Abstract
MITF-M and PAX3 are proteins central to the establishment and transformation of the melanocyte lineage. They control various cellular mechanisms, including migration and proliferation. BRN2 is a POU domain transcription factor expressed in melanoma cell lines and is involved in proliferation and invasion, at least in part by regulating the expression of MITF-M and PAX3. The T361 and S362 residues of BRN2, both in the POU domain, are conserved throughout the POU protein family and are targets for phosphorylation, but their roles in vivo remain unknown. To examine the role of this phosphorylation, we generated mutant BRN2 in which these two residues were replaced with alanines (BRN2TS→BRN2AA). When expressed in melanocytes in vitro or in the melanocyte lineage in transgenic mice, BRN2TS induced proliferation and repressed migration, whereas BRN2AA repressed both proliferation and migration. BRN2TS and BRN2AA bound and repressed the MITF-M promoter, whereas PAX3 transcription was induced by BRN2TS but repressed by BRN2AA. Expression of the BRN2AA transgene in a Mitf heterozygous background and in a Pax3 mutant background enhanced the coat color phenotype. Our findings show that melanocyte migration and proliferation are controlled both through the regulation of PAX3 by nonphosphorylated BRN2 and through the regulation of MITF-M by the overall BRN2 level.
INTRODUCTION
POU family transcription factors are expressed in a wide variety of cell types. They are involved in diverse functions, such as cell type determination, proliferation, renewal, invasion, and migration. The members of the POU domain family of transcription factors share the POU DNA-binding domain (DBD) called the POU domain. The POU domain consists of two DNA-binding units (POUs for POU specific and POUh for POU homeodomain) connected by a flexible linker (3). This molecular structure allows POU proteins to recognize a large set of DNA targets and also to bind different transactivator proteins, depending on the spacing and the positioning adopted by the two subdomains of the POU DBD (50).
POU transcription factor function can be modulated by posttranslational modifications, including sumoylation, oxidation, ubiquitinylation, glycosylation, and particularly phosphorylation (5, 20, 32). Two residues in the DBD domain, a threonine and a serine, are conserved in all mammalian POU domains (see Table S1 at http://umr3347.curie.fr/fr/quipes-de-recherche/d-veloppement-normal-et-pathologie-des-m-lanocytes/differential-function-non-pho). These serine and/or threonine residues in Oct-1, Pit-1, and BRN2 are phosphorylated by protein kinase A (PKA) (31, 46, 55).
Several lines of evidence suggest that PKA is involved in melanocyte lineage proliferation. Forskolin stimulates the proliferation of human melanocytes (54). Proliferation of human melanocytes is induced in a dose-dependent manner by alpha-melanocyte-stimulating hormone (1, 57, 58). Dibutyryl adenosine cyclic AMP induces the proliferation of epidermal melanocytes in culture (29). PKA phosphorylates claudin-1 and allows its translocation to the nucleus in melanoma cells, a process associated with the induction of migration (21). Lastly, PKA phosphorylates serine 362 of BRN2 in vitro (46).
BRN2 and other POU proteins (OCT1, OCT4, and BRN1) are expressed in melanocytes. BRN2, also known as NOCT3 and POU3F2, was originally found to be strongly expressed in neuronal cells. It is involved in the commitment of neurons and central nervous system development and is found in adult brain (26, 44, 52, 61). BRN2 is also expressed in melanocytes, which are derived from the neural crest, and is overexpressed in melanoma cell lines. BRN2 induces proliferation and invasion of the melanocyte lineage (23–25, 48, 59).
The physiological role of BRN2 phosphorylation remains unknown. However, phosphorylation of BRN2 Ser362 by PKA modifies its DNA-binding properties in vitro (46). Three distinct types of BRN2 DNA recognition elements have been characterized: PORE (palindromic Oct recognition element), MORE (more palindromic Oct recognition element) (50), and NORE (N-OCT-3 recognition element) (2). POU family proteins bind these elements in different ways. A common feature of BRN2 binding to PORE and NORE motifs is that the POUh domain adopts a rigid arm anchoring in the minor groove of the DNA (2). Conversely, when the BRN2 DBD binds to a MORE motif, the POUh arm does not bind in the DNA minor groove. The protein-DNA interfaces are different in these three types of complexes. A phosphate group on the Ser362 residue is expected to prevent insertion of the rigid arm of POUh into the DNA minor groove. Indeed, in vitro, phosphorylated BRN2 is unable to bind to either PORE or NORE motifs but is still able to bind to MORE elements (46).
BRN2 positively regulates the transcription of the GADD45a, DELTA1 (NOTCHL), and PAX3 genes and represses the transcription of MITF-M (14, 23, 37, 48, 64). GADD45a, DELTA1, PAX3, and MITF-M are all involved in the proliferation of the melanocyte lineage. This proliferation is regulated by GADD45a only after UVB irradiation. DELTA1/NOTCH signaling plays a crucial role in melanocyte stem cell survival (43, 53). However, DELTA1 has not been demonstrated to be a direct target of BRN2 in the melanocyte lineage. Note that in melanoma cell lines, BRN2 is an activator and, conversely, MITF-M is a repressor of the NOTCH pathway (60). PAX3 is involved in melanoblast proliferation and migration (9, 62) and is expressed at high levels in human cutaneous malignant melanomas but at low level in mature melanocytes (6, 9, 40, 51). PAX3Sp/+ and PAX3IRESnLacZ/+ mice display a white belly spot phenotype and have white tails and paws, implicating PAX3 in melanoblast proliferation and migration in vivo (49). MITF-M is a critical regulator of proliferation and migration, and optimal activity of this protein leads to the most favorable rate of proliferation and migration in a particular molecular context (12, 13). Phosphorylation has differential effects on BRN2 interactions with the various classes of binding element in the promoters of the PAX3 and MITF genes, and both of these genes are involved in the proliferation and migration of melanocytes; these observations provide clues to the mechanisms underlying the phenotypes observed (13, 23, 34, 62).
We report a series of in vitro, in cellulo, and in vivo experiments demonstrating that BRN2 and its phosphorylation are involved in the migration and proliferation of cells of the melanocyte lineage. We show that BRN2 represses melanocyte migration and, depending on its phosphorylation status, represses or activates proliferation. These effects involve BRN2-mediated regulation of MITF-M and PAX3. MITF-M transcription is repressed similarly and in a phosphorylation-independent manner by both BRN2TS and BRN2AA, whereas PAX3 transcription is induced by BRN2TS but repressed by BRN2AA.
MATERIALS AND METHODS
BRN2 phosphorylation.
Recombinant full-length BRN2 and DBD were produced and purified as described in reference 11. Nuclear extracts were obtained as described in reference 45. Purified proteins were incubated for 60 min at 30°C at a concentration of 10 μM in the presence of 100 μM ATP and either 50 U/ml of the catalytic subunit of PKA from bovine heart (Sigma) or 1 μg of a nuclear extract from Lyse human melanoma cells in buffer B (25 mM Tris-HCl [pH 8.0], 5% glycerol, 100 mM NaCl, 125 μg/ml bovine serum albumin [BSA], 1 mM EGTA, 5 mM MgCl2). For radiolabeling experiments, 1 μCi of [γ-32P]ATP was added.
Plasmid constructs.
Murine BRN2 cDNA was obtained by PCR with a genomic clone (28) using primers that generate BamHI sites at each end. BRN2AA cDNA was prepared using an in vitro site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Around nucleotides 1084 (a) and 1087 (t), the sequence of BRN2 is GAAAAAGCGGACCTCCATCGAGGTG and that of BRN2AA is GAAAAAGCGGGCCGCAATTGAGGTG (changes are underlined). The oligonucleotides used resulted in the replacement of threonine and serine with alanines (T361A and S362A). BamHI fragments carrying BRN2TS (wild type [WT]) and BRN2AA were inserted into the BamHI site of the multiple cloning site in pEGFP-C1 (a C-terminal fusion vector; Clontech) to produce pEGFP-BRN2TS (no. 896) and pEGFP-BRN2AA (no. 898). A BglII fragment encoding the 13 amino acids of the SV5 epitope (27) was inserted into the BamHI site at the 5′ end of the BRN2TS and BRN2AA fragments. These cDNAs were used to generate constructs for obtaining transgenic mice. The mouse tyrosinase gene enhancer (Enh) was fused to the promoter region (Tyr prom) to produce a 6.1-kb regulatory element driving the expression of a cDNA encoding a WT form (BRN2TS) or a mutated form (BRN2AA) of BRN2. A simian virus 40 (SV40) small T antigen splice site and polyadenylation sequence were added to the 3′ end of the constructs to produce Tyr::BRN2TS (no. 705) and Tyr::BRN2AA (no. 706). BRN2TS, BRN2AA, and BRN2 DBD were introduced into appropriate plasmids to produce recombinant proteins as previously described (11). The MITF-M and PAX3 promoters were provided by R. Ballotti and J. Epstein and have been described previously (36, 41).
Cell culture and luciferase assays.
The mouse melanocyte cell line melan-a was kindly provided by D. Bennett (7). These cells were maintained in F12 medium supplemented with 10% fetal calf serum, 5 mM l-glutamine, and 200 nM tetradecanoyl phorbol acetate (Sigma). Lyse human melanoma cells were cultured in RPMI 1640 medium containing 10% fetal calf serum and 5 mM l-glutamine (42). The method for culturing primary melanocytes is described elsewhere (18, 35). BRN2AA, BRN2TS, and WT melanocytes were established in culture, and the number of melanocytes was estimated weekly under a microscope. All cells were grown in the presence of antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) and incubated at 37°C in a humidified atmosphere with 5% carbon dioxide. Cells at 70% confluence were transiently transfected in six-well plates, using 3 μl of Lipofectamine 2000 (Invitrogen), Opti-MEM medium (Gibco), and 1.5 μg (melan-a cells) or 2 μg (Lyse cells) of total plasmid DNA. Cells were cotransfected with the thymidine kinase::Renilla luciferase construct as a control. The amount of DNA was equalized with pBluescript. We determined luciferase activity and Renilla luciferase activity 48 h after transfection. Luciferase activity was normalized against Renilla luciferase activity.
Cell motility.
melan-a murine melanocytes were seeded into six-well plates (3 × 105/well) on day zero. On day 1, cells were transfected with 250 ng of the expression vector Brn2TS-GFP or Brn2AA-GFP or the empty green fluorescent protein (GFP) expression vector (equimolar) as a control. On day 2, the cells were seeded sparsely (5 × 104/well) to minimize cell-cell interactions. On day 3, single-cell migration analysis was performed using an inverted fully automated microscope (Leica DM IRB; Leica Microsystems) equipped for fluorescence imaging with a 20× lens; the cells were placed in an environmental chamber with 5% CO2 in a humidified atmosphere at 37°C (Life Imaging Services). Time-lapse recording started 48 h after transfection. Fluorescence and phase-contrast images were collected at 4-min intervals for 12 h with a charge-coupled device camera (CoolSNAP fx; Photometrics) operated by Metamorph software (Molecular Devices). Cell migration was analyzed by manually tracking individual cells in fluorescence frames using the ImageJ software and a manual tracking plugin (http://rsb.info.nih.gov/ij/plugins/track/track.html).
Proliferation test.
Cells were seeded onto coverslips at 7 × 104/cm2 and cultured overnight. On day 1, cells were transfected with 25 ng of the expression vector Brn2TS-GFP or Brn2AA-GFP or the empty GFP vector (equimolar) as a control. On day 3, cells were pulsed with 10 μM bromodeoxyuridine (BrdU; BD Biosciences) for 2 h. The cells were then fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 10 min at room temperature. Samples were blocked with 5% (vol/vol) donkey serum in 1× PBS for 30 min at room temperature and treated with 300 μg/ml DNase I (Sigma-Aldrich) for 1 h at 37°C. Cells were probed with a mouse polyclonal anti-BrdU antibody diluted 1/150 (BD Biosciences) and a rabbit monoclonal anti-GFP antibody diluted 1/500 (Abcam) for 12 h at 4°C. Alexa Fluor 555–anti-mouse and Alexa Fluor 488–anti-rabbit antibodies, respectively, were added, and the samples were incubated for 1 h (22).
Transgenic mouse generation and analysis.
Transgenic mice were generated with Tyr::BRN2TS and Tyr::BRN2AA constructs as described previously (19, 38). Tyr::BRN2TS and Tyr::BRN2AA mice were identified by PCR analysis of DNA prepared from tail biopsy specimens collected at weaning (see Table S2 at the URL mentioned in the introduction). The primers used were LL1463, corresponding to a 3′ part of BRN2, and LL1165, corresponding to the SV40 fragment; they generate a fragment of 215 bp of DNA from both Tyr::BRN2TS and Tyr::BRN2AA mice. LL1477, corresponding to the sequence around the TS→AA mutation, and LL1165 were used to screen Tyr::BRN2AA mice and gave a fragment of 212 bp. Transgene expression was detected in several founder mice by reverse transcription (RT)-PCR with RNA isolated from skin biopsy samples. Mice were crossed with Dct::LacZ mice (39), and the resulting embryos were collected at embryonic day 13.5 (E13.5) and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as previously described (8, 19). The transgenic Mitfmivga9, Pax3IRESnLacZ, Tyr::Cre, and ZEG mice used were described previously (16, 19, 30, 47, 49). Transgenic mice were repeatedly backcrossed to C57BL/6 mice and maintained for up to 18 months. We determined the number of melanoblasts and melanocytes in vivo and ex vivo as reported elsewhere (8, 18, 38). All animals were housed under specific-pathogen-free conditions at the Institut Curie in conformity with French and European Union laws.
RNA extraction and semiquantitative real-time PCR.
Isolation of embryonic melanoblasts and extraction of their RNA were done as described elsewhere (15). mRNA was analyzed by semiquantitative real-time PCR using an iCycler (Bio-Rad). For each sample, 2 μl of cDNA was used in a reaction volume of 25 μl containing 2× IQ SYBR green Supermix (Bio-Rad) and 300 nM primers for hypoxanthine phosphoribosyltransferase (HPRT), MITF-M, PAX3, BRN2, and LacZ (see Table S2 at the URL mentioned in the introduction). All samples were analyzed in triplicate. The thermal profile was 95°C for 1 min 30 s and 40 cycles of 95°C for 30 s and 60°C for 1 min. Results were standardized to the results for HPRT mRNA. RNA extraction from WT and mutant skin biopsy specimens was done as described elsewhere (42). Real-time quantitative RT-PCR (qRT-PCR) analyses for the mRNAs for the transgenes Tyr::BRN2TS and Tyr::BRN2AA (LL1463 and LL302) and HPRT (LL1615 and LL1616) (see Table S2) were performed with a Bio-Rad iCycler iQ multicolor real-time PCR detection system. Each 25-μl reaction mixture consisted of 2 μl of cDNA, 1× iQ SYBR green Supermix (Bio-Rad), 600 nM Tyr::BRN2 transgene, and HPRT primers. The amount of the target transcript was related to that of a reference gene (HPRT) by the ΔCT (threshold cycle) method. Each sample was assayed in triplicate. For each mouse line, at least two skin biopsy specimens or three to five independent melanoblast preparations were tested.
Electrophoretic mobility shift assays (EMSAs).
EMSAs for BRN2 were done as described in reference 46. The sense DNA oligonucleotides used in the assays are reported in Table S2 at the URL mentioned in the introduction.
DNase I footprinting.
Two vector plasmids carrying the −2135 to +136 region of the MITF promoter or the kb −1.6 region of the PAX3 promoter were used in PCR experiments to amplify a 236-bp fragment of MITF (−226 to +10, MITF-M promoter region) and two fragments of 146 bp (−1397 to −1251, PAX3 promoter region) and 148 bp (−944 to −796, PAX3 promoter region) of PAX3. For each fragment, two PCRs were performed in parallel using both sense and antisense primers. In the first PCR, the sense primer was 32P end labeled, whereas in the second PCR, the antisense primer was labeled. The radiolabeled DNA fragments were incubated at 28°C for 30 min with a series of amounts of BRN2 in 20 μl binding buffer (25 mM Tris-HCl [pH 8], 100 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2 2 mM dithiothreitol, 5% glycerol, 125 μg/ml BSA, 0.004% Tween 20). DNase I (Roche) was added to a concentration of 1 U/ml, and the incubation was continued for 2 min at 28°C. Next, 5 μl of DNase I stop solution (1.5 M sodium acetate, 250 mM EDTA [pH 8.0], 120 μg/ml tRNA) was added. The mixture was treated with phenol, and DNA-protein complexes were precipitated with ethanol. The products were analyzed by 10% PAGE containing 7 M urea. The gel was vacuum dried and autoradiographed.
Chromatin immunoprecipitation (ChIP) experiments.
Lyse melanoma cell lines were treated for 2 h with 10 μM forskolin or 0.5 μM staurosporine. ChIP experiments were performed with a Diagenode Transcription factor ChIP kit according to the manufacturer's protocol. Anti-BRN2 antibody (sc-6029) was from Santa Cruz. The nonrelevant antibody used was an anti-Flag antibody (29674)from AnaSpec. Anti-TATA-binding protein (anti-TBP) antibody and c-fos and exon Myo oligonucleotides were provided in the ChIP kit. The Mitf and Pax3 oligonucleotides used are shown in Table S2 at the URL mentioned in the introduction. PCR was performed as described above.
RESULTS
Phosphorylation of BRN2 differentially affects migration and proliferation in cellulo.
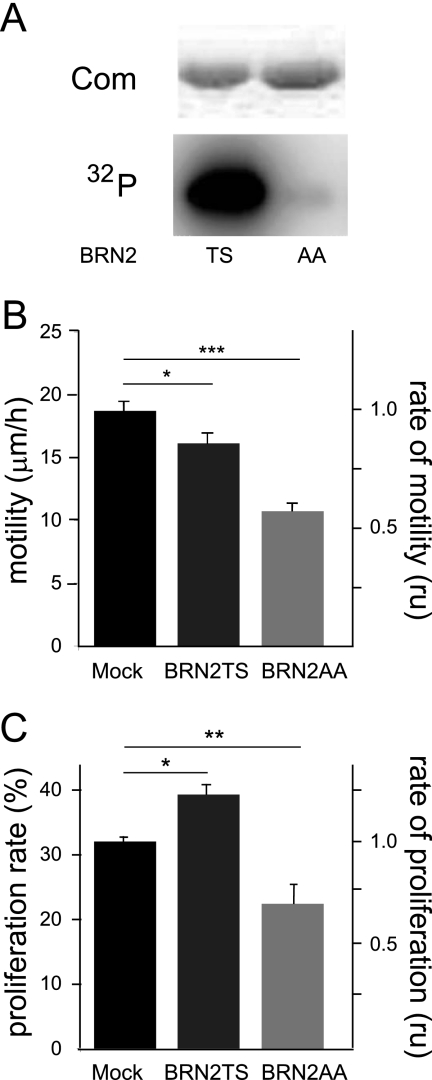
To study the role of the phosphorylation of residues Thr361 and Ser362 in BRN2 in the melanocyte lineage, these two residues were replaced with alanines (BRN2TS→BRN2AA). The WT protein (here named BRN2TS) and mutated protein (here named BRN2AA) produced in bacteria were incubated in the presence of [γ-32P]ATP and purified PKA. The phosphorylated form of BRN2TS was detectable, but BRN2AA was not phosphorylated (Fig. 1A); the mutation of these two residues is thus sufficient to inhibit BRN2 phosphorylation by PKA.
Fig 1.
Phosphorylation of BRN2 affects cell migration and proliferation in cellulo. (A) Analysis of BRN2 phosphorylation profile. BRN2TS and BRN2AA were produced in bacteria, purified, and incubated with [γ-32P]ATP in the presence of purified PKA. The proteins were analyzed by SDS-PAGE. Com, Coomassie staining. (B) In cellulo motility of murine melan-a melanocytes. Cells were transfected with GFP-coupled BRN2TS and BRN2AA expression vectors. The empty GFP expression vector (Mock) was used as a control. Forty-eight hours after transfection, the cells were subjected to a single-cell migration assay using two-dimensional video microscopy for 12 h and cell motility was determined. Each experiment was performed at least three times. The migration speeds were calculated as means ± standard deviations using Excel (Microsoft). Student's t test was used to compare migration speeds between groups. *, P < 0.01; ***, P < 0.001. (C) The cell proliferation rates of Lyse melanoma cells after 48 h of BRN2TS and BRN2AA expression were estimated by following BrdU incorporation into transfected cells over 2 h. Experiments were performed at least three times. The proliferation rates were calculated as means ± standard deviations using Excel (Microsoft). Student's t test was used to compare migration speeds. *, P < 0.01; **, P < 0.005. Similar results were obtained with all of the cell lines tested.
To determine the effects of BRN2TS and BRN2AA on migration and proliferation in the melanocyte lineage, two murine melanocyte cell lines (melan-a and 9v) and Lyse human melanoma cells were transfected with expression vectors encoding GFP (mock transfection), BRN2TS-GFP, or BRN2AA-GFP. Forty-eight hours later, the motility of transfected melan-a and 9v cells was determined using video microscopy and the average migration speed was recorded (Fig. 1B; see Fig. S1 at the URL mentioned in the introduction). The migration speed of cells expressing BRN2TS was slightly, but significantly, slower than that of mock-transfected cells; the motility of BRN2AA-transfected cells was substantially lower than that of mock-transfected cells. The effects of BRN2TS and BRN2AA on the proliferation of Lyse melanoma cell lines were assessed. Forty-eight hours after transfection, cells were labeled with BrdU for 2 h. The cells were fixed and immunostained, and GFP and BrdU staining was assessed. The rate of proliferation was evaluated from the proportions of GFP-positive cells that were BrdU positive and negative (Fig. 1C). These results were confirmed in two murine melanocyte cell lines (see Fig. S2 at the URL mentioned in the introduction). Under these conditions, BRN2TS significantly enhanced, and BRN2AA reduced, cell proliferation. Thus, BRN2TS induced proliferation and BRN2AA reduced proliferation and migration of melanocytes and melanoma cells in cellulo.
BRN2 is not expressed during melanoblast development.
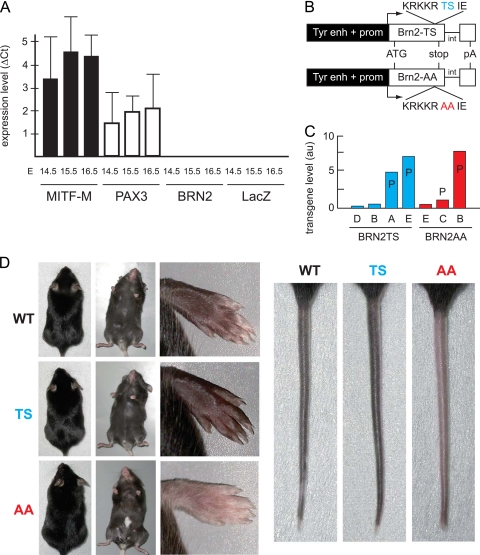
To evaluate the effects of BRN2TS and BRN2AA, we looked for a cell type that (i) belongs to the melanocyte lineage, (ii) does not produce endogenous BRN2, and (iii) is PKA sensitive. We could not find such cells among murine or human melanoma cell lines, as all express BRN2, albeit at a low level in some cases. We previously showed that BRN2 was not detectable before E11.5 in mouse embryos but is present in the roof plate at the time of delamination of neural crest cells (24). To evaluate BRN2 expression in melanoblasts in vivo, we isolated a pure population of melanoblasts. In this respect, we generated transgenic (Tyr::Cre/°; ZEG/ZEG) mice producing fluorescent melanoblasts after specific recombination of the ZEG locus in cells expressing tyrosinase (Tyr) and LacZ in all other cells, including keratinocytes (16). GFP-positive cells were isolated from the epidermis of Tyr::Cre/°; ZEG/ZEG embryos (15). qRT-PCR analysis demonstrated that MITF-M and PAX3 were expressed in melanoblasts at E14.5 to E16.5, but no BRN2 was detected (Fig. 2A). In situ hybridization experiments using BRN2 probes confirmed these observations (data not shown). These findings indicate that BRN2 is not expressed in melanoblasts during embryogenesis, at least until E16.5.
Fig 2.
BRN2TS mice are hyperpigmented, and BRN2AA mice are hypopigmented. (A) BRN2 is not produced in melanoblasts during embryonic development. Skin melanoblasts were isolated, as enhanced-GFP-positive cells, by fluorescence-activated cell sorter from Tyr::Cre/°; ZEG/ZEG mice from E14.5 to E16.5. RNA was extracted, and qRT-PCR was used to assay the mRNAs of endogenous MITF-M, PAX3, BRN2, and LacZ using HPRT as an internal reference. The experiments were performed four or five times for each embryonic stage. For each stage, three Tyr::Cre/°; ZEG/ZEG embryos were used. Tyr::Cre/°; °/° and °/°; ZEG/ZEG embryos do not produce any enhanced GFP (not shown). Note that LacZ was used as internal control and nondefloxed cells produce this reporter mRNA. qRT-PCR with C57BL/6 mouse brain samples was used as a positive control for BRN2 (not shown). (B) Map of the Tyr::BRN2TS and Tyr::BRN2AA transgenes. int, intron of SV40; enh, enhancer; prom, promoter; pA, polyadenylation site of SV40. (C) Expression of BRN2TS or BRN2AA in various Tyr::BRN2 transgenic mouse lines. P indicates that the mice of these lines present a phenotype. The transgenes were similarly strongly expressed in Tyr::BRN2TS-E and Tyr::BRN2AA-B mice. These two lines are directly compared with each other. au, arbitrary unit. (D) Phenotype of WT, Tyr::BRN2TS-E (TS), and Tyr::BRN2AA-B (AA) mice. The mice do not present any phenotype on the dorsal side. On the ventral side, AA mice present a white belly spot. The paws and tails of TS mice are darker and those of AA mice are lighter than those of WT mice. The gray intensity of paws and tails was estimated (arbitrary relative scale) as 1 and 1 in WT mice, as 1.2 and 1.1 in Tyr::BRN2TS mice, and as 0.6 and 0.8 in Tyr::BRN2AA mice, respectively.
BRN2 expression in melanocytes affects coat color.
To determine the effects of BRN2TS and BRN2AA in the melanocyte lineage in vivo, we generated transgenic mice producing the WT form of BRN2 (Tyr::BRN2TS mice) and a mutated form of BRN2 (Tyr::BRN2AA mice) in committed cells of the melanocyte lineage (Fig. 2B). The tyrosinase (Tyr) promoter becomes active at around E10.5 (19). The expression of BRN2TS and BRN2AA at this stage has the advantage of evaluating the effects of these proteins in the absence of endogenous BRN2. BRN2TS and BRN2AA transgenes were detected in several founder mice by RT-PCR.
Mouse lines weakly expressing the BRN2TS (bars B and D) and BRN2AA (bar E) transgenes did not present any observable phenotype (Fig. 2C). In contrast, mouse lines strongly expressing the BRN2TS (bars A and E) and BRN2AA (bars C and B) transgenes displayed a coat color phenotype; Tyr::BRN2TS (TS) animals showed significant overall hyperpigmentation of the paws and tail (Fig. 2D). This phenotype was seen in around 30% of animals, and therefore penetrance was not complete. Tyr::BRN2AA (AA) animals presented a white belly spot (50% penetrance) and lower-than-WT pigmentation of the paws and tail (fully penetrant) (Fig. 2D). Tyr::BRN2TS and Tyr::BRN2AA mice did not show any hair greying when adult and did not develop melanoma.
BRN2AA and BRN2TS affect melanoblast proliferation.
To study the effects of BRN2AA and BRN2TS further, we compared mouse lines producing similar amounts of mRNA from the transgenes (Tyr::BRN2TS-E and Tyr::BRN2AA-B) in the same genetic background (C57BL/6).
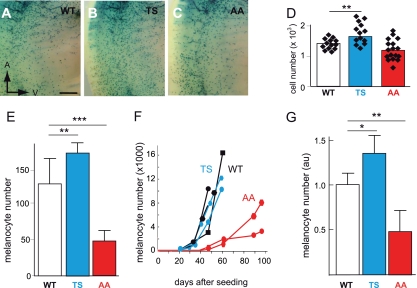
The number of melanoblasts in the trunk region of embryos at E13.5 was determined by crossing Dct::LacZ/° mice (LacZ), which carry a β-galactosidase reporter under the control of the melanocytic-specific Dct promoter (39), with Tyr::BRN2TS-E/° (BRN2TS), Tyr::BRN2AA-B/° (BRN2AA), and control mice to produce Tyr::BRN2TS-E/°; Dct::LacZ/° (BRN2TS-LacZ), Tyr::BRN2AA-B/°; Dct::LacZ/° (BRN2AA-LacZ), and °/°; Dct::LacZ/° (WT-LacZ) mice. The number of melanoblasts in BRN2TS-LacZ embryos was slightly higher than that in WT-LacZ embryos; that in BRN2AA-LacZ embryos was lower than that in the controls (Fig. 3A to D). The standard errors for the BRN2TS-LacZ and BRN2AA-LacZ groups were larger than that for the WT-LacZ group, with the higher and lower limits, respectively, differing from those for WT-LacZ. Such results may explain the partial penetrance of the phenotype. The more numerous melanoblasts in BRN2TS than in control embryos was due to a higher proliferation rate and was independent of apoptosis (see Fig. S3 at the URL mentioned in the introduction).
Fig 3.
In vivo melanoblast proliferation is induced by BRN2TS and repressed by BRN2AA. (A to D) Number of melanoblasts in WT-Lac, BRN2TS-LacZ (TS), and BRN2AA-LacZ (AA) embryos at E13.5 in the trunk region between the front and back limbs, from somites 13 to 25. Melanoblasts were genetically labeled using Dct::LacZ and identified by eye as X-Gal-positive cells. The number of melanoblasts was estimated to be 1,388 ± 183 for WT embryos (14 independent embryo sides), 1,750 ± 350 for TS embryos (14 independent embryo sides), and 1,201 ± 340 for AA embryos (17 independent embryo sides). The P value is 0.003 for WT versus TS mice (**) and 0.08 for WT versus AA mice. The anterior (A) and ventral (V) orientations of embryos are indicated in panel A. Note that the intensity of X-Gal staining is similar in WT and mutant melanoblasts and that the level of expression of Dct is slightly affected, but it is not for Tyr and Tyrp1, by the expression of BRN2TS or BRN2AA (see Fig. S7 at the URL mentioned in the introduction) (E) Numbers of WT, TS, and AA melanocytes 5 days after the explantation of newborn skin. Similar numbers of cells isolated from the skin of 3-day-old pups were grown in culture. The number of melanocytes was estimated in six independent cultures of cells from WT pups, four independent cultures of cells from TS pups, and eight independent cultures of cells from AA pups. StatView software was used for statistical analysis. **, P < 0.01; ***, P < 0.001. (F) Primary melanocyte growth curves. Melanocytes were established in culture from two independent WT (black), two independent TS (blue), and two independent AA (red) new born pups, and growth curves were determined from the numbers of pigmented cells. Each point is derived from the mean count of melanocytes from duplicate dishes. On day zero, 106 cells containing a mixture of melanocytes, keratinocytes, and fibroblasts were seeded for each WT, TS, and AA mouse. (G) Numbers of X-Gal-positive cells estimated from at least three independent WT-LacZ, TS-LacZ, and AA-LacZ tails. au, arbitrary units. StatView software was used for statistical analysis: *, P < 0.05; **, P < 0.01.
Primary melanocytes were established in culture from truncal skin collected after birth from WT, BRN2TS, and BRN2AA mice. The growth of WT, BRN2TS, and BRN2AA melanocytes was evaluated after various times. After the short period of growth, the number of BRN2AA melanocytes was about one-third and the number of BRN2TS melanocytes was slightly higher (30%) than for WT melanocytes (Fig. 3E). After a longer period, the overall growth of BRN2AA melanocytes was delayed relative to that of WT melanocytes (Fig. 3F); in contrast, there was no significant difference in growth between WT and BRN2TS melanocytes.
The BRN2TS phenotype may be the result of more melanocytes in the tail and/or more melanin per melanocyte. Similarly, the BRN2AA phenotype may reflect fewer melanocytes, poorer migration, and/or less melanin per melanocyte than in controls. The amount of melanin in hair is a marker of the production of melanin per melanocyte. We did not observe any difference in the amount of melanin in BRN2TS or BRN2AA hairs from that in hairs from WT littermates (see Fig. S4A at the URL mentioned in the introduction). The density of melanocytes in the tail was higher in BRN2TS mice than in WT mice (30% higher) and in WT mice than in BRN2AA mice (50% higher) (Fig. 3G; see Fig. S4B to D at the URL mentioned in the introduction). Therefore, the differences in pigmentation in the paws and tails are due mainly to the different numbers of melanocytes. These experiments show, both in mice and in culture, that BRN2TS induced proliferation, whereas BRN2AA repressed proliferation. Phosphorylation of BRN2 on Thr361 and/or Ser362 thus appears to be required for BRN2 to exert a positive effect on the proliferation of melanoblasts and melanocytes.
BRN2TS and BRN2AA have different actions on PAX3 transcription but not on MITF-M transcription.
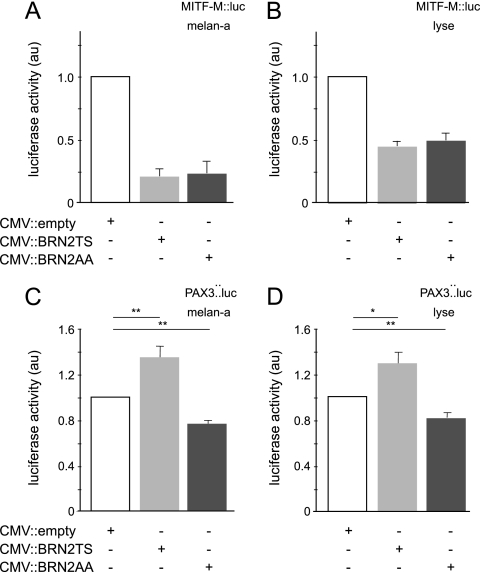
To understand the differential regulation of proliferation by BRN2TS and BRN2AA in the melanocyte lineage, we investigated four known BRN2 targets: GADD45a, DELTA1, MITF-M, and PAX3 (14, 23, 37, 64). Murine melan-a melanocytes and human Lyse melanoma cells were cotransfected with an expression vector encoding either BRN2TS or BRN2AA or mock transfected (cytomegalovirus [CMV]::empty) and reporter constructs containing the promoters of interest driving the luciferase reporter gene. Neither BRN2TS nor BRN2AA acted as an activator or a repressor of the GADD45a or the DELTA1 promoter (data not shown). However, in both melan-a and Lyse cells, luciferase assays revealed that BRN2TS and BRN2AA similarly repressed the MITF-M promoter (Fig. 4A and B). In contrast, BRN2TS induced the transcription of PAX3, whereas this promoter was repressed by BRN2AA (Fig. 4C and D). The replacement of the TS residues in BRN2 with AA residues therefore did not affect the protein's ability to repress MITF-M transcription. However, phosphorylation appears to be required for activation of PAX3 transcription, whereas in the absence of phosphorylation, repression is observed. We confirmed the previous observation (23) that mutation of the BRN2 target associated with MITF-M abolished BRN2 regulation. Similarly, mutations in the PAX3 promoter slightly affected activation by BRN2 (see Fig. S5A and B at the URL mentioned in the introduction).
Fig 4.
BRN2TS and BRN2AA have different effects on the PAX3 promoter, but not on the MITF-M promoter. melan-a (A and C) and Lyse (B and D) cells were transfected with a control (CMV::empty) or a CMV::BRN2TS or CMV::BRN2AA expression vector and a MITF-M::luciferase (A and B) or a PAX3::luciferase (C and D) reporter vector. StatView software was used for statistical analysis. *, P < 0.05; **, P < 0.01. au, arbitrary unit.
Phosphorylation regulates BRN2 binding to the MITF-M and PAX3 promoters.
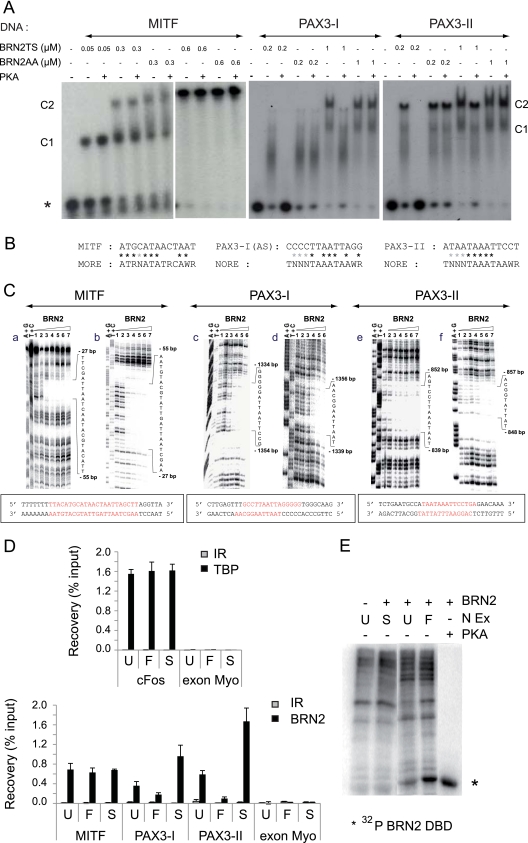
BRN2 directly regulates the MITF-M promoter by interacting with a sequence in the proximal promoter (23). To determine whether BRN2 phosphorylation modifies its interaction with this element, we performed in vitro DNA-binding assays; the assays involved an oligonucleotide probe spanning this element and BRN2TS and BRN2AA proteins produced and purified from Escherichia coli. Aliquots of the BRN2TS and BRN2AA preparations were incubated with purified PKA and ATP, and then EMSAs were carried out using 32P-radiolabeled oligonucleotides corresponding to the WT and mutated MITF-M promoter sequences and phosphorylated or nonphosphorylated BRN2TS and BRN2AA.
There were no significant differences in BRN2-DNA complex formation between the phosphorylated and nonphosphorylated forms and the WT MITF-M promoter sequence (Fig. 5A, lanes 1 to 11). Mutation of the interaction site abolished the binding of BRN2 (see Fig. S5C at the URL mentioned in the introduction), as previously shown (23). Phosphorylation of BRN2 on the Thr361 and/or Ser362 residues therefore does not modify the interaction between BRN2 and the MITF-M target. The BRN2-binding site 5′-ATGCATAACTAAT-3′ (located at −51 to −39) in the MITF-M promoter (23) matches a degenerate MORE motif (Fig. 5B). To confirm the nature of the BRN2 recognition site, DNase I footprinting experiments were carried out. The binding of the first monomer (Fig. 5C, parts a and b) protected both strands, with a clear footprint visible over the recognition sequence, confirming that this recognition site is a MORE-type target. These findings are in good agreement with the similar regulation of the MITF-M promoter by BRN2TS and BRN2AA.
Fig 5.
The nonphosphorylated form of BRN2 binds NORE elements in the PAX3 promoter, and phosphorylated/nonphosphorylated forms of BRN2 bind MORE elements in the MITF-M promoter. (A) BRN2 phosphorylation reduced its affinity for PAX3 but not for MITF-M targets in vitro. Radioactively labeled probes (10 nM) MITF-M, PAX3-I, and PAX3-II were incubated with various concentrations of recombinant BRN2TS or BRN2AA protein pretreated with PKA or not pretreated. Samples were analyzed by nondenaturing 8% acrylamide gel electrophoresis. Results are representative of at least three independent experiments. Under our conditions, the protein was bound to NORE and MORE sequences as monomers (C1) and as dimers (C2). The phosphorylation of BRN2TS and BRN2AA by PKA was evaluated for each independent experiment by mass spectrometry (not shown) and indirectly as shown in Fig. 1A. The asterisk indicates the labeled free probe. (B) The BRN2-binding sequences in the MITF-M and PAX3 promoters match degenerate MORE and NORE sequences. Asterisks indicate matches. (C) DNase I footprinting analysis of the BRN2-DNA interaction indicates that the MITF-M target is a MORE sequence whereas the PAX3 targets are NORE sequences. Autoradiograms of 12% polyacrylamide denaturing gels showing the DNase I footprints on the sense (a, c, and e) and complementary (b, d, and f) strands of one MITF-M and two PAX3 promoter fragments. Lanes A+G and T+C, Maxam-Gilbert chemical sequencing references. Lane 1, free DNA cleavage products. Lanes 2 to 7, purified BRN2 protein at 1, 5, 20, 50, 75, and 225 nM, respectively. Sequences of the MITF-M and PAX3 promoters with the BRN2-binding sites in red. (D) ChIP assays of BRN2 binding to the Mitf and Pax3 promoters. Lyse melanoma cells were either left untreated (U) or treated with forskolin to induce PKA (F) or with staurosporine to repress PKA (S). The samples were subjected to ChIP assays with antibodies against BRN2 or TBP or irrelevant (IR) antibodies and analyzed by qRT-PCR using primers specifically encompassing the BRN2-binding site. The c-fos promoter was used to normalize the assay, and a myoglobin exon was used as a negative control. All of the data shown are representative of a minimum of three independent assays. (E) Forskolin (F) and staurosporine (S) control the level of phosphorylation of BRN2. In the presence of [γ-32P]ATP, the BRN2 DBD produced from E. coli (BRN2) was incubated with nuclear extracts (N Ex) obtained from Lyse melanoma cells, which were either left untreated (U) or treated with forskolin or staurosporine. Phosphorylation of the BRN2 DBD by purified PKA was used as a positive control. Untreated N Ex lacking the BRN2 DBD was used as a negative control. The radiolabeled proteins were analyzed by 12% SDS-PAGE and autoradiography. Phosphorylation of the BRN2 DBD was highest when the cells were treated with forskolin and lowest when the cells were treated with staurosporine.
Two putative BRN2-binding sites (PAX3-I and PAX3-II) have been mapped in the PAX3 promoter (64). These sites match the NORE consensus sequence imperfectly (Fig. 5B). The interaction between BRN2 and sequences spanning the putative binding sites in the PAX3 promoter was tested by EMSA and footprinting experiments analogous to those described above for MITF-M. Mutation of these two interaction sites abolished the binding of BRN2 (see Fig. S5C at the URL mentioned in the introduction). EMSA showed that PKA phosphorylation significantly reduced the affinity of BRN2TS for the PAX3-I and PAX3-II sequences (Fig. 5A, lanes 12 to 29) but had no effect on the binding of BRN2AA to these sites. Footprinting experiments indicated that, for both PAX3 target elements, the binding of the first monomer protected mainly the sense strand (Fig. 5C, parts c to f) and the binding of a second molecule was necessary for complete protection of the complementary strand; these findings are characteristic of a NORE motif (2).
To confirm the involvement of BRN2 phosphorylation in selective DNA binding in cellulo, ChIP assays were performed using Lyse melanoma cells (Fig. 5D). Significant occupancy of the MITF-M and both of the PAX3 target sites was detected using an anti-BRN2 antibody, whereas no signal was observed using nonspecific IgG as a control. In an additional negative-control experiment, no BRN2 was detected bound to the irrelevant myoglobin exon region. We tested whether phosphorylation by PKA affected promoter occupancy by BRN2; cells were treated with forskolin, a stimulator of PKA activity, or staurosporine, an inhibitor of PKA activity, before ChIP assays. ChIP with anti-TBP antibody and amplification with c-fos core promoter oligonucleotides were done to normalize the assay (Fig. 5D; see Fig. S6 at the URL mentioned in the introduction). As predicted from the EMSAs, BRN2 occupancy of the PAX3 promoter was increased in staurosporine-treated cells, compared to untreated cells, whereas occupancy was reduced by forskolin treatment. These treatments did not significantly affect the occupancy of the MITF promoter site. To test the efficacy of these treatments to modify phosphorylation of BRN2, the purified recombinant BRN2 DBD was incubated with nuclear extracts from untreated cells and forskolin- or staurosporine-treated cells in the presence of [γ-32P]ATP (Fig. 5E). BRN2 DBD was used rather than full-length BRN2 protein because several sites in the N-terminal domain of BRN2 are phosphorylated by nuclear extracts (data not shown). As expected, BRN2 DBD phosphorylation by forskolin-treated cell extracts was greater and that by staurosporine-treated cell extracts was weaker than that by control extracts. These various findings confirm that BRN2 binding to its sites in the PAX3 promoter is modulated by the DBD phosphorylation status, probably due to PKA, whereas its binding to the MITF-M site is unaffected by phosphorylation.
In conclusion, BRN2 phosphorylation does not modify its interaction with, or its ability to repress, the MITF-M promoter but modifies its interaction with the PAX3 promoter, leading to differential regulation of PAX3 by BRN2TS and BRN2AA.
Enhancement of the coat color phenotypes of BRN2TS and BRN2AA mice on PAX3- or MITF-M-deficient backgrounds.
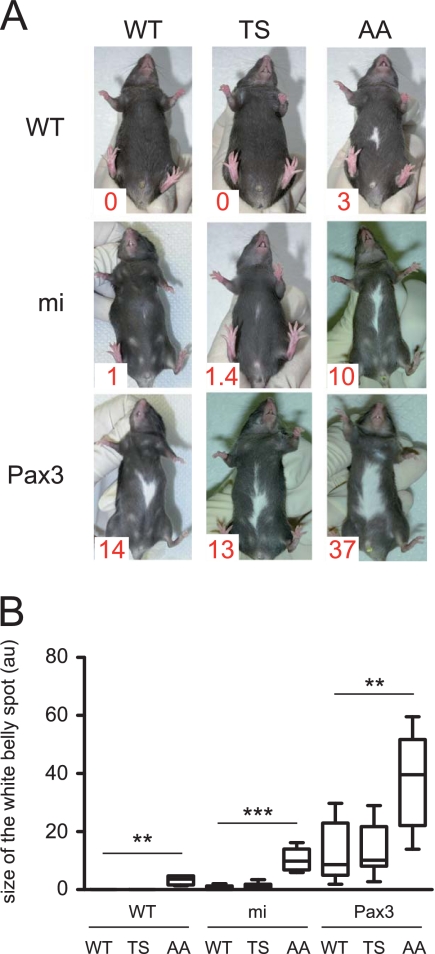
The phenotypes of Tyr::BRN2AA mice (mild hypopigmentation of the paws and tail and a white belly spot) may be a consequence of the repression of the expression of PAX3 and MITF. The phenotypes of Tyr::BRN2TS mice (mild hyperpigmentation of the paws and tail and no white belly spot phenotypes) may reflect the results of antagonistic signaling through PAX3 and MITF. To evaluate the importance of these signals and the potential genetic cooperation of BRN2 with PAX3 and MITF in melanocytes, we crossed Tyr::BRN2TS/° (TS) and Tyr::BRN2AA/° (AA) mice with Mitfvga9/+ (mi) and Pax3IRESnLacZ/+ (Pax3) mice and evaluated white belly spot size as a measure of the migration and proliferation of melanoblasts.
The size of the white belly spot of BRN2AA mice in the mi and Pax3 backgrounds was larger than in the parental controls (10, 37, and 3 arbitrary units, respectively) (Fig. 6; see Table S3 at the URL mentioned in the introduction). These results are in good agreement with the molecular (direct repression of MITF and PAX3 expression) and cellular (repression of proliferation and migration) results presented above. In contrast, the size of the white belly spot of BRN2TS mice in the mi and Pax3 backgrounds is not greater. These results confirm the antagonistic function found for BRN2TS at the molecular (direct transcriptional repression of MITF and activation PAX3) and cellular (activation of proliferation and repression of migration) levels.
Fig 6.
BRN2 cooperates with MITF and PAX3 to determine the size of the white belly spot in vivo. (A) WT, Tyr::BRN2TS/° (TS), and Tyr::BRN2AA/° (AA) mice were crossed with Mitfmiva9/+ (mi) or Pax3IRESnLacZ/+ (Pax3) mice. The area of the white spot was determined as a percentage of the total area of the belly with ImageJ software and is expressed in arbitrary units (au); 100 au means that the entire belly is white, and 0 au means that the entire belly is black. The mean area of the white belly spot is shown in red at the bottom left corner of each photograph. (B) Box-and-whisker plot of white belly spot sizes. Each horizontal bar represents the median, and the limits of each rectangle correspond to the first and third quartiles. The ends of the error bars represent the 5th and 95th centiles. **, P < 0.01; ***, P < 0.001.
Overall, these diverse approaches provide several lines of evidence to suggest that (i) MITF-M involved in proliferation and migration is regulated in vivo by the total amount of BRN2 and (ii) PAX3 involved in proliferation and migration is controlled by both the total amount and the phosphorylation status of BRN2.
DISCUSSION
Appropriate control of the level and activity of MITF-M and PAX3 is essential for regulating proliferation and migration of the melanocyte lineage. It has been suggested that BRN2 regulates the expression of these two genes in cells in vitro. Here we addressed the role of BRN2, its relative level, and the phosphorylation of its DBD, on the regulation of proliferation and migration of melanocytes both in vitro and in vivo. We report differential effects of DBD phosphorylation on the interactions with, and regulation of, the PAX3 and MITF-M promoters. These findings provide a mechanism to explain the effects of BRN2 on melanocyte proliferation and migration.
BRN2 is regulated at the transcriptional and posttranslational levels.
A combination of BRAF, β-catenin, and MITF/miR-211 regulates the amount of BRN2 mRNA and protein (10, 24, 25, 56). In this study, we evaluated the significance of posttranslational modification of conserved residues (Thr361 and Ser362) in the BRN2 DBD for cell migration and proliferation. POU transcription factor functions are modulated by posttranslational modifications and particularly by phosphorylation (5, 20, 32). Knowing that PKA can affect proliferation and migration, we showed that mutation of these two residues in alanine was sufficient to inhibit BRN2 POU domain phosphorylation by PKA. Obviously, Thr361 and Ser362 may also be targets of other kinases, and additional sites in BRN2 may also be phosphorylated by other kinases. Nevertheless, Thr361 and Ser362 are of particular interest with respect to the role of BRN2 in these cellular mechanisms because their modification modulates the DNA-binding properties of this transcription factor.
BRN2TS and BRN2AA mice present coat color phenotypes.
The phenotypes observed in vivo are the result of various actions at the cellular and molecular levels. The expression of a nonphosphorylatable form of BRN2 (BRN2AA) in melanoblasts leads to hypopigmentation of the tail and paws and a white belly spot; these features indicate proliferation and migration defects. The white belly spots were larger in a PAX3 or MITF heterozygous background. The expression of a phosphorylatable form of BRN2 (BRN2TS) in melanoblasts led to mild hyperpigmentation of the tail and paws without any white belly spot, revealing slightly increased proliferation, which was not amplified in a PAX3 or MITF background. Some BRN2TS molecules may be phosphorylated (depending of the context), and one possible interpretation of these findings is that the absence of the white belly spot in BRN2TS mice is due to compensation of the reduced migration by increased proliferation.
Molecular mechanism associated with the expression of BRN2TS and BRN2AA.
During the establishment of the melanocyte lineage, when BRN2 is not produced in melanoblasts, MITF and PAX3 are transcribed and are presumably regulated by exogenous BRN2 in transgenic mice. The phosphorylated and nonphosphorylated forms of BRN2 bind the MORE element of the MITF-M promoter and repress its activity. Moreover, the nonphosphorylated form of BRN2 represses PAX3 transcription and binds the two NORE elements of the PAX3 promoter more efficiently than the phosphorylated form of BRN2 does. Thus, the differential functions of nonphosphorylated and phosphorylated BRN2 on migration and proliferation can be explained on the basis of the idea that MITF and PAX3 are the main targets. However, the involvement of other BRN2 targets cannot be excluded from any explanation of the white belly spot of BRN2AA mice. A large series of potential targets, including Steel (= KitL), were identified by a genome-wide approach using ChIP-chip experiments and 501Mel melanoma cells (33). Other targets, such as the cGMP-specific phosphodiesterase PDE5A, have been identified as BRN2 targets and are involved in melanoma invasion (4).
Our experiments indicate that BRN2 is mainly a repressor of transcription; we observed repression of MITF-M by BRN2TS and BRN2AA and repression of PAX3 by BRN2AA. However, the mechanism by which BRN2TS activates or represses transcription remains unknown. The binding of the phosphorylated form of BRN2 to its DNA target is weaker than the binding of the nonphosphorylated BRN2. This may explain the lack of repression of PAX3 by BRN2TS, but it does not explain its activation. BRN2 binds DNA as a monomer, homodimer, or heterodimer and interacts with the PAX3, SOX10, and POU proteins, including OCT1 (17). Induction of PAX3 may depend on the type of dimer, the nature of the coactivator recruited, and/or the presence of appropriate binding sites in the promoter.
Brn2 and melanomagenesis.
BRN2 is overexpressed in some melanoma cell lines and plays a role in controlling the proliferation and invasion of many (10, 17, 23, 48, 63). The production of BRN2 is variable in human melanoma biopsy specimens, with some cells producing more BRN2 than others (10, 23). The induction of expression of BRN2 can be associated with the induction of the Wnt/β-catenin pathway in a subset of the melanoma cell population (24) and/or the induction of the mitogen-activated protein kinase using BRAF specifically (4, 25). In tumors, proliferative potential (and invasiveness) may well differ between BRN2-positive and -negative cells (48). The importance and relevance of posttranslational modification, including phosphorylation, of BRN2 in human melanoma biopsy specimens cannot be addressed for the time being; we do not have access to and were not able to raise an antibody of quality directed against phosphorylated Thr361 and Ser362 of BRN2. For the time being, we show in this article that the overexpression of phosphorylatable and nonphosphorylatable forms of BRN2 is not sufficient to induce melanomagenesis in mice. Although BRN2-positive cells may be more invasive than other cells, our findings in vivo and in cellulo show that these cells migrate less. The proliferative capacity of BRN2-positive cells presumably depends on the kinases (including PKA) activated and certainly the environment; nevertheless, phosphorylated BRN2-positive cells are likely to proliferate more actively than nonphosphorylated BRN2-positive cells. These two types of cells may, however, migrate less than BRN2-negative cells.
Work in several laboratories clearly implicates BRN2 in melanomagenesis, but its role in the normal life of the melanocyte remains elusive. We have shown that BRN2 is not produced (or is undetectable) in melanoblasts until fetal development during development and is produced at a low level in melanocytes in culture in the absence of WNT or/and BRAF induction (15, 24). Possibly, BRN2 (N-OCT3) is involved in melanocyte renewal. This notion is certainly of interest, in view of the function of OCT4 in embryonic stem cells and stem cells and its potential role during melanomagenesis. Appropriate molecular genetic experiments should bring some insight into the functions, if any, of BRN2 during the renewal of melanocytes and melanomagenesis.
ACKNOWLEDGMENTS
We are grateful to F. Relaix (Pax3IRESnLacZ mice), I. Jackson (Dct::LacZ mice), and C. Lobe (Z/EG mice) for providing mouse strains, to D. Bennett (melan-a cells) for providing melanocyte lines, and to R. Ballotti (MITF-M, DCT, and TYRP1 promoters) and J. Epstein (PAX3 promoter) for providing constructs. We thank the teams caring for the animal colony and imaging facilities of the Institut Curie, especially Y. Bourgeois, F. Cordelières, and H. Harmange.
I.B. and I.P. were supported by fellowships from the Institut Curie and the ARC. L.D. was supported by a fellowship from the Ligue Contre le Cancer (Comité de l'Oise). This work was supported by the Ligue Nationale Contre le Cancer (Equipe labelisée), INCa, and Cancéropole IdF.
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Abdel-Malek ZA, et al. 2001. The melanocortin 1 receptor is the principal mediator of the effects of agouti signaling protein on mammalian melanocytes. J. Cell Sci. 114:1019–1024 [DOI] [PubMed] [Google Scholar]
- 2. Alazard R, et al. 2005. Identification of the ‘NORE’ (N-Oct-3 responsive element), a novel structural motif and composite element. Nucleic Acids Res. 33:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersen B, Rosenfeld MG. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 22:2–35 [DOI] [PubMed] [Google Scholar]
- 4. Arozarena I, et al. 2011. Oncogenic BRAF Induces Melanoma Cell Invasion by Downregulating the cGMP-Specific Phosphodiesterase PDE5A. Cancer Cell 19:45–57 [DOI] [PubMed] [Google Scholar]
- 5. Augustijn KD, et al. 2002. Structural characterization of the PIT-1/ETS-1 interaction: PIT-1 phosphorylation regulates PIT-1/ETS-1 binding. Proc. Natl. Acad. Sci. U. S. A. 99:12657–12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barr FG, et al. 1999. Predominant expression of alternative PAX3 and PAX7 forms in myogenic and neural tumor cell lines. Cancer Res. 59:5443–5448 [PubMed] [Google Scholar]
- 7. Bennett DC, et al. 1989. Cloned mouse melanocyte lines carrying the germline mutations albino and brown: complementation in culture. Development 105:379–385 [DOI] [PubMed] [Google Scholar]
- 8. Berlin I, et al. 2012. General strategy to analyse coat colour phenotypes in mice. Pigment Cell Melanoma Res. 25:117–119 [DOI] [PubMed] [Google Scholar]
- 9. Blake JA, Ziman MR. 2005. Pax3 transcripts in melanoblast development. Dev. Growth Differ. 47:627–635 [DOI] [PubMed] [Google Scholar]
- 10. Boyle GM, et al. 2011. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 24:525–537 [DOI] [PubMed] [Google Scholar]
- 11. Cabos-Siguier B, et al. 2009. Expression and purification of human full-length N Oct-3, a transcription factor involved in melanoma growth. Protein Expr. Purif. 64:39–46 [DOI] [PubMed] [Google Scholar]
- 12. Carreira S, et al. 2005. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 433:764–769 [DOI] [PubMed] [Google Scholar]
- 13. Carreira S, et al. 2006. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 20:3426–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Castro DS, et al. 2006. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell 11:831–844 [DOI] [PubMed] [Google Scholar]
- 15. Colombo S, Champeval D, Rambow F, Larue L. 2012. Transcriptomic analysis of mouse embryonic skin cells reveals previously unreported genes expressed in melanoblasts. J. Invest. Dermatol. 132:170–178 [DOI] [PubMed] [Google Scholar]
- 16. Colombo S, Kumasaka M, Lobe C, Larue L. 2010. Genomic localization of the Z/EG transgene in the mouse genome. Genesis 48:96–100 [DOI] [PubMed] [Google Scholar]
- 17. Cook AL, Sturm RA. 2008. POU domain transcription factors: BRN2 as a regulator of melanocytic growth and tumourigenesis. Pigment Cell Melanoma Res. 21:611–626 [DOI] [PubMed] [Google Scholar]
- 18. Delmas V, et al. 2007. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 21:2923–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delmas V, Martinozzi S, Bourgeois Y, Holzenberger M, Larue L. 2003. Cre-mediated recombination in the skin melanocyte lineage. Genesis 36:73–80 [DOI] [PubMed] [Google Scholar]
- 20. Diamond SE, Chiono M, Gutierrez-Hartmann A. 1999. Reconstitution of the protein kinase A response of the rat prolactin promoter: differential effects of distinct Pit-1 isoforms and functional interaction with Oct-1. Mol. Endocrinol. 13:228–238 [DOI] [PubMed] [Google Scholar]
- 21. French AD, et al. 2009. PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int. J. Med. Sci. 6:93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallagher SJ, et al. 2011. General strategy to analyse melanoma in mice. Pigment Cell Melanoma Res. 24:987–988 [DOI] [PubMed] [Google Scholar]
- 23. Goodall J, et al. 2008. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Res. 68:7788–7794 [DOI] [PubMed] [Google Scholar]
- 24. Goodall J, et al. 2004. Brn-2 expression controls melanoma proliferation and is directly regulated by beta-catenin. Mol. Cell. Biol. 24:2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodall J, et al. 2004. The Brn-2 transcription factor links activated BRAF to melanoma proliferation. Mol. Cell. Biol. 24:2923–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hagino-Yamagishi K, et al. 1997. Predominant expression of Brn-2 in the postmitotic neurons of the developing mouse neocortex. Brain Res. 752:261–268 [DOI] [PubMed] [Google Scholar]
- 27. Hanke T, Szawlowski P, Randall RE. 1992. Construction of solid matrix-antibody-antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J. Gen. Virol. 73(Pt. 3):653–660 [DOI] [PubMed] [Google Scholar]
- 28. Hara Y, Rovescalli AC, Kim Y, Nirenberg M. 1992. Structure and evolution of four POU domain genes expressed in mouse brain. Proc. Natl. Acad. Sci. U. S. A. 89:3280–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirobe T, Abe H. 1999. Genetic and epigenetic control of the proliferation and differentiation of mouse epidermal melanocytes in culture. Pigment Cell Res. 12:147–163 [DOI] [PubMed] [Google Scholar]
- 30. Hodgkinson CA, et al. 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74:395–404 [DOI] [PubMed] [Google Scholar]
- 31. Kapiloff MS, Farkash Y, Wegner M, Rosenfeld MG. 1991. Variable effects of phosphorylation of Pit-1 dictated by the DNA response elements. Science 253:786–789 [DOI] [PubMed] [Google Scholar]
- 32. Kasibhatla S, et al. 1999. Jun kinase phosphorylates and regulates the DNA binding activity of an octamer binding protein, T-cell factor beta1. Mol. Cell. Biol. 19:2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kobi D, et al. 2010. Genome-wide analysis of POU3F2/BRN2 promoter occupancy in human melanoma cells reveals Kitl as a novel regulated target gene. Pigment Cell Melanoma Res. 23:404–418 [DOI] [PubMed] [Google Scholar]
- 34. Kubic JD, Young KP, Plummer RS, Ludvik AE, Lang D. 2008. Pigmentation PAX-ways: the role of Pax3 in melanogenesis, melanocyte stem cell maintenance, and disease. Pigment Cell Melanoma Res. 21:627–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larue L, Dougherty N, Porter S, Mintz B. 1992. Spontaneous malignant transformation of melanocytes explanted from Wf/Wf mice with a Kit kinase-domain mutation. Proc. Natl. Acad. Sci. U. S. A. 89:7816–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. 2000. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J. Biol. Chem. 275:37978–37983 [DOI] [PubMed] [Google Scholar]
- 37. Lefort K, Rouault JP, Tondereau L, Magaud JP, Dore JF. 2001. The specific activation of gadd45 following UVB radiation requires the POU family gene product N-oct3 in human melanoma cells. Oncogene 20:7375–7385 [DOI] [PubMed] [Google Scholar]
- 38. Luciani F, et al. 2011. Biological and mathematical modeling of melanocyte development. Development 138:3943–3954 [DOI] [PubMed] [Google Scholar]
- 39. Mackenzie MA, Jordan SA, Budd PS, Jackson IJ. 1997. Activation of the receptor tyrosine kinase Kit is required for the proliferation of melanoblasts in the mouse embryo. Dev. Biol. 192:99–107 [DOI] [PubMed] [Google Scholar]
- 40. Medic S, Rizos H, Ziman M. 2011. Differential PAX3 functions in normal skin melanocytes and melanoma cells. Biochem. Biophys. Res. Commun. 411:832–837 [DOI] [PubMed] [Google Scholar]
- 41. Milewski RC, et al. 2004. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development 131:829–837 [DOI] [PubMed] [Google Scholar]
- 42. Moore R, et al. 2004. Involvement of cadherins 7 and 20 in mouse embryogenesis and melanocyte transformation. Oncogene 23:6726–6735 [DOI] [PubMed] [Google Scholar]
- 43. Moriyama M, et al. 2006. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J. Cell Biol. 173:333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakai S, et al. 1995. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9:3109–3121 [DOI] [PubMed] [Google Scholar]
- 45. Nakatani Y, Ogryzko V. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430–444 [DOI] [PubMed] [Google Scholar]
- 46. Nieto L, et al. 2007. Differential effects of phosphorylation on DNA binding properties of N Oct-3 are dictated by protein/DNA complex structures. J. Mol. Biol. 370:687–700 [DOI] [PubMed] [Google Scholar]
- 47. Novak A, Guo C, Yang W, Nagy A, Lobe CG. 2000. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28:147–155 [PubMed] [Google Scholar]
- 48. Pinner S, et al. 2009. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res. 69:7969–7977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Relaix F, et al. 2003. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 17:2950–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reményi A, et al. 2001. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell 8:569–580 [DOI] [PubMed] [Google Scholar]
- 51. Scholl FA, et al. 2001. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 61:823–826 [PubMed] [Google Scholar]
- 52. Schonemann MD, et al. 1995. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9:3122–3135 [DOI] [PubMed] [Google Scholar]
- 53. Schouwey K, et al. 2007. Notch1 and Notch2 receptors influence progressive hair graying in a dose-dependent manner. Dev. Dyn. 236:282–289 [DOI] [PubMed] [Google Scholar]
- 54. Schwahn DJ, Xu W, Herrin AB, Bales ES, Medrano EE. 2001. Tyrosine levels regulate the melanogenic response to alpha-melanocyte-stimulating hormone in human melanocytes: implications for pigmentation and proliferation. Pigment Cell Res. 14:32–39 [DOI] [PubMed] [Google Scholar]
- 55. Segil N, Roberts SB, Heintz N. 1991. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254:1814–1816 [DOI] [PubMed] [Google Scholar]
- 56. Strub T, et al. 2011. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 30:2319–2332 [DOI] [PubMed] [Google Scholar]
- 57. Swope VB, Medrano EE, Smalara D, Abdel-Malek ZA. 1995. Long-term proliferation of human melanocytes is supported by the physiologic mitogens alpha-melanotropin, endothelin-1, and basic fibroblast growth factor. Exp. Cell Res. 217:453–459 [DOI] [PubMed] [Google Scholar]
- 58. Tamura A, et al. 1987. Normal murine melanocytes in culture. In Vitro Cell Dev. Biol. 23:519–522 [DOI] [PubMed] [Google Scholar]
- 59. Thomson JA, et al. 1995. The Brn-2 gene regulates the melanocytic phenotype and tumorigenic potential of human melanoma cells. Oncogene 11:691–700 [PubMed] [Google Scholar]
- 60. Thurber AE, et al. 2011. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene 30:3036–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vierbuchen T, et al. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463:1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Q, Kumar S, Slevin M, Kumar P. 2006. Functional Analysis of Alternative Isoforms of the Transcription Factor PAX3 in Melanocytes In vitro. Cancer Res. 66:8574–8580 [DOI] [PubMed] [Google Scholar]
- 63. Wellbrock C, et al. 2008. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS One 3:e2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu BK, Pruitt SC. 2005. Determination of transcription factors and their possible roles in the regulation of Pax3 gene expression in the mouse B16 F1 melanoma cell line. Melanoma Res. 15:363–373 [DOI] [PubMed] [Google Scholar]