Abstract
Herpesvirus proteins pUL34 and pUL31 form a complex at the inner nuclear membrane (INM) which is necessary for efficient nuclear egress. Pseudorabies virus (PrV) pUL34 is a type II membrane protein of 262 amino acids (aa). The transmembrane region (TM) is predicted to be located between aa 245 and 261, leaving only one amino acid in the C terminus that probably extends into the perinuclear space. It is targeted to the nuclear envelope in the absence of other viral proteins, pointing to intrinsic localization motifs, and shows structural similarity to cellular INM proteins like lamina-associated polypeptide (Lap) 2ß and Emerin. To investigate which domains of pUL34 are relevant for localization and function, we constructed chimeric proteins by replacing parts of pUL34 with regions of cellular INM proteins. First the 18 C-terminal amino acids encompassing the TM were exchanged with TM regions and C-terminal domains of Lap2ß and Emerin or with the first TM region of the polytopic lamin B receptor (LBR), including the nine following amino acids. All resulting chimeric proteins complemented the replication defect of PrV-ΔUL34, demonstrating that the substitution of the TM and the extension of the C-terminal domain does not interfere with the function of pUL34. Complementation was reduced but not abolished when the C-terminal 50 aa were replaced by corresponding Lap2ß sequences (pUL34-LapCT50). However, replacing the C-terminal 100 aa (pUL34-LapCT100) resulted in a nonfunctional protein despite continuing pUL31 binding, pointing to an important functional role of this region. The replacement of the N-terminal 100 aa (pUL34-LapNT100) had no effect on nuclear envelope localization but abrogated pUL31 binding and function.
INTRODUCTION
During herpesvirus morphogenesis, nucleocapsids are assembled in the host cell nucleus and have to cross the nuclear membranes to gain access to the cytosol, where final tegumentation and envelopment occurs. To this end, nucleocapsids bud at the inner nuclear membrane (INM), which subsequently encloses the nucleocapsid, thereby forming a primary enveloped virion located in the perinuclear space. This primary envelope is lost after fusion with the outer nuclear membrane (ONM), releasing the nucleocapsid into the cytosol (reviewed in references 25, 38, 39, and 40). To gain access to the budding sites at the INM, the nuclear lamina, a filamentous meshwork consisting mainly of lamin types A/C and B which underlies and supports the nuclear membrane, has to be softened and/or dissolved at least locally (reviewed in references 25 and 40). This partial dissolution is thought to be accomplished by the nuclear egress complex (NEC), which is highly conserved throughout the herpesviruses (reviewed in references 25 and 40). It consists of viral proteins homologous to herpes simplex virus type 1 (HSV-1) pUL34 and pUL31 and functions via the recruitment of cellular and viral protein kinases which phosphorylate lamins, thereby triggering their dissolution (3, 43, 49). In the absence of either pUL31 or pUL34, nucleocapsids are trapped in the nucleus and only a few infectious particles are released (6, 10, 13, 18, 28, 44, 50, 51, 52, 59). However, pUL31 and pUL34 not only are required for efficient nuclear egress but also are sufficient for the formation of vesicles from the INM, resembling primary envelopes (31), indicating that these two proteins form the core budding machinery.
Pseudorabies virus (PrV) pUL34, which exhibits a smooth nuclear rim staining in infected and transfected cells, is a predicted type II membrane protein (28) with the hydrophobic domain located between amino acids (aa) 245 and 261 (according to PSort II [http://www.psort.org/]) (24), leaving only one amino acid in the C terminus to extend into the perinuclear lumen either from the INM or from the primary virion envelope. The deduced amino acid sequence does not contain a typical nuclear localization motif (NLS) (24) but contains an RXR (RQR) sequence at amino acids 173 to 175 which has been defined as an efficient INM-sorting motif for human cytomegalovirus glycoprotein B (41, 42). However, the significance of this motif in PrV pUL34 has not been investigated, and pUL34 might be small enough to diffuse passively into the INM. Although it has been speculated that pUL34 is retained in the nuclear membrane by interaction with nucleoplasmic pUL31, this is not the case in PrV, where pUL34 shows distinct nuclear rim localization even in the absence of other viral proteins, although the presence of pUS3 kinase might enhance nuclear membrane targeting (29).
pUL31 is a small, soluble nuclear protein which forms a complex with pUL34 and colocalizes with pUL34 at the nuclear rim in infected or transfected cells (13, 31). PrV pUL31 contains a predicted bipartite NLS between amino acids 4 and 20 (24) and is efficiently targeted to the nucleoplasm in infected and transfected cells (13, 31), most probably by using the cellular nuclear import machinery. The pUL31 interaction domain of pUL34 was mapped to the N-terminal 162 amino acids in PrV (13), to amino acids 1 to 178 in murine cytomegalovirus MCMV (6), and to amino acids 137 to 181 in HSV-1 (34), indicating the conservation of this domain across herpesvirus pUL34 homologs.
The nuclear envelope consists of an inner and an outer membrane which are connected at the nuclear pores. While the outer nuclear membrane and the perinuclear space are contiguous with the rough endoplasmic reticulum (RER), sharing a common set of proteins, the INM is enriched for a specific subset of proteins. Approximately 80 different integral membrane proteins of the INM have been identified by a proteomic approach (54). Many of those proteins bind either directly or indirectly to lamins and DNA and provide structural as well as functional support for the lamina and chromatin. Three different types of INM integral membrane proteins have been identified: proteins which span the INM once and extend with most of their mass into the nucleoplasm, like Lap2 and Emerin; proteins with two transmembrane (TM) domains, like Man1; and proteins with multiple hydrophobic domains, such as the lamin B receptor (LBR) and Nurim (53). pUL34 and its homologues in other herpesviruses share structural similarities to the cellular INM proteins with a single transmembrane region, and it is likely that they are localized by pathways similar to those of the cellular proteins.
Newly synthesized INM proteins are inserted into the RER membrane and move laterally along the RER and ONM to the INM at nuclear pores. Their targeting to the INM has long been explained solely by the diffusion-retention model (23, 56), suggesting that nuclear interaction partners arrest the laterally diffusing membrane proteins. However, as a general mechanism, this model has been challenged by the observation that energy-dependent and receptor-mediated targeting mechanisms exist using either classical nuclear localization signals or INM-specific motifs (5, 27, 36, 41, 47).
However, the exact targeting mechanisms of herpesviral pUL34 homologs is not understood, nor are the molecular details of primary envelopment and the mechanism of fusion of the primary virion envelope with the outer nuclear membrane. In HSV-1, glycoproteins B (gB) and H (gH), which are essential for the penetration of virions during entry, have been reported to be involved in fusion during nuclear egress (11). However, this is not the case in PrV, since PrV mutants lacking either gB, gH, gD, or gL singly or in different combinations are fully competent in the nuclear egress of nucleocapsids as well as virus release into the supernatant (32). On the other hand, the simultaneous expression of pUL31 and pUL34 leads to the efficient formation of vesicles from the inner nuclear membrane, which resemble primary envelopes in size and form. Putative fusion intermediates with the outer nuclear membrane also were observed in ultrastructural studies (31), indicating that pUL34 and pUL31 are sufficient for vesicle formation, fission, and fusion. Interestingly, only one C-terminal amino acid of pUL34 is predicted to be exposed from the primary envelope into the perinuclear cleft, and it is difficult to imagine how this could trigger the fusion event. Thus, we wanted to assay functional domains by replacing different regions of PrV pUL34 with corresponding domains of cellular INM proteins.
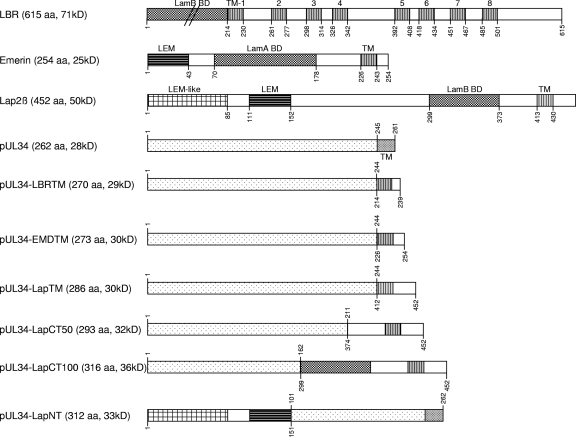
Lamina-associated polypeptide 2 (Lap2) is a group of six alternatively spliced proteins known as Lap2α, Lap2β, Lap2γ, Lap2δ, Lap2ε, and Lap2ζ (2). The β, γ, δ, and ε isoforms are integral proteins of the inner nuclear membrane, while Lap2α has no transmembrane region and is present in the nucleoplasm (reviewed in reference 23). Lap2β comprises 452 aa and is the most ubiquitous isoform, with the TM predicted to be between aa 413 and 430 and a C-terminal domain of 22 aa (Fig. 1). It is able to bind lamin B via a C-terminally located domain (aa 299 to 373) (14, 15). The chromatin binding domain, which binds to barrier to autointegration factor (BAF), is localized in the N terminus, and an LEM motif is present between amino acids 111 and 152 (16, 55). The LEM domain is an ∼40-residue motif found in nuclear membrane-associated proteins, including Lap2, Emerin, MAN1, Otefin, and Lem-3 (35). An additional LEM-like motif responsible for chromosome binding is located within the first 85 amino acids of the N terminus (16). Lap2β binding to lamins and chromatin is inhibited by mitotic phosphorylation (12).
Fig 1.
Construction of pUL34 chimeras. Chimeric genes were generated by fusion PCR with overlapping primers and cloned into the eukaryotic expression vector pcDNA3. Indicated are expressed open reading frames, including the transmembrane domains in Lap2β, Emerin, LBR, and pUL34. The LEM-like domain, the LEM domain, and the lamin A and/or B binding regions (BD) are also shown. Amino acid positions are given above for pUL34 amino acid sequences and below for LBR, Emerin, and Lap2β. The sizes of the generated chimeras are shown in amino acids and predicted molecular masses. The N-terminal domain of LBR is not drawn to scale.
Emerin has limited homology to Lap2ß in structure and function. It consists of 254 aa, and its TM is predicted between aa 226 and 243. An LEM domain is located between aa 1 and 43 (Fig. 1). In contrast to Lap2ß, which binds only B-type lamins, Emerin binds A- and B-type lamins (7, 9). The 615-aa LBR is characterized by 8 predicted transmembrane domains (Fig. 1) (48). The hydrophilic N terminus reaches into the nucleoplasm, where it interacts with lamin B (aa 1 to 216).
To test which domains in PrV pUL34 are necessary and/or sufficient for nuclear envelope localization and for function during nuclear egress, we generated chimeras between pUL34 and Lap2ß, Emerin, and LBR and tested the hybrid proteins for their intracellular localization and for the functional complementation of a UL34-deleted PrV mutant.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney cells (RK13) were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Laboratory strain PrV Kaplan (PrV-Ka) (26) was grown on RK13 cells and PrV-ΔUL34 on RK13-UL34 cells as described previously (28, 29).
Construction of UL34 chimeras.
Chimeric genes were generated by a modified fusion PCR (22, 46) using primers shown in Table 1. In a first PCR, the corresponding regions of either PrV UL34 or rat Lap2β, human Emerin, and human LBR were amplified with Pfx DNA polymerase (Invitrogen) separately with specific primers comprising overlapping extensions. pcDNA-UL34 (28) or pEGFP-Lap2β (1), pNP07 (kindly provided by Peter O'Hare), or pLBR1TMgfp (8) were used as the template. In a second PCR, the overlapping regions of the generated PCR products were annealed and used as a template with forward and reverse primers (Table 1). The fusion PCR products for UL34-LapTM, UL34-EMDTM, UL34-LBRTM, and UL34-LapCT50 were cleaved with EcoRI/HindIII, for which recognition sites were added with the primers (underlined in Table 1), and cloned into vector pcDNA3 (Invitrogen). UL34-LapCT100 and UL34-LapNT were first cloned blunt ended into the SmaI site of pBluescript SK+ (Stratagene) and recloned into pcDNA3 after digestion with HindIII. Correct amplification, in-frame fusion, and cloning were verified by sequencing.
Table 1.
Primers used for construction of chimeric genesa
| Name | Sequence 5′ to 3′ | Location (bp) in: |
|||
|---|---|---|---|---|---|
| pEGFP-Lap2β (Euroscarf P30463) | pLBR1TM-GFP (Euroscarf P30453) | NP07 | PrV-Ka (GenBank no. BK001744) | ||
| UL34for | CACAAAGCTTACCATGAGCGGCACCCT | 31395–31411 | |||
| UL34rev | CACAGGATCCGCCGCCTTTAACGCATGTTCAG | 32193–32712 | |||
| Lap2βfor | CACAAAGCTTGGGACGAGATGCCGGAGTTC | 1370–1389 | |||
| Lap2βrev | GAGAGAATTCGAGATCGAGTCGGATGTGCC | 2768–2749 | |||
| UL34-LapTMfor | CGGCGCCTCGCCGGCTACTGGATAAAAATGCTGCTGTTTG | 2614–2635 | 32112–32129 | ||
| UL34-LapTMrev | CAAACAGCAGCATTTTTATCCAGTAGCCGGCGAGGCGCCG | 2635–2614 | 32129–32112 | ||
| UL34-LapCT50 for | CGCCCGTGCCCGGGCACGCGCCGCTCGAGCTCAGTGACTT | 2494–2514 | 32011–32020 | ||
| UL34-LapCT50 rev | AAGTCACTGAGCTCGAGCGGCGCGTGCCCGGGCACGGGCG | 2514–2494 | 32020–32011 | ||
| UL34-LapCT100 for | GGCCCCGACGACGCCTCG AGGTTGACTGGAAATTTCAAGC | 2263–2284 | 31863–31880 | ||
| UL34-LapCT100 rev | GCTTCAAATTTCCAGTCAACCTCGAGGCGTCGTCGGGGCC | 2284–2263 | 31880–31863 | ||
| UL34-LapNTfor | CTGTTGAAGCTGAGGGAACAGCACAACAACGTGATCCTGGCC | 1810–1830 | 31698–31718 | ||
| UL34-LapNTrev | GGCCAGGATCACGTTGTTGTGCTGTTCCCTCAGCTTCAACAG | 1830–1810 | 31718–31698 | ||
| UL34-LBRTMfor | CGGCGCCTCGCCGGCTACGGTGTGTTTCTCATCATGTTTG | 1395–1416 | 32112–32129 | ||
| UL34-LBRTMrev | CAAACATGATGAGAAACACACCGTAGCCGGCGAGGCGCCG | 1416–1395 | 32129–32112 | ||
| LBRrev | CACAGAATTCTTATGGATCTTTCTGTTTAC | 1453–1472 | |||
| UL34-EMDTMfor | CGGCGCCTCGCCGGCTACTGGGGCCAGCTGCTGCTTTTCC | 5372–5391 | 32112–32129 | ||
| UL34-EMDTMrev | GGAAAAGCAGCAGCTGGCCCCAGTAGCCGGCGAGGCGCCG | 5391–5372 | 32129–32112 | ||
| EMDrev | CACAGAATTCTTAGAAGGGGTTGCCTTC | 5459–5442 | |||
Recognition sequences are underlined.
Generation of stable cell lines.
Stable cell lines expressing pUL34-LBRTM, pUL34-EMDTM, pUL34-LapTM, pUL34-LapCT50, pUL34-LapCT100, or pUL34-LapNT were isolated after transfection of RK13 cells with the corresponding pcDNA3 constructs using calcium phosphate precipitation (17) and selection of transfected cells in medium containing 0.5 mg/ml G418 (Invitrogen). Resistant cell clones were picked and tested by indirect immunofluorescence with the anti-pUL34 serum (28).
Laser-scanning confocal microscopy.
RK13 cells were either transfected singly with pcDNA-UL34, pcDNA-UL34-LBRTM, pcDNA-UL34-EMDTM, pcDNA-UL34-LapTM, pcDNA-UL34-LapCT50, pcDNA-UL34-LapCT100, and pcDNA-UL34-LapNT or cotransfected with these constructs and pcDNA-UL31 (13). Twenty-four h after transfection, cells were fixed with 3% paraformaldehyde for 20 min and subsequently permeabilized with 3% paraformaldehyde–0.3% Triton X-100 and immunostained for pUL34 and/or pUL31. To this end, polyclonal rabbit anti-pUL31 (13) and polyclonal mouse anti-pUL34 serum (31) were diluted 1:500 in phosphate-buffered saline (PBS) and incubated for 1 h at room temperature. Bound antibody was detected by Alexa Fluor 555 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) diluted 1:1,000. Nuclei were stained with TO-PRO-3 (Invitrogen). Fluorescence images were acquired using a confocal laser-scanning microscope (LSM510; Zeiss, Germany).
Western blot analysis.
RK13, RK13-UL34, RK13-UL34-LBRTM, RK13-UL34-EMDTM, RK13-UL34-LapTM, RK13-UL34-LapCT50, RK13-UL34-LapCT100, and RK13-UL34-LapNT cells were harvested and lysed, and proteins were separated on SDS-10% polyacrylamide gels. After electrotransfer onto a nitrocellulose membrane, the membrane was incubated with the monospecific anti-pUL34 serum (28) at a dilution of 1:50,000. Bound antibody was detected by peroxidase-coupled goat anti-rabbit antibodies and visualized by enhanced chemiluminescence, which was recorded by an image analyzer (ChemiDoc XRS+; Bio-Rad).
One-step growth kinetics and plaque size.
For one-step growth kinetics, cells were infected at a multiplicity of infection (MOI) of 3. After 1 h at 4°C, the inoculum was replaced by prewarmed medium and cells were incubated for an additional hour at 37°C to allow virus entry. Nonpenetrated virus was inactivated by low pH treatment (37), and cells and supernatant were harvested either immediately or after incubation for an additional 4, 8, 12, 24, or 36 h. Progeny virus titers were determined on RK13-UL34 cells. Mean values from three independent experiments and corresponding standard deviations were plotted.
For the determination of plaque sizes, cells were infected with either PrV-Ka or PrV-ΔUL34 under plaque assay conditions and stained with 1% crystal violet 2 days postinfection. For each virus-cell combination, 50 plaques were measured microscopically. The plaque size for PrV-Ka on a given cell line was set as 100%, and corresponding values for the mutant viruses were calculated. Mean values from three independent experiments are given with the corresponding standard deviations.
Electron microscopy.
Cell lines were infected with PrV-ΔUL34 at an MOI of 1 and processed for electron microscopy 14 h postinfection as described previously (28).
RESULTS
Construction of pUL34 chimeric proteins.
pUL34 and the INM proteins Lap2ß, Emerin, and LBR are efficiently targeted to the nuclear envelope. To investigate which domains of pUL34 are necessary and/or sufficient for the targeting and function of pUL34 during herpesvirus nuclear egress, we constructed chimeras between these proteins. First, the functional role of the pUL34 transmembrane domain, including the penultimate amino acid, which might constitute the short periplasmic domain, was tested. To this end, the hydrophobic C-terminal domain of pUL34 (aa 245 to 262) was replaced by (i) the first hydrophobic region of LBR located between aa 214 and 230 and nine additional C-terminal amino acids, (ii) the TM region of Emerin (aa 226 to 243) including the 11 C-terminal amino acids, and (iii) the Lap2β TM (aa 413 to 452) including the 22 C-terminal amino acids (Fig. 1).
In a second approach, the altered region was enlarged toward the N terminus of pUL34. To this end, aa 1 to 211 of pUL34 were fused to the Lap2ß C-terminal region (aa 374 to 452), resulting in pUL34-LapCT50. Furthermore, aa 1 to 162 of pUL34, which have been shown to be sufficient for interaction with pUL31 (13), were fused to the 153 C-terminal Lap2β amino acids, including the lamin B binding domain (aa 299 to 373) (15), giving rise to pUL34-LapCT100 (Fig. 1). In the reverse construct, the N-terminal 100 aa of pUL34 were replaced by aa 1 to 152 of Lap2β, comprising the LEM motif, generating pUL34-LapNT (Fig. 1). This construct was chosen because it should retain the pUL31 interaction domain which has been predicted at aa 137 to 181 in HSV-1 pUL34 (34), corresponding to aa 123 to 167 of PrV pUL34. Correct in-frame cloning was verified by sequencing, and expression was tested by coupled in vitro transcription-translation (data not shown) and immunofluorescence after transient transfection (see below).
Localization and complex formation with pUL31 of pUL34-INM chimeric constructs.
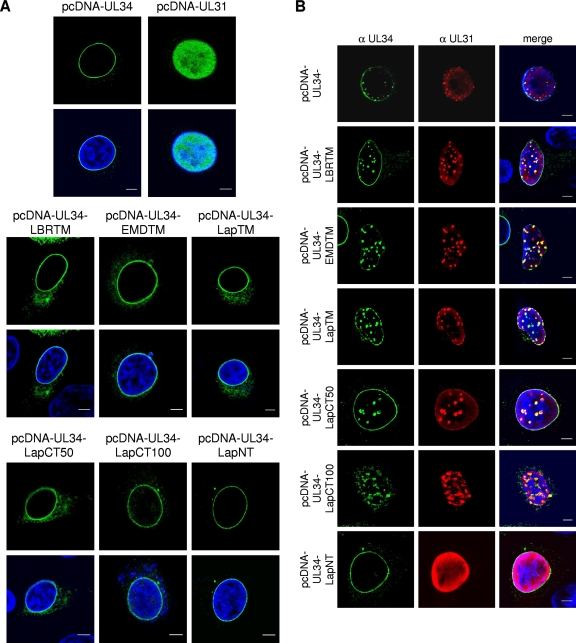
To analyze the intracellular localization of the chimeric protein RK13, cells were transfected with the different expression vectors. Cells were fixed and processed for indirect immunofluorescence with the anti-pUL34-specific serum 2 days after transfection. All chimeric proteins exhibited smooth nuclear rim staining similar to that of PrV pUL34 (Fig. 2A) or cellular Lap2β, Emerin, and LBR (12, 20, 28, 45), while pUL31 in transfected cells shows a diffuse nucleoplasmic pattern (Fig. 2A) (13, 31). Due to the high expression levels, some cytoplasmic staining also was observed. To test for interaction with pUL31, plasmids expressing the chimeric proteins were cotransfected with pcDNA-UL31 (Fig. 2B). The cotransfection of pcDNA-UL31 and pcDNA-UL34 resulted in the relocalization of pUL31 to speckles near the nuclear rim, where both complex partners colocalize, as described previously (31). A similar staining pattern was detectable after the cotransfection of pcDNA-UL34-LBRTM, pcDNA-UL34-EMDTM, pcDNA-UL34-LapTM, pcDNA-UL34-LapCT50, and pcDNA-UL34-LapCT100 with pcDNA-UL31, demonstrating that these chimeric proteins still were able to recruit nucleoplasmic pUL31. However, the cotransfection of pcDNA-UL31 with pcDNA-UL34-LapNT did not result in speckle formation or the relocalization of pUL31, indicating that this protein is unable to interact with and/or relocate its complex partner.
Fig 2.
Localization of pUL34 and pUL34 chimeras. RK13 cells were transfected either singly with pcDNA-UL34, pcDNA-UL34-LBRTM, pcDNA-UL34-EMDTM, pcDNA-UL34-LapTM, pcDNA-UL34-LapCT50, pcDNA-UL34-LapCT100, or pcDNA-UL34-LapNT, (A) or cotransfected with pcDNC-UL31, (B). Cells were fixed, and expressed proteins were detected with monospecific anti-pUL31 rabbit and anti-pUL34 mouse serum. Nuclei were counterstained with TO-PRO-3 (Invitrogen). Images were recorded with a laser-scanning microscope (LSM 510; Zeiss). Scale bars, 5 μm.
Complementation of PrV-ΔUL34 by pUL34-INM chimeras.
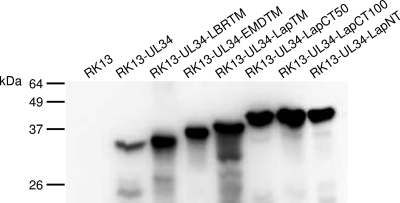
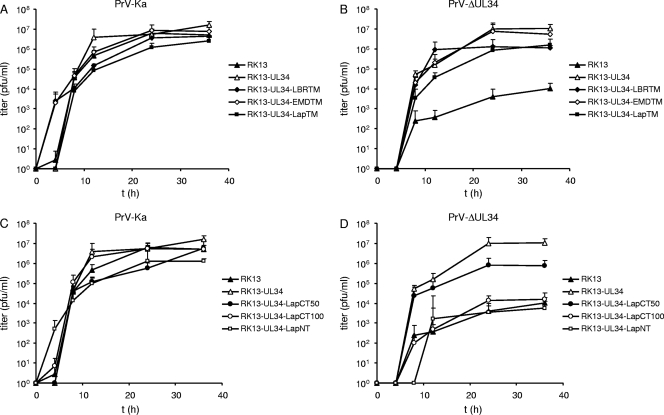
Since all chimeric proteins localized to the nuclear rim, we tested whether they were also able to complement the replication defect of PrV-ΔUL34. Thus, RK13 cells were transfected with the corresponding expression constructs and selected in medium containing G418. Cell clones were picked and tested with the pUL34 monospecific rabbit serum by indirect immunofluorescence and immunoblotting. Several cell clones were tested, and one of each was used for further studies. Western blot analyses of the selected cell lines showed the expected electrophoretic mobility of the corresponding chimeras (Fig. 3). To assay for transcomplementation, cell lines were infected at an MOI of 3 with either PrV-Ka or PrV-ΔUL34 (Fig. 4). After 0, 4, 8, 12, 24, and 36 h, cells were harvested and infectious progeny titrated on RK13-UL34 cells. While PrV-Ka replicated to similar titers on all cell lines, indicating that the chimeras do not exert general dominant-negative effects on virus replication (Fig. 4A and C), all cell lines expressing chimeras carrying heterologous transmembrane and C-terminal domains complemented the defect of the UL34 deletion mutant to titers similar to that of the wild-type pUL34-expressing cells (Fig. 4B). The enlargement of the replaced region toward the N terminus of pUL34 in RK13-UL34-LapCT50 resulted in a ca. 10-fold decrease in viral titers (Fig. 4D). In contrast, further extension of the exchanged region in RK13-UL34-LapCT100 resulted in the abrogation of complementation despite continuing pUL31 recruitment (Fig. 2). RK13-UL34-LapNT cells were unable to support virus replication to titers exceeding those on noncomplementing RK13 cells (Fig. 4D).
Fig 3.
Western blot analysis. Lysates of RK13, RK13-UL34, RK13-UL34-LBRTM, RK13-UL34-EMDTM, RK13-UL34-LapTM, RK13-UL34-LapCT50, RK13-UL34-LapCT100, and RK13-UL34-LapNT cells were separated on SDS-10% polyacrylamide gels. After transfer to nitrocellulose, the membrane was incubated with the polyclonal monospecific rabbit anti-pUL34 serum. The location of molecular mass markers is indicated on the left.
Fig 4.
One-step growth kinetics. RK13, RK13-UL34, RK13-UL34-LBRTM, RK13-UL34-EMDTM, RK13-UL34-LapTM, RK13-UL34-LapCT50, RK13-UL34-LapCT100, and RK13-UL34-LapNT cells were infected at an MOI of 3 with either PrV-Ka (A and C) or PrV-ΔUL34 (B and D). Cells were harvested after 0, 4, 8, 12, 24, and 36 h, and progeny virus titers were determined on RK13-UL34 cells. Given are mean values from three independent experiments with corresponding standard deviations.
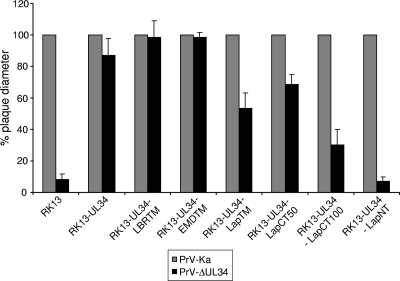
Plaque diameters of PrV-ΔUL34 reached wild-type-like values on RK13-UL34, RK13-UL34-LBRTM, and RK13-UL34-EMDTM (Fig. 5). In contrast, although RK13-UL34-LapTM cells produced titers similar to those of RK13-UL34 cells, plaque size was decreased and reached only approximately 50% of that of PrV-Ka, while plaque size decreased to approximately 70% on RK13-UL34-LapCT50 (Fig. 5). Surprisingly, although RK13-UL34-LapCT100 did not enhance virus production (Fig. 4D), compared to that of noncomplementing RK13 cells the plaque size was increased from only tiny foci to measurable plaques, reaching 30% of the size of those of PrV-Ka. RK13-UL34-LapNT did not support efficient cell-cell spread beyond that observed on RK13 cells (Fig. 5).
Fig 5.
Plaque sizes. RK13, RK13-UL34, RK13-UL34-LBRTM, RK13-UL34-EMDTM, RK13-UL34-LapTM, RK13-UL34-LapCT50, RK13-UL34-LapCT100, and RK13-UL34-LapNT cells were infected with PrV-Ka (gray columns) and PrV-ΔUL34 (black columns). Cells were fixed 2 days postinfection, and plaque diameters were measured microscopically. Plaque sizes of PrV-Ka on the corresponding cell line were set as 100%, and diameters of plaques or foci were calculated accordingly. Given are mean values from three independent experiments and corresponding standard deviations.
Ultrastructural analyses.
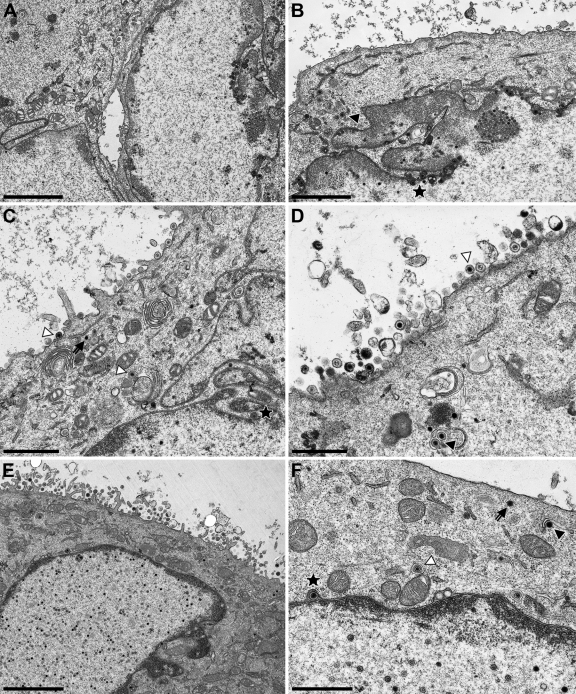
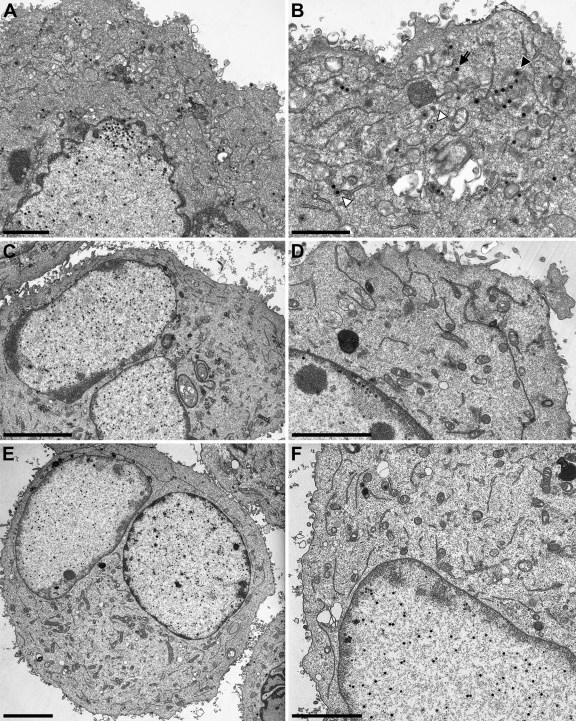
To investigate the complementation of PrV-ΔUL34 on cell lines expressing the pUL34 chimeric proteins in more detail, cells were infected in parallel at an MOI of 1 with PrV-ΔUL34 and processed for high-resolution transmission electron microscopy after 14 h. Ultrastructural analyses of infected cells expressing transmembrane domain substitutions (pUL34-LBRTM, pUL34-EMDTM, and pUL34-LapTM) showed immature capsids as well as numerous nucleocapsids in the nucleus, primary enveloped virions in the perinuclear cleft, budding stages into membranous vesicles in the cytoplasm, and mature virions on the cell surface (Fig. 6). This pattern is similar to that observed in cells infected by wild-type PrV-Ka (19). After infection of RK13-UL34-LapCT50 cells, wild-type-like replication stages were observed (Fig. 7A and B). In contrast, extranuclear capsids were not detectable in infected RK13-UL34-LapCT100 or RK13-UL34-LapNT cells, although the nuclei were filled with DNA-containing capsids, a phenotype resembling that of RK13 cells infected with PrV-ΔUL34 (Fig. 7C to F) (28).
Fig 6.
Ultrastructural analysis of TM substitutions. RK13-UL34-LBRTM (A and B), RK13-UL34-EMDTM (C and D), and RK13-UL34-LapTM (E and F) were infected with PrV-ΔUL34 at an MOI of 1 and processed for electron microscopy 14 h after infection. On the left overviews of infected cells are shown, and higher magnifications are given on the right. Primary enveloped virions are marked by asterisks, intracytoplasmic nucleocapsids by arrows, intracytoplasmic nucleocapsids undergoing secondary envelopment by closed triangles, and enveloped intracytoplasmic virions in vesicles by open triangles. Bars correspond to 3 (A), 2 (B), 1.4 (C), 1 (D), 3 (E), and 1 μm (F).
Fig 7.
Ultrastructural analysis of Lap2ß-Chimera. RK13-UL34-LapCT50 (A and B), RK13-UL34-LapCT100 (C and D), and RK13-UL34-LapNT (E and F) were infected with PrV-ΔUL34 at an MOI of 1 and processed for electron microscopy 14 h after infection. On the left overviews of infected cells are shown, and higher magnifications are given on the right. Bars correspond to 2 (A), 1.4 (B), 5 (C), 3 (D), 5 (E), and 3 μm (F). In panel B, the primary enveloped virion is marked by an asterisk, an intracytoplasmic nucleocapsid by an arrow, an intracytoplasmic nucleocapsid undergoing secondary envelopment by a closed triangle, and an enveloped intracytoplasmic virion in a vesicle by an open triangle. No extranuclear virus particles are present in panels C to F, whereas intranuclear DNA-filled capsids are clearly visible.
DISCUSSION
In this study, we aimed at further delineating the targeting and functional motifs in PrV pUL34. To this end, different parts of pUL34 were replaced by corresponding regions of the cellular proteins Emerin, LBR, and Lap2β, thereby generating chimeric proteins.
pUL34-LBRTM, pUL34-EMDTM, and pUL34-LapTM.
The replacement of the TM domain, including the C-terminal amino acids of pUL34 by TM domains of LBR, Emerin, and Lap2β, resulted in chimeric proteins which efficiently complemented the replication of PrV-ΔUL34. Thus, the role of this region of the protein in nuclear egress, including primary envelope formation and fusion, is not virus specific but can be supplied by heterologous sequences. Even the extension of the C-terminal domain, which could protrude into the perinuclear space, did not affect primary virion formation, nuclear egress, or the release of infectious virions, which shows that any hypothetical recruitment of cellular proteins to mediate primary envelope formation and/or fusion must either be effected by regions outside the altered area or be a property of both TM and C-terminally extended areas.
The full complementation of plaque formation was observed for cell lines expressing pUL34-LBRTM and pUL34-EMDTM. In contrast, pUL34-LapTM-expressing cells complemented the plaque formation of PrV-ΔUL34 to only approximately 50% compared to that of PrV-Ka, although formation of infectious progeny comparable to that of infection with PrV-Ka could be observed. Recently, a critical role for HSV-1 pUL34 in cell-to-cell spread has been suggested (21), and it had been speculated that HSV-1 pUL34 influences cell-to-cell spread indirectly by targeting of proteins toward the cell periphery. Thus, our chimeric pUL34-LapTM may have a defect in this function which could be due to the extension of the C-terminal domain, since smaller C-terminal extensions represented by pUL34-LBRTM and pUL34-EMDTM had no effect on plaque formation.
pUL34-LapCT50.
Another chimeric protein was based on pUL34-LapTM but carried an extension of the alteration encompassing the 50 C-terminal pUL34 residues with Lap2β aa 374 to 452. In cotransfection experiments, the colocalization of pUL34-LapCT50 with pUL31 confirmed that pUL34 aa 212 to 262 are not essential for interaction with pUL31. In one-step growth kinetics with the PrV-ΔUL34 deletion mutant, a ca. 10-fold reduction in the production of infectious progeny was observed. However, replication still was significantly better than that on noncomplementing RK13 cells. Plaque size was reduced to 70% of wild-type levels, which correlates with the one-step replication phenotype. Thus, aa 1 to 211 are sufficient for pUL34 function despite producing reduced titers and plaque sizes.
pUL34-LapCT100.
We also extended the altered region preceding the transmembrane domain toward the N terminus of pUL34, retaining only aa 1 to 162 of authentic PrV pUL34. This region has been shown to be sufficient to interact with pUL31 in yeast two-hybrid analyses (13). The corresponding chimeric pUL34-LapCT100 still was targeted to the nuclear envelope and was able to colocalize with and relocate pUL31 to the nuclear rim, demonstrating that the pUL31 interaction domain was intact and correctly folded. The formation of speckles as observed during coexpression of wild-type pUL34 and pUL31 (31) also was evident, indicating that the chimeric protein can induce sufficient membrane curvature to form primary capsid-less (L) particles. After infection with PrV-ΔUL34, RK13-UL34-LapCT100 cells exhibited numerous DNA-containing capsids, sometimes close to the INM. However, neither intracytoplasmic virus particles nor primary envelopment stages were observed. This result suggests that correct membrane anchoring and interaction with pUL31 is not sufficient for pUL34 function in primary envelopment and nuclear egress. It can be speculated that the region adjacent to the pUL31 interaction domain and preceding the transmembrane domain (aa 163 to 211) is required for either direct or indirect interaction with the nucleocapsid. Interaction between HSV-1 pUL34 and the major capsid protein VP5 has been described (60). However, since the major substrates for primary envelopment are DNA-filled capsids, it is more likely that a C capsid-specific component, either pUL25 alone (30) or a complex of pUL25/pUL17 (57), is targeted. Direct interactions between the NEC and the C capsid have been suggested recently for PrV and HSV-1 but with contradictory results (33, 58). The absence of primary L particles in PrV-ΔUL34-infected RK13-pUL34-LapCT100 cells likely is due to the presence of pUS3, which allows for the efficient fusion of these vesicles with the outer nuclear membrane (31).
The amino acid sequences of herpesvirus pUL34 homologs exhibit high conservation from the N terminus to approximately amino acid position 170 in PrV pUL34, comprising the region shown to interact with pUL31 in PrV, HSV-1, and MCMV (6, 13, 34), while homology is low beyond that part. Despite the low conservation, this region is of functional importance. A deletion mutant of the MCMV pUL34 homolog lacking amino acids 178 to 277 (of 316 residues) showed behavior similar to that of pUL34-LapCT100. Complex formation with the pUL31 homolog was unimpaired, but the corresponding viral deletion mutant could not be complemented. In contrast, a mutant lacking only aa 207 to 277 fully complemented the defect (6), delineating this essential region between aa 178 (aa 1 to 178 have been shown to be sufficient to interact with the pUL31 homolog) and 207, which contains a polyproline stretch (6). However, charged cluster mutants in this region of HSV-1 pUL34 which should disrupt protein-protein interactions had no effect in complementation assays (4). Thus, further studies are required to define in more detail this region of interest.
pUL34-LapNT.
The chimeric protein in which the N-terminal 100 amino acids of pUL34 were replaced by the 151 N-terminal amino acids of Lap2β, including the LEM and a LEM-like motif, still showed nuclear rim staining indicative of proper targeting to the nuclear membrane. However, interaction with coexpressed pUL31 was not detectable. In HSV-1 pUL34, aa 137 to 181, corresponding to aa 123 to 167 in PrV pUL34, were shown to be sufficient for complex formation with pUL31 (34), whereas in our yeast two-hybrid studies pUL34 clones interacting with the pUL31 bait contained pUL34 sequences from aa 1 up to at least aa 162 (13). Due to the lack of pUL31-pUL34 interaction, it was not surprising that this chimera did not support virus replication, since NEC formation is required for efficient nuclear egress (reviewed in references 25 and 38–40).
In summary, our data show that the alteration of the transmembrane domain of pUL34 as well as the addition of more C-terminal, putatively periplasmic amino acids do not interfere with the intracellular localization or function of pUL34 in nuclear egress. In contrast, both the N-terminal and the C-terminal domains contain essential functions necessary for interaction with the complex partner pUL31 for intrinsic pUL34 function, respectively.
ACKNOWLEDGMENTS
We thank D. Werner, C. Meinke, and P. Meyer for expert technical assistance, K. Grimm for help with the infection of cell lines, and M. Jörn for photographic help.
This study was supported by the Deutsche Forschungsgemeinschaft Priority Program SPP 1175, grant Me 854/8-2.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108:83–96 [DOI] [PubMed] [Google Scholar]
- 2. Berger R, et al. 1996. The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res. 6:361–370 [DOI] [PubMed] [Google Scholar]
- 3. Bjerke SL, Roller RJ. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bjerke SL, et al. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601–7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braunagel SC, Williamson ST, Ding Q, Wu X, Summers MD. 2007. Early sorting of inner nuclear membrane proteins is conserved. Proc. Natl. Acad. Sci. U. S. A. 104:9307–9312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bubeck A, et al. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clements L, Manilal S, Love DR, Morris GE. 2000. Direct interaction between emerin and lamin A. Biochem. Biophys. Res. Commun. 267:709–714 [DOI] [PubMed] [Google Scholar]
- 8. Ellenberg J, et al. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fairley EA, Kendrick-Jones J, Ellis JA. 1999. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J. Cell Sci. 112:2571–2582 [DOI] [PubMed] [Google Scholar]
- 10. Farina A, et al. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farnsworth A, et al. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foisner R, Gerace L. 1993. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 73:1267–1279 [DOI] [PubMed] [Google Scholar]
- 13. Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furukawa K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112:2485–2492 [DOI] [PubMed] [Google Scholar]
- 15. Furukawa K, Kondo T. 1998. Identification of the lamina-associated-polypeptide-2-binding domain of B-type lamin. Eur. J. Biochem. 251:729–733 [DOI] [PubMed] [Google Scholar]
- 16. Furukawa K, Fritze CE, Gerace L. 1998. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J. Biol. Chem. 273:4213–4219 [DOI] [PubMed] [Google Scholar]
- 17. Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467 [DOI] [PubMed] [Google Scholar]
- 18. Granato M, et al. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82:4042–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Granzow H, et al. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haraguchi T, et al. 2000. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113:779–794 [DOI] [PubMed] [Google Scholar]
- 21. Haugo AC, Szpara ML, Parsons L, Enquist LW, Roller RJ. 2011. Herpes simplex virus type 1 pUL34 plays a critical role in cell-to-cell spread of virus in addition to its role in virus replication. J. Virol. 85:7203–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 23. Holmer L, Worman HJ. 2001. Inner nuclear membrane proteins: functions and targeting. Cell. Mol. Life Sci. 58:1741–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horton P, Nakai K. 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5:147–152 [PubMed] [Google Scholar]
- 25. Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 26. Kaplan AS, Vatter AE. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407 [DOI] [PubMed] [Google Scholar]
- 27. King MC, Lusk CP, Blobel G. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442:1003–1007 [DOI] [PubMed] [Google Scholar]
- 28. Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klupp BG, Granzow H, Mettenleiter TC. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363–2371 [DOI] [PubMed] [Google Scholar]
- 30. Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klupp BG, et al. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U. S. A. 104:7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klupp B, Altenschmidt J, Granzow H, Fuchs W, Mettenleiter TC. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 82:6299–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leelawong M, Guo D, Smith GA. 2011. A physical link between the pseudorabies virus capsid and the nuclear egress complex. J. Virol. 85:11675–11684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang L, Baines JD. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J. Virol. 79:3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin F, et al. 2000. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 275:4840–4847 [DOI] [PubMed] [Google Scholar]
- 36. Lusk CP, Blobel G, King MC. 2007. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8:414–420 [DOI] [PubMed] [Google Scholar]
- 37. Mettenleiter TC. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625 [DOI] [PubMed] [Google Scholar]
- 38. Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mettenleiter TC, Klupp BG, Granzow H. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423–429 [DOI] [PubMed] [Google Scholar]
- 40. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 41. Meyer GA, Radsak KD. 2000. Identification of a novel signal sequence that targets transmembrane proteins to the nuclear envelope inner membrane. J. Biol. Chem. 275:3857–3866 [DOI] [PubMed] [Google Scholar]
- 42. Meyer G, Gicklhorn D, Strive T, Radsak K, Eickmann M. 2002. A three-residue signal confers localization of a reporter protein in the inner nuclear membrane. Biochem. Biophys. Res. Commun. 291:966–971 [DOI] [PubMed] [Google Scholar]
- 43. Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 44. Neubauer A, Rudolph J, Brandmüller C, Just FT, Osterrieder N. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189–204 [DOI] [PubMed] [Google Scholar]
- 45. Nili E, et al. 2001. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J. Cell Sci. 114:3297–3307 [DOI] [PubMed] [Google Scholar]
- 46. Nixdorf R, Klupp BG, Mettenleiter TC. 2001. Role of the cytoplasmic tails of pseudorabies virus glycoproteins B, E and M in intracellular localization and virion incorporation. J. Gen. Virol. 82:215–226 [DOI] [PubMed] [Google Scholar]
- 47. Ohba T, Schirmer EC, Nishimoto T, Gerace L. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olins AL, Rhodes G, Welch DB, Zwerger M, Olins DE. 2010. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus 1:53–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park R, Baines JD. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Popa M, et al. 2010. Dominant negative mutants of the murine cytomegalovirus M53 gene block nuclear egress and inhibit capsid maturation. J. Virol. 84:9035–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reynolds AE, et al. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schirmer EC, Gerace L. 2005. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 30:551–558 [DOI] [PubMed] [Google Scholar]
- 54. Schirmer EC, Florens L, Guan T, Yates JR, III, Gerace L. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301:1380–1382 [DOI] [PubMed] [Google Scholar]
- 55. Shumaker DK, Lee KK, Tanhehco YC, Craigie R, Wilson KL. 2001. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20:1754–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith S, Blobel G. 1993. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J. Cell Biol. 120:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trus BL, et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc. Natl. Acad. Sci. U. S. A. 108:14276–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ye GJ, Roizman B. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. U. S. A. 97:11002–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ye GJ, Vaughan KT, Vallee RB, Roizman B. 2000. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]