Abstract
Starvation, like many other catabolic conditions, induces loss of skeletal muscle mass by promoting fiber atrophy. In addition to the canonical processes, the starvation-induced response employs many distinct pathways that make it a unique atrophic program. However, in the multiplex of the underlying mechanisms, several components of starvation-induced atrophy have yet to be fully understood and their roles and interplay remain to be elucidated. Here we unveiled the role of tumor necrosis factor receptor-associated factor 6 (TRAF6), a unique E3 ubiquitin ligase and adaptor protein, in starvation-induced muscle atrophy. Targeted ablation of TRAF6 suppresses the expression of key regulators of atrophy, including MAFBx, MuRF1, p62, LC3B, Beclin1, Atg12, and Fn14. Ablation of TRAF6 also improved the phosphorylation of Akt and FoxO3a and inhibited the activation of 5′ AMP-activated protein kinase in skeletal muscle in response to starvation. In addition, our study provides the first evidence of the involvement of endoplasmic reticulum stress and unfolding protein response pathways in starvation-induced muscle atrophy and its regulation through TRAF6. Finally, our results also identify lysine 63-linked autoubiquitination of TRAF6 as a process essential for its regulatory role in starvation-induced muscle atrophy.
INTRODUCTION
Skeletal muscle atrophy is a debilitating consequence of starvation and many other catabolic conditions, such as aging, immobilization, denervation, and chronic disease states (22). Muscle atrophy that occurs in response to fasting/nutritional deprivation has several common, as well as distinct, features (29). Like other atrophic programs, the loss of muscle mass upon starvation involves activation of the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal system (ALS). However, a degree of distinction in starvation-induced muscle atrophy is introduced by the fact that in the case of nutrient deprivation, muscle proteins are degraded and mobilized for amino acid production, which is further used for gluconeogenesis. In addition, other evidence suggests the involvement of distinct stimuli in starvation-induced catabolic changes, such as altered levels of insulin growth factors and glucocorticoids (13, 49). Recently, activating transcription factor 4 (ATF4) has been reported to be involved in starvation-induced loss of skeletal muscle mass (15). Furthermore, while muscle wasting in response to cancer cachexia or disuse conditions (e.g., denervation, unloading, immobilization, etc.) involves the activation of the transcription factor nuclear factor kappa B (NF-κB), there has been no evidence regarding the activation or involvement of NF-κB in the loss of skeletal muscle in response to starvation (1, 32).
TRAFs are a family of conserved adaptor proteins which, through their association with cytoplasmic domains of different receptors, mediate the activation of various intracellular signaling pathways (55). Distinct from other TRAFs, TRAF6 is an E3 ubiquitin ligase, and through association with the dimeric ubiquitin-conjugating enzyme Ubc13/Uev1A, it catalyzes lysine 63 (K63)-linked polyubiquitination of several target proteins (26, 43, 55). Although it remains enigmatic whether the E3 ubiquitin ligase activity of TRAF6 is essential for its signaling function under all conditions, recent studies have shown that TRAF6 functions as a central regulator in multiple signaling pathways, such as NF-κB, mitogen-activated protein kinase, and phosphatidylinositol 3-kinase/Akt, in response to various cytokines and microbial products (8, 26, 28, 55, 61, 63). In addition to its association with cytoplasmic domains of various cell surface receptors, such as Toll-like receptors and the interleukin-1 receptor (IL-1R), TRAF6 has also been found to interact with multiple components of the UPS and/or the ALS in some cell types (27, 38, 39, 54). We have also recently reported that TRAF6 regulates skeletal muscle mass and activation of ALS and UPS in denervated skeletal muscle (42). However, the role and mechanisms of action of TRAF6 and whether the E3 ubiquitin ligase activity of TRAF6 is required for muscle atrophy in response to fasting remain unknown.
Several cellular stress conditions, such as starvation and alterations in glycosylation status, lead to the accumulation of unfolded and/or misfolded proteins in the endoplasmic reticulum (ER) lumen and cause ER stress (18, 19). The ER responds by activation of a range of signaling pathways that are collectively termed the ER stress response or the unfolded protein response (UPR), which is essentially a cytoprotective response, but an excessive or prolonged UPR can produce deleterious effects, including cell death (25). The UPR has three distinct arms, which have their specific transducers. Activation of these arms is mediated by PERK (protein kinase RNA-like ER kinase), IRE1 (inositol-requiring protein 1), and ATF6 (activating transcription factor 6) (12, 18, 19). Until recently, there was no evidence that ER stress-related proteins are involved in skeletal muscle metabolism or that there is any regulatory mediation of ER chaperones in skeletal muscle atrophy. Recently, it has been demonstrated that ATF4, a transcription factor involved in the cellular responses to starvation, is involved in muscle atrophy through mechanisms independent of the induction of muscle-specific E3 ubiquitin ligases MAFBx (also known as atrogin-1) and MuRF1 (15). It is notable that the expression of ATF4 is increased through activation of the PERK arm of the UPR in addition to other activating kinases, such as general control derepressible 2 kinase and heme-regulated inhibitor kinase (23). Intriguingly, ER stress-responsive transcription factor ATF6 has been implicated in muscle adaptation in response to acute exercise (60). However, direct evidence regarding UPR activation in skeletal muscle under atrophic conditions is still lacking.
Proinflammatory cytokines have been traditionally suggested as the mediators of skeletal muscle wasting in various chronic disease conditions. Recently, we discovered that inflammatory cytokine tumor necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) is also a mediator of skeletal muscle atrophy under disuse conditions (36). The expression of TWEAK receptor Fn14 was found to be increased about 6- to 7-fold in skeletal muscle 4 days after denervation. The role of the TWEAK-Fn14 system in disuse atrophy was supported by the findings that denervation-induced loss of skeletal muscle mass, activation of the NF-κB signaling pathway, and expression of MuRF1 were significantly inhibited in TWEAK-null mice compared to those in wild-type mice (36). More recently, Wu et al. have reported the increased expression of Fn14 in skeletal muscles of mice in a hind limb unloading model of disuse atrophy (59). However, the role of the TWEAK-Fn14 dyad in starvation-induced muscle atrophy remains unknown. Moreover, signaling mechanisms governing the expression of Fn14 in skeletal muscle under atrophic conditions have not yet been investigated.
In this study, using skeletal-muscle-specific TRAF6 knockout mice, we have investigated the role of TRAF6 and the mechanisms by which it regulates starvation-induced muscle atrophy. Our results show that muscle-specific depletion of TRAF6 inhibits starvation-induced activation of the UPS and the ALS and muscle atrophy in mice. Intriguingly, TRAF6 is essential for the inducible expression of several ER stress response-related genes, including ATF4, in response to fasting. Our results also suggest that the TWEAK-Fn14 system is involved in fasting-induced muscle atrophy and that inducible expression of Fn14 in response to starvation requires TRAF6. Finally, our experiments demonstrate that the E3 ubiquitin ligase activity of TRAF6 is essential for induction of the atrophic program in skeletal muscle in response to starvation.
MATERIALS AND METHODS
Animals.
Floxed TRAF6 (TRAF6f/f) and muscle-specific TRAF6 knockout (TRAF6mko) mice have been described previously (42). TWEAK knockout (TWEAK-KO) mice (described previously [33]) were provided by Avi Ashkenazi (Genentech, South San Francisco, CA). All of the mice were in the C57BL/6 background, and their genotype was determined by PCR assay of tail DNA. For starvation studies, mice were provided water but kept unfed for 6 h, 12 h, 24 h, or 48 h. All experimental protocols with mice were approved in advance by the Institutional Animal Care and Use Committee of the University of Louisville.
Cell culture.
C2C12 cells (a myoblastic cell line) were obtained from the American Type Culture Collection. These cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). Myoblasts were transfected with different plasmids using Effectene transfection reagent (Qiagen). To induce differentiation, the cells were incubated in differentiation medium (2% horse serum in DMEM) for 96 h as described previously (42). For acute starvation studies, the medium of the differentiated myotubes was replaced with sterile phosphate-buffered saline (PBS) for 3 h or 6 h of incubation and the myotubes were examined by morphometric or biochemical assays. TRAF6+/+ and TRAF6−/− mouse embryonic fibroblasts (MEF) were grown in DMEM containing 10% FBS.
Total RNA extraction and QRT-PCR assay.
RNA isolation and quantitative real-time PCR (QRT-PCR) were performed with gene-specific primers as described previously (36).
Analysis of spliced form of XBP-1.
Total RNA was extracted from the cells using TRIzol reagent. cDNA was prepared with the SuperScript II first-strand synthesis system (Invitrogen, Carlsbad, CA). The sequences of the primers were 5′-TTA CGG GAG AAA ACT CAC GGC-3′ (forward) and 5′-GGG TCC AAC TTG TCC AGA ATG C-3′ (reverse). The primer annealing temperature was 56°C, and reaction mixtures containing 100 ng of cDNA proceeded for 35 cycles. The PCR products were run on a 2% agarose gel to identify the presence of unspliced and spliced XBP-1 cDNA.
AMPK assay.
5′ AMP-activated protein kinase (AMPK) enzymatic activity in skeletal muscle tissue extracts was measured using a commercially available kit (MBL International) as previously described (42).
In vivo gene delivery.
The injection of plasmid DNA into tibial anterior (TA) muscles of mice and electroporation were performed as previously described (52). In brief, pcDNA3, FLAG-TRAF6 C70A, and pEGFPC1 were prepared using an endotoxin-free kit (Qiagen) and suspended in sterile saline solution in a 1:10 ratio. Mice were anesthetized, and a small portion of the TA muscles of both hind limbs was surgically exposed and injected with 30 μl of 0.5 U/μl hyaluronidase (EMD Biosciences). After 2 h, plasmid DNA (30 μg in 25 μl saline) was injected into the TA muscle; 1 min after plasmid DNA injection, a pair of platinum plate electrodes were placed against the closely shaved skin on both sides and electric pulses were delivered. Three 20-ms square-wave pulses of 75 V/cm at a 1-Hz frequency were generated using a stimulator (model S88; Grass Technologies) and delivered to the muscle. The polarity was then reversed, and a further three pulses were delivered to the muscle. After electroporation, mice were returned to their cages and fed a standard diet. Mice were used 10 days after plasmid electroporation for starvation studies.
Histology and morphometric analysis.
Hind limb (soleus and TA) muscles of mice were removed, frozen in isopentane cooled in liquid nitrogen, and sectioned using a microtome cryostat. For the assessment of tissue morphology, 10-μm-thick transverse sections of muscles were stained with hematoxylin and eosin (H&E) and staining was visualized (without any imaging medium) at room temperature on a microscope (Eclipse TE 2000-U) using a Plan 10× numerical aperture (NA) 0.25 PH1 DL or Plan-Fluor ELWD 20× NA 0.45 Ph1 DM objective lens, a digital camera (Digital Sight DS-Fi1), and NIS Elements BR 3.00 software (all from Nikon). The images were stored as JPEG files, and image levels were equally adjusted using Photoshop CS2 software (Adobe). Fiber cross-sectional area (CSA) was analyzed in H&E-stained soleus or TA muscle sections. For each muscle, the distribution of fiber CSA was calculated by analyzing 200 to 250 myofibers using NIS Elements BR 3.00 software (Nikon) as described previously (36).
Immunoprecipitation and Western blot assay.
Levels of different proteins in skeletal muscle were determined by performing immunoblotting as described previously (36). In brief, tissues were washed with PBS and homogenized in Western blot assay lysis buffer A (50 mM Tris-Cl [pH 8.0], 200 mM NaCl, 50 mM NaF, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 0.3% IGEPAL, protease inhibitors). Approximately 100 μg of protein was resolved per lane by 10 to 12% SDS-PAGE; electrotransferred onto nitrocellulose membrane; probed using anti-TRAF6 (1:1,000; Millipore), anti-TRAF3 (1:1,000; Santa Cruz Biotechnology, Inc.), anti-TRAF5 (1:1,000; Santa Cruz Biotechnology, Inc.), antiubiquitin (1:1,000; Santa Cruz Biotechnology, Inc.), antiphospho-FoxO3a (1:1,000; Cell Signaling Technology), anti-FoxO3a (1:1,000; Cell Signaling Technology); anti-phospho-Akt (1:200; Cell Signaling Technology), anti-Akt (1:1,000; Cell Signaling Technology), anti-phospho-eukaryotic translation initiation factor-2 (anti-phospho-eIF2; 1:1,000; Cell Signaling Technology), anti-eIF2 (1:1,000; Cell Signaling Technology), anti-ATF3 (1:1,000; Santa Cruz Biotechnology), anti-ATF4 (1:1,000; Santa Cruz Biotechnology), anti-protein disulfide isomerase (anti-PDI; 1:1,000; Santa Cruz Biotechnology), anti-CHOP (1:1,000; Santa Cruz Biotechnology), anti-LC3B (1:1,000; Cell Signaling Technology), anti-p62 (1:1,000; MBL International), anti-Fn14 (1:1,000; Cell Signaling Technology), and MF-20 (1:1,000; Development Studies Hybridoma Bank, University of Iowa) antibodies; and detected by chemiluminescence. The bands were quantified using ImageQuant TL software (GE Healthcare). To study the autoubiquitination of TRAF6, muscle extract (400 μg protein) was incubated overnight with 1 μg of anti-TRAF6 antibody (MBL) in 600 μl of lysis buffer, protein A-Sepharose beads were added, and the mixture was incubated at 4°C for an additional 2 h. The beads were washed four times with lysis buffer and finally suspended in 2× Laemmli sample buffer. Proteins were resolved on 10% SDS-PAGE gel and immunoblotted using Lys-63-specific ubiquitin antibody (1:1,000; Millipore).
Statistical analysis.
Results are expressed as mean ± standard deviation (SD). The Student t test or analysis of variance was used to compare quantitative data populations with normal distributions and equal variance. A P value of <0.05 was considered statistically significant unless otherwise specified.
RESULTS
TRAF6 mediates starvation-induced muscle atrophy in mice.
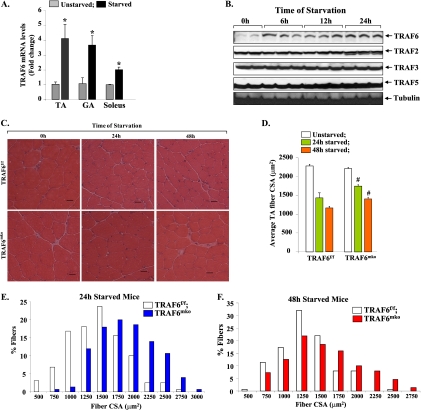
We have previously reported that the expression of TRAF6 is increased in skeletal muscle under multiple catabolic conditions, including denervation and cancer cachexia (42). We first investigated how the expression of TRAF6 is affected in skeletal muscles of mice in response to fasting. Wild-type mice were given access to normal water but deprived of food for 24 h. TRAF6 transcript levels in hind limb muscle were measured by QRT-PCR assay. As shown in Fig. 1A, mRNA levels of TRAF6 were significantly increased in the TA, gastrocnemius (GA), and soleus muscles of fasted mice compared to those of controls. We also performed a Western blot assay for a few TRAFs. In the TA muscles of unstarved mice, the level of TRAF6 protein was much lower than that of TRAF2, TRAF3, or TRAF5. However, the levels of TRAF6 protein were considerably increased within 6 h and remained elevated even after 24 h of fasting. In contrast, fasting did not affect TRAF2, TRAF3, or TRAF5 protein levels in the TA muscles of mice (Fig. 1B). To identify the physiological significance of the increased expression of TRAF6 in fasted mouse muscle, we investigated the effects of deletion of TRAF6 on muscle atrophy in response to starvation. Skeletal muscle-specific TRAF6 knockout (here TRAF6mko) mice and their corresponding littermates (i.e., TRAF6f/f; described previously [42]) were fasted for 24 h or 48 h. Isolated hind limb muscles were used for the preparation of transverse cryosections, followed by H&E staining and quantification of fiber CSA. Fasting for 24 h or 48 h caused a significant reduction in fiber CSA in TA muscles of TRAF6f/f mice (Fig. 1C). However, fasting-induced loss of fiber CSA was significantly inhibited in TA muscles of TRAF6mko mice (Fig. 1C and D). Furthermore, the proportion of fibers with a larger CSA was notably higher in TA muscles of TRAF6mko mice than in those of TRAF6f/f littermates evaluated after 24 h (Fig. 1E) or 48 h (Fig. 1F) of fasting. TA muscles of mice contain predominately fast-type fibers. In contrast, soleus muscles contain almost equal proportions of fast- and slow-type fibers (36). We also performed H&E staining and quantified fiber CSA in soleus muscles of TRAF6f/f and TRAF6mko mice. Similar to that in TA muscles, fasting-induced loss of fiber CSA was significantly inhibited in the soleus muscles of TRAF6mko mice compared to that in TRAF6f/f mice (see Fig. S1A and B in the supplemental material). These results demonstrate that TRAF6 mediates starvation-induced muscle atrophy in vivo.
Fig 1.
Involvement of TRAF6 in starvation-induced muscle atrophy. (A) TRAF6 transcript levels in TA, GA, and soleus muscles of wild-type control mice and those fasted for 24 h. Error bars represent SD. n = 3 in each group. *, P < 0.01 (values significantly different from those for unstarved muscle). (B) Western blot analyses of TA muscle extracts for TRAF2, TRAF3, TRAF5, and TRAF6 proteins at different time points after food deprivation. (C) Twelve-week-old TRAF6f/f and TRAF6mko mice were starved for 24 h or 48 h. Shown are representative photomicrographs of H&E-stained sections of TA muscles of TRAF6f/f and TRAF6mko mice 24 h and 48 h after starvation. Scale bars, 20 μm. (D) Average fiber CSA in TA muscle sections from TRAF6f/f and TRAF6mko mice. #, P < 0.05 (values significantly different from those of TRAF6f/f mice at corresponding times of starvation). Frequency distribution histograms representing CSA of fibers in TA muscles from TRAF6f/f and TRAF6mko mice after 24 h (E) and 48 h (F) of starvation (n = 8 in each group).
TRAF6 is required for the activation of the UPS and the ALS in skeletal muscle in response to starvation.
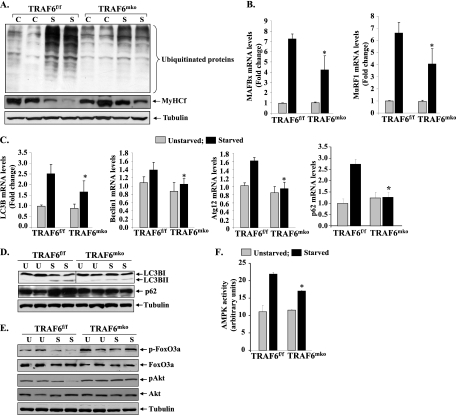
The UPS is responsible for the breakdown of the majority of the proteins in mammalian cells (45, 53). Under distinct catabolic conditions, the difference between the ubiquitination of all proteins and that of specific proteins can affect the development of pathology (9). To understand the mechanisms by which TRAF6 causes muscle atrophy and to determine whether TRAF6 is involved in UPS activation, we first investigated how the conjugation of ubiquitin to muscle proteins is affected in skeletal muscles of fasted TRAF6f/f and TRAF6mko mice. GA muscle extracts prepared from TRAF6f/f and TRAF6mko mice unstarved or starved for 24 h were immunoblotted using antibody against ubiquitin. As shown in Fig. 2A, fasting augmented the conjugation of ubiquitin to muscle proteins. However, the fasting-induced increase in protein ubiquitylation was significantly lower in TA muscles of TRAF6mko mice than in those of TRAF6f/f mice (increases in ubiquitination upon starvation: 3.41-fold ± 1.09-fold [TRAF6f/f] versus 1.64-fold ± 0.02-fold [TRAF6mko]; P < 0.05), indicating that TRAF6 is required for UPS activation in skeletal muscle upon starvation (Fig. 2A). While muscle atrophy involves the enhanced degradation of several contractile proteins, myosin heavy chain fast type (MyHCf) is one such protein which undergoes rapid degradation under many catabolic conditions, potentially through the UPS (32, 36). To further verify that TRAF6 is involved in the degradation of muscle proteins, we measured levels of MyHCf protein by Western blot assay. As shown in Fig. 2A (middle panel), levels of MyHCf were considerably higher in muscles of fasted TRAF6mko mice than in those of TRAF6f/f littermates (decreases in MyHCf levels: 41.90% ± 5.01% [TRAF6f/f] versus 17.9% ± 2.7% [TRAF6mko]; P < 0.05).
Fig 2.
Role of TRAF6 in fasting-induced activation of the UPS and autophagy in skeletal muscle. TRAF6f/f and TRAF6mko mice were starved for 24 h, and isolated GA and TA muscles were analyzed. (A) Representative immunoblots showing levels of ubiquitinated proteins (top), MyHCf (middle), and the unrelated protein tubulin (bottom) in TA muscle. C, control unstarved; S, starved. (B) MAFBx and MuRF1 transcript levels measured by QRT-PCR assay were significantly lower in the GA muscles of starved TRAF6mko mice than in those of starved TRAF6f/f mice. (C) Fold changes in transcript levels of autophagy-related genes for LC3B, Beclin1, Atg12, and p62 in GA muscles of starved TRAF6mko mice compared with those of starved TRAF6f/f mice (n = 4). (D) Immunoblots demonstrating reduced p62 protein levels and inhibition of conversion of LC3BI protein into LC3BII. The vertical black line indicates that intervening lanes have been spliced out. (E) Levels of phosphorylation of FOXO3a and Akt in TA muscles of unstarved TRAF6f/f and TRAF6mko mice (U) and mice starved for 24 h (S). (F) Enzymatic activity of AMPK in TA muscle extracts of TRAF6f/f and TRAF6mko mice. Error bars represent SD. *, P < 0.05 (values significantly different from TA muscles of starved TRAF6f/f mice).
UPS activation under multiple atrophic conditions involves the increased expression of two muscle-specific E3 ubiquitin ligase genes, MuRF1 and MAFBx (6, 7, 16). To further verify the role of TRAF6 in UPS activation in response to starvation, we measured MuRF1 and MAFBx transcript levels in skeletal muscles of TRAF6f/f and TRAF6mko mice. Fasting significantly increased the expression of both MAFBx and MuRF1 in skeletal muscles of mice (Fig. 2B). However, the increase in both MAFBx and MuRF1 transcript levels in response to fasting was found to be significantly lower in TRAF6mko mice than in TRAF6f/f mice (Fig. 2B).
It has been previously reported that fasting also induces the activation of ALS in skeletal muscle (37, 66). Furthermore, there is evidence that the ALS coordinates with the UPS to stimulate muscle atrophy under diverse conditions, including starvation (34, 50, 66). To evaluate the role of TRAF6 in the activation of autophagy, we measured mRNA levels of the autophagy-related genes for LC3B, Beclin1, and Atg12, which have been previously reported to be upregulated in skeletal muscle in response to fasting (34). As shown in Fig. 2C, fasting-induced expression of LC3B, Beclin1, and Atg12 was significantly inhibited in skeletal muscles of TRAF6mko mice in comparison with that in TRAF6f/f mice. We further measured mRNA levels of p62/SQSTM1, a multidomain adaptor protein, which interacts with and is activated by TRAF6 (38, 39). p62 binds to LC3 through a LiR (LC3-interacting region) motif and tethers protein to autophagosomes (5). Interestingly, the starvation-induced increase in the level of p62 mRNA in skeletal muscle was also significantly inhibited in TRAF6mko mice in comparison with that in TRAF6f/f mice (Fig. 2C).
Conversion of LC3BI to the active form LC3BII is a critical event in autophagy and considered one of the most reliable markers of autophagosome formation (31). We performed Western blot assays to determine whether TRAF6 affects the conversion of LC3BI into LC3BII in skeletal muscle upon starvation. As shown in Fig. 2D, starvation significantly increased the levels of the LC3BII form in skeletal muscles of TRAF6f/f mice but not in TRAF6mko mice (increases in LC3BII levels upon starvation: 15.24-fold ± 4.24-fold [TRAF6f/f] versus 4.52-fold ± 2.24-fold [TRAF6mko]; P < 0.01). We also found that the levels of p62 were increased in skeletal muscles of TRAF6f/f mice in response to starvation. However, the starvation-induced increase in p62 protein levels was not observed in skeletal muscles of TRAF6mko mice (increases in p62 levels: 1.87-fold ± 0.02-fold [TRAF6f/f] versus 1.02-fold ± 0.01-fold [TRAF6mko]; P < 0.05) (Fig. 2D). These results are in concert with previous findings suggesting that TRAF6 also acts through p62/LC3 binding to activate autophagy (39).
We next sought to determine whether ablation of TRAF6 intervenes with upstream regulators of the UPS and the ALS such as p62, FOXO3a, Akt, and AMPK in skeletal muscle. FOXO3a is a member of the forkhead family of transcription factors and is known to induce the expression of MAFBx and MuRF1 in atrophying skeletal muscle (51). Akt phosphorylates the FOXO family of transcription factors, including FOXO3a, which leads to their inactivation through retention in cytosol (56). Using a constitutively active form of FOXO3 with three mutated Akt phosphorylation sites, it has been previously found that FOXO3 induces atrophy by accelerating proteolysis through activation of autophagy (66). Our results showed that starvation led to a marked decrease in the basal level of phosphorylation of both Akt and FOXO3a in TA muscles of TRAF6f/f mice (Fig. 2E). In contrast, no noticeable decrease in the phosphorylation of Akt or FOXO3a was observed in TRAF6mko mice upon starvation (Fig. 2E).
AMPK, which is activated in response to an increased AMP/ATP ratio, is linked to muscle atrophy under multiple conditions, including fasting (46). As shown in Fig. 2F, fasting induced the activation of AMPK in GA muscles of mice. Furthermore, fasting-induced activation of AMPK was significant inhibited in TA muscles of TRAF6mko mice compared to that in TRAF6f/f mice (Fig. 2F). Taken together, these results suggest that under conditions of starvation, TRAF6 mediates the activation of both the UPS and the ALS in skeletal muscle potentially through the upstream activation of p62, Akt, FOXO3a, and AMPK.
TRAF6 augments the expression of ER stress response-related genes in skeletal muscle.
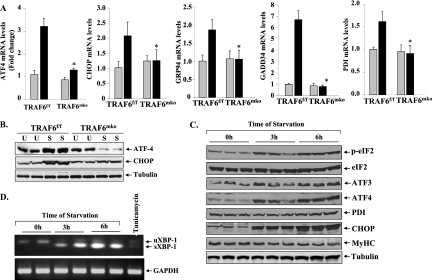
The ER stress and UPR pathways are employed by cells as a corrective measure to avoid an increase in the unfolded-protein load. In general, it has been observed that the PERK and IRE1 arms of the UPR are involved in mediating deleterious effects whereas the ATF6 arm mediates adaptive responses. We investigated whether starvation augments the expression of various genes involved in UPR pathways and whether TRAF6 plays a role in their induction in skeletal muscle. Interestingly, a drastic increase in the mRNA levels of several UPR markers, such as transcription factors ATF4 and CCAAT/enhancer-binding protein homologous protein (CHOP), the ER stress-inducible enzyme PDI, and the ER-resident chaperons glucose-regulated protein 94 (GRP94) and growth arrest and DNA damage-inducible protein (GADD34), was observed in skeletal muscles of TRAF6f/f mice (Fig. 3A). More importantly, we found that starvation-induced expression of UPR genes was almost completely blunted in skeletal muscles of TRAF6mko mice. Western blot analysis also demonstrated that starvation augments ATF4 and CHOP protein levels in skeletal muscles of TRAF6f/f mice. In contrast, there was no noticeable increase in ATF4 or CHOP protein levels in GA muscles of fasted TRAF6mko mice (differences in ATF4 levels upon starvation, 1.52-fold ± 0.07-fold [TRAF6f/f] versus 0.32-fold ± 0.01-fold [TRAF6mko]; differences in CHOP levels upon starvation, 1.89-fold ± 0.21-fold [TRAF6f/f] versus 1.17-fold ± 0.04-fold [TRAF6mko]) (Fig. 3B). To further confirm that TRAF6 s involved in the induction of ER stress, we subjected cultured TRAF6+/+ and TRAF6−/− MEF to serum starvation for 24 h and measured the levels of various ER stress-related genes. Interestingly, the starvation-induced increases in ATF4, GRP94, GADD34, and CHOP transcript levels were found to be significantly inhibited in TRAF6−/− MEF compared to those in TRAF6+/+ MEF (see Fig. S2A in the supplemental material). Overexpression of ATF6 in cells causes ER stress response-related increased expression of various ER chaperons. Interestingly, ATF6-induced expression of GRP78 and GRP94 was completely blunted in TRAF6−/− MEF compared to that in TRAF6+/+ MEF (see Fig. S2B in the supplemental material). Taken together, these data provide the first evidence that TRAF6 is involved in the induction of the ER stress response.
Fig 3.
Role of TRAF6 in the induction of ER stress-responsive genes in skeletal muscle upon fasting. Three-month-old TRAF6f/f and TRAF6mko mice were fasted for 24 h, and isolated GA muscles were used for biochemical analysis. (A) ATF4, CHOP, GRP94, GADD34, and PDI transcript levels were found to be significantly lower in fasted GA muscles of TRAF6mko mice than in those of TRAF6f/f mice. n = 4 in each group. Error bars represent SD. *, P < 0.05 (values significantly different from those for muscles of starved TRAF6f/f mice). (B) Representative immunoblots demonstrating reduced protein levels of ATF4 and CHOP in TA muscles of starved TRAF6mko mice compared with those of TRAF6f/f mice. C2C12 myoblasts were differentiated into myotubes and starved for 3 and 6 h in PBS. U, unstarved; S, starved. (C) Representative immunoblots from two independent experiments performed in triplicate showing increased phosphorylation of eIF2α protein and elevated levels of ER stress-responsive proteins ATF3, ATF4, PDI, and CHOP in myotubes starved for 3 h or 6 h. Levels of MyHCf were reduced upon incubation of myotubes in PBS. (D) Splicing of XBP-1 increased upon fasting or the ER stressor tunicamycin in C2C12 myotubes measured by QRT-PCR assays using primers that detected both unspliced and spliced forms of XBP-1. uXBP-1, unspliced XBP-1; sXBP-1, spliced XBP-1.
Although fasting induced the expression of ER stress response genes in skeletal muscle, we could not detect any change in the level of phosphorylation of eIF2α (a downstream phosphorylation target and marker of activation of the PERK arm of the UPR) or splicing of XBP-1 mRNA (a marker of activation of the IRE1 arm of the UPR) in skeletal muscle in vivo. A recently published study also reported no changes in the level of phosphorylation of eIF2α in rat skeletal muscles measured after 1, 2, or 3 days of fasting (40). While there is a possibility that the expression of many of these genes (i.e., ATF4, CHOP, GADD34, and PDI) is governed through mechanisms independent of the ER stress response, it is also a possibility that eIF2α is activated transiently only at specific time point after starvation. To further examine whether fasting induces the ER stress response in skeletal muscle, we employed cultured myotubes and studied the phosphorylation of eIF2α and splicing of XBP-1 mRNA. As a model of acute starvation, culture medium of C2C12 myotubes was replaced with sterile PBS for 3 h or 6 h as described previously (51, 66). Protein extracts prepared from these myotubes were subjected to Western blotting. As shown in Fig. 3C, the phosphorylation of eIF2α was significantly increased at both 3 h and 6 h after the incubation of myotubes with PBS. Similarly, ATF3, ATF4, PDI, and CHOP protein levels were considerably increased in starved myotubes (Fig. 3C). Moreover, the levels of MyHCf protein were reduced, indicating that 6 h of starvation was sufficient to cause significant atrophy in vitro (Fig. 3C). XBP-1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor causing UPR induction (64). We studied the splicing of XBP-1 mRNA by performing semi-QRT-PCR using primers that detect both unspliced and spliced mRNAs. As shown in Fig. 3D, acute starvation of C2C12 myotubes by incubation with PBS dramatically increased the levels of the spliced form of XBP-1 (sXBP-1), indicating that starvation also activates the IRE1 arm of the ER stress response in myotubes.
Although ER stress has been studied in a plethora of homeostasis-rectifying mechanisms, its involvement in skeletal muscle atrophy has not been explored. To understand the potential role of ER stress in the induction of the atrophic program, we studied the effects of pharmacological inducers of ER stress on the expression of various components of the UPS and the ALS. C2C12 myotubes were treated with the ER stressor tunicamycin or thapsigargin for 18 h, and then the mRNAs of various genes were measured by QRT-PCR assay. As shown in Fig. S3A in the supplemental material, the expression of MAFBx and MuRF1 (important components of the UPS) and LC3B and Beclin1 (important components of the ALS) was significantly increased in tunica-mycin- or thapsigargin-treated myotubes compared to that in myotubes treated with the vehicle alone. Furthermore, myosin heavy chain II (i.e., MYH4) mRNA levels were found to be significantly reduced in C2C12 myotubes upon treatment with tunicamycin or thapsigargin (see Fig. S3B in the supplemental material). The increased expression of CHOP, ATF4, and GADD34 in tunicamycin- or thapsigargin-treated C2C12 myotubes confirmed that these agents activated the UPR (see Fig. S3C in the supplemental material). Collectively, these results suggest that ER stress-responsive pathways could be involved in the induction of muscle atrophy through both acceleration of proteolysis and inhibition of the expression of specific muscle genes such as that for MYH4.
TRAF6 is involved in the increased expression of Fn14 in skeletal muscle upon starvation.
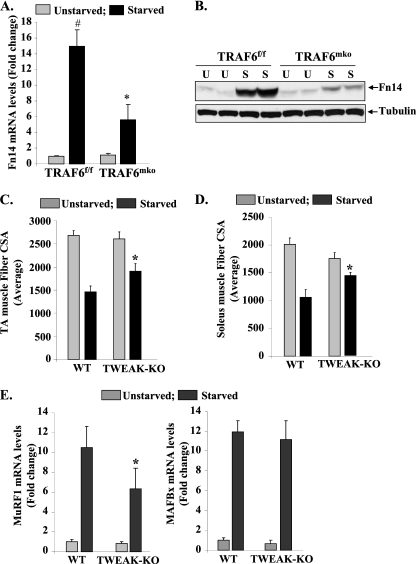
The TWEAK-Fn14 system has emerged as one of the important regulators of skeletal muscle atrophy, especially under disuse conditions (36). However, activation of the TWEAK-Fn14 system does not occur under all atrophy conditions (36). We first investigated whether fasting affects the expression of TWEAK or its receptor Fn14 in skeletal muscle. The expression of Fn14 (but not TWEAK itself) was dramatically induced in skeletal muscle in response to fasting. Intriguingly, fasting-induced expression of Fn14 was significantly inhibited in skeletal muscles of TRAF6mko mice compared to that in TRAF6f/f mice. Western blotting using muscle extracts also verified that the expression of Fn14 is increased in skeletal muscle in response to fasting and TRAF6 is required for fasting-induced expression of Fn14 in skeletal muscle (increases in Fn14 protein levels upon starvation: 5.04-fold ± 0.52-fold [TRAF6f/f] versus 1.72-fold ± 0.08-fold [TRAF6mko]; P < 0.01) (Fig. 4B). Fasting did not affect IL-1R, TNF receptor I (TNFRI), or TNFRII expression in skeletal muscles of either TRAF6f/f or TRAF6mko mice (our unpublished observation). These results suggest that the conditions of fasting specifically augment the expression of the Fn14 receptor through TRAF6-dependent mechanisms.
Fig 4.
Activation of the TWEAK-Fn14 system by TRAF6 in skeletal muscles of mice. TRAF6f/f and TRAF6mko mice were fasted for 24 h, and isolated GA muscles were analyzed for expression of Fn14. (A) Fn14 transcript levels were significantly reduced in GA muscles of starved TRAF6mko mice compared with those of TRAF6f/f mice. (B) Representative immunoblot demonstrating reduced levels of Fn14 protein but not the unrelated protein tubulin in TA muscles of starved TRAF6mko mice compared to those of TRAF6f/f mice. Three-month-old wild-type TWEAK-KO mice were starved for 24 h. Quantification of mean fiber CSA in H&E-stained sections of TA (C) and soleus (D) muscles of wild-type and TWEAK-KO mice (n = 8 in each group). (E) MuRF1 but not MAFBx transcript levels were found to be significantly reduced in muscles of fasted TWEAK-KO mice compared to those in wild-type mice. Error bars represent SD. #, P < 0.01 (values significantly different from those of GA muscles of unstarved TRAF6f/f mice); *, P < 0.05 (values significantly different from those of GA muscles of starved TRAF6f/f mice).
While it was interesting to learn that TRAF6 regulates the expression of Fn14 in skeletal muscle, it was not clear whether the TWEAK-Fn14 system also contributes to muscle atrophy upon starvation. To answer this question, we employed TWEAK-KO mice, which have been previously found to be resistant to muscle atrophy in response to denervation (36). Three-month-old wild-type and TWEAK-KO mice were provided normal food or fasted for 24 h and then euthanized, and then their hind limb muscles were isolated for analyses. Transverse sections prepared from TA and soleus muscles were subjected to H&E staining, and the average fiber CSA was quantified by morphometric methods. Interestingly, starvation-induced loss of fiber CSA was significantly inhibited in the TA (Fig. 4C) and soleus (Fig. 4D) muscles of TWEAK-KO mice compared to that in wild-type mice.
To evaluate the mechanisms, we also studied whether the increased activity of TWEAK-Fn14 augments the expression of the components of the UPS, ALS, and/or UPR in skeletal muscle. UPS activation was evaluated by measuring MAFBx and MuRF1 mRNA levels by QRT-PCR. As shown in Fig. 4E, the starvation-induced increase in MuRF1 was significantly inhibited in GA muscles of TWEAK-KO mice compared to that in wild-type mice. In contrast, there was no significant difference in MAFBx mRNA levels between wild-type and TWEAK-KO mice upon starvation (Fig. 4E). QRT-PCR analysis also revealed that there was no significant difference in mRNA levels of LC3B, Beclin1, and Atg12 in skeletal muscles of wild-type and TWEAK-KO mice in response to starvation (see Fig. S4 in the supplemental material). Furthermore, there was also no significant difference in the starvation-induced expression of the ER stress-related genes for ATF4, GADD34, CHOP, HERPES, PDI, and GRP94 in the muscles of fasted wild-type and TWEAK-KO mice (see Fig. S5 in the supplemental material). Taken together, these results indicate that TWEAK-Fn14 might be contributing to starvation-induced muscle loss through activation of the components of the UPS pathway but not those of the ALS or UPR pathway.
E3 ubiquitin ligase activity of TRAF6 is essential for starvation-induced muscle atrophy.
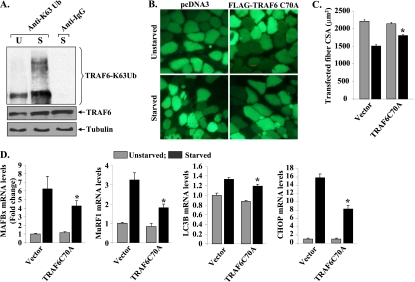
TRAF6 functions both as an adaptor protein and as an E3 ubiquitin ligase catalyzing K63-linked autoubiquitination, as well as ubiquitination of target proteins, which stimulates protein trafficking (26). While the E3 ubiquitin ligase activity of TRAF6 has been found to be critical for many of its cellular functions, it has also been reported that the ubiquitin ligase activity of TRAF6 may be dispensable for the activation of several downstream signaling pathways in response to specific stimuli (57). We first studied whether starvation increases TRAF6 autoubiquitination in skeletal muscle in vivo. Protein extracts prepared from skeletal muscles of unstarved or starved wild-type mice were immunoprecipitated with anti-TRAF6 and then immunoblotted with anti-K63 ubiquitin antibody, which recognizes only K63-linked ubiquitinated proteins. As shown in Fig. 5A, TRAF6 ubiquitination was significantly increased in fasted mouse skeletal muscle. Mutation of cystine to alanine (C70A) in the TRAF6 zinc finger domain renders it unable to interact and bind with the Ubc13-Uev complex (26). The TRAF6C70A mutant form has previously been used to study the role of TRAF6 E3 ubiquitin ligase activity in various cellular responses (26, 28). To understand whether the E3 ubiquitin ligase activity of TRAF6 is required for fasting-induced muscle atrophy, TA muscles of wild-type mice were electroporated with the vector alone or with TRAF6C70A cDNA. Efficiency of gene delivery was monitored by coelectroporation with an enhanced green fluorescent protein (EGFP)-expressing vector as described previously (44). After 10 days of plasmid electroporation, the mice were given normal food or fasted for 24 h and isolated TA muscles were used to make sections and for measurement of fiber CSA. Interestingly, overexpression of the TRAF6C70A mutant protein significantly inhibited starvation-induced fiber atrophy in the TA muscles of mice (Fig. 5B and C).
Fig 5.
TRAF6 E3 ubiquitin ligase activity is required for starvation-induced muscle atrophy. Three-month-old mice were fasted for 24 h. (A) TA muscle extracts prepared from unstarved (U) and starved (S) mice were immunoprecipitated with TRAF6 antibody and then Western blotted using an antibody that recognizes only K63-linked ubiquitinated (Ub) substrates (top). Western blotting of total muscle extracts using anti-TRAF6 antibody (middle) and anti-α-tubulin (bottom). (B) TA muscles of C57BL6 mice were electroporated with plasmids expressing EGFP and FLAG-TRAF6C70A mutant protein. Ten days later, mice were fasted for 24 h and myofibers expressing EGFP in combination with pCDNA3 or FLAG-TRAF6C70A were analyzed by fluorescence microscopy. (C) Quantification of fiber CSA of transfected TA muscles of TRAF6f/f and TRAF6mko mice. (D) Cultured C2C12 myoblasts were transfected with the vector alone or the TRAF6C70A mutant protein, differentiated into myotubes, and starved for 6 h in PBS. The starvation-induced increase in MAFBx, MuRF1, LC3B, and CHOP mRNA levels was significantly inhibited in TRAF6C70A-transfected cultures compared to that in cultures treated with the vector alone. Error bars represent SD. *, P < 0.05 (values significantly different from starved myotubes transfected with the vector alone).
To further evaluate the role of the ubiquitin ligase activity of TRAF6 in starvation-induced muscle atrophy, we investigated whether overexpression of the TRAF6C70A mutant protein can affect the activation of the atrophic program in cultured myotubes. C2C12 myoblasts were stably transfected with the vector alone or TRAF6C70A; this was followed by their differentiation into myotubes. To induce starvation, the myotubes were incubated in PBS for different length of time. As shown in Fig. S6 in the supplemental material, overexpression of TRAF6C70A inhibited starvation-induced atrophy in cultured myotubes. Quantitative estimation of myotube diameters also revealed that the TRAF6C70A mutant protein inhibits atrophy in cultured myotubes (see Fig. S7 in the supplemental material). Finally, we investigated whether overexpression of the TRAF6C70A mutant protein can block the expression of the markers of the UPS, the ALS, and the ER stress response in myotubes. Interestingly, fasting-induced increases in MAFBx, MuRF1, LC3B, and CHOP mRNA levels was found to be significantly inhibited in TRAF6C70A-transfected myotubes compared to that in myotubes transfected with the vector alone (Fig. 5D). Together, these experiments provide convincing evidence that the E3 ubiquitin ligase activity of TRAF6 is essential for starvation-induced skeletal muscle atrophy.
DISCUSSION
This study identified TRAF6 as a novel regulator of a complex array of intracellular mechanisms that underlie the starvation-induced atrophic response in skeletal muscle. This investigation is also the first one to recognize potential roles for the TWEAK-Fn14 system and UPR pathways in the development of skeletal muscle atrophy in response to starvation.
It has been consistently observed that under almost all atrophic conditions, the activation of the UPS and the ALS is a common denominator of the loss of skeletal muscle mass. Rapid muscle atrophy might stem from the degradation of thick and thin filament proteins, some of which are targeted by muscle-specific E3 ubiquitin ligases MuRF1 and MAFBx (6, 10, 11, 16). As also noticed under other atrophic conditions (42), one of the mechanisms by which TRAF6 mediates starvation-induced muscle atrophy is activation of the UPS because fasting-induced overall protein ubiquitination and the expression of both MAFBx and MuRF1 were significantly lower in skeletal muscles of TRAF6mko mice than in those of control littermates (Fig. 2A and B).
The contribution of ALS to the regulation of skeletal muscle mass under both physiological and pathological conditions has gained increasing attention. It has also been found that along with the UPS, activation of the ALS also induces myofiber degradation in atrophying skeletal muscle (4, 50, 66). Although autophagy is a homeostasis maintenance mechanism and physiological autophagy is necessary for the removal of protein aggregates and defunct cellular compartments (35, 50), the hyperactivated ALS can contribute to muscle proteolysis under various conditions, including fasting (50). Consistent with previously published reports (34, 66), our results indicate that fasting causes increased expression of several autophagy genes (Fig. 2C and D) and activation of the FOXO3a transcription factor (Fig. 2E) and AMPK (Fig. 2F). While AMPK is known to stimulate the expression of LC3B, the FOXO3a transcription factor is purported to upregulate MAFBx and MuRF1 and regulates the ALS in skeletal muscle (34, 66). All of these markers and effectors of macroautophagy were found to be significantly inhibited in TRAF6mko mice compared to those in TRAF6f/f mice (Fig. 2), providing further evidence of the role of TRAF6 and autophagy in starvation-induced atrophy. TRAF6 interacts with LC3B through p62 (39) and ubiquitinylates (Lys-63-linked) Beclin-1 and thus further regulates autophagosome formation in response to Toll-like receptor 4 signaling (54). Our results also indicate a similar interaction in starvation-induced muscle atrophy because levels of p62, LC3B, and Beclin-1 were found to be induced by fasting in TRAF6f/f mice and were rescued in TRAF6mko mice (Fig. 2C and D). Though it has been suggested that proteasomal degradation dominates over lysosomal breakdown of proteins during muscle atrophy (34, 66), our results indicate that, irrespective of their relative contributions, TRAF6 recruits both the UPS and ALS in starvation-induced muscle atrophy.
The ER is a site for and an important regulator of protein folding, trafficking, targeting, and quality control. Under the conditions of increased unfolded protein load/stress, ER stimulates an elaborate corrective response known as the UPR (47), which is mediated by three ER membrane-associated proteins, PERK, IRE1, and ATF6. UPR pathways have been found to be involved in several pathological and metabolic disorders, such as impaired glucose metabolism (3, 41), adipocyte stress (17, 48), and inflammatory response and metabolic abnormalities (20, 62, 65). However, there has been no published evidence yet suggesting direct involvement of the UPR in skeletal muscle atrophy. In an attempt to study the involvement of ER stress in disuse atrophy, it was previously reported that while the genes that maintain sarcoplasmic reticulum calcium levels are induced in disuse atrophy, there is no such increase in the markers of ER stress or the UPR (21). However, a recent report suggests that in response to starvation, the expression of ATF4 is increased, which promotes myofiber atrophy (15). Interestingly, it was found that ATF4 affects the expression of a few genes involved in muscle growth; however, it does not affect the expression of MAFBx or MuRF1, suggesting that ATF4 can induce muscle atrophy through mechanisms independent of activation of the UPS (15). Although ATF4 can be activated by multiple mechanisms under fasting conditions, it belongs to the PERK/eIF2α branch of the UPR and thus there is a possibility that UPR pathways are activated and mediate starvation-induced muscle atrophy.
Recent investigations have highlighted the role the ER stress and UPR pathways play in energy and glucose metabolism and have shown ER sensitivity to glucose availability. Transcriptional networks activated by ER stress regulate the expression of several genes involved in glucose metabolism. GADD34, XBP-1, and ATF6 have been found to be involved in glucose output, glycogen synthesis and gluconeogenesis (2, 30, 41, 58), implying that all three of the arms of the UPR play roles in energy and glucose metabolism. Muscles, being the largest reservoir of proteins, are the first ones to be mobilized as a source of amino acids for gluconeogenesis. Although UPR activation and its role in glucose metabolism in atrophying muscle have not yet been elucidated, there is a possibility that fasting activates the UPR, which in turn induces secondary mechanisms regulating muscle atrophy. Our results provide initial evidence that the UPR is activated in skeletal muscle both in vitro and in vivo (Fig. 3) and underline the possibility that through transcriptional activation of downstream effectors, UPR pathways could be instrumental in the regulation of skeletal muscle mass. This inference is also supported by our finding that treatment of myotubes with the ER stressor tunicamycin or thapsigargin augmented the expression of components of the UPS and the ALS (see Fig. S3 in the supplemental material). Interestingly, all of the markers of the UPR were significantly inhibited in TRAF6mko mice (Fig. 3), suggesting that TRAF6 plays a critical role in the induction of the UPR in starvation-induced skeletal muscle atrophy. Previously published reports bolster this observation by implicating TRAF2 in the activation of the UPR and proteolytic degradation and cell death (24). TRAF2 is another E3 ubiquitin ligase of the TRAF family and has many features in common with TRAF6 (55). Therefore, it is possible that functional redundancy exists between TRAF2 and TRAF6 in the activation of UPR pathways in response to different stimuli. Future research will reveal an interaction and/or mechanisms through which TRAF6 regulates the ER stress response in fasted mouse muscle.
The TWEAK-Fn14 axis is another recently discovered regulator of skeletal muscle atrophy (14, 36). Increased expression of the Fn14 receptor appears to be a key determinant in TWEAK-mediated skeletal muscle wasting, especially under disuse conditions (36). In the present study, we observed that the levels of Fn14 were dramatically induced in fasted mouse skeletal muscle and that the TWEAK-Fn14 system contributed to starvation-induced muscle atrophy in mice (Fig. 4). It is noteworthy that although Fn14 is a cell surface receptor and TRAF6 is an intracellular protein, signaling through TRAF6 induces the expression of Fn14 in fasted mouse muscles. Our previous investigations showed that TRAF6 does not play a role in the inducible expression of Fn14 in denervated muscle (42), further suggesting that TRAF6 mediates different types of muscle atrophy through the activation of distinct mechanisms. The observation that Fn14 is one of the downstream targets of TRAF6 in starvation-induced atrophy is also supported by our finding that inhibition of the TWEAK-Fn14 system did not affect the activation of either the ALS (see Fig. S4 in the supplemental material) or the UPR (see Fig. S5 in the supplemental material) in skeletal muscle, whereas ablation of TRAF6 inhibited both of these pathways in response to starvation (Fig. 2). On the similar lines, we found that while MAFBx mRNA levels in TRAF6mko mice were lower than those in TRAF6f/f mice (Fig. 2B), there was no significant difference in the levels of expression of MAFBx in the skeletal muscles of TWEAK-KO and wild-type mice in response to starvation (Fig. 4E).
In the recent years, significant progress has been made in understanding the mechanisms by which TRAF6 propagates downstream signaling. The most accepted model suggests that upon activation, the RING finger ubiquitin E3 ligase domain makes complexes with E2 conjugating enzyme Ubc13/Uev1a to mediate the conjugation of K63-linked ubiquitin chains to TRAF6 substrates, including TRAF6 itself. These chains recruit several factors, including adaptor proteins TAB2/3, which contain atypical zinc finger domains with an affinity for K63-linked ubiquitin chain binding, resulting in activation of the transforming growth factor beta-activated kinase 1 (TAK1) complex. However, Walsh et al. have previously reported that while the RING finger domain of TRAF6 is essential for the activation of TAK1, it is not required for interaction between TRAF6 and the TAK1 complex in vitro (57). Furthermore, this study suggested that TRAF6 autoubiquitination is dispensable for both its interaction with and activation of the TAK1 complex and also for RANKL-induced osteoclastogenesis (57). In contrast, our experiments demonstrate that TRAF6 undergoes K63-linked autoubiquitination in skeletal muscle in response to starvation (Fig. 5A). Furthermore, using mutant TRAF6 lacking ubiquitin ligase activity (i.e., TRAF6C70A) (26), we found that the ligase activity of TRAF6 is essential for orchestrating the activation of the ALS, UPS, and UPR upon acute starvation (Fig. 5D). The requirement of E3 ubiquitin ligase for muscle atrophy in response to starvation (Fig. 5; see Fig. S6 and S7 in the supplemental material) but not during osteoclastogenesis (57) suggests that TRAF6 induces the activation of different signaling complexes in a context-dependent manner. It is also notable that starvation-induced muscle atrophy may not even involve activation of the TAK1–IKK–NF-κB pathway, for which the autoubiquitination of TRAF6 or its E3 ligase activity has been found to be dispensable during osteoclastogenesis (57).
In conclusion, the results of the present study identify TRAF6 as a novel regulator of starvation-induced atrophy. Since molecular pathways regulated by TRAF6 are also implicated in many other cell types and physiological responses, a better understanding of its regulatory role will be of significant clinical importance for developing new therapeutic strategies for the treatment of muscle disorders and other diseases.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to A. Ashkenazi for providing TWEAK-KO mice.
This study was supported by funding from National Institutes of Health grants AG029623 and AR059810 to A.K.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 30 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Acharyya S, Guttridge DC. 2007. Cancer cachexia signaling pathways continue to emerge yet much still points to the proteasome. Clin. Cancer Res. 13:1356–1361 [DOI] [PubMed] [Google Scholar]
- 2. Acosta-Alvear D, et al. 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27:53–66 [DOI] [PubMed] [Google Scholar]
- 3. Back SH, et al. 2009. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 10:13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bechet D, Tassa A, Taillandier D, Combaret L, Attaix D. 2005. Lysosomal proteolysis in skeletal muscle. Int. J. Biochem. Cell Biol. 37:2098–2114 [DOI] [PubMed] [Google Scholar]
- 5. Bjørkøy G, et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bodine SC, et al. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704–1708 [DOI] [PubMed] [Google Scholar]
- 7. Cao PR, Kim HJ, Lecker SH. 2005. Ubiquitin-protein ligases in muscle wasting. Int. J. Biochem. Cell Biol. 37:2088–2097 [DOI] [PubMed] [Google Scholar]
- 8. Chen ZJ. 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7:758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciechanover A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarke BA, et al. 2007. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 6:376–385 [DOI] [PubMed] [Google Scholar]
- 11. Cohen S, et al. 2009. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J. Cell Biol. 185:1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox JS, Shamu CE, Walter P. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197–1206 [DOI] [PubMed] [Google Scholar]
- 13. Dehoux M, et al. 2004. Role of the insulin-like growth factor I decline in the induction of atrogin-1/MAFbx during fasting and diabetes. Endocrinology 145:4806–4812 [DOI] [PubMed] [Google Scholar]
- 14. Dogra C, et al. 2007. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 21:1857–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ebert SM, et al. 2010. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol. Endocrinol. 24:790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. 2001. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U. S. A. 98:14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregor MG, Hotamisligil GS. 2007. Thematic review series: adipocyte biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J. Lipid Res. 48:1905–1914 [DOI] [PubMed] [Google Scholar]
- 18. Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274 [DOI] [PubMed] [Google Scholar]
- 19. Haze K, Yoshida H, Yanagi H, Yura T, Mori K. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH. 2006. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol. Cell. Biol. 26:3071–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunter RB, Mitchell-Felton H, Essig DA, Kandarian SC. 2001. Expression of endoplasmic reticulum stress proteins during skeletal muscle disuse atrophy. Am. J. Physiol. Cell Physiol. 281:C1285–C1290 [DOI] [PubMed] [Google Scholar]
- 22. Kandarian SC, Jackman RW. 2006. Intracellular signaling during skeletal muscle atrophy. Muscle Nerve 33:155–165 [DOI] [PubMed] [Google Scholar]
- 23. Kilberg MS, Shan J, Su N. 2009. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 20:436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim I, Xu W, Reed JC. 2008. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 7:1013–1030 [DOI] [PubMed] [Google Scholar]
- 25. Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462–464 [DOI] [PubMed] [Google Scholar]
- 26. Lamothe B, et al. 2007. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J. Biol. Chem. 282:4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lamothe B, et al. 2008. The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J. Biol. Chem. 283:24871–24880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lamothe B, et al. 2007. TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochem. Biophys. Res. Commun. 359:1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lecker SH, et al. 2004. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 18:39–51 [DOI] [PubMed] [Google Scholar]
- 30. Lee AH, Scapa EF, Cohen DE, Glimcher LH. 2008. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320:1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Malhotra S, Kumar A. 2008. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J. Mol. Med. 86:1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maecker H, et al. 2005. TWEAK attenuates the transition from innate to adaptive immunity. Cell 123:931–944 [DOI] [PubMed] [Google Scholar]
- 34. Mammucari C, et al. 2007. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6:458–471 [DOI] [PubMed] [Google Scholar]
- 35. Masiero E, et al. 2009. Autophagy is required to maintain muscle mass. Cell Metab. 10:507–515 [DOI] [PubMed] [Google Scholar]
- 36. Mittal A, et al. 2010. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J. Cell Biol. 188:833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15:1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moscat J, Diaz-Meco MT, Wooten MW. 2007. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32:95–100 [DOI] [PubMed] [Google Scholar]
- 39. Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. 2010. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J. Biol. Chem. 285:2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ogata T, Oishi Y, Higuchi M, Muraoka I. 2010. Fasting-related autophagic response in slow- and fast-twitch skeletal muscle. Biochem. Biophys. Res. Commun. 394:136–140 [DOI] [PubMed] [Google Scholar]
- 41. Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. 2008. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7:520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paul PK, et al. 2010. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J. Cell Biol. 191:1395–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533 [DOI] [PubMed] [Google Scholar]
- 44. Rana ZA, Ekmark M, Gundersen K. 2004. Coexpression after electroporation of plasmid mixtures into muscle in vivo. Acta Physiol. Scand. 181:233–238 [DOI] [PubMed] [Google Scholar]
- 45. Rock KL, et al. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761–771 [DOI] [PubMed] [Google Scholar]
- 46. Romanello V, et al. 2010. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 29:1774–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ron D, Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 [DOI] [PubMed] [Google Scholar]
- 48. Rutkowski DT, et al. 2008. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell 15:829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. 2004. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am. J. Physiol. Endocrinol. Metab. 287:E591–E601 [DOI] [PubMed] [Google Scholar]
- 50. Sandri M. 2010. Autophagy in skeletal muscle. FEBS Lett. 584:1411–1416 [DOI] [PubMed] [Google Scholar]
- 51. Sandri M, et al. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117:399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schertzer JD, Plant DR, Lynch GS. 2006. Optimizing plasmid-based gene transfer for investigating skeletal muscle structure and function. Mol. Ther. 13:795–803 [DOI] [PubMed] [Google Scholar]
- 53. Schwartz AL, Ciechanover A. 1999. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 50:57–74 [DOI] [PubMed] [Google Scholar]
- 54. Shi CS, Kehrl JH. 2010. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3:ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Silke J, Brink R. 2010. Regulation of TNFRSF and innate immune signalling complexes by TRAFs and cIAPs. Cell Death Differ. 17:35–45 [DOI] [PubMed] [Google Scholar]
- 56. Tran H, Brunet A, Griffith EC, Greenberg ME. 2003. The many forks in FOXO's road. Sci. STKE 2003:RE5. [DOI] [PubMed] [Google Scholar]
- 57. Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. 2008. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS One 3:e4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Y, Vera L, Fischer WH, Montminy M. 2009. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 460:534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu CL, Kandarian SC, Jackman RW. 2011. Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS One 6:e16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu J, et al. 2011. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 13:160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamashita M, et al. 2008. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell 31:918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamazaki H, et al. 2009. Activation of the Akt-NF-kappaB pathway by subtilase cytotoxin through the ATF6 branch of the unfolded protein response. J. Immunol. 183:1480–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang WL, et al. 2009. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325:1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891 [DOI] [PubMed] [Google Scholar]
- 65. Zhang K, et al. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587–599 [DOI] [PubMed] [Google Scholar]
- 66. Zhao J, et al. 2007. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6:472–483 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.