Abstract
TREX is a conserved multiprotein complex that is necessary for efficient mRNA export to the cytoplasm. In Saccharomyces cerevisiae, the TREX complex is additionally implicated in RNA quality control pathways, but it is unclear whether this function is conserved in mammalian cells. The Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein binds and recruits the TREX component REF/Aly to viral mRNAs. Here, we demonstrate that REF/Aly is recruited to the KSHV noncoding polyadenylated nuclear (PAN) RNA by ORF57. This recruitment correlates with ORF57-mediated stabilization of PAN RNA, suggesting that REF/Aly promotes nuclear RNA stability. Further supporting this idea, tethering REF/Aly to PAN RNA is sufficient to increase the nuclear abundance and half-life of PAN RNA but is not sufficient to promote its export. Interestingly, REF/Aly appears to protect the poly(A) tail from deadenylation, and REF/Aly-stabilized transcripts are further adenylated over time, consistent with previous reports linking poly(A) tail length with nuclear RNA surveillance. These studies show that REF/Aly can stabilize nuclear RNAs independently of their export and support a broader conservation of RNA quality control mechanisms from yeast to humans.
INTRODUCTION
Like many RNA processing events in the eukaryotic cell, mRNA export from the nucleus to the cytoplasm is coupled with other steps in mRNA synthesis (56, 59, 65). In mammalian cells, the protein complex TREX associates with the 5′ end of transcripts as a result of splicing, and TREX subsequently promotes the export of the mature mRNA (6, 51). The mammalian TREX complex contains REF/Aly, UAP56, hTEX1, and CIP29 as well as components of the THO complex (hHpr1, hTho2, fSAP35, fSAP79, and fSAP24), and many of these proteins are conserved from Saccharomyces cerevisiae to humans (17, 51, 60, 70, 71, 82). However, in yeast, TREX recruitment is linked with transcription and 3′-end formation rather than splicing (21, 36, 39). TREX deposition subsequently recruits TAP/NXF1, and the mRNA is “handed off” from REF/Aly to TAP/NXF1 (24). TAP/NXF1, with its partner p15/NXT1, is the receptor responsible for bulk polyadenylated RNA export (18, 31).
A role for the TREX component REF/Aly in mRNA export is supported by several lines of evidence. The human REF/Aly protein is conserved from yeast to humans, and the yeast homolog, Yra1, is essential for bulk mRNA export (69, 71). REF/Aly interacts directly with both UAP56 and TAP/NXF1, two factors required for bulk mRNA export (17, 51, 69–71). REF/Aly increases mRNA export efficiency in Xenopus oocyte systems (45, 61), and artificial tethering of REF/Aly increases the export efficiency of otherwise inefficiently exported transcripts (13, 17, 25, 80). Small interfering RNA (siRNA)-mediated knockdown of REF family members decreases bulk mRNA export to various degrees in metazoan cells. In some cases, little or no nuclear poly(A) accumulation is observed, while other studies report an accumulation of poly(A) RNA in the nucleus (15, 19, 25, 37, 43, 55). These distinct phenotypes may be due to redundancy with other adaptors (78), compensatory changes in gene expression (25), and/or differences in experimental procedures. Knockdown of REF is toxic to both Drosophila and human cells (19, 25), but in Drosophila cells, no bulk poly(A) accumulation is observed. This observation suggests that Drosophila REF is necessary for export of only a subset of essential mRNAs, or that it may have an additional essential function(s) (19). Indeed, REF/Aly has previously been implicated in transcriptional control (3, 77), and here we propose a role for REF/Aly in nuclear RNA stability.
RNA surveillance or RNA quality control pathways are the processes that destroy transcripts that are misprocessed or unfolded and/or do not assemble into a suitable ribonucleoprotein particle (RNP) (16, 65). In yeast, Yra1 is linked to the RNA quality control machinery. Iglesias and colleagues demonstrated that Yra1 ubiquitination leads to its release from the nuclear messenger RNP (mRNP) and proposed that this is part of a nuclear RNA surveillance mechanism that selectively promotes export of mature mRNPs (30). More generally, export factors are physically or genetically linked to the nuclear RNA decay machinery involved in transcript surveillance (14, 26, 33, 41, 44, 76, 81). For example, defects in mRNA export factors lead to hyperadenylation and retention of transcripts at the site of transcription. Moreover, this retention depends on Rrp6, an exonuclease that otherwise degrades aberrant RNAs, but the precise mechanism of retention remains unknown (27, 34, 62). Significantly less is known about the interrelationships between mRNA export, polyadenylation, and RNA surveillance in mammalian nuclei, but recent work has shown that inhibition of mRNA export by TAP/NXF1 knockdown leads to a hyperadenylation phenotype similar to that observed in yeast (58), and one factor, ZC3H3, has been proposed to link regulation of polyadenylation with export in Drosophila and human cells (29).
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a multifunctional regulator of gene expression called ORF57 (Mta) that has been implicated in transcriptional control, splicing regulation, translation, and mRNA export (2, 10, 48, 73). ORF57 interacts directly with both REF/Aly and viral mRNAs which, in some cases, increases the export efficiency of the mRNA (1, 47, 49, 54). Because most KSHV mRNAs are transcribed from single-exon genes, it has been proposed that ORF57 recruits the TREX complex to viral mRNAs to promote splicing-independent export (1, 49). In addition, we have recently shown that ORF57 binds and stabilizes the KSHV polyadenylated nuclear (PAN) RNA (63), a 5′-capped, RNA polymerase II-transcribed, polyadenylated noncoding RNA that is retained in the nucleus (63, 72, 84). Thus, ORF57 promotes the nuclear stability of transcripts independently of its role in mRNA export.
Because of its mRNA-like characteristics and nuclear localization, PAN RNA serves as a useful tool for examining nuclear events in gene expression uncoupled from downstream processes such as mRNA export or translation. The present studies of ORF57 and PAN RNA reveal that REF/Aly stabilizes RNA in a fashion that is separable from its role in mRNA export. ORF57 recruits REF/Aly to PAN RNA where it binds directly to the transcript. The REF/Aly association with PAN RNA displays a 5′ bias, reminiscent of the placement of REF/Aly on spliced mRNAs (6, 51). Deletion of the REF/Aly binding domain from ORF57 abolishes its stabilization function, supporting the model that REF/Aly is an essential cofactor for ORF57-mediated nuclear RNA stabilization. Artificial tethering of REF/Aly to PAN RNA in the absence of ORF57 leads to higher PAN RNA levels by increasing the PAN RNA half-life and maintaining longer poly(A) tail lengths. Perhaps surprisingly, REF/Aly tethering is not sufficient to promote PAN RNA export from the nucleus. Taken together, our results strongly support a role for REF/Aly in nuclear RNA stability and suggest that binding of export factors actively protects nuclear transcripts from degradation by the RNA quality control machinery in the mammalian cell nucleus.
MATERIALS AND METHODS
Plasmids.
With the exception of TetRP-PANΔENE-6MS2, described below, all PAN RNA and ORF57 expression plasmids were described previously, as well as the MS2-only (pcNMS2-NLS-Flag) construct (11–13, 63). To create TetRP-PANΔENE-6MS2, six MS2 binding sites were cut from MS2-PANΔ79 using NcoI and ligated to Trp-PANΔ79 cut with NcoI. The Fl-ΔReBD expression plasmid, pc-Flag-ORF57II ΔReBD, was created using SOEing PCR techniques (28). Twenty-eight of the 35 amino acids in the putative REF-binding domain were deleted. Primers NC576 (5′ GAGCAATTGTCCGAACCCGC 3′) and NC577 (5′ GGGACGTGGGATGGTGGGGCGCTCGCAGGAGTCTGAGTT 3′) were used to generate the 5′ fragment, and primers NC578 (5′ AACTCAGACTCCCTGCGAGCGCCCCACCATCCCACGTCCC 3′) and NC579 (5′ TGCTCTTATGAGAGCGGTGA 3′) were used to create the 3′ fragment. pc-Flag-ORF57II was used as the template. The resulting PCR amplicons were used as templates for PCR using primers NC576 and NC579 to generate the ORF57 fragment containing the REF-binding domain deletion. The resulting PCR product was cut with KpnI and EcoRV and inserted into pc-Flag-ORF57II cut with the same restriction enzymes.
Cell culture, transfection, and RNA procedures.
HEK293 and 293TOA cells were grown in Dulbecco's modified Eagle's medium (Sigma) containing 10% fetal bovine serum (FBS), 1× penicillin-streptomycin (Sigma), and 2 mM l-glutamate. Tetracycline-free FBS (Clontech) was used for 293TOA medium. 293TOA medium was supplemented with 100 μg/ml G418.
HEK293 and 293TOA cells were transfected using TransIT-293 (Mirus) according to the manufacturer's instructions. UV cross-linking experiments were performed in 10-cm plates, with a total of 12 μg plasmid DNA. A typical transfection mixture consisted of 1.5 μg PAN-WT, 3.0 μg ORF50, 3.0 μg ORF57, and 3.0 μg Fl-REF. Because ORF57 increases PAN RNA levels, we increased the amount of PAN-WT plasmid to 6.0 μg in the samples lacking ORF57 expression to yield roughly equivalent PAN RNA expression levels. All control samples were balanced with matched empty vector plasmids so that all samples contained equal amounts of transfected DNA. MS2-REF and ORF57 titrations were performed in 12-well tissue culture plates with 0.7 to 0.8 μg plasmid DNA. Transfections were set up as previously described (63). Total RNA was harvested 18 to 24 h after transfection using TRI reagent (Molecular Research Center). RNA was analyzed by standard Northern blotting techniques (7, 13). In situ hybridization experiments were performed as previously described (13, 63).
For Northern blots with oligonucleotide probes (see Fig. 6D), the probes were 5′-end labeled with T4 polynucleotide kinase using standard procedures. Oligonucleotides NC29 (5′ ATCGGCGGCACCAATGAAAACCAGAAGCGGCAAGAAGGCA 3′) and NC31 (GCACGTTAAATTGTCAAAAGTATAACATGTTTTTCCAATA 3′) were used to detect the PAN RNA 5′ and 3′ ends, respectively. Hybridization was performed for 4 to 18 h in ExpressHyb (Clontech) at 42°C.
RNase H digestion.
To remove poly(A) tails, total RNA was incubated for 1 h at 37°C with 0.375 units of RNase H (Promega), 10 U RNasin (Promega), and 1.25 μM oligonucleotide dT40, in RNase H Buffer (20 mM Tris [pH 7.5], 100 mM KCl, 10 mM dithiothreitol [DTT], 10 mM MgCl2) in a final volume of 20 μl. To cleave PAN RNA and resolve changes in poly(A) tail length (see Fig. 6), RNase H digests were identical, except that NC581 (5′ AATCCAATGCAATAACCCGCAAGG 3′) replaced the dT40 oligonucleotide. Differences in poly(A) tail length were determined by running a 0.1- to 2-kb RNA ladder (Invitrogen) next to treated samples. A standard curve was generated by measuring the migration of each band from a fixed point and plotting the mobility by the log10 of the molecular weight of the ladder. Mean poly(A) tail lengths, as defined by the midpoint of the band, were determined based on the standard curve.
Cross-linking experiments.
UV cross-linking experiments were performed as previously described (9). However, two different wash conditions were used. Radioimmunoprecipitation assay (RIPA) washes were performed by washing beads five times in 1 ml RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 150 mM sodium chloride, 50 mM Tris-HCl [pH 8.0], 2 mM EDTA) and nutating at room temperature for 3 min. In some cases, we used a more stringent washing procedure, as follows: (i) 1 ml RIPA, (ii) 500 μl RIPA-U Plus [RIPA supplemented with 1 M urea, 0.2 mg/ml poly(U) RNA, and 1 mg/ml torula yeast RNA], (iii) 500 μl RIPA-U Plus, (iv) 1 ml low-salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8], 150 mM NaCl), (v) 1 ml high-salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8], 500 mM NaCl), and (vi) 1 ml Tris-EDTA. To ensure that both conditions abolished the ORF57-REF interaction, protein was taken from inputs and pellets and analyzed by Western blotting (see Fig. S1 in the supplemental material).
To determine the position of REF binding (see Fig. 2), UV cross-linking assays were modified and real-time PCR was performed as previously described (57, 66). Primer amplification efficiencies for the indicated amplicons (see Fig. 2C) were determined to be 92% (50 to 124), 91% (293 to 372), 82% (642 to 728), and 83% (994 to 1064). Threshold cycle (CT) values and amplification efficiencies were used to determine the relative quantities (RQ) of inputs and pellets. RQ values of the no-RT controls were subtracted from those of RT-containing samples, and percent immunoprecipitation was determined by calculating the pellet/input ratio. These values were then normalized to those of the +UV +Fl-REF samples to compare between experiments, and all values are presented relative to those for the 5′-most amplicon.
Decay assays.
Decay assays were performed as previously described (11) with the following exceptions. 293TOA cells were plated on 12-well plates and transfected with 0.3 μg TetRP-PANΔENE-6MS2 and 0.4 μg of the appropriate MS2 expression construct per well. At given time points, total RNA was harvested using TRI reagent. The endogenous 7SK RNA served as a loading control.
RESULTS
REF/Aly binds to PAN RNA in an ORF57-dependent manner.
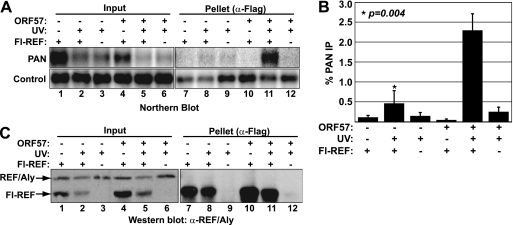
ORF57 binds viral mRNAs and recruits REF/Aly to the mRNP, presumably to facilitate viral mRNA export. However, ORF57 also binds to PAN RNA but does not lead to its export. It is possible that ORF57 binding does not promote PAN RNA export because REF/Aly is not recruited to PAN RNA. Alternatively, REF/Aly recruitment by ORF57 may not be sufficient for PAN RNA export. To distinguish between these possibilities, we employed a UV irradiation cross-linking RNA immunoprecipitation approach (9). We transfected HEK293 cells with expression constructs for PAN RNA, ORF57, and/or an N-terminally Flag-tagged version of the murine REF2-I protein (Fl-REF), which is 74% identical to human REF/Aly but has shorter variable regions (71). The following day, transfected cells were exposed to UV light to covalently cross-link protein with RNA. Immediately following UV treatment, the cells were lysed under stringent conditions, Fl-REF was immunoprecipitated with anti-Flag antibodies, and the presence of PAN RNA in the immunoprecipitates was examined by Northern blot. Figure 1A shows that PAN RNA was coimmunoprecipitated with Fl-REF in the presence of ORF57 (lane 11), but it was nearly undetectable in the absence of ORF57 (lane 8). Importantly, neither the no-cross-linking (Fig. 1A, lanes 7 and 10) nor the no-Fl-REF controls (lanes 9 and 12) showed significant immunoprecipitation of PAN RNA. Quantification of the data demonstrated a statistically significant increase in Fl-REF cross-linking to PAN RNA when ORF57 was expressed (Fig. 1B). Western blot analysis of the proteins verified that Fl-REF was efficiently immunoprecipitated under the stringent conditions used for UV cross-linking experiments (Fig. 1C, lanes 7 to 12). Additionally, inspection of the REF/Aly levels in the cell lysate using a REF/Aly antibody showed that the exogenously expressed Fl-REF levels were only slightly above the endogenous levels. Moreover, there was no dramatic upregulation of Fl-REF upon coexpression of ORF57 (Fig. 1C, compare lanes 1 and 2 to lanes 4 and 5). As an additional control, we verified that the immunoprecipitation conditions used were stringent enough to disrupt the REF/Aly interaction with ORF57 (see Fig. S1 in the supplemental material). From these data, we conclude that ORF57 promotes a direct interaction between REF/Aly and PAN RNA. Because live cells were exposed to UV in this experiment, the absence of signal in the no-UV controls (Fig. 1C, lanes 7 and 10) indicates that the interaction occurs in cells and is not due to reassortment of protein-RNA complexes postlysis (9, 53). Furthermore, UV irradiation is a zero-length cross-linker, so UV-dependent cross-linking can be interpreted as the result of a direct interaction between a protein and an RNA (9). Thus, these data show that Fl-REF is recruited to the PAN RNP in an ORF57-dependent fashion and that, upon recruitment, Fl-REF interacts directly with the RNA.
Fig 1.
REF/Aly binds PAN RNA in an ORF57-dependent fashion. (A) UV cross-linking shows that Fl-REF binds PAN RNA in an ORF57-dependent manner. Cells transfected with PAN RNA, Fl-REF, and ORF57 expression constructs as indicated were exposed to UV light ∼24 h after transfection or were left untreated. Lysates from the transfected cells were subjected to immunoprecipitation with anti-Flag–agarose. Northern blotting was performed to detect PAN RNA in the input and pellets; different exposures of the input and pellets are displayed. The control panels show an exogenously added β-globin transcript used to normalize RNA recovery and loading. (B) Quantification of UV cross-linking experiments. PAN RNA signals were first normalized to the β-globin control; then the pellet values were divided by input values to determine the percent PAN RNA immunoprecipitation. Average values were plotted for each of the samples; error bars show standard deviations (n = 3). The P value is from an unpaired Student's t test of the data sets for the +Fl-REF +UV samples. (C) Western blot analysis to confirm immunoprecipitation in UV cross-linking experiments. Input and pellet protein samples were run on a gel, transferred to nitrocellulose, and probed with a monoclonal anti-REF/Aly antibody that recognizes Fl-REF as well as the endogenous REF/Aly. The murine Fl-REF migrates at a higher mobility due to its shorter nonconserved variable regions (71). The input and pellet panels had different exposure times.
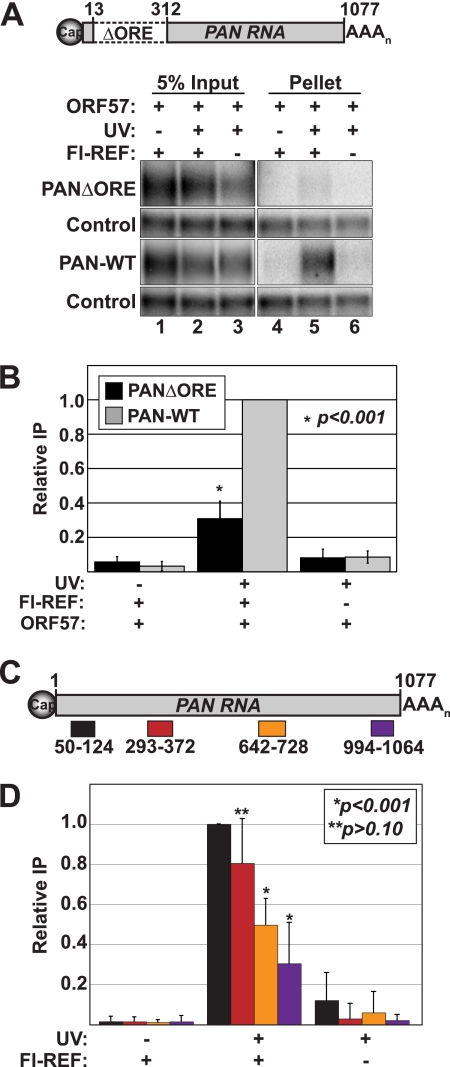
PAN RNA contains an ORF57-responsive element (ORE) in its 5′-most 300 nucleotides (nt) that is sufficient for ORF57 binding (50, 63, 66). In addition, deletion of this element from PAN RNA dramatically decreases the efficiency of ORF57 binding to PAN RNA. Therefore, it seems likely that ORF57 binding to the ORE recruits REF/Aly to the PAN RNP. Alternatively, it is possible that ORF57 activates REF/Aly binding independent of the ORF57 association with PAN RNA. Indeed, accessibility of the REF/Aly RNA-binding domain (RBD) to RNA is regulated by interactions with other proteins (20). To distinguish between these models, we performed UV cross-linking experiments in which we expressed an ORE deletion (PANΔORE) (Fig. 2A) instead of full-length PAN RNA. Because ORF57 binding to PANΔORE is significantly reduced (63, 66), the recruitment model predicts that the loss of ORF57 binding to PAN RNA will lead to loss of Fl-REF recruitment in the presence of ORF57. Consistent with this prediction, PANΔORE RNA was coimmunoprecipitated significantly less efficiently than wild-type PAN RNA (Fig. 2A and B), even though ORF57 was coexpressed. Some residual binding was still observed, but this was also observed in the case of ORF57 (66). Thus, efficient REF/Aly binding to PAN RNA depends on the presence of both ORF57 (Fig. 1) and the ORE (Fig. 2), strongly supporting the model that ORF57-binding to PAN RNA recruits REF/Aly to the PAN RNP.
Fig 2.
REF/Aly recruitment is enhanced by the ORE, and its binding is enriched at the PAN RNA 5′ end. (A) (Top) Diagram of PANΔORE RNA. The numbers refer to the PAN RNA nucleotide sequence relative to its transcription start site (68). (Bottom) Representative Northern blots from a UV cross-linking coimmunoprecipitation experiment with a wild-type PAN (PAN-WT) and PANΔORE RNAs. Details are as in Fig. 1. (B) Quantification of data from PANΔORE cross-linking experiments. Error bars represent standard deviations, and P values are from an unpaired Student's t test of the PAN-WT and PANΔORE data sets from the +UV +Fl-REF +ORF57 samples (n = 4). IP, immunoprecipitation. (C) Diagram of the amplicons used for qRT-PCR. (D) Results from UV-cross-linking qRT-PCR experiments. The data are plotted for each of the indicated samples and controls. The colors used in the bar graphs match the depiction of the amplicons in panel C. The bars are the mean percent immunoprecipitation values relative to that of the 5′-most amplicon, and the error bars show standard deviations (n = 3). Determination of primer efficiencies was described previously (57, 66). P values are from a two-tailed, unpaired Student's t test comparing each data set to that of the 5′-most amplicon.
REF/Aly binds to the 5′ ends of spliced cellular RNAs in a splicing and cap-binding complex (CBC)-dependent fashion (6). The data above show that ORF57 recruits REF/Aly to PAN RNA independently of splicing, but efficient recruitment depends on a 5′ RNA element. To determine whether REF/Aly binds to the 5′ end of PAN RNA directly, we modified the UV cross-linking protocol to include a partial RNase digestion prior to immunoprecipitation. The immunoprecipitated fragments were then analyzed by quantitative reverse transcription-PCR (qRT-PCR) to approximate the region of REF/Aly interaction. Four primer sets spanning PAN RNA were used for the qRT-PCR analysis (Fig. 2C). As expected, we observed a UV-dependent and Fl-REF-dependent enrichment for PAN RNA in the immunoprecipitates. In addition, we saw higher immunoprecipitation efficiency at the 5′ end of the molecule. However, the 5′ bias is not absolute; REF/Aly associated with fragments further downstream on PAN RNA. Importantly, using the same assay, ORF57 was shown to bind to PAN RNA with a nearly identical trend (66). The observed downstream binding may be due to insufficient RNase treatment, or it is possible that ORF57 and REF/Aly bind at more than one site on PAN RNA. Because these data mirror those seen with ORF57, they strongly support the conclusion that ORF57 recruits REF/Aly to PAN RNA at its 5′ end, but the interaction may occur at other sites on the transcript as well.
The ORF57 REF/Aly interaction domain is necessary for its nuclear stabilization function.
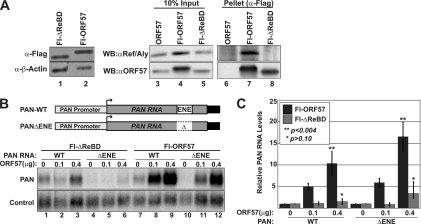
To determine if REF/Aly is an essential cofactor for ORF57-mediated increases in RNA stability, we generated a Flag-tagged ORF57 expression construct that lacks 28 amino acids essential for the interaction of REF/Aly with ORF57 (Fl-ΔReBD) (49, 54). This deletion does not alter the localization of ORF57 (47, 54), nor does it have a substantial effect on ORF57 protein levels (Fig. 3A, lanes 1 and 2). To verify the loss of interaction between Fl-ΔReBD and REF/Aly, we immunoprecipitated Fl-ORF57 or Fl-ΔReBD using anti-Flag–agarose beads and looked for coimmunoprecipitation of endogenous REF/Aly (Fig. 3A, lanes 3 to 8). While REF/Aly was coimmunoprecipitated with the wild-type Fl-ORF57, coimmunoprecipitation of REF/Aly with Fl-ΔReBD was undetectable (Fig. 3A, lanes 7 and 8). No immunoprecipitation was observed when ORF57 was untagged (lane 6).
Fig 3.
REF/Aly-binding correlates with ORF57 activity. (A) Immunoprecipitation of REF/Aly is abolished in the Fl-ΔReBD mutant. (Lanes 1 and 2) Western analysis demonstrating similar expression levels of the Fl-ORF57 and Fl-ΔReBD proteins. Endogenous β-actin served as a loading control. (Lanes 3 to 8) Western blot analysis of 10% input and 100% pellets from a coimmunoprecipitation experiment. Whole-cell extracts from HEK293 cells transiently expressing either Fl-ORF57, Fl-ΔReBD, or an untagged ORF57 were used for immunoprecipitation with anti-Flag–agarose in the presence of RNase. Western blotting (WB) was performed with REF/Aly or Flag antibodies as indicated. (B) (Top) Schematic diagram of the PAN-WT and PANΔENE expression constructs. Transcription is driven by the PAN RNA promoter, and constructs contain the PAN RNA polyadenylation signal (black box). (Bottom) Northern analysis comparing the effects of Fl-ORF57 or Fl-ΔReBD on the accumulation of full-length PAN RNA (WT) or PANΔENE (ΔENE). Membranes were probed with PAN RNA or control probes, as indicated. The loading control detects RNA from a cotransfected plasmid that expresses the murine mgU2-19/30 small Cajal body RNA (scaRNA) (11, 75) (C) Quantification of the Northern blot data. Each PAN RNA value was first normalized to the loading control, and all values are relative to the corresponding no-ORF57 sample. The error bars show standard deviations (n = 3); the P values are from a two-tailed, unpaired Student's t test of the indicated result with the matching no-ORF57 data set.
We next tested whether deletion of the REF-binding domain abolished the ability of ORF57 to increase PAN RNA levels. Consistent with previously published results (54), Fl-ΔReBD was strongly compromised in its ability to upregulate PAN RNA (Fig. 3B, compare lanes 1 to 3 with 7 to 9, and 3C). We previously showed that ORF57's ability to stabilize PAN RNA was at least partially redundant with the cis-acting element called the ENE (63). The ENE is a 79-nt element near the 3′ end of PAN RNA that interacts with the poly(A) tail to stabilize PAN RNA in cis. Stabilization by the ENE is not dependent on ORF57, nor does ORF57 sequence-specifically bind this region (63, 66). However, the ENE and ORF57 likely protect PAN RNA from the same decay pathway. As a result, deletion of the ENE causes significant decreases in PAN RNA levels due to its destabilization in the absence of ORF57 (11–13). As a result, an ENE deletion mutant of PAN RNA (PANΔENE) provides us with a broader dynamic range to detect increases in stabilization by Fl-ΔReBD. Even in this case, Fl-ΔReBD did not significantly increase the levels of PANΔENE RNA (Fig. 3B, lanes 4 to 6 and lanes 10 to 12, and 3C). These data show that when ORF57 is compromised for REF/Aly binding, it is unable to contribute to PAN RNA stability. One caveat to these experiments is that the deletion may affect interactions with other proteins or activities of ORF57 unrelated to REF/Aly function. Optimally, we would assess ORF57 activity in cells in which REF/Aly is knocked down with RNAi. Unfortunately, when we knock down REF/Aly using RNAi in HEK293 cells, we concomitantly decrease exogenous ORF57 expression, so it is technically not feasible to assay for loss of ORF57 stabilizing function in the absence of REF/Aly. Even so, these data are consistent with the model that ORF57 recruits REF/Aly to PAN RNA, where it functions to stabilize the transcript.
REF/Aly is sufficient to increase nuclear PAN RNA levels.
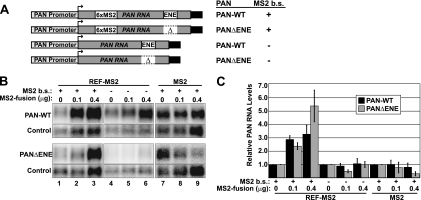
REF/Aly is recruited to PAN RNA by ORF57, and it appears to be a necessary cofactor for ORF57-mediated increases in PAN RNA stability. We next tested whether REF/Aly recruitment was sufficient to confer increased accumulation of PAN RNA. To address this experimentally, we employed a tethering assay to promote an interaction between REF and PAN RNA in HEK293 cells (8). We expressed PAN RNA from plasmids containing six binding sites for the bacteriophage MS2 coat protein that either included or excluded the ENE (Fig. 4A). We transiently coexpressed these transcripts with a fusion protein containing REF2-I fused to the MS2 coat protein at its C terminus (REF-MS2) and examined the effects of tethering REF-MS2 on PAN RNA accumulation (Fig. 4B, lanes 1 to 3). As controls, we tested PAN RNA lacking MS2 binding sites or we replaced REF-MS2 expression with MS2 coat protein alone (Fig. 4B, lanes 4 to 9). The MS2 protein contains a simian virus 40 (SV40) nuclear localization sequence to ensure its translocation to the nucleus. Quantification of the data showed that tethering of REF-MS2 to PAN RNA increases its accumulation in a dose-dependent fashion (Fig. 4C), and this increase is not observed in the absence of MS2-binding sites or when MS2 coat protein alone is expressed. Thus, recruitment of REF-MS2 to PAN RNA independently of ORF57 is sufficient to increase PAN RNA accumulation.
Fig 4.
REF/Aly is sufficient to increase PAN RNA abundance. (A) Diagram of the PAN RNA expression constructs used in the tethering experiments. PAN RNA is driven by its own promoter and has its own polyadenylation signals (black rectangles). Δ, absence of the ENE; b.s., MS2 binding sites. (B) Representative Northern blots from tethering experiments. The presence or absence of MS2 binding sites and the amounts of cotransfected MS2 construct are shown above the panels. The probe and PAN RNA expression construct are indicated on the left. Panels have different exposures, to best display the given signal, but relative quantification is shown in panel C. The loading control detects RNA from a cotransfected mgU2-19/30 plasmid that expresses a previously described murine scaRNA (11, 75). (C) Quantification of the data from multiple REF-MS2 tethering assays. The data are mean values relative to the matched no-REF-MS2 or no-MS2 control. The error bars show standard deviations (n = 3).
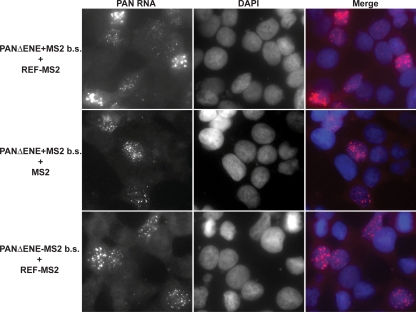
The decay machinery in the nucleus differs from that in the cytoplasm, so it is possible that the observed increases in PAN RNA accumulation were an indirect result of REF-mediated export of PAN RNA to the cytoplasm. In addition, because PAN RNA is a nuclear transcript, the hypothesis that ORF57 recruits REF/Aly to PAN RNA predicts that REF/Aly recruitment is insufficient for PAN RNA export. Therefore, we performed fluorescence in situ hybridization (FISH) to determine the localization of PANΔENE RNA upon REF-MS2 tethering (Fig. 5). As expected, in the controls lacking either REF-MS2 (Fig. 5, middle row) or MS2-binding sites (bottom row), PANΔENE RNA displayed punctate nuclear staining (63). Most importantly, PANΔENE remains localized exclusively to the nucleus in the presence of REF-MS2 (Fig. 5, top row). In this case, both the number of cells showing signal above background and the intensity of the signal in individual cells were increased compared to controls. We conclude that the REF-dependent increases in PAN RNA levels reflect an increase in nuclear RNA abundance, demonstrating that REF/Aly is sufficient to increase the nuclear accumulation of PAN RNA but is not sufficient to promote its export.
Fig 5.
REF/Aly tethering does not promote PAN RNA export. In situ hybridization of PANΔENE RNA containing MS2 binding sites (top and middle) or lacking MS2 binding sites (bottom) is shown. MS2 and REF-MS2 were cotransfected as indicated. (Left) PAN RNA probe; (middle) DAPI-stained nuclei; (right) merged signal. The PAN RNA plus REF-MS2 panels are shown at a lower exposure to avoid saturation of the signal.
REF/Aly increases the half-life of PAN RNA and affects poly(A) tail lengths.
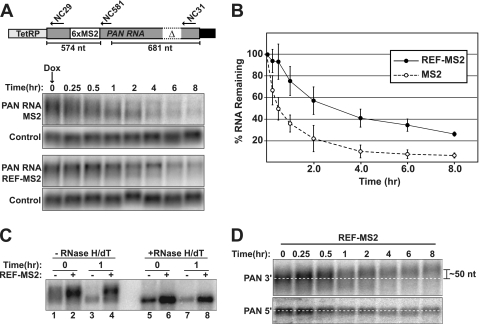
Next, we addressed the cellular mechanism underlying the REF-mediated increases in nuclear PAN RNA abundance. To directly test whether REF/Aly stabilizes PAN RNA in the nucleus, we determined the half-life of PANΔENE RNA in the presence or absence of REF-MS2 tethering. PAN RNA lacking the ENE but containing six MS2 binding sites was cloned behind the tetracycline-responsive promoter (TetRP) (Fig. 6A). We cotransfected HEK293 Tet-Off Advanced (293TOA) cells with this construct and either the REF-MS2 or the MS2 coat protein expression construct. 293TOA cells allow us to inhibit TetRP-driven transcription by adding doxycycline (Dox) to the medium. Approximately 18 h after transfection, doxycycline was added to the medium, RNA was harvested at given intervals, and expression levels were compared by Northern blotting (Fig. 6A). Quantification of the Northern blotting data showed that the apparent half-life was extended from ∼0.5 h to ∼3 h when REF-MS2 was tethered to PAN RNA (Fig. 6B). Taking these results with those reported above, we conclude that REF/Aly association with PAN RNA is sufficient to increase the nuclear half-life of PAN RNA.
Fig 6.
REF/Aly promotes PAN RNA stability. (A) (Top) Schematic diagram of the TetRP-PANΔENE-6xMS2 construct. Positions of oligonucleotides (arrows) for RNase H-targeted cleavage (NC581) as well as probes for detection of the 5′ (NC29) and 3′ (NC31) ends of PAN RNA are shown. The schematic is not to scale. (Bottom) Northern blots from an in vivo RNA decay experiment. 293TOA cells were cotransfected with TetRP-PANΔENE-6MS2 and MS2 (upper panels) or REF-MS2 (lower panels). After transcription shutoff by addition of Dox to the medium, RNA was collected at the indicated intervals. Blots were probed first for PAN RNA and then for an endogenous loading control (7SK RNA). (B) Decay curve of PANΔENE with or without REF-MS2. PAN RNA signals were first normalized to the loading control and expressed as a percentage of the time-zero samples. Each data point is the average of three experiments, with the error bars representing the standard deviation. (C) Northern blot of 0- and 1-h samples from an RNA decay experiment with either MS2 alone (−) or REF-MS2 (+). The same samples were treated with RNase H and oligo(dT) to remove the poly(A) tails and run on the same gel. (D) REF-MS2 promotes PAN RNA hyperadenylation subsequent to transcription shutoff. Total RNA from a REF-MS2 transcription shutoff experiment was cleaved with RNase H and NC581 and subjected to Northern blotting. (Top) The 3′ end of the cleaved PAN RNA was detected with probe NC31. (Bottom) The 5′ PAN RNA fragment was detected on the same blot using probe NC29. The dashed white lines serve as guides to show that the mean length of the 3′ end increases while the 5′ end remains constant. The mean poly(A) tail lengths were estimated to be ∼150 nt at 0 h and ∼200 nt at 8 h (data not shown). The positions of the oligonucleotides used for RNase H cleavage and RNA detection are depicted in panel A.
Inspection of the Northern blot data revealed that in the absence of REF-MS2, the length of PAN RNA shortened over time. In contrast, little or no shortening was observed when REF-MS2 was tethered to the RNA (Fig. 6A). The differences in mobility were more readily seen when samples from the 0- and 1-h time points were run on the same gel (Fig. 6C). Upon treatment of these samples with RNase H and oligo(dT) to remove poly(A) tails, each of the RNAs had the same mobility (Fig. 6C, lanes 5 to 8), which was slightly higher than that for the 1-h MS2-only samples (compare lane 3 with lanes 5 to 8). Thus, the shortened forms of PAN RNA were due to decreases in poly(A) tail length, consistent with the previous report that PAN RNA undergoes deadenylation prior to its destruction (11). We interpret these data as indicating that REF/Aly recruitment protects the transcripts from deadenylation and/or promotes ongoing readenylation that counterbalances nuclear deadenylation. Interestingly, we noticed a subtle but reproducible trend that PAN RNA mobility decreases over time in the presence of REF-MS2 after transcription shutoff (Fig. 6A and C). To better resolve the relative lengths of the transcripts, we cleaved samples from a REF-MS2 transcription shutoff experiment with RNase H and the DNA oligonucleotide NC581 (Fig. 6A, top) prior to Northern blot analysis. This treatment shortens the transcripts, making changes in gel mobility due to poly(A) tail extension more obvious. We then detected the 3′ or 5′ fragments using an oligonucleotide probe specific for each fragment (Fig. 6A, top). This analysis showed that the 3′ fragments lengthened over time after transcription shutoff, while the 5′ fragment mobility remained constant (Fig. 6D). We estimated the increase over the 8-h period to be ∼50 nt, extending an ∼150-nt poly(A) tail to ∼200 nt. Thus, when REF-MS2 is recruited to PAN RNA in the nucleus, the transcript is subject to poly(A) extension subsequent to the initial cleavage and polyadenylation step.
It is worth noting that when we performed the decay experiments using short (2-h) transcription pulses, we saw no differences between PAN RNA decay rates of tethered MS2 and REF-MS2. Previous studies of PAN RNA decay kinetics reported diminution of the effects of stabilizing factors using shorter transcription pulses that are due to the multiple pathways involved in PAN RNA destruction (11, 63). An additional contributing factor here may be that the levels of transcript are not sufficient to drive binding to REF-MS2 after only a short transcription pulse. Alternatively, it remains formally possible that REF/Aly may play a role in transcription, as has been previously proposed (3, 77), and that some of the effects on steady-state levels are transcriptional. Even so, the decay data presented in Fig. 6 as well as the changes in poly(A) tail length clearly indicate a posttranscriptional role for REF/Aly in PAN RNA accumulation.
DISCUSSION
The data presented here are consistent with two primary conclusions. First, we conclude that the cellular REF/Aly protein stabilizes transcripts in the nucleus in a manner that is separable from its role in nuclear export. We show that recruitment of REF/Aly is sufficient to increase the abundance of PAN RNA in the nucleus (Fig. 4 and 5), so the effect cannot be attributed to its previously defined role in RNA export. Rather, binding of REF/Aly to PAN RNA leads to an increase in transcript stability, apparently by directly or indirectly protecting the poly(A) tail (Fig. 6). Second, we conclude that the KSHV ORF57 protein recruits REF/Aly to PAN RNA to promote the stabilization of the transcript. KSHV ORF57 binds an element in PAN RNA near its 5′ end, increasing the PAN RNA half-life (63, 66). Here, using a stringent UV cross-linking technique, we showed that ORF57 recruits REF/Aly to PAN RNA (Fig. 1 and 2), resulting in a direct interaction between REF/Aly and the 5′ end of PAN RNA in vivo. Three observations support the conclusion that ORF57-mediated stabilization is a result of REF/Aly activity: (i) REF-MS2 is sufficient to stabilize PAN RNA in the nucleus when recruited to the transcript independent of ORF57 (Fig. 4 to 6), (ii) the REF interaction domain of ORF57 is necessary for its stabilization function (Fig. 3) (54), and (iii) both REF-MS2 tethering and ORF57 lead to stabilization, apparently by affecting poly(A) tail metabolism (Fig. 6) (63). Thus, these studies have uncovered a cellular cofactor for the KSHV ORF57 protein's RNA stability function and have identified a novel function for a cellular export factor.
A role for REF/Aly in nuclear RNA surveillance?
Our data indicate that the stabilization function of REF/Aly can be separated from its role in nuclear export. When REF/Aly is recruited to PAN RNA either by ORF57 or by direct tethering, PAN RNA is not exported from the nucleus; however, its half-life increases. This observation has interesting implications for RNA biogenesis. At least in some cases, intronless transcripts are less stable than their spliced counterparts (11, 13, 83), and inefficiently exported transcripts tend to be less abundant presumably due to nuclear decay (for example, see reference 25). Given the promotion of export by splicing, it is reasonable to propose that stabilization is an indirect consequence of a shorter exposure to nuclear decay factors. However, our data support a more direct role for REF/Aly in stabilizing nuclear transcripts. Specifically, transcripts that associate with REF/Aly resist nuclear decay even upon increased nuclear dwell times. Further supporting this idea, a recent report showed that TREX components lead to the nuclear stabilization of naturally occurring intronless mRNAs (40). These data are consistent with the idea that assembly of export factors protects transcripts from a nuclear RNA surveillance machinery that degrades “nonexportable” mRNPs.
We propose that the upstream events in mRNA export, specifically REF/Aly recruitment, retain an mRNP that is not fully export competent in a state in which it is protected from nucleolytic attack. We can imagine two broad non-mutually exclusive models for protection of the RNA. First, the changes in stability may result from alterations in the mRNP that render it refractory to the exonucleases. For example, the ENE stabilizes transcripts by limiting access of the poly(A) tail to the degradation machinery (11, 12). Perhaps TREX similarly protects the poly(A) tail by promoting interactions between the 3′ and 5′ ends of the transcript. Indeed, Yra1 interacts with the nuclear poly(A)-binding protein in yeast (30), and one can imagine that interactions between REF/Aly at the 5′ end of a transcript with factors at the 3′ end may promote circularization and stabilization of the transcripts in the context of the mRNP. Second, we can envision a model in which the REF/Aly complex maintains transcripts at a particular subnuclear locale that is devoid of nuclease activity. Similar models, in which hyperadenylated transcripts accumulate at the sites of transcription, where they are relatively stable, have been proposed for yeast (62). Further studies are necessary to distinguish between these models.
Another observation consistent with the idea that REF/Aly protects PAN RNA from the nuclear RNA surveillance machinery is that upon REF/Aly binding, poly(A) tails appear to be protected from deadenylation and subject to further adenylation. In yeast and mammalian cells, hyperadenylation occurs as a consequence of compromised Mex67 or TAP/NXF1 function, respectively. While one can imagine that this hyperadenylation is an indirect consequence of export blockage, recent work suggests that this may be due to a direct role of Mex67 in poly(A) tail metabolism (58). In cleavage and polyadenylation assays, extracts from Mex67 temperature-sensitive strains generated significantly longer poly(A) tails than their wild-type counterparts (58). Moreover, the changes in length were due primarily to the lack of poly(A) trimming in the Mex67-deficient extracts. Yeast TREX components, including Yra1, are necessary to release transcripts from the cleavage and polyadenylation machinery prior to export (30, 58). Dias and colleagues showed that in mammalian cells, REF/Aly and UAP56 are necessary for release of RNAs from nuclear speckle domains (15) and that TREX components are recruited to naturally occurring intronless transcripts to stabilize them (40). Taking these observations with our data, we propose that REF/Aly is recruited early in mRNA biogenesis and that this recruitment renders the transcript resistant to deadenylation and decay by affecting mRNP structure or localization. After REF/Aly recruitment, further maturation of the mRNP leads to release from retention, poly(A) tail trimming, and export. Interestingly, REF/Aly primarily binds to polyadenylated transcripts that are in insoluble nuclear material (52), consistent with the proposed role for REF/Aly upstream of transcript release.
What is the role of REF/Aly in KSHV RNA metabolism?
We used the unique PAN RNA to probe the mechanisms of ORF57 and REF/Aly in nuclear RNA metabolism. However, the present study also provides mechanistic information regarding ORF57-mediated stabilization of PAN RNA. While the function of PAN RNA remains unknown, ORF57 promotes its accumulation in both transfected and infected cells (23, 38, 46, 54, 63). It seems reasonable to propose that its function depends on its extraordinarily high abundance: nearly 70 to 80% of the poly(A)+ RNA in lytically reactivated cells is PAN RNA (68, 72). Several factors contribute to this abundance. First, the PAN RNA promoter has robust activity in the lytic phase, driven by the KSHV RTA protein (79). Second, the ENE stabilizes PAN RNA in cis. Third, ORF57 binds to PAN RNA and promotes its stability. Our present results strongly suggest that the cellular REF/Aly protein is a necessary cofactor for ORF57-mediated stabilization. Thus, assuming the high abundance of a PAN RNA is necessary for its function in KSHV, REF/Aly may promote this function by increasing PAN RNA half-life.
ORF57 is a member of the herpes simplex virus (HSV) ICP27 family of herpesvirus proteins that have been implicated in nearly every step of RNA biogenesis from transcription through decay (10, 22, 48, 64, 67, 74). While the mechanisms of these proteins differ in some respects, one common theme is that each family member interacts with the cellular mRNA export machinery. For example, ICP27 binds to REF/Aly and TAP/NXF1 directly (4, 5), while human cytomegalovirus (hCMV) UL69 interacts directly with UAP56 (42). KSHV ORF57 binds to REF/Aly directly, but only weak direct binding to TAP/NXF1 has been observed (49). In all cases, the prevailing model is that ICP27 family members provide a link between viral mRNAs and the cellular export machinery. Herpesvirus mRNAs are presumed to require this function because their single-exon genes are not subject to splicing-coupled mechanisms for export factor recruitment. However, the extent to which this mechanism is used by each herpesvirus remains an area of significant interest, as is the determination of whether this export mechanism is utilized by the majority of viral mRNAs or only a subset.
In nearly every case examined, ICP27 homologs are multifunctional and essential for viral replication. However, whether the functions driven by interaction with export factors are essential for viral replication remains unclear. In HSV-infected cells, TAP/NXF1 is essential for viral mRNA export, whereas REF/Aly is dispensable (35). This may be due to REF/Aly redundancy with other cellular factors like UIF, which has recently been reported to be redundant with REF/Aly in both KSHV-infected and uninfected mammalian cells (25, 32). Interestingly, while REF/Aly may not be required for HSV mRNA export, ICP27 recruits REF/Aly, but not TAP/NXF1, to sites of viral transcription (4). Therefore, consistent with our observations with ORF57, ICP27 appears to recruit REF/Aly to transcripts early in their biogenesis to perform a function independent of its role in mRNA export. In light of these results, it will be interesting to determine whether ORF57 and REF/Aly are cotranscriptionally transferred to nascent transcripts to promote herpesvirus evasion of host RNA quality control pathways.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julie Pfeiffer and Ivan D'Orso for critical review of the manuscript. This work was funded by the NIH-NIAID grant AI081710 and by Welch Foundation research grant I-1732. N.K.C. is a Southwestern Medical Foundation Scholar in Biomedical Research. S.H.S. was supported by an NIH Molecular Microbiology training grant (5T32AI007520-12). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
S.H.S. performed experiments, wrote the paper, and designed experiments, O.V.H. and A.H. performed experiments, and N.K.C. designed experiments and wrote the paper.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 30 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Boyne JR, Colgan KJ, Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 29:1851–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruhn L, Munnerlyn A, Grosschedl R. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 11:640–653 [DOI] [PubMed] [Google Scholar]
- 4. Chen IH, Li L, Silva L, Sandri-Goldin RM. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen IH, Sciabica KS, Sandri-Goldin RM. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng H, et al. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127:1389–1400 [DOI] [PubMed] [Google Scholar]
- 7. Church GM, Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coller J, Wickens M. 2002. Tethered function assays using 3′ untranslated regions. Methods 26:142–150 [DOI] [PubMed] [Google Scholar]
- 9. Conrad NK. 2008. Co-immunoprecipitation techniques for assessing RNA-protein interactions in vivo. Methods Enzymol. 449:317–342 [DOI] [PubMed] [Google Scholar]
- 10. Conrad NK. 2009. Posttranscriptional gene regulation in Kaposi's sarcoma-associated herpesvirus. Adv. Appl. Microbiol. 68:241–261 [DOI] [PubMed] [Google Scholar]
- 11. Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. 2006. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell 24:943–953 [DOI] [PubMed] [Google Scholar]
- 12. Conrad NK, Shu MD, Uyhazi KE, Steitz JA. 2007. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc. Natl. Acad. Sci. U. S. A. 104:10412–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conrad NK, Steitz JA. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 24:1831–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das B, Butler JS, Sherman F. 2003. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 23:5502–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dias AP, Dufu K, Lei H, Reed R. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 1:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doma MK, Parker R. 2007. RNA quality control in eukaryotes. Cell 131:660–668 [DOI] [PubMed] [Google Scholar]
- 17. Dufu K, et al. 2010. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 24:2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erkmann JA, Kutay U. 2004. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp. Cell Res. 296:12–20 [DOI] [PubMed] [Google Scholar]
- 19. Gatfield D, Izaurralde E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159:579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Golovanov AP, Hautbergue GM, Tintaru AM, Lian LY, Wilson SA. 2006. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA 12:1933–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gomez-Gonzalez B, et al. 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30:3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han Z, et al. 2007. Multiple roles of Epstein-Barr virus SM protein in lytic replication. J. Virol. 81:4058–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han Z, Swaminathan S. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. 2008. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. U. S. A. 105:5154–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hautbergue GM, et al. 2009. UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 19:1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hieronymus H, Yu MC, Silver PA. 2004. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 18:2652–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hilleren P, Parker R. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7:753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horton RM. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93–99 [DOI] [PubMed] [Google Scholar]
- 29. Hurt JA, et al. 2009. A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. J. Cell Biol. 185:265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iglesias N, et al. 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24:1927–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Izaurralde E. 2002. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur. J. Cell Biol. 81:577–584 [DOI] [PubMed] [Google Scholar]
- 32. Jackson BR, et al. 2011. An interaction between KSHV ORF57 and UIF provides mRNA-adaptor redundancy in herpesvirus intronless mRNA export. PLoS Pathog. 7:e1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jensen TH, Boulay J, Rosbash M, Libri D. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11:1711–1715 [DOI] [PubMed] [Google Scholar]
- 34. Jensen TH, Patricio K, McCarthy T, Rosbash M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887–898 [DOI] [PubMed] [Google Scholar]
- 35. Johnson LA, Li L, Sandri-Goldin RM. 2009. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83:6335–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson SA, Cubberley G, Bentley DL. 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell 33:215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katahira J, Inoue H, Hurt E, Yoneda Y. 2009. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 28:556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirshner JR, Lukac DM, Chang J, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lei EP, Silver PA. 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 16:2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lei H, Dias AP, Reed R. 2011. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl. Acad. Sci. U. S. A. 108:17985–17990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Libri D, et al. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26:1631–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Longman D, Johnstone IL, Caceres JF. 2003. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luna R, et al. 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell 18:711–722 [DOI] [PubMed] [Google Scholar]
- 45. Luo ML, et al. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644–647 [DOI] [PubMed] [Google Scholar]
- 46. Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8α, and K8.1 and the production of infectious virus. J. Virol. 81:1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365–28378 [DOI] [PubMed] [Google Scholar]
- 48. Majerciak V, Zheng ZM. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 14:1516–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Malik P, Blackbourn DJ, Clements JB. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001–33011 [DOI] [PubMed] [Google Scholar]
- 50. Massimelli MJ, et al. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 7:1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masuda S, et al. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mili S, Shu HJ, Zhao Y, Pinol-Roma S. 2001. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 21:7307–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mili S, Steitz JA. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10:1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okada M, Jang SW, Ye K. 2008. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. U. S. A. 105:8649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pandit S, Wang D, Fu XD. 2008. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell Biol. 20:260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qu X, et al. 2009. Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol. Cell. Biol. 29:5327–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reed R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15:326–331 [DOI] [PubMed] [Google Scholar]
- 60. Rehwinkel J, et al. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11:558–566 [DOI] [PubMed] [Google Scholar]
- 61. Rodrigues JP, et al. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. U. S. A. 98:1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rougemaille M, et al. 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 26:2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sahin BB, Patel D, Conrad NK. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 6:e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sandri-Goldin RM. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241–5256 [DOI] [PubMed] [Google Scholar]
- 65. Schmid M, Jensen TH. 2008. Quality control of mRNP in the nucleus. Chromosoma 117:419–429 [DOI] [PubMed] [Google Scholar]
- 66. Sei E, Conrad NK. 2011. Delineation of a core RNA element required for Kaposi's sarcoma-associated herpesvirus ORF57 binding and activity. Virology 419:107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sergeant A, Gruffat H, Manet E. 2008. The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front. Biosci. 13:3798–3813 [DOI] [PubMed] [Google Scholar]
- 68. Song MJ, Brown HJ, Wu TT, Sun R. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Strasser K, Hurt E. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Strasser K, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304–308 [DOI] [PubMed] [Google Scholar]
- 71. Stutz F, et al. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sun R, Lin SF, Gradoville L, Miller G. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 93:11883–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Swaminathan S. 2005. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell Biochem. 95:698–711 [DOI] [PubMed] [Google Scholar]
- 74. Toth Z, Stamminger T. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939–2949 [DOI] [PubMed] [Google Scholar]
- 75. Tycowski KT, Aab A, Steitz JA. 2004. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 14:1985–1995 [DOI] [PubMed] [Google Scholar]
- 76. Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24:813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Virbasius CM, Wagner S, Green MR. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell 4:219–228 [DOI] [PubMed] [Google Scholar]
- 78. Walsh MJ, Hautbergue GM, Wilson SA. 2010. Structure and function of mRNA export adaptors. Biochem. Soc. Trans. 38:232–236 [DOI] [PubMed] [Google Scholar]
- 79. West JT, Wood C. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150–5163 [DOI] [PubMed] [Google Scholar]
- 80. Wiegand HL, Lu S, Cullen BR. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. U. S. A. 100:11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang M, Green MR. 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 15:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao C, Hamilton T. 2007. Introns regulate the rate of unstable mRNA decay. J. Biol. Chem. 282:20230–20237 [DOI] [PubMed] [Google Scholar]
- 84. Zhong W, Wang H, Herndier B, Ganem D. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. U. S. A. 93:6641–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.