Abstract
Nr2e1 encodes a stem cell fate determinant of the mouse forebrain and retina. Abnormal regulation of this gene results in retinal, brain, and behavioral abnormalities in mice. However, little is known about the functionality of human NR2E1. We investigated this functionality using a novel knock-in humanized-mouse strain carrying a single-copy bacterial artificial chromosome (BAC). We also documented, for the first time, the expression pattern of the human BAC, using an NR2E1-lacZ reporter strain. Unexpectedly, cerebrum and olfactory bulb hypoplasia, hallmarks of the Nr2e1-null phenotype, were not fully corrected in animals harboring one functional copy of human NR2E1. These results correlated with an absence of NR2E1-lacZ reporter expression in the dorsal pallium of embryos and proliferative cells of adult brains. Surprisingly, retinal histology and electroretinograms demonstrated complete correction of the retina-null phenotype. These results correlated with appropriate expression of the NR2E1-lacZ reporter in developing and adult retina. We conclude that the human BAC contained all the elements allowing correction of the mouse-null phenotype in the retina, while missing key regulatory regions important for proper spatiotemporal brain expression. This is the first time a separation of regulatory mechanisms governing NR2E1 has been demonstrated. Furthermore, candidate genomic regions controlling expression in proliferating cells during neurogenesis were identified.
INTRODUCTION
During embryogenesis, the proper development of an organism relies on orchestration between proliferation, differentiation, and death of different cell populations. The resulting dynamic balance depends on both cell intrinsic regulators and environmental factors. One such intrinsic regulator is Nr2e1 (also known as Tlx, Tll, and Tailless), which encodes a highly conserved transcription factor known to be a key stem cell fate determinant in both the developing mouse forebrain and retina (18, 24, 29, 39, 59). In mouse, abnormal regulation of this gene results in blindness, behavior abnormalities, and brain tumor initiation and progression (27, 35, 36, 41, 56, 58). In humans, candidate regulatory mutations have been found in patients suffering from microcephaly, bipolar disorder, schizophrenia, and aggression (22, 23). Upregulation of NR2E1 expression was also found in cancer, and a somatic-protein-coding mutation was found in glioblastoma (27, 35–38, 47). These data argue in favor of strengthening our knowledge regarding the regulation and function of human NR2E1 and thereby the potential for this gene to have a role in disease. To date, the main information regarding human NR2E1 expression comes from assembled analyses done on homogenized tissue samples and information from a humanized mouse model (1, 18, 23). This model demonstrated complete correction of the null-brain phenotype while only ameliorating the eye phenotype. The uncorrected eye phenotype was proposed to be due to gene dosage sensitivity during eye development; a hypothesis that has not been verified to date.
During development, the mammalian forebrain can be divided into two regions, the telencephalon and diencephalon. The telencephalon will become the cerebrum, comprising the cerebral cortex and basal ganglia, and the diencephalon will give rise to the brain thalamus regions and optic vesicles (OV); the latter will mature into the optic nerve and eye. The mouse Nr2e1 gene is first detected at embryonic day 8 (E8) in the ventricular zone (VZ) of the neuroepithelium layer and later spreads posteriorly into the diencephalon (E10.5) (32, 59). At E12.5, the expression is detected in the VZ region of the lateral telencephalon, comprising the dorsal and lateral pallium (DP and LP), the medial pallium (MP), the lateral ganglionic eminence (LGE), and the medial ganglionic eminence (MGE) (48). Mice lacking the Nr2e1 gene (Tlx−/−, Nr2e1frc/frc, also called fierce, referred to here as Nr2e1-null) show forebrain defects, leading to cerebrum and olfactory bulb (OB) hypoplasia (31, 56). The cerebrum and OB defects are mainly attributed to the role of Nr2e1 in controlling the proliferation rate of neural stem cells in the VZ of the telencephalon during development (24, 40, 49). The role of Nr2e1 in maintaining the neural stem cell population also appears to be important in the adult brain. Expression studies using lacZ reporter mice demonstrate a positive signal for the Nr2e1 gene in proliferative cells lining the subventricular zone (SVZ), rostral migratory stream (RMS), and subgranular layer of the dentate gyrus (DG) (24, 26, 45). Functional studies of the adult brain, using conditional knockout and transgenic mouse models, revealed that Nr2e1 is involved in controlling the number of proliferative cells in both the SVZ and DG (26, 60). This control in the SVZ appears to be highly dependent on the copy number found in the genome (27). Surprisingly, nonproliferative cells populating the cornu Ammonis regions (CA1 and CA3) and DG regions of the hippocampus, as well as the striatum and cortex, have also been shown to express sparse to strong levels of Nr2e1 in the adult brain (60).
In the developing mouse eye, Nr2e1 is detected in the optic processes of the developing embryo as early as E9 (32). At E11.5, the expression becomes restricted to the innermost surface of the retina, corresponding to the end feet of retinal progenitor cells (RPC) found in the neuroblastic layer (NBL) of the developing embryo (29). Nr2e1 expression in the NBL peaks at E15.5, suggesting a role for this gene in an early phase of retinogenesis during embryonic development (29, 61). Nr2e1-null mice have retinal and optic nerve dystrophy, leading to blindness (56, 58, 61). During development, the null animals display a deregulation in the proliferation rate of RPC and an increase in apoptotic levels in the ganglion cell layer (GCL), which results in a marked reduction in thickness of the distinct layers in the adult retina (29, 56). Nr2e1-null animals also suffer from retinal vasculature defects which are explained by the role of this gene in the proper assembly of fibronectin matrices secreted by proangiogenic astrocytes (53). An absence of Nr2e1 expression in these cells during development has been attributed to defects in normal vasculature formation in the null mice. In the adult retina, Nr2e1 expression is restricted to the Müller cells, a retinal glia subtype population that is immunoreactive for cellular retinaldehyde-binding protein (CRALBP) (29, 61).
With the advancement of technologies allowing easy modification of large DNA constructs and the use of novel docking sites to transfer these large constructs to the mouse genome, it is now possible to generate multiple humanized mouse strains in a relatively short time (9, 16). In the present study, we sought to investigate the functionality of the human gene in both brain and eye, using a novel humanized mouse strain carrying a single-copy bacterial artificial chromosome (BAC) insert of NR2E1 knocked in at the Hprt locus. The high level of sequence similarity at the NR2E1 locus between mouse and human and the fact that mice carrying a single copy of the mouse Nr2e1 gene (Nr2e1frc/+) have virtually no phenotype were elements justifying the choice of our approach (1, 2, 18, 31, 56). Thus, we tested the hypothesis that a single-copy human gene is functionally equivalent to a single-copy mouse gene and will correct both the brain and eye phenotypes of the Nr2e1-null mice. We also documented, for the first time, the expression pattern of the human gene using an NR2E1-lacZ reporter strain generated with the same technology. Finally, employing the site-specific, single-copy, docking technology allowed us to develop a platform for future testing of candidate human mutations in NR2E1 (44).
MATERIALS AND METHODS
BAC retrofitting.
Two BAC constructs were generated: first, bEMS223, which is a modified version of BAC RP11-144P8 (http://bacpac.chori.org/hmale11.htm; accessed 6 June 2011) containing the human NR2E1 gene and retrofitted to contain Hprt homologous recombination targeting arms, second, bEMS86, which was a further retrofitting of bEMS223 to carry a lacZ reporter cassette inserted at ATG of NR2E1. Both BAC constructs were generated using the lambda recombination system (57).
To generate the bEMS223 construct, the HPRT region in pJDH8A/246b (16) was modified by NotI digestion, resulting in a 1.3-kb fragment that contained the Ori site. This fragment was extracted and amplified using the following primers: 5′-AATTGCGGCCGCTGTTCACTGATTCACGCGGTTCAAAAATGACGATCGATGGTATTAACTCAAACGATATTTAAATCGCTCTTCCGCTTCCTCG-3′ and 5′-AATTGCGGCCGCTCAGCGTTTTGCAGCGGCCAGCTGTCCCACACATCAAGTCTTTTGCAGACTCAATATTTAAATTGGATGGAGGCGGATAAA-3′. These primers added 60-bp targeting arms for integration into the sacB gene and NotI (bold) and SwaI (italics) restriction sites to the Ori linear DNA cassette. The modified Ori cassette replaced the original Ori site in pJDH8A/246b through NotI digestion and ligation. This produced the modified pJDH8A/246b construct pEMS1907, which contained an HPRT sequence flanked by the sacB gene and SwaI sequence. The HPRT cassette was removed from pEMS1907 using SwaI and recombineered into the sacB gene of the NR2E1 parental BAC (RP11-144P8), using the lambda recombination system in SW102 cells.
The bEMS86 construct was generated by adding a lacZ reporter cassette in the first exon starting at the first codon of NR2E1 and creating a downstream 25-bp deletion of genomic DNA. A custom synthesized lacZ/kanamycin cassette, pEMS1908 (GeneArt, Regensburg, Germany), was PCR amplified to contain BAC homology arms using the following primers: 5′-GCCGGGACTCGGGCAGCGCCCACCAACCGCTCCGCCCCGGGACAGCCAGCATGGCGGATCCCGTCGTTTT-3′ and 5′-TCGCCCCAGGCTGCGCGCCTAGGCCCCACGGCGGCCCGAGAGGTACCCACGAAGTTCCTATACTTTCTAG-3′.
The resulting fragment was retrofitted into bEMS223 as described above and transformed into SW105 cells. The kanamycin gene adjacent to the lacZ gene was designed with flanking full frt sites (28), which were used to excise the kanamycin gene via induction of FLPe recombinase (6, 43). The resulting construct, bEMS86, contained a lacZ reporter gene under the influence of the NR2E1 human regulatory regions.
Strain generation, husbandry, and breeding.
Three different strains were generated: B6.129P2(Cg)-Hprttm86(NR2E1,bEMS223)Ems from embryonic stem cell (ESC) clone mEMS2044 harbored the BAC bEMS223; B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems from mEMS4751 and B6.129P2(Cg)-Hprttm87(Ple142-lacZ)Ems from mEMS4749 both harbored the BAC bEMS86. These strains were generated using a variation of the previously described strategy to insert constructs 5′ of Hprt on the mouse X chromosome (5, 16, 55). Briefly, BAC DNA was purified using the Nucleobond BAC 100 kit (Clontech Laboratories, Mountain View, CA) and linearized with I-SceI as described in the literature (16). bEMS223 was electroporated into 4 × 107 mEMS1204 ESCs (55), and bEMS86 similarly was electroporated into mEMS1202 (55) using the following conditions: voltage, 190 V; capacitance, 500 μF; resistance, none; a BTX ECM 630 Electro cell manipulator (BTX, San Diego, California) (55) was used. ESC clones were selected in hypoxanthine aminopterin thymidine (HAT) and isolated, and DNA was purified as described previously (55). Note that B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems and B6.129P2(Cg)-Hprttm87(Ple142-lacZ)Ems were obtained from two independent ESC clones from the same electroporation but different plates. PCR assays used for BAC DNA integrity characterization are listed elsewhere (see Table 1 at http://cisreg.ca/MCB-NR2E1/). These human-specific PCR assays were designed to span the entire BAC construct inserted in the mouse genome; they had a maximum of 11 kb between them, with an average of 6 kb apart. The PCR assays were performed as previously described (55). PCR-positive ESC clones for bEMS223 were subsequently tested in Southern blot assays to verify the integrity of the insertion site using a 5′ Hprt probe and RSA probe (16). Copy number quantification for bEMS223 ESC clones, which had passed both the PCR and Southern blot screening, was performed on genomic DNA extracted with the Qiagen DNeasy blood and tissue kit (Qiagen Inc., Toronto, Ontario, Canada). A custom TaqMan copy number assay was designed in an intronic region with identical sequences in mouse and human NR2E1 genes and used in accordance with the manufacturer's instructions (Life Technologies, Carlsbad, CA). The custom assay employed the following primers: 5′-AAGCTCTGGAAAGTAGTGTTATGAA-3′, 5′-TAATAGGCATCCCAAACACAAA-3′, and 5′-TGGGAATGCTCTGTGAATGA-3′ (probe sequence).
Copy number evaluation was done using raw threshold cycle (CT) value comparisons between the custom assay and two different TaqMan copy number mouse reference assays, Tfrc and Tert (Life Technologies, Carlsbad, CA). The quantitative PCR (Q-PCR) results from three technical replicates were pooled together and compared to wild-type ESC CT values. Correctly targeted ESC clones were microinjected into B6(Cg)-Tyrc-2/J (JAX stock no. 000058) blastocysts to generate chimeras that were bred to C57BL/6J (B6) (JAX stock no. 000664) to obtain offspring carrying the BAC insert. Backcrossing to B6 mice continued such that mice used in this study were N5 or higher. The Mutant Mouse Regional Resource Center (MMRRC) at The Jackson Laboratory is distributing strain B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems/Mmjax (MMRRC stock no. 032962).
Male animals were used in all studies to avoid any variability due to random X inactivation of the knock-in alleles at Hprt. Experimental animals for studying the functional NR2E1 BAC (bEMS223) were generated through a breeding scheme similar to that used previously (1). Briefly, B6 females heterozygous for the BAC insert and for the fierce deletion [B6.Cg-Hprttm86(NR2E1,bEMS223)Ems/X, Nr2e1frc/+], were crossed to 129 males heterozygous for the fierce mutation (129S1/SvImJ.Cg-Nr2e1frc/+). This produced first-generation hybrid offspring (B6129F1), abbreviated here as Wt (wild type) (X/Y, Nr2e1+/+), NR2E1/fierce [Hprttm86(NR2E1,bEMS223)Ems/Y, Nr2e1frc/frc], fierce (X/Y, Nr2e1frc/frc), and NR2E1 [Hprttm86(NR2E1,bEMS223)Ems/Y, Nr2e1+/+]. Males harboring the NR2E1 lacZ reporter BAC (bEMS86) were used to evaluate the expression pattern of the human NR2E1 gene [B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems/Y and B6.129P2(Cg)-Hprttm87(Ple142-lacZ)Ems/Y], abbreviated here as NR2E1-lacZ. The age of adult animals used in the different experiments ranged from 8 to 20 weeks.
All mice were maintained in the pathogen-free Centre for Molecular Medicine and Therapeutics animal facility on a 6 am-to-6 pm light cycle, 20 ± 2°C, with 50% ± 5% relative humidity, and had food and water ad libitum. All procedures involving animals were in accordance with the Canadian Council on Animal Care (CCAC) and UBC Animal Care Committee (ACC) (Protocol no. A07-0435).
Embryo and adult tissue preparation.
Timed-pregnant mice were euthanized by cervical dislocation, and embryos at E12.5 or E15.5 were dissected and then fixed in 4% paraformaldehyde (PFA) with 0.1 M Na2HPO4 (PO) buffer (pH 8.0) for 1 h at 4°C. Whole embryos were incubated in lacZ staining solution overnight at 37°C [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (1 mg/ml), MgCl2 (2 mM), K3Fe(CN)6 (4 mM), and K4Fe(CN)6 (4 mM) in 1× phosphate-buffered saline (PBS)] before being postfixed in 4% PFA for an additional 4 h. The embryos were then cryoprotected as described in the literature and embedded in optimal-cutting-temperature (OCT) compound (Tissue-tek, Torrance, CA) on dry ice (24). Embryos were then sectioned at 16 μm using a Cryo Star HM550 cryostat (Microm International, Kalamazoo, MI) and mounted for imaging or cleared as described in the literature (42), and pictures were taken in 100% glycerol solution.
Intracardial perfusions were performed on avertin-anesthetized mice with 4% PFA with 0.1 M PO buffer (pH 8.0). For lacZ expression analysis, brain and eye tissues were collected and postfixed in 4% PFA for an additional 30 min at 4°C and then transferred to 20% sucrose with 0.05 M PO buffer overnight at 4°C and embedded the next day in OCT compound on dry ice. For luxol fast blue/cresyl violet staining and immunofluorescence, brains were collected, postfixed in PFA overnight at 4°C, and then transferred to 20% sucrose with 0.05 M PO buffer overnight at 4°C and embedded the next day in OCT compound on dry ice. For hematoxylin-and-eosin staining, eyes were collected and transferred to Davison fixative for postfixing overnight at 4°C and then subsequently washed in 1× PBS and 50 to 70% ethanol solution before being processed for paraffin embedment.
Histology.
For lacZ staining, 20 to 25 μm cryosections from adult brains (floating sections) or adult eyes (sections on slides) were rehydrated in sequential washes of PBS and permeabilized in PBS with 0.1% Triton before being incubated in lacZ solution overnight at 37°C [X-Gal (1 mg/ml), MgCl2 (2 mM), K3Fe(CN)6 (4 mM), and K4Fe(CN)6 (4 mM) in 1× PBS–0.1% Triton, deoxycholate (0.01%), and NP-40 (0.02%)].
For antibody staining, 20- to 25-μm cryosections from adult brains (floating sections) or E15.5 embryos and adult eyes (sections on slides) were rehydrated in sequential washes of PBS, permeabilized in PBS with 0.1% Triton (adults) or 0.3% Triton (embryos), and quenched in 0.1 M glycine-PBS solution. The cryosections were blocked with 1% bovine serum albumin (BSA) in PBS with 0.1% Triton (adults) or 0.3% Triton (embryos) for 1 h at room temperature before applying the primary antibodies. Colocalization experiments were performed using chicken anti-β-galactosidase (β-Gal) antibody (ab9361; Abcam, San Francisco, CA) 1:5,000), rabbit anti-Sox2 antibody (ab97959; Abcam) (1:1,000), mouse anti-CRALBP antibody (ab15051; Abcam) (1:1,200), rabbit anti-Ki67 antibody (ab15580; Abcam) (1:1,000), mouse anti-glial fibrillary acidic protein (anti-GFAP) antibody (VP-G805; Vector, Burlington, Ontario, Canada) (1:1,000), or mouse anti-NeuN antibody (MAB377; Chemicon, Billerica, Massachusetts) (1:1,000) and incubated overnight at 4°C. Corresponding secondary antibodies coupled to Alexa 488 or Alexa 594 (Invitrogen, Burlington, Ontario, Canada) were incubated at room temperature for 2 h in the dark (1:1,000). Hoechst 33342 was used for nuclear staining on all sections. Ki67-positive cell counting was performed as described in the literature (54).
For brain histology, coronal floating sections of 25 μm were mounted on slides and dehydrated with 95% ethyl alcohol before being left overnight at 56°C in a luxol fast blue solution (0.1% luxol fast blue in 95% ethyl alcohol). Excess stain was then rinsed with 95% ethyl alcohol, and the slides were rinsed in distilled water before being differentiated in a lithium carbonate solution for 30 s (0.05% lithium carbonate in distilled water). The differentiation was then continued using 70% ethyl alcohol for 15 s, and sections were counterstained in cresyl echt violet solution (0.1% cresyl violet in distilled water) for 6 min before being dehydrated and mounted for microscopy.
For retinal histology, 5-μm paraffin sections were rehydrated, incubated in hematoxylin solution for 5 min, and then washed in tap water and incubated in 1% lithium carbonate solution for 30 s before being washed in tap water again and incubated in acid alcohol (1%) for 5 s. The sections were then washed again in tap water before being incubated in eosin Y solution for 5 min and finally washed in tap water and dehydrated in 95 to 100% ethanol and xylene before mounting for microscopy.
Funduscopy.
Direct funduscopy was performed as described in the literature (15). Eyes were dilated with 1% atropine 30 min before examination. The animals were restrained from moving without sedation.
Electroretinograms.
Electroretinogram responses were recorded as previously described (14). Briefly, animals were dark adapted, anesthetized with xylazine (13 mg/ml) and ketamine (87 mg/ml), and maintained on a heating pad. The corneas were locally anesthetized with 0.5% proparacaine hydrochloride, and the pupils were dilated with 2.5% phenylephrine and 1% atropine. Experiments were conducted using an Espion E2 system with a Colordome mini-Ganzfeld stimulator (Diagnosys LLC, Lowell, MA). DTL Plus corneal electrodes were used (Diagnosys LLC, Lowell, MA). Dark-adapted responses were recorded by averaging 15 responses for each stimulus intensity (0.01 and 3.16 cd · s/m2).
Comparative genomic and transcription factor binding site (TFBS) overrepresentation analysis.
For the comparative genomic analysis, relative sequence coordinates were retrieved for the BAC construct used in this and three other studies (1, 12, 26) and then visualized using the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/index.html; assembly in February 2009).
For the TFBS overrepresentation analysis of the highly conserved regions, phylogenetic footprinting was performed by extracting a phastCons profile to identify regions with significant identity (default parameters for multispecies analysis were used). TFBS predictions were performed using the ORCA toolkit system (http://burgundy.cmmt.ubc.ca/cgi-bin/ORCAtk/orca; version 1.0.0). TFBSs were predicted using the “all JASPAR CORE vertebrate” profiles (default parameters). A gene ontology (GO) term overrepresentation analysis of the enriched transcription factor population was performed using “GOHyperGAll function” of “GOstats library” from the “R package” (11, 17). Terms with a Bonferroni-corrected P value of <0.05 were considered significantly enriched in the analysis.
Nucleotide shuffling analysis.
Sequences from the highly conserved regions were used in a random mononucleotide shuffling analysis. The nucleotides were shuffled to obtain random sequences, conserving the original mononucleotide distribution among the conserved regions. TFBS prediction analysis and GO term overrepresentation analysis were performed as described in the previous paragraph with the obtained random sequences. The following terms were retained for the shuffling analysis: “nervous system development,” “central nervous system development,” “neurogenesis,” “eye development,” and “camera-type eye development,” for both the 5′ and 3′ conserved regions. The analysis compiled for each GO term the number of times (n) a Bonferroni-corrected P value lower than or equal to the original Bonferroni-corrected P value was found on the shuffled sequences (10,000 times). The associated P value obtained for each GO term (randomized P value) correspond to n divided by 10,000.
Mouse statistical analysis.
All analyses were performed using the STATISTICA 6.0 software program (Statsoft, Inc., Tulsa, OK). Nonparametric analysis using the Kruskal-Wallis test was performed as described in the literature (1).
RESULTS
Human NR2E1-lacZ embryos displayed an unexpected absence of expression in the dorsal pallium in the brain while retaining appropriate region-specific expression in the eyes.
To understand the functionality, and document the expression pattern, of the human NR2E1 gene in brain and eye development, we generated novel humanized mouse strains for NR2E1 as described in Material and Methods. The frequency of properly targeted ESC clones varied between 24% for bEMS223 (NR2E1 functional construct) and 21% for bEMS86 (NR2E1-lacZ construct).
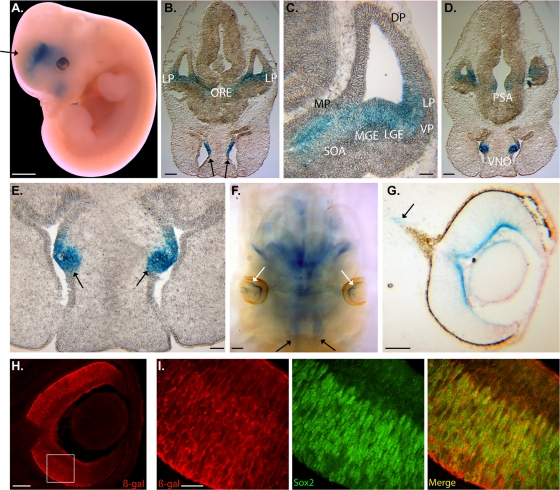
Two independent human NR2E1-lacZ strains, B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems and B6.129P2(Cg)-Hprttm87(Ple142-lacZ)Ems, were generated and yielded similar embryonic expression results; all the data presented here were obtained using B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems. The mouse Nr2e1 gene expression at E12.5 is detected at high levels in the VZ of the dorsal and lateral pallium (DP and LP), as well as the lateral ganglionic eminence (LGE). Lower expression levels are also detected in the medial pallium (MP) and the medial ganglionic eminence (MGE) at this time point (48). As expected, whole mounts of E12.5 NR2E1-lacZ embryos showed expression in the nasal cavities (NC), eyes, and diencephalon. Expression in the telencephalon was distributed in a rostral caudal manner, where low expression was apparent at the rostral/basal level and extended strongly to the caudal/lateral regions. Unexpectedly, no expression in the dorsal telencephalon was apparent in these animals (Fig. 1A, black arrow). Cryosections of E12.5 NR2E1-lacZ embryos revealed a staining pattern that extended from the LP to the optic recess (ORE) in the caudal regions of the developing telencephalon (Fig. 1B). On this section, it was also possible to see staining in the innermost surface of the NC (Fig. 1B, black arrows). Higher magnification focusing on the lateral telencephalon demonstrated that the staining extended from the LP to the ventral pallium (VP), the LGE, the MGE, and the supra optic area (SOA), as well as the MP (Fig. 1C). Unexpectedly, no lacZ staining was found in the DP, a region that has been reported to strongly express mouse Nr2e1 at this time point (48). Further caudally, the staining became more restricted to the postoptic area (PSA) in the diencephalon and the vomeronasal organ (VNO) in the NC (Fig. 1D and E, arrows). Staining in E12.5 cleared embryos showed strong expression in the NC (black arrows) and the innermost surface of the neural retina (white arrows), two regions known to express mouse Nr2e1 at this time point (Fig. 1F) (29, 32, 58). The eye expression at E12.5 was restricted to the innermost surface of the retina and extended toward the periphery (Fig. 1F and G). Lower levels of expression were present in the developing optic nerve (Fig. 1G, arrow). This staining pattern in eye development from NR2E1-lacZ mimicked that obtained from a mouse Nr2e1-lacZ reporter strain previously reported (29, 61). From E15.5 onward, the mouse Nr2e1 gene expression becomes uniformly localized in RPC populating the NBL (29). Sox2, a known marker of RPC in the NBL of the developing retina, was used to perform colocalization experiments on retinal cryosections from E15.5 NR2E1-lacZ embryos with a β-Gal-specific antibody (52). The results revealed that the human NR2E1-lacZ reporter gene was also expressed uniformly along the NBL, with strong staining within the outer NBL region (Fig. 1H). Higher magnification revealed that the staining was mainly found in the cytoplasm of Sox2-positive cells in the NBL of the developing retina (Fig. 1I). Overall, these results suggested proper expression of the human NR2E1 gene in the developing eye and NC but also demonstrated an absence of expression in the VZ of the DP in the developing telencephalon, a region critical for the development of the neocortex (7, 8).
Fig 1.
NR2E1-lacZ targeted at the Hprt locus demonstrated an unexpected absence of expression in the dorsal pallium in the developing brain while retaining appropriate region-specific expression in the developing eye. (A) BAC NR2E1-lacZ was expressed in the lateral and caudal regions of the telencephalon, the diencephalon, nasal cavities (NC), and developing eyes as shown in embryonic day 12.5 (E12.5) embryos. The arrow highlights the absence of expression in the dorsal telencephalon. (B) Expression was restricted to ventral structures of the caudal telencephalon and extended from the lateral pallium (LP) to the optic recess (ORE). Strong expression in the NC was also observed (arrows). (C) Higher magnification demonstrated an absence of lacZ expression in the ventricular/subventricular zone (VZ/SVZ) of the dorsal pallium (DP). The medial pallium (MP), LP, ventral pallium (VP), lateral ganglionic eminence (LGE), medial ganglionic eminence (MGE), and supra-optic area (SOA) showed staining for lacZ. (D) Staining in the diencephalon was restricted to the postoptic area (PSA). (E) The vomeronasal organ (VNO) also displayed strong lacZ staining (arrows). (F) Eye expression was restricted to the innermost surface of the developing retina (white arrows). Expression was also in the NC (black arrows). (G) Strong expression was observed in the innermost surface of the developing retina, which extended to the periphery of the developing eye. Staining was also in the developing optic nerve (arrow). (H) Immunofluorescence analysis performed with E15.5 embryo retina using an anti-β-Gal antibody (red) revealed staining along the innermost surface of the retina as well as staining in cells found in the neuroblastic layer. The boxed region is shown in panel I. (I) Colocalization performed between β-Gal (red) and Sox2 (green), focusing on the NBL, revealed β-Gal-positive staining in the cytoplasm of Sox2-positive cells, suggesting expression in the retinal progenitor cells. Panels A and F show whole mounts; panels B, C, D, E, G, H, and I show cryosections. (Scale bar, 2 mm [A, B, D, and F], 100 μm [C, E, G, and H], or 20 μm [I].)
Human NR2E1-lacZ displayed an unexpected absence of expression in neurogenic regions in the adult brain while retaining appropriate cell type-specific expression in the retina.
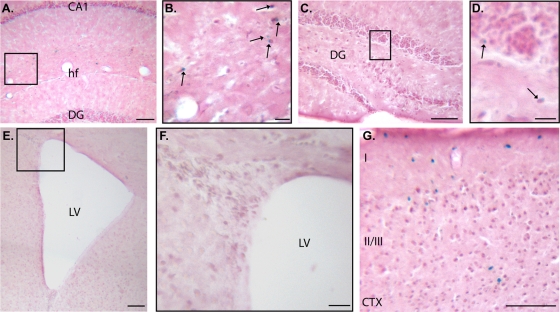
Mouse Nr2e1 gene expression in the adult brain is found in proliferative cells lining the SVZ, RMS, and subgranular layer of the dentate gyrus (DG), as well as neurons populating the hippocampal formation (HPF), striatum, and cortex (24, 26, 45, 60). Thus, as expected, adult brain sections stained from the BAC NR2E1-lacZ reporter animals demonstrated sparse expression in the HPF. The staining was restricted to the region bordered by the CA1, CA2, and CA3 dorsally and the hippocampal fissure (hf) ventrally (Fig. 2A and arrows in panel B). Very few lightly stained cells were found at the border of the subgranular layer in the DG (Fig. 2C and arrows in panel D), but there was an unexpected absence of staining in the SVZ of the lateral ventricles (Fig. 2E and F). As expected, the cortex contained lacZ-positive cells in the upper layers (layers I and II/III) (Fig. 2G). Overall, these results unexpectedly showed an absence of expression of the human BAC NR2E1-lacZ reporter in the SVZ of the lateral ventricles but an appropriate expression in the subgranular layer of the DG, the HPF, and superficial layer of the cortex.
Fig 2.
NR2E1-lacZ targeted at the Hprt locus demonstrated an unexpected absence of expression in neurogenic regions of the adult forebrain. (A and B) BAC NR2E1-lacZ was sparsely expressed in the hippocampal formation (arrows). The boxed region in panel A is shown in panel B. CA1, cornu ammonis 1; hf, hippocampal fissure; DG, dentate gyrus. (C and D) Very few lacZ-positive cells were in the subgranular layer of the DG (arrows). The boxed region in panel C is shown in panel D. (E and F) lacZ staining demonstrated an absence of expression in the subventricular zone of the lateral ventricles (LV). The boxed region in panel E is shown in panel F. (G) lacZ staining was in layers I, II, and III of the cortex (CTX). (Cryosection scale bar, 100 μm [A, C, E, and G] or 20 μm [B, D, and F].)
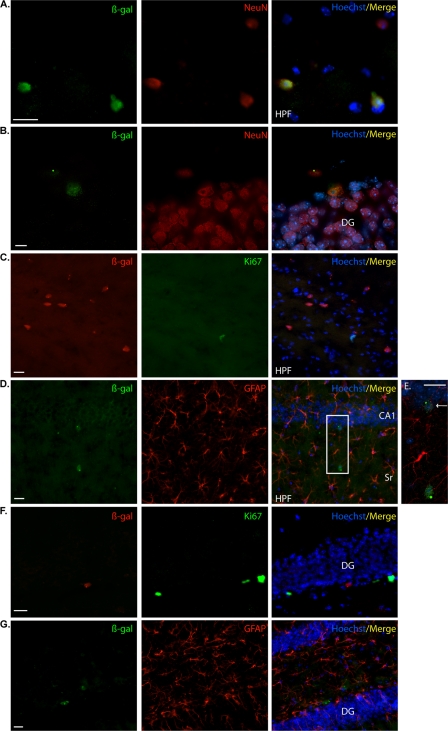
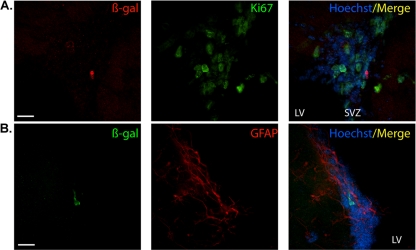
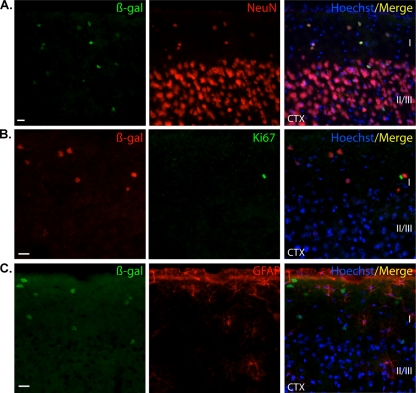
We sought to investigate the nature of these lacZ-positive cells using colocalization analyses with a β-Gal antibody. Colocalization with NeuN and β-Gal revealed that the reporter gene was expressed in neurons populating the HPF, including the subgranular layer of the DG (Fig. 3A and B). These results were confirmed by an absence of colocalization with either Ki67 or GFAP antibodies in these regions (HPF; Fig. 3C, D, and E and DG in 3F and G). In contrast to direct staining, immunolocalization revealed very few β-Gal-positive cells in the SVZ. However, these cells were not positive for Ki67, indicating that they were nonproliferative (Fig. 4A). The mouse Nr2e1 gene has been shown to be expressed in astrocyte-like type B cells, a population immunoreactive for GFAP in the mouse SVZ (26). The results obtained in our study demonstrated that the few β-Gal-positive cells surrounding the SVZ were not immunoreactive for GFAP, confirming the absence of staining of our reporter gene in astrocyte-like type B cells (Fig. 4B). The β-Gal-positive cells in the cortex were immunoreactive for NeuN and not immunoreactive for Ki67 or GFAP, together implying that the BAC NR2E1-lacZ reporter was expressed only in mature neurons in the cortex (Fig. 5A, B, and C). Finally, both the lacZ staining and the β-Gal antibody revealed an absence of expression in the striatum and RMS in the BAC NR2E1-lacZ animals (data not shown). Overall, our results showed that like the mouse gene, human NR2E1 was sparsely expressed in neurons in the cortex and hippocampus (60), but the results also demonstrated that the human BAC NR2E1-lacZ reporter was not expressed in proliferative cells in regions where neurogenesis normally occurs in adult mice.
Fig 3.
NR2E1-lacZ targeted at the Hprt locus demonstrated an unexpected absence of expression in adult proliferative cells of the dentate-gyrus-subgranular layer. (A) In BAC NR2E1-lacZ mice, immunofluorescence using an anti-β-Gal antibody (green) revealed positive cells in the hippocampal formation (HPF) that colocalized with NeuN (red), suggesting expression in mature neurons. (B) Colocalization revealed few β-Gal-immunoreactive cells (green) in the subgranular layer of the dentate gyrus (DG) that colocalized with NeuN (red), suggesting expression in mature neurons. (C) β-Gal-immunopositive cells (red) in the HPF did not colocalize with Ki67 (green). (D) β-Gal-immunopositive cells (green) in the HPF did not colocalize with GFAP (red). The boxed region in panel D is shown in panel E. (E) Higher magnification of the colocalization between β-Gal (green) and GFAP (red) revealed β-Gal-positive cells in cornu ammonis 1 (CA1) of the hippocampus (arrow). These cells did not colocalize with GFAP. (F) β-Gal-immunopositive cells (red) in the DG did not colocalize with Ki67 (green). (G) β-Gal-immunopositive cells (green) in the DG did not colocalize with GFAP (red). Sr, stratum radiatum. (Cryosection scale bars, 20 μm.)
Fig 4.
NR2E1-lacZ targeted at the Hprt locus demonstrated an unexpected absence of expression in astrocyte-like type B cells in the subventricular zone of the lateral ventricle. (A) In BAC NR2E1-lacZ mice, colocalization revealed the presence of few β-Gal-immunoreactive cells (red) in the subventricular zone (SVZ) of the lateral ventricle (LV). These cells were not positive for Ki67 (green). (B). β-Gal-immunopositive cells (green) in the SVZ of the LV did not colocalize with GFAP (red). (Cryosection scale bars, 20 μm.)
Fig 5.
NR2E1-lacZ targeted at the Hprt locus demonstrated expected expression in neurons populating the upper cortical layers I and II/III. (A) In BAC NR2E1-lacZ mice, colocalization revealed the presence of β-Gal-positive cells (green) in the cortex (CTX) that were positive for NeuN (red), suggesting expression in mature neurons. (B) β-Gal-immunopositive cells (red) in the CTX did not colocalize with Ki67 (green). (C) β-Gal-immunopositive cells (green) in the CTX did not colocalize with GFAP (red). I, II/III, cortex layers. (Cryosection scale bars, 20 μm.)
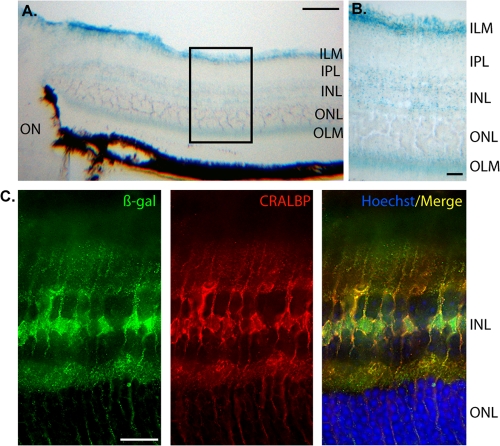
In the adult eye, the mouse Nr2e1 gene is known to express in Müller cells populating the retina (29). We thus examined where NR2E1-lacZ was expressed in adult retina sections using both the staining method and colocalization experiments. The stained sections revealed characteristic expression in Müller cells, with the body of cells located in the inner nuclear layer (INL) and fibers extending radially throughout the inner plexiform layer (IPL) and outer nuclear layer (ONL) (Fig. 6A). The end feet of these stained cells were located in the inner limiting membrane (ILM) and the outer limiting membrane (OLM) (Fig. 6B). Colocalization with the β-Gal antibody and CRALBP was shown; the latter is a known marker of Müller cells (Fig. 6C). Thus, as expected, these results demonstrated Müller cell-type-specific expression of the human BAC NR2E1-lacZ reporter in the adult mouse retina.
Fig 6.
NR2E1-lacZ targeted at the Hprt locus demonstrated appropriate cell type-specific adult retina expression. (A and B) In BAC NR2E1-lacZ mice, lacZ expression in the adult eye was strong in the inner nuclear layer (INL) and extended radially to the inner limiting membrane (ILM) and outer limiting membrane (OLM). This staining pattern was consistent with Müller cells in the adult retina. IPL, inner plexiform layer; ON, optic nerve; ONL, outer nuclear layer. (C) Colocalization, using an anti-β-Gal antibody (green) and anti-CRALBP antibody (red), revealed cell appropriate expression in Müller cells in the adult retina. The β-Gal expression pattern was localized to the large nucleus and perinucleus structure in the INL. CRALBP staining was mainly in the cytoplasm of these cells as expected. (Cryosection scale bar, 100 μm [A] or 20 μm [B and C].)
NR2E1/fierce animals displayed adult forebrain abnormalities and neurogenesis defects.
The results obtained with the BAC NR2E1-lacZ reporter mouse strain showed an absence of expression in key neurogenic regions and cells in both the developing and adult brain. To understand the importance of this absence of expression, we used the same human BAC but without the lacZ reporter and thus containing a functional NR2E1 gene, knocked in to the same chromosomal Hprt locus. This single-copy human knock-in allele (abbreviated here as NR2E1) was bred onto the fierce (Nr2e1frc/frc) background, which is null for mouse Nr2e1, to generate the experimental animals referred to here as NR2E1/fierce. In these resulting animals, brain and eye development relied solely on single-copy functional human NR2E1. Comparison analyses were performed with three different controls: wild-type (Wt) animals, fierce animals, and NR2E1 animals with the human BAC on the Wt background.
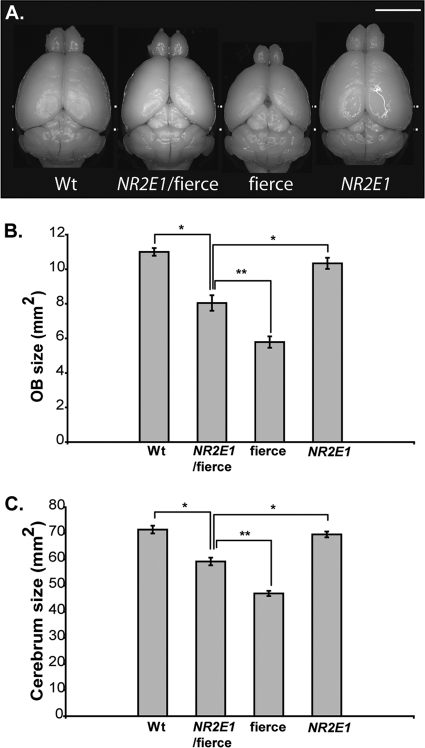
Fierce mice are named for their aggressive behavior and display gross brain abnormalities, such as hypoplasia of the OB and cerebrum (56). Our results demonstrated that a single copy of the human NR2E1 gene can only partially correct the fierce brain phenotype. NR2E1/fierce animals were difficult to handle and aggressive toward other mice. They also displayed OB and cerebrum hypoplasia with exposed colliculi, phenotypes that were absent in Wt and NR2E1 animals (Fig. 7A). Quantitative analyses of the surface area from the OB (Fig. 7B) and cerebrum (Fig. 7C) demonstrated significant reduction of the forebrain structures in NR2E1/fierce animals compared to those of Wt and NR2E1 animals (P < 0.001). However, NR2E1/fierce animals displayed larger OB and cerebrum sizes than the fierce animals, showing that the NR2E1 BAC can partially correct the fierce phenotype (P value for OB size, <0.01; P value for cerebrum size, <0.001). Nevertheless, no significant difference was observed when comparing the OB and cerebrum sizes of Wt and NR2E1 animals, indicating no effects of an additional human copy on the developing brain of Wt animals.
Fig 7.
NR2E1/fierce mouse brains were partially corrected for reduced olfactory bulb and cerebrum size. (A) Gross brain dissection revealed reduced olfactory bulb (OB) and cerebrum size for fierce mice, a known characteristic of the null phenotype. To a lesser extent, NR2E1/fierce mice also displayed these abnormalities, suggesting incomplete correction of the fierce brain phenotype by the human gene. (Scale bar, 5 mm.) (B) Surface quantification revealed significant reduction of the OB in NR2E1/fierce mice compared to that in Wt and NR2E1 mice (∗, P < 0.001). The NR2E1/fierce OB was also significantly bigger than the fierce OB (∗∗, P < 0.01). No significance was found when comparing Wt and NR2E1 mice. (C) Similarly, cerebrum area was significantly reduced in NR2E1/fierce mice compared to that in Wt and NR2E1 mice (∗, P < 0.001). The NR2E1/fierce cerebrum was also significantly bigger than the fierce cerebrum (∗∗, P < 0.001). No significance was found when comparing Wt and NR2E1 mice. (B and C) The Kruskal-Wallis H test was performed on 6 to 10 mice for all genotypes. Error bars represent standard errors of the means.
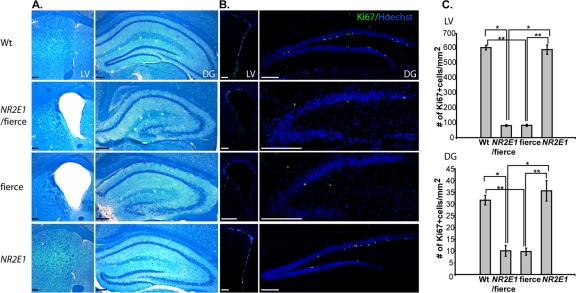
To understand the importance of an absence of NR2E1 expression in the adult brain, we looked at the morphology of the SVZ of the lateral ventricles and the DG of the hippocampus. Fierce mice display enlarged ventricles and reduced and poorly defined DG due in part to neurogenesis defects in these areas (31, 56). Histological analyses demonstrated no difference in morphology between NR2E1/fierce and fierce mice, suggesting similar neurogenesis defects in both brain regions (Fig. 8A). No morphological differences were observed between Wt and NR2E1 animals in either brain region, again showing no effect of an additional human copy (Fig. 8A). Nr2e1 has been shown to be a critical regulator of the adult neural stem cell population in both the SVZ and DG in adult mice (26, 60). We used Ki67 immunolabeling to understand the cortical hypoplasia defects found in NR2E1/fierce adult mice. Initial results obtained from both brain regions suggested similar numbers of proliferative cells in NR2E1/fierce animals and in fierce animals (Fig. 8B). No apparent differences in proliferative cell numbers were found when comparing Wt and NR2E1 animal sections (Fig. 8B). These results were confirmed by Ki67-positive cell counting in both the SVZ and DG regions of these animals (Fig. 8C). No statistical differences were found in the number of Ki67-positive cells in both the SVZ and DG between NR2E1/fierce and fierce animals, whereas significance was obtained between NR2E1/fierce and Wt or NR2E1 animals (P < 0.05). Also, no statistical differences were obtained when comparing the number of Ki67-positive cells between Wt and NR2E1 animals in both neurogenic regions, again showing no effect of an additional human copy. Overall, the results correlated with the expression data obtained with the BAC NR2E1-lacZ animals and strongly suggested that an absence of NR2E1 expression in proliferative cells of both the SVZ and DG led to an inability to correct proliferative and structural defects in these brain regions. This also suggests that the partial correction of both the OB and cerebrum size occurred during embryonic development rather than in the adult brain.
Fig 8.
NR2E1/fierce mice brains were not corrected for adult neurogenesis defects. (A) Luxol fast blue cresyl violet-stained cryosection analysis revealed enlarged lateral ventricles (LV) and reduced and poorly defined dentate gyrus (DG) in NR2E1/fierce mice, a phenotype indistinguishable from that found in fierce mice. No differences were found when comparing Wt and NR2E1 mice. (B) Ki67-immunopositive cell numbers were reduced in NR2E1/fierce mice in the LV and DG compared to those for Wt and NR2E1 mice. (C) Quantitative analysis showed a significant reduction of Ki67-immunopositive cell numbers in NR2E1/fierce mice compared to those in Wt and NR2E1 mice in both the LV (∗, P < 0.05) and DG (∗, P < 0.05). No significance was found when comparing Ki67-immunopositive cell numbers from NR2E1/fierce and fierce mice in either the LV or DG. Also, no significance was found when comparing Ki67-immunopositive cell numbers from Wt and NR2E1 mice in either the LV or DG. As expected, there was a significant reduction in Ki67-immunopositive cell numbers in fierce mice compared to those in Wt and NR2E1 mice in both the LV (∗∗, P < 0.05) and DG (∗∗, P < 0.05). (Scale bars, 200 μm.) (C) The Kruskal-Wallis H test was performed on 3 mice for all genotypes for both the LV and DG. Error bars represent standard errors of the means.
NR2E1/fierce animals displayed appropriate retinal architecture.
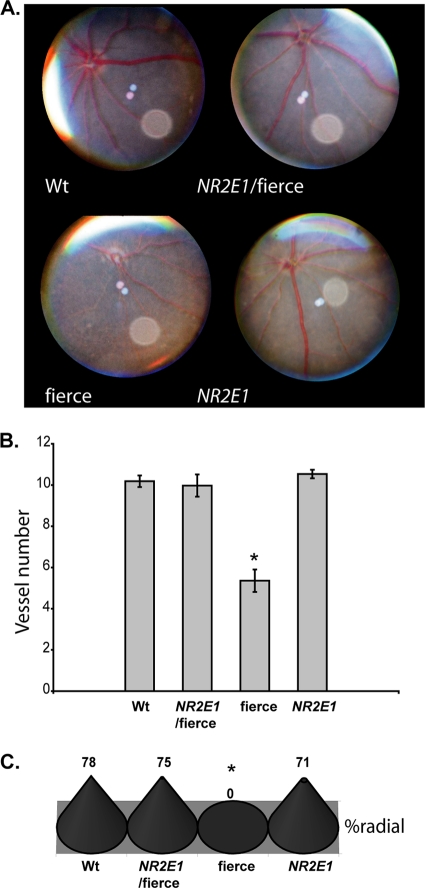
We looked at the eyes of the NR2E1/fierce mice, which exhibited an appropriate expression pattern in the BAC NR2E1-lacZ mice. We began our investigation by evaluating the retinal architecture of the NR2E1/fierce animals using fundus microscopy. Fierce mice characteristically display a reduced and abnormal vascular patterning of the major retinal arteries and veins (1, 56). Our investigation revealed that the blood vessels populating the retina of the NR2E1/fierce animals showed structural characteristics comparable to those of Wt animals, suggesting a correction of this aspect of the fierce phenotype (Fig. 9A). Quantitative analysis of the vessel numbers for NR2E1/fierce animals showed no differences from those for Wt or NR2E1 animals, whereas a significant reduction in blood vessel count was observed when comparing fierce to the other genotypes (P = 0.001) (Fig. 9B). As described in the literature, radial asymmetry was only observed in fierce (P < 0.001) (Fig. 9C) (1, 56). These results render the eyes from the NR2E1/fierce animals indistinguishable from those of the Wt animals, demonstrating complete correction of the vasculature defects of the fierce phenotype.
Fig 9.
NR2E1/fierce mice were corrected for retinal blood vessel defects. (A) Eye fundus photos showed normal blood vessel organization in the Wt, NR2E1/fierce, and NR2E1 retinal surface. The expected blood vessel abnormalities were seen for fierce mice. (B) No significant difference was found in the blood vessel numbers of Wt, NR2E1/fierce, and NR2E1 mice. The blood vessel number was significantly reduced in fierce eyes compared to the other mice (∗, P = 0.001). Error bars represent standard errors of the means. (C) No significant differences were found between Wt, NR2E1/fierce, and NR2E1 mice for asymmetry; only fierce mice showed asymmetry of the blood vessels (∗, P < 0.001). The Kruskal-Wallis H test was performed on 6 to 9 mice for all genotypes (B and C).
The eye defects in the fierce animals also include thinning of the INL and ONL in comparison to Wt animals (56). Adult retinas from NR2E1/fierce animals revealed normal thickness of the INL and ONL compared to Wt and NR2E1 retinas (Fig. 10A). The fierce animal INL measured 23.3 ± 2.40 μm, demonstrating a significant reduction compared to 36.4 ± 0.844 μm (Wt), 35.6 ± 4.48 μm (NR2E1/fierce), and 33.6 ± 3.84 μm (NR2E1) (P < 0.01) (Fig. 10B). The fierce animal ONL also revealed a marked reduction of thickness, measuring 32.3 ± 2.87 μm compared to 56.0 ± 2.72 μm (Wt), 55.6 ± 4.88 μm (NR2E1/fierce), and 50.4 ± 6.23 μm (NR2E1) (P < 0.05) (Fig. 10C). The fierce animal GCL also demonstrated incomplete differentiation, a phenomenon characterized by the presence of remnant retinal ganglion cells (RGC) in the IPL (56). We observed a significant increase in displaced RGC only in fierce retinas, with an average of 20.4 ± 2.44 displaced ganglion cells in comparison to 8.20 ± 0.970 cells (Wt), 8.50 ± 0.957 cells (NR2E1/fierce), and 7.75 ± 1.18 cells (NR2E1) (P < 0.01) (Fig. 10D). These results, combined with the expression pattern analysis of the BAC NR2E1-lacZ, revealed that the genomic fragment used to generate these animals contained all of the elements for proper temporal and spatial expression of the human gene in the eye. An additional human copy of NR2E1 on the Wt background had no effect and corrected the retinal architecture deleterious phenotype of the fierce background. Overall, these results suggested an appropriate functional role of the NR2E1 human protein in the developing eyes, resulting in a correction of the blood vessels and retinal defects in adult animals.
Fig 10.
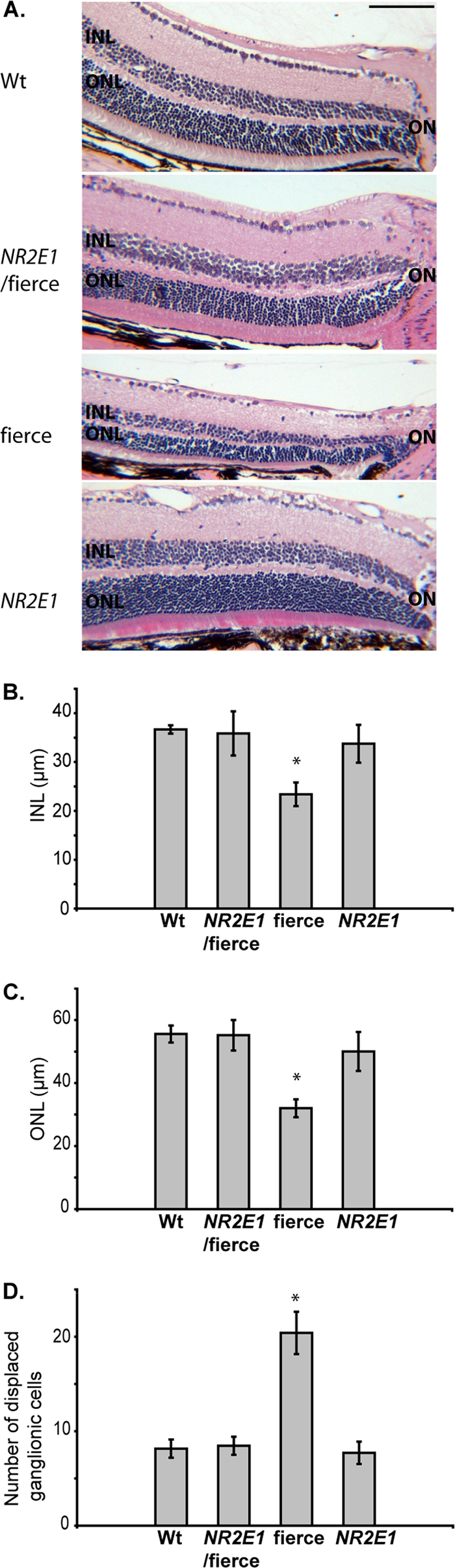
NR2E1/fierce mice were corrected for retinal architecture defects. (A) Hematoxylin-and-eosin-stained paraffin section analysis revealed that the retina of Wt, NR2E1/fierce, and NR2E1 mice appeared similar when the inner nuclear layer (INL) and outer nuclear layer (ONL) thicknesses were examined. Only the fierce retina demonstrated a reduction in the INL and ONL compared to results for the three other genotypes. ON, optic nerve. (Scale bar, 100 μm.) (B) Quantitative analysis showed no significant difference when comparing the INL thicknesses of Wt, NR2E1/fierce, and NR2E1 mice. Only the fierce retina demonstrated a reduction in INL thickness compared to results for the three other genotypes (∗, P < 0.01). (C) No significant difference was found when comparing the ONL thicknesses of Wt, NR2E1/fierce, and NR2E1 mice. Only the fierce retina demonstrated a reduction in ONL thickness compared to the three other genotypes (∗, P < 0.05). (D) No significant difference was found when comparing the number of displaced ganglionic cells in the inner plexiform layer (IPL) of Wt, NR2E1/fierce, and NR2E1 mice. Only the fierce retina demonstrated increased displaced retinal ganglion cells in the IPL compared to results for the other three genotypes (∗, P < 0.01). (B, C, and D) The Kruskal-Wallis H test was performed on 4 to 6 mice for all genotypes. Error bars represent standard errors of the means.
NR2E1/fierce animals displayed functional retinas.
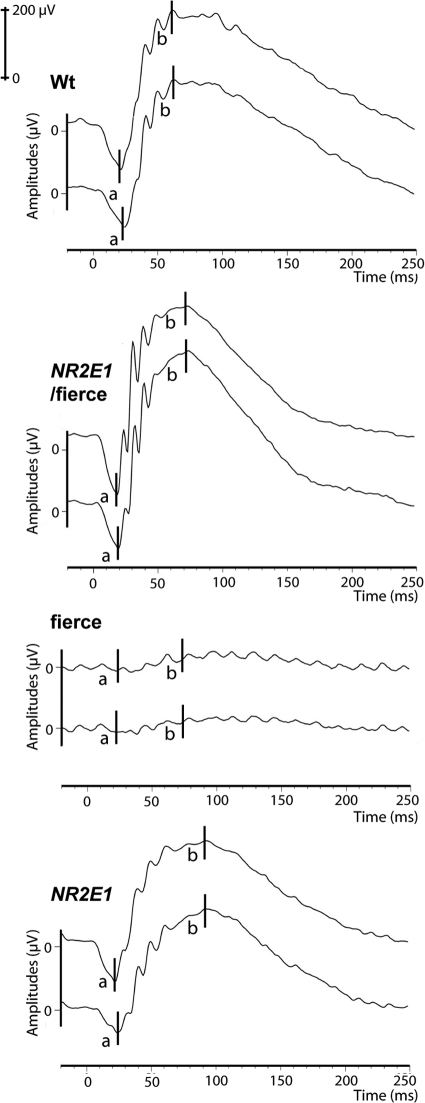
To confirm the functionality of the NR2E1/fierce retinas, we performed electroretinogram (ERG) experiments. In the literature, fierce mice subjected to an ERG demonstrate reduced to absent a-wave and b-wave signals in adult animals (56, 58). We obtained a similar pattern for fierce animals, with an average a-wave of 11.3 ± 3.91 μV and b-wave of 29.3 ± 9.00 μV at 3 cd · s/m2 (Fig. 11). These results demonstrated a significant reduction of amplitude for both the a-wave and b-wave compared to findings for animals of the three other genotypes at the same intensity (P < 0.01) (Fig. 11). No significant difference was found between Wt, NR2E1/fierce and NR2E1 amplitude values, suggesting proper functionality of their retinas as well as no effect of the additional copy of NR2E1 and complete correction of the retina-null phenotype.
Fig 11.
NR2E1/fierce mice were corrected for retinal functional defects. Electroretinogram (ERG) experiments demonstrated normal a-wave and b-wave amplitudes at 3 cd · s/m2 in Wt, NR2E1/fierce, and NR2E1 mice. Only the fierce ERG values differed significantly compared to the other three genotypes (a-wave, P < 0.01; b-wave, P < 0.01). Each line presents measurements from one eye; each graph presents two eyes from one mouse. The Kruskal-Wallis H test was performed on 3 mice for all genotypes.
Comparative genomic analysis revealed candidate brain-specific stem cell-regulatory elements.
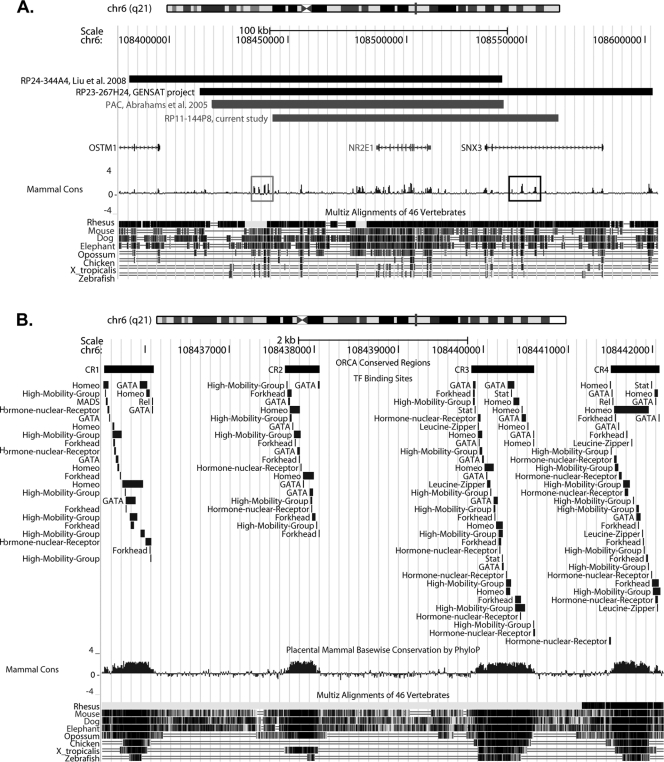
To understand the discrepancy between the results obtained from the human BAC derived mice used in this work to those obtained with a previous humanized mouse model for NR2E1, which demonstrated complete correction of the brain phenotype while only ameliorating the eye phenotype, we decided to undertake a comparative genomic analysis (Fig. 12A) (1). Sequence alignment comparison between our current mouse model and three others, which were proven to be successful in either functionally correcting the brain phenotype using the human gene or conferring proper expression in the brain using the mouse gene, revealed that our sequence was the shortest at the 5′ end (Fig. 12A) (1, 12, 26). Our construct was missing 25 kb of sequence which contained four highly conserved regions, located in the intragenic region and spanning a distance of approximately 6 kb. These regions correspond to the following coordinates according to the UCSC genome browser (assembly February 2009): conserved region (CR) 1, chr6, 108,435,521 to 108,436,101; CR 2, chr6, 108,437,653 to 108,438,062; CR 3, chr6, 108,439,859 to 108,440,601; CR 4, chr6, 108,441,507 to 108,442,084 (Fig. 12A, gray box). The construct used in the current study also contained additional 3′ sequence that was not found in the previously humanized mouse model, which demonstrated partial correction of the eye phenotype (Fig. 12A) (1). Within this additional sequence of 24 kb, a series of five conserved regions were found, located within intron two of the neighboring SNX3 gene and spanning a distance of approximately 11 kb. These regions correspond to the following coordinates according to the UCSC genome browser (assembly February 2009): CR 5, chr6, 108544148 to 108544255; CR 6, chr6, 108547452 to 108547478; CR 7, chr6, 108547888 to 108548739; CR 8, chr6, 108550671 to 108550708; CR 9, chr6, 108553391 to 108554246 (Fig. 12A, black box).
Fig 12.
Comparative genomic analysis revealed candidate regulatory elements. (A) Relative sequence coordinates from constructs used in four different mouse models were retrieved and visualized using the UCSC genome browser (http://genome.ucsc.edu/). The results showed that the bacterial artificial chromosome (BAC) used in the current study model terminated prior to a 5′ conserved element, composed of four conserved regions (gray box). This element was present in all three other mouse models generated. An additional 3′ conserved element, composed of five conserved regions (black box), was also present in the BAC construct used in this study. (B) The four conserved regions found in the 5′ conserved element and described for panel A contained an enriched population of binding sites for transcription factors involved in neurogenesis. The transcription factor binding sites are grouped by family for easier representation.
We used TFBS prediction analysis to identify, and evaluate the overrepresentation of, transcription factors that could bind the four 5′ intragenic conserved regions. The results revealed an enriched presence for TFBSs predicted for transcription factors with gene ontology (GO) terms involved in biological processes such as “nervous system development” (P = 9.23 × 10−12), “central nervous system development” (P = 8.45 × 10−10), “neurogenesis” (P = 2.63 × 10−07), “eye development” (P = 9.45 × 10−06), and “camera-type eye development” (P = 1.18 × 10−04) (Table 1; the complete GO term list is in Table 2 at http://cisreg.ca/MCB-NR2E1/). A randomized statistical analysis performed on these four conserved regions also demonstrated that these GO terms were significant and were not due to chance (Table 1, randomized P value). The transcription factors corresponding to the most significantly enriched GO term, “neurogenesis” (Table 1; randomized P = 0.0122), were particularly relevant to this study. This category contains 25 transcription factors, including the “high mobility group” family (HMG) (Sox2, Sox5, Sox9, and Sox10), the “homeobox” family (including Pax2 and Pax6), and the “hormone nuclear receptor” family (Nr2f1, Nr2e3, and Nr4a2), all of which are known to have important roles in brain development (Table 2, family binding site distribution; Fig. 12B, exact coordinates; see also Table 3 at http://cisreg.ca/MCB-NR2E1/) (13, 20, 34, 50).
Table 1.
Relevant GO termsa
| GO term | GO ID | Transcription factors | 5′ conserved regions analysis |
3′ conserved regions analysis |
||
|---|---|---|---|---|---|---|
| Bonferroni-corrected P value | Randomized P value | Bonferroni-corrected P value | Randomized P value | |||
| Nervous system development | GO:0007399 | STAT3, NR2F1, ESR2, SOX2, MEF2A, PAX5, FEV, PAX2, CREB1, PAX6, TLX1, ZEB1, NHLH1, FOXC1, FOXL1, SOX10, FOXD1, FOXD3, RELA, TFAP2A, FOXA2, MAFB, SOX5, GATA2, NR3C1, NFATC2, E2F1, EN1, HNF1B, RXRA, PBX1, NKX2-5, FOXA1, NR2E3, SRF, SOX9, PPARG, NR4A2 | 9.23094119467758e−12 | 0.0327 | 1.23502929422786e−11 | 0.0220 |
| Central nervous system development | GO:0007417 | PAX6, TLX1, ZEB1, NHLH1, FOXC1, FOXL1, ESR2, FOXA2, MAFB, SOX2, SOX5, CREB1, GATA2, NR3C1, NFATC2, NR2F1, E2F1, EN1, HNF1B, SOX9, SOX10, PPARG | 8.4507010705991e−10 | 0.0147 | 2.99022615274573e−09 | 0.0356 |
| Neurogenesis | GO:0022008 | STAT3, NR2F1, ESR2, SOX2, PAX2, CREB1, RELA, FOXA2, SOX5, GATA2, TLX1, RXRA, PBX1, NKX2-5, FOXA1, NR2E3, PAX6, SRF, EN1, SOX9, SOX10, PPARG, FOXD1, FOXD3, NR4A2 | 2.63390159936542e−07 | 0.0122 | 5.7376060069322e−07 | 0.0344 |
| Eye development | GO:0001654 | PAX6, SOX2, FOXC1, STAT3, FOXL1, YY1, SP1, ZEB1, PAX4, MAX, NR2E3 | 9.45330733200263e−06 | 0.0300 | 6.69632064726976e−05 | 0.2595 |
| Camera-type eye development | GO:0043010 | FOXC1, FOXL1, YY1, SP1, ZEB1, PAX4, MAX, NR2E3, SOX2 | 1.18317513845212e−04 | 0.0473 | 9.3334459117566e−04 | 0.4437 |
The most relevant Gene Ontology (GO) terms that have been found in the overrepresentation analysis are presented. GO terms are given in the first column, with their GO identifier (ID) in the second column. Gene symbols for the transcription factors that belong to those GO terms and have at least one binding site predicted within the conserved regions are given in the third column. A Bonferroni-corrected P value, associated with the overrepresentation of the corresponding GO term, is given in the fourth and sixth columns. The fifth and seventh columns contain the randomized P values obtained after the nucleotide shuffling analysis (see Materials and Methods).
Table 2.
Neurogenesis transcription factor familiesa
| Family | Transcription factor(s) |
|---|---|
| Stat | Stat3 |
| Hormone nuclear receptor | NR2F1, Esr2, RXRA, Nr2e3, PPARG, Nr4a2 |
| High-mobility group | Sox2, SOX5, SOX9, Sox10 |
| Homeo | PAX2, Tlx1, Pbx1, NKX2-5, PAX6 EN1 |
| Leucine zipper | CREB1 |
| Rel | RELA |
| Forkhead | FOXA2, Foxa1, FOXD1, FOXD3 |
| GATA | GATA2 |
| MADS | SRF |
The family classification used in Fig. 12B is presented. The first column gives the family classification of the different transcription factors (listed in the second column) that are associated with the neurogenesis Gene Ontology (GO) term. Structural classification came from JASPAR (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl?rm=struct_browse).
Similarly, we found enrichment for TFBSs prediction in the five 3′ SNX3 intronic conserved regions for all the previously reported GO terms: “nervous system development” (P = 1.24 × 10−11), “central nervous system development” (P = 2.99 × 10−09), “neurogenesis” (P = 5.74 × 10−07), “eye development” (P = 6.70 × 10−05), and “camera-type eye development” (P = 9.33 × 10−04) (Table 1, complete GO term list; see also Table 4 at http://cisreg.ca/MCB-NR2E1/). However, only the terms “nervous system development,” “central nervous system development,” and “neurogenesis” remained significant after the randomized analysis was performed (Table 1). In addition, since the SNX3 gene is also expressed in the central nervous system (30), these regions may be present to regulate the gene in which they reside, and there is no requirement to hypothesize that they regulate NR2E1. In contrast, the location proximal to the NR2E1 promoter supports the involvement of the four 5′ conserved regions in NR2E1 regulation. This allows us to predict that the incomplete NR2E1 expression and function observed in the brains of our animals were most likely due to the absence of these four 5′ conserved regions.
DISCUSSION
In the current study, we have demonstrated for the first time the expression resulting from the insertion of a BAC carrying the human NR2E1 gene in both developing and adult mice. We have shown that human NR2E1-lacZ expression mimics that of the mouse homolog in both developing and adult eyes, resulting in normal retina in the animals harboring only one functional allele of the human gene, thereby correcting the retina-null phenotype. These results show that the BAC construct used in this study contains all of the elements for appropriate spatial and temporal eye expression of the human gene. Further, the results support the hypothesis that the human gene is functionally equivalent to the mouse gene in the mouse eye and that the previous incomplete correction of the fierce eye phenotype may indeed be attributed to excess copy number (1). Finally, the functional conservation of NR2E1 highlights the potential role of this gene in human eye diseases of unknown etiology.
In stark contrast, our results also show that the BAC used in this study demonstrates an unexpected absence of expression of the human NR2E1 gene in key neurogenic regions of both the developing and adult brain. The Nr2e1 mouse gene has been previously shown to regulate the proliferation rate of neural stem cell populations in the developing telencephalon, and the absence of expression of the Nr2e1 gene results in premature neuronal differentiation, leading to depletion of the neural stem cell populations (40). This affects the development of subsequent forebrain structures, such as the upper layer of the cortex, the DG, and the OB. In this study, animals harboring the functional NR2E1 allele generated from the human BAC display a fierce-like phenotype in adult neurogenic brain regions. This phenotype includes morphological defects in the DG of the hippocampus and SVZ of the lateral ventricles, which correlate with an absence of expression of the BAC NR2E1-lacZ reporter gene in proliferating cells in these regions. These results provide a distinct separation of the regulatory mechanisms governing the NR2E1 gene, leading to an abnormal brain expression pattern while retaining normal cell-specific retinal expression.
In addition to the phenotype found in adult neurogenic brain regions, the NR2E1/fierce brain displays an attenuated version of cortex and OB hypoplasia, a hallmark of the fierce phenotype. We conclude that this is due to a partial correction of the fierce phenotype during embryonic development as the formation of these structures involves the radial migration of neurons from the VZ of the DP but also the tangential migration of neurons from the VZ of the subpallium (3, 4, 10, 33, 51). In our current study, the expression results obtained in the BAC NR2E1-lacZ embryos demonstrate an absence of expression in the DP of the telencephalon while preserving expression in the MP, LP/VP, and subpallium regions. This suggests that the partial correction found in the adult animals harboring the current human BAC construct is due to expression of the gene in the LP/VP and subpallium regions during development.
Our hypothesis that the human gene is capable of completely correcting the fierce brain phenotype was based primarily on previous published results using a random-insertion multiple-copy mouse model (1). Thus, the current incomplete correction could be due to the single-copy insert or location on the X chromosome. X inactivation makes it impossible for us to test two expressed copies of the human BAC in female mice (16, 25). However, taking into consideration the complete absence of expression of the human gene in the mouse DP, we favor an explanation involving missing key regulatory regions. Following this path led us to the identification of 5′ conserved regulatory regions that are statistically enriched for binding sites of transcription factors involved in neurogenesis. Among these, Pax6 has been shown to genetically interact with Nr2e1 in the establishment of the pallio-subpallial boundary (48), and Sox2 has been shown to play an important role in controlling Nr2e1 transcription in cultured neural stem cells (46). Furthermore, Pax6 and Sox2 can form a co-DNA binding partner that regulates initiation of lens development (19). Thus, we hypothesize that we have identified important regulatory regions that may interact with Pax6 and Sox2 to control specific expression of NR2E1 in both the developing and adult brain.
The findings from the current study are also of critical importance for future human genetic disease studies. The candidate novel regulatory regions found in the current study were not included in previous association studies and deep sequencing analysis of NR2E1 (21–23). Two of these studies have reported the finding of candidate regulatory mutations in patients with various brain disorders, and some of these mutations were predicted to affect the binding sites of specific regulators involved in brain development (22, 23). Hence, we argue that future patient studies, screening for mutations at the NR2E1 locus, should include these new candidate regulatory regions in their analysis. In addition, NR2E1 is now a very strong candidate to be involved in human eye disease, a role never examined in patients to date. Importantly, the docking technology used to generate the animals in this study can be reapplied using BAC recombineered to carry candidate human mutations, including regulatory mutations, to test their ability to correct the fierce phenotype. Finally, this work serves as a paradigm and can be generalized to the study of other human genes for which a BAC construct can be derived and a mutant mouse phenotype exists.
ACKNOWLEDGMENTS
We thank Elodie Portales-Casamar for the design of bEMS86, Jacek Mis and Kristi Hatakka for microinjection, Baoping Song for tissue paraffin expertise, Jingsong Wang for microscopy expertise, Christopher Roach for his technical help with the experiments, and Ximena Corso-Diaz, Charles N. de Leeuw, Katrina Bepple, and Bibiana K. Y. Wong for aid in manuscript preparation. We also thank the entire Pleiades Promoter Project team for their pipeline work, which directly and indirectly facilitated the generation of B6.129P2(Cg)-Hprttm86(NR2E1,bEMS223)Ems, B6.129P2(Cg)-Hprttm73(Ple142-lacZ)Ems, and B6.129P2(Cg)-Hprttm87(Ple142-lacZ)Ems.
This work was funded by the U.S. National Institute for Mental Health (4R33MH083515-03 to E.M.S.), Genome Canada, Genome British Columbia, GlaxoSmithKline R&D Ltd., BC Mental Health and Addiction Services, Child and Family Research Institute, University of British Columbia (UBC) Institute of Mental Health, and UBC Office of the Vice President Research (048PLE to E.M.S.), Canadian Research Chairs (950-202735 Tier II to E.M.S.), and Canadian Institutes of Health Research (MSH-691153 to W.W.W. and VIH-105440 to K.G.-E. and C.Y.G-E.).
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Abrahams BS, et al. 2005. Pathological aggression in “fierce” mice corrected by human nuclear receptor 2E1. J. Neurosci. 25:6263–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abrahams BS, et al. 2002. Novel vertebrate genes and putative regulatory elements identified at kidney disease and NR2E1/fierce loci. Genomics 80:45–53 [DOI] [PubMed] [Google Scholar]
- 3. Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. 1997. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278:474–476 [DOI] [PubMed] [Google Scholar]
- 4. Angevine JB, Jr, Sidman RL. 1961. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192:766–768 [DOI] [PubMed] [Google Scholar]
- 5. Bronson SK, et al. 1996. Single-copy transgenic mice with chosen-site integration. Proc. Natl. Acad. Sci. U. S. A. 93:9067–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchholz F, Angrand PO, Stewart AF. 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16:657–662 [DOI] [PubMed] [Google Scholar]
- 7. Butler AB. 1994. The evolution of the dorsal pallium in the telencephalon of amniotes: cladistic analysis and a new hypothesis. Brain Res. Rev. 19:66–101 [DOI] [PubMed] [Google Scholar]
- 8. Butler AB. 1994. The evolution of the dorsal thalamus of jawed vertebrates, including mammals: cladistic analysis and a new hypothesis. Brain Res. Rev. 19:29–65 [DOI] [PubMed] [Google Scholar]
- 9. Copeland NG, Jenkins NA, Court DL. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769–779 [DOI] [PubMed] [Google Scholar]
- 10. de Carlos JA, Lopez-Mascaraque L, Valverde F. 1996. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J. Neurosci. 16:6146–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23:257–258 [DOI] [PubMed] [Google Scholar]
- 12. Gong S, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425:917–925 [DOI] [PubMed] [Google Scholar]
- 13. Graham V, Khudyakov J, Ellis P, Pevny L. 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39:749–765 [DOI] [PubMed] [Google Scholar]
- 14. Guerin K, et al. 2008. Systemic aminoglycoside treatment in rodent models of retinitis pigmentosa. Exp. Eye Res. 87:197–207 [DOI] [PubMed] [Google Scholar]
- 15. Hawes NL, et al. 1999. Mouse fundus photography and angiography: a catalogue of normal and mutant phenotypes. Mol. Vis. 5:22. [PubMed] [Google Scholar]
- 16. Heaney JD, Rettew AN, Bronson SK. 2004. Tissue-specific expression of a BAC transgene targeted to the Hprt locus in mouse embryonic stem cells. Genomics 83:1072–1082 [DOI] [PubMed] [Google Scholar]
- 17. Horan K, et al. 2008. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147:41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson A, Panayiotidis P, Foroni L. 1998. The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21. Genomics 50:34–43 [DOI] [PubMed] [Google Scholar]
- 19. Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. 2001. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 15:1272–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitambi SS, Hauptmann G. 2007. The zebrafish orphan nuclear receptor genes nr2e1 and nr2e3 are expressed in developing eye and forebrain. Gene Expr. Patterns 7:521–528 [DOI] [PubMed] [Google Scholar]
- 21. Kumar RA, et al. 2007. Absence of mutations in NR2E1 and SNX3 in five patients with MMEP (microcephaly, microphthalmia, ectrodactyly, and prognathism) and related phenotypes. BMC Med. Genet. 8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar RA, et al. 2007. Mutation and evolutionary analyses identify NR2E1-candidate-regulatory mutations in humans with severe cortical malformations. Genes Brain Behav. 6:503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar RA, et al. 2008. Initial association of NR2E1 with bipolar disorder and identification of candidate mutations in bipolar disorder, schizophrenia, and aggression through resequencing. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B:880–889 [DOI] [PubMed] [Google Scholar]
- 24. Li W, et al. 2008. Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol. Endocrinol. 22:56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liskay RM, Evans RJ. 1980. Inactive X chromosome DNA does not function in DNA-mediated cell transformation for the hypoxanthine phosphoribosyltransferase gene. Proc. Natl. Acad. Sci. U. S. A. 77:4895–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu HK, et al. 2008. The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev. 22:2473–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu HK, et al. 2010. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev. 24:683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lyznik LA, Mitchell JC, Hirayama L, Hodges TK. 1993. Activity of yeast FLP recombinase in maize and rice protoplasts. Nucleic Acids Res. 21:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyawaki T, et al. 2004. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J. Neurosci. 24:8124–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mizutani R, et al. 2011. Developmental expression of sorting nexin 3 in the mouse central nervous system. Gene Expr. Patterns 11:33–40 [DOI] [PubMed] [Google Scholar]
- 31. Monaghan AP, et al. 1997. Defective limbic system in mice lacking the tailless gene. Nature 390:515–517 [DOI] [PubMed] [Google Scholar]
- 32. Monaghan AP, Grau E, Bock D, Schutz G. 1995. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development 121:839–853 [DOI] [PubMed] [Google Scholar]
- 33. Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. 2001. Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 4:143–150 [DOI] [PubMed] [Google Scholar]
- 34. Naka H, Nakamura S, Shimazaki T, Okano H. 2008. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat. Neurosci. 11:1014–1023 [DOI] [PubMed] [Google Scholar]
- 35. Park HJ, et al. 2011. Erratum to: the neural stem cell fate determinant TLX promotes tumorigenesis and genesis of cells resembling glioma stem cells. Mol. Cells 31:199. [DOI] [PubMed] [Google Scholar]
- 36. Park HJ, et al. 2010. The neural stem cell fate determinant TLX promotes tumorigenesis and genesis of cells resembling glioma stem cells. Mol. Cells 30:403–408 [DOI] [PubMed] [Google Scholar]
- 37. Parsons DW, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phillips HS, et al. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173 [DOI] [PubMed] [Google Scholar]
- 39. Pignoni F, et al. 1990. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62:151–163 [DOI] [PubMed] [Google Scholar]
- 40. Roy K, et al. 2004. The Tlx gene regulates the timing of neurogenesis in the cortex. J. Neurosci. 24:8333–8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roy K, Thiels E, Monaghan AP. 2002. Loss of the tailless gene affects forebrain development and emotional behavior. Physiol. Behav. 77:595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schatz O, Golenser E, Ben-Arie N. 2005. Clearing and photography of whole mount X-gal stained mouse embryos. Biotechniques 39:650–656 [DOI] [PubMed] [Google Scholar]
- 43. Schlake T, Bode J. 1994. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33:12746–12751. [DOI] [PubMed] [Google Scholar]
- 44. Schmouth JF, Bonaguro RJ, Corso-Diaz X, Simpson EM. Modeling human-regulatory variation in mouse: finding the function in GWAS and whole genome sequencing. PLoS Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shi Y, et al. 2004. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427:78–83 [DOI] [PubMed] [Google Scholar]
- 46. Shimozaki K, et al. 22 December 2011. Sex determining region Y-box 2 (SOX2) regulation of nuclear receptor tailless (TLX) transcription in adult neural stem cells. J. Biol. Chem. doi/10.1074/jbc.M111.290403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sim FJ, et al. 2006. Neurocytoma is a tumor of adult neuronal progenitor cells. J. Neurosci. 26:12544–12555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stenman J, Yu RT, Evans RM, Campbell K. 2003. Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development 130:1113–1122 [DOI] [PubMed] [Google Scholar]
- 49. Stenman JM, Wang B, Campbell K. 2003. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J. Neurosci. 23:10568–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stoykova A, Treichel D, Hallonet M, Gruss P. 2000. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 20:8042–8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tamamaki N, Fujimori KE, Takauji R. 1997. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J. Neurosci. 17:8313–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taranova OV, et al. 2006. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 20:1187–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uemura A, Kusuhara S, Wiegand SJ, Yu RT, Nishikawa S. 2006. Tlx acts as a proangiogenic switch by regulating extracellular assembly of fibronectin matrices in retinal astrocytes. J. Clin. Invest. 116:369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wong BK, et al. 2010. Hyperactivity, startle reactivity and cell-proliferation deficits are resistant to chronic lithium treatment in adult Nr2e1(frc/frc) mice. Genes Brain Behav. 9:681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang GS, et al. 2009. Next generation tools for high-throughput promoter and expression analysis employing single-copy knock-ins at the Hprt1 locus. Genomics 93:196–204 [DOI] [PubMed] [Google Scholar]
- 56. Young KA, et al. 2002. Fierce: a new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav. Brain Res. 132:145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu D, et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu RT, et al. 2000. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc. Natl. Acad. Sci. U. S. A. 97:2621–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu RT, McKeown M, Evans RM, Umesono K. 1994. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370:375–379 [DOI] [PubMed] [Google Scholar]
- 60. Zhang CL, Zou Y, He W, Gage FH, Evans RM. 2008. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451:1004–1007 [DOI] [PubMed] [Google Scholar]
- 61. Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. 2006. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 20:1308–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]