Abstract
The general stress response of Bacillus subtilis can be activated by a wide range of signals, including low intensities of visible light. It is regulated by a dedicated σ factor via a complex signal transduction pathway that makes use of stressosomes: hetero-oligomeric complexes that include one or more of the RsbR proteins (RsbRA, RsbRB, RsbRC, and RsbRD). The response to blue light is mediated by the photoreceptor YtvA. We show here which of the four RsbR proteins are necessary for the activation of the σB response by blue light. Experiments performed with single-, double-, and triple-deletion strains in the rsbR genes show that RsbRB and RsbRA function antagonistically, with the former being a negative regulator and the latter a positive regulator of the YtvA-dependent light activation of the stress response. A strain with RsbRB as the only RsbR protein is unable to respond to light-activation of σB. Furthermore, RsbRC and RsbRD can replace RsbRA's function only in the absence of RsbRB. This differentiation of function is confined to light stress, since strains with RsbRA or RsbRB as the only RsbR protein behave similarly in our experimental conditions in response to physicochemical stresses. Interestingly, RsbRB's absence is sufficient to result in light activation of the general stress response at wild-type expression levels of ytvA, while it was previously reported that YtvA could only activate σB when overproduced, or when cells are supplemented with an additional environmental stress.
INTRODUCTION
The general stress response (GSR) of Bacillus subtilis, mediated by the alternative sigma factor B (σB), is activated when cells encounter “energy” (or nutritional) stresses, such as carbon, phosphorus, or oxygen limitation, or “environmental” (or physicochemical) stresses, such as acid, ethanol, heat, or NaCl shock (6, 7, 38). In addition, it was recently discovered that illumination with blue light can activate the GSR (3). In B. subtilis, σB regulates the expression of approximately 150 genes that are involved in processes such as carbon metabolism, macromolecular turnover, and envelope function and in counteracting stresses, either directly or indirectly (18, 33, 34). Several important pathogens, such as Staphylococcus aureus and Listeria monocytogenes, have a GSR-like regulatory network that is involved in regulation of virulence (20).
The activity of σB is regulated by a partner switch mechanism, which in turn is regulated by two convergent signal transduction pathways. In unstressed cells, σB is held inactive by the anti-sigma factor RsbW. Upon exposure to stress, the phosphorylated form of the anti-anti-sigma factor RsbV (RsbV-P) is dephosphorylated, whereupon RsbV binds to RsbW and σB is released, leaving it free to associate with RNA polymerase and activate the GSR. When the stress signal has decayed, RsbV-P is no longer actively dephosphorylated, and the balance shifts back toward rephosphorylation of RsbV by the kinase activity of RsbW. This results in the liberation of RsbW from RsbV, which switches off the GSR (for a review, see Hecker et al. [17]).
Dephosphorylation of RsbV-P is catalyzed by two phosphatases. RsbP, one of the two phosphatases that contains an N-terminal PAS (Per-ARNT-Sim) domain, is required for the response to energy stress (37) and, as shown recently, to red light (4). Although the activation mechanism of RsbP remains unclear, a correlation with a drop in ATP levels has been reported (41), and the activity of an α/β-hydrolase (RsbQ) is required for this response (9).
More is known about RsbU, the other phosphatase, which is activated by environmental stresses (40). RsbU activation is based on interaction between its N-terminal domain and the kinase RsbT (13, 40). RsbT is normally held inactive by a high-molecular-weight complex called the stressosome, which is speculated to function as a signal integration hub (12, 28). The stressosome contains at least three classes of proteins: one or more of the RsbR proteins (see below), RsbS, and the aforementioned kinase RsbT (10, 12, 23). Both RsbS and the RsbR proteins contain a conserved STAS (sulfate transporter and anti-sigma factor antagonist) domain, which can be phosphorylated by RsbT upon activation by environmental stress (1, 11, 16, 22). This results in release of RsbT, which activates RsbU. When the stress signal is extinguished, the prestress condition is restored by the phosphatase RsbX, which dephosphorylates RsbS and RsbRA, resulting in a stressosome capable of recapturing RsbT (11, 40). Genome sequence analyses have shown that the stressosome is not unique to B. subtilis and that it can likely be coupled to a variety of output functions (32).
Initially only one RsbR protein was known, but the availability of the genome sequence of B. subtilis led to the identification of three paralogs. These four proteins have been renamed RsbRA (previously RsbR), RsbRB (YkoB), RsbRC (YojH), and RsbRD (YqhA) (1, 23). All have a highly conserved C-terminal STAS domain, which forms the core of the stressosome, together with the STAS domain of RsbS (27). Their N-terminal domains, in contrast, are diverse (1). For clarity, these four proteins are referred to here as “RsbR proteins.” Two additional paralogs, each having a C-terminal STAS domain, were identified: the split paralog YetI/YezB, which does not appear to be involved in the regulation of the GSR and will not be considered here, and YtvA (1). YtvA was demonstrated to sense blue light with its N-terminal LOV (light, oxygen, voltage) domain in vitro (26). The loss or overproduction of YtvA abolishes or enhances, respectively, the environmental σB response induced by moderate intensities of blue light (3, 4, 36). However, the light enhancement of the GSR by YtvA only becomes apparent when YtvA is overproduced or when light is combined with an additional environmental stress, such as NaCl shock (3, 36). YtvA has been shown to act upstream of RsbU and RsbT (15), suggesting that it interacts with the stressosome. However, its role in or on the stressosome is not yet resolved. YtvA, RsbRB, RsbRC, and RsbRD can all be copurified with RsbRA from B. subtilis cell extracts (11, 12, 15, 23), strongly suggesting that YtvA is associated with the stressosome. However, RsbT is able to phosphorylate all four RsbR proteins, but not YtvA (1), and stressosomes can be made in vitro by mixing RsbS with any of the four RsbR proteins, but this has not yet been reported for YtvA (10, 12). In addition, a quadruple knockout of the RsbR proteins leads to a constitutive and high level of activation of the GSR, presumably due to the inability to form (sufficient) stressosomes to capture RsbT, even though YtvA is still present in such strains (1, 23).
The specific role of each individual RsbR protein in the stressosome has remained unclear. The most intuitive explanation would be that each paralog senses a different (set of) stress(es) via its N-terminal domain. However, this appears unlikely, since mutants with any three of the four RsbR proteins deleted respond in a very similar way to NaCl and ethanol shock (1, 23). An alternative explanation is that all four RsbR proteins respond to a common intracellular signal(s), presumably created by all environmental stresses, but with a different sensitivity or threshold. This would allow the cell to fine-tune the sensitivity of the stressosome as required. This hypothesis is supported by the fact that each rsbR gene is contained in a different transcription unit, implying that differential regulation of expression may be important. Indeed, transcriptional analyses have shown that rsbRD is upregulated by σB (33), whereas rsbRA and rsbRB are upregulated by the antimicrobials 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on, respectively (30). Additional studies have shown that transcription of ytvA increases in strains that overproduce Spx and σE: important regulators involved in disulfide stress and sporulation, respectively (14, 15). Intriguingly, many other bacteria that contain the genes for a stressosome also contain more than one RsbR paralog (32).
Because neither the separate input signals for each of the four RsbR proteins nor the upstream components of the environmental stress pathway are known, it is difficult to determine their individual functions in vivo. Therefore, we studied the roles of the RsbR proteins with respect to YtvA-mediated light stress. To this end, we performed a systematic study of the activation of the GSR in strains with all possible combinations of null mutations in the four rsbR genes. The photoreceptor YtvA was overproduced in these strains, and the GSR activation was measured in the presence or absence of light. Our results show a differentiation of function, most notably between RsbRB and the other RsbR proteins. In addition, we show that a single deletion of rsbRB is sufficient for a light-sensitive GSR, without the need for overproduction of YtvA or an additional environmental stress.
MATERIALS AND METHODS
Bacterial strains and genetic manipulation.
The bacterial strains and plasmids used in this investigation are listed in Table 1. DNA manipulations and molecular genetic techniques were carried out according to standard procedures. All B. subtilis strains used in the present study are derivatives of the wild-type PB2 (8).
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Genotypea | Reference or constructionb |
|---|---|---|
| Plasmids | ||
| pDG148-Stu | Pspac-MCS bla kan ble (cloning vector) | 19 |
| pYtvA | Pspac-ytvA bla kan ble (replicating plasmid) | 3 |
| Strains | ||
| PB2 | trpC2 (168 wild type) | 8 |
| PB198 | amyE::(Pctc-lacZ cat) trpC2 | 8 |
| PB491 | ΔrsbRA amyE::(Pctc-lacZ cat) trpC2 | 2 |
| PB528 | ΔrsbRB::kan amyE::(Pctc-lacZ cat) trpC2 | 1 |
| PB565 | ΔytvA::ery amyE::(Pctc-lacZ cat) trpC2 | 1 |
| PB574 | ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 | 1 |
| PB520 | ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | 1 |
| PB198/pYtvA | amyE::(Pctc-lacZ cat) trpC2 pYtvA | 3 |
| PB491/pYtvA | ΔrsbRA amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→PB491 |
| PB528/pYtvA | ΔrsbRB::kan amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→PB528 |
| PB574/pYtvA | ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→PB574 |
| PB520/pYtvA | ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2pYtvA | pYtvA→PB520 |
| JBS10011 | ΔrsbRB::kan ΔytvA::ery amyE::(Pctc-lacZ cat) trpC2 | PB565→PB528 |
| MA1 | ΔrsbRA ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 | PB574→PB491 |
| MA2 | ΔrsbRA ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB520→PB491 |
| MA3 | ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB520→PB574 |
| MA4 | ΔrsbRA ΔrsbRB::kan amyE::(Pctc-lacZ cat) trpC2 | PB528→PB491 |
| MA5 | ΔrsbRB::kan ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 | PB528→PB574 |
| MA6 | ΔrsbRB::kan ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB528→PB520 |
| MA7 | ΔrsbRB::kan ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB528→MA3 |
| MA8 | ΔrsbRA ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB520→MA1 |
| MA9 | ΔrsbRA ΔrsbRB::kan ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB520→MA4 |
| MA10 | ΔrsbRA ΔrsbRB::kan ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 | PB528→MA1 |
| MA11 | ΔrsbRA ΔrsbRB::kan ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 | PB520→MA10 |
| MA1/pYtvA | ΔrsbRA ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA1 |
| MA2/pYtvA | ΔrsbRA ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA2 |
| MA3/pYtvA | ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA3 |
| MA4/pYtvA | ΔrsbRA ΔrsbRB::kan amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA4 |
| MA5/pYtvA | ΔrsbRB::kan ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA5 |
| MA6/pYtvA | ΔrsbRB::kan ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA6 |
| MA7/pYtvA | ΔrsbRB::kan ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA7 |
| MA8/pYtvA | ΔrsbRA ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA8 |
| MA9/pYtvA | ΔrsbRA ΔrsbRB::kan ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA9 |
| MA10/pYtvA | ΔrsbRA ΔrsbRB::kan ΔrsbRC::ery amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA10 |
| MA11/pYtvA | ΔrsbRA ΔrsbRB::kan ΔrsbRC::ery ΔrsbRD::spc amyE::(Pctc-lacZ cat) trpC2 pYtvA | pYtvA→MA11 |
Genes for antibiotic selection: bla, ampicillin; ble, phleomycin; cat, chloramphenicol; erm, erythromycin; kan, kanamycin; spc, spectinomycin.
Arrows indicate the direction of transformation. Purified DNA (left) was transformed into a specific strain (right).
Plasmid and strain constructions.
The construction of pYtvA, a plasmid for overexpression of ytvA based on pDG148-Stu (19), has been reported elsewhere (3). This plasmid features the ytvA gene under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible spac promoter.
For strain constructions, transformants were selected on Bacto tryptic soy broth (TSB; 30 g/liter) agar plates, containing, when appropriate, 10 μg of kanamycin/ml, 5 μg of chloramphenicol/ml, 0.5 μg of erythromycin/ml, 100 μg of spectinomycin/ml, or 10 μg of phleomycin/ml, after overnight incubation at 37 or 30°C. Transformations of B. subtilis were carried out according to a previously described protocol (24). The double, triple, and quadruple null mutant strains MA1 to MA11, as well as JBS10011, were obtained by transforming the parent strain with chromosomal DNA of the strain bearing the desired mutation (see Table 1 for details). The pYtvA plasmid was also introduced into a number of strains by standard transformation. The only exception to this was strain MA11, which we could not transform using natural competence. For this strain, electrotransformation was used instead, according to a previously described protocol (39).
The identities of all transformants were checked by the presence of the appropriate antibiotic resistance(s) and by various PCRs to check for the presence of the correct knockout(s) and/or plasmid(s).
Growth conditions and β-galactosidase assays.
The medium used for the β-galactosidase assays was TSB supplemented with 0.5% glucose, 5 μg of chloramphenicol/ml, and 10 μg of kanamycin/ml or 10 μg of phleomycin/ml, where applicable, to keep selective pressure on the replicating plasmids. The antibiotics did not result in a reduction in growth rate in our conditions. TSB medium was inoculated with a single colony from a fresh plate of the particular strain and incubated overnight in a shaking water bath at 37°C and approximately 250 rpm. The overnight cultures were diluted and allowed to grow until they had reached the exponential growth phase, rediluted, and distributed over different Erlenmeyer flasks to initiate an experiment. No volume larger than 10 ml in a 100-ml Erlenmeyer flask was used to assure sufficient aeration. The Erlenmeyer flasks were placed in the shaking water bath at 37°C and approximately 250 rpm in the light or in the dark. Dark controls were wrapped tightly in aluminum foil. When the cultures had reached an optical density at 600 nm (OD600) of 0.3 to 0.6 (early exponential growth phase) sampling was started. Depending on the specific assay conditions, described in the text and figure legends, a stress was usually induced at this point. YtvA overproduction was induced by adding IPTG to a final concentration of 1 mM (from a 1 M stock solution). Environmental stresses were induced by adding the stress factor (NaCl or ethanol [for amounts see the text]) or light (by turning on a lamp). Whenever a stress factor was added an equal amount of water was added to controls without stress induction to correct for any dilution effects. Samples of the cultures were taken at various time points in complete darkness or with a minimal red background light when necessary (see the illumination conditions below). The samples were immediately transferred to an ice/water mixture, flash-frozen with liquid nitrogen, and stored at −80°C. In all of the experiments reported here, sampling was stopped before the cells reached stationary phase. β-galactosidase activities were measured and expressed in Miller units to correct for cell growth according to a previously described protocol (21). To compare the many different strains in the experiments with YtvA overproduction, each strain was assayed together with the “wild-type” strain (PB198/pYtvA, i.e., the strain with intact copies of all four rsbR genes) under the same conditions (light and dark). The relative GSR activation was then calculated by dividing the actual activation by the activation in the presence of light of the “wild-type” strain at the same time point. In all graphs, error bars represent the variation of two biological replicates in the same experiment. Error bars are only shown when they exceed the size of the symbols. The data shown are from representative experiments, which were repeated at least two times.
Western blots.
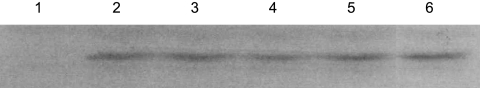
Samples for Western blots were taken from cultures grown and treated exactly as the cultures for the β-galactosidase assays (see above). Instead of inducing a stress at an OD600 of 0.3 to 0.6, samples were taken at an OD600 of 0.8 to 1.0, i.e., in the middle of the growth phase where the reporter-enzyme experiments were performed. Cells were harvested by centrifugation and lysed with lysozyme and boiling prior to loading an amount corrected for small variations in OD600 on a SDS–15% PAGE gel. Proteins were subsequently transferred to Immobilon-P transfer membrane (Millipore). Complete transfer was ensured by monitoring the transfer efficiency of a prestained protein ladder (Fermentas). Immunoblotting was performed with polyclonal anti-N-terminal-RsbRA antibodies (10), which were kindly provided by Jon Marles-Wright (Newcastle University), in 5% (wt/vol) milk powder in phosphate-buffered saline (PBS; 80 mM Na2PO4, 20 mM NaH2PO4, 100 mM NaCl [pH 7.5]) for 2 h. After a thorough washing in PBS, the blots were incubated with goat anti-rabbit peroxidase antibodies (Thermo Fisher Scientific) in 5% (wt/vol) milk powder in PBS for 1 h, after which they were thoroughly washed again prior to visualization using Amersham Hyperfilm ECL and an Amersham ECL Plus Western blot detection system (GE Healthcare). The blot shown is from a representative experiment, repeated three times. Minimal processing was done in Adobe Photoshop CS5: the image was cropped, an irrelevant lane was removed, and the contrast was enhanced equally on the entire image for printability.
Illumination conditions.
Without further specification “light” refers to white light from a compact fluorescent lamp. Moderate white light intensities were set in the range of 25 to 35 micro-einstein m−2 s−1—more than enough to saturate the YtvA-dependent light response (4). Illumination with this intensity of visible light does not significantly affect growth rate or growth yield of B. subtilis. Dark controls were wrapped tightly in aluminum foil. Occasionally, a low-intensity red LED that emits wavelengths unable to activate YtvA (λmax 632 nm with a spectral width at half maximum of ∼25 nm) was used as a background light for sampling in the dark.
RESULTS
Previous studies have shown that YtvA is a photoreceptor that acts in the environmental signaling pathway of the GSR, on (or upstream of) the main structural hub in this pathway: the stressosome (3, 15, 27, 36). The stressosome can contain one or more proteins of the RsbR family—RsbRA, RsbRB, RsbRC, and RsbRD—which appear to be redundant (1, 23). As a first step toward elucidation of the role of the individual RsbR proteins, the requirement of each RsbR protein in the YtvA-dependent light activation of the GSR was systematically analyzed. All possible single, double, triple, and quadruple null mutant combinations of the four RsbR proteins were constructed and the plasmid pYtvA, which has the ytvA gene under the control of the IPTG-inducible spac promoter, was introduced in all of these strains. In addition, all strains contained a fusion between a σB-dependent promoter (Pctc) and a promoter-less gene encoding a reporter-enzyme (lacZ) in their genome, of which the β-galactosidase activity (expressed in Miller units) was determined as a measure for the level of σB (and thus GSR) activation. The ability of YtvA to activate the GSR was assayed by comparing light- and dark-grown cultures at different time points after the induction of overexpression of YtvA by the addition of IPTG (for details, see Materials and Methods).
Due to the large number of strains that this procedure required, not all could be assayed at the same time. To allow for a comparison of the results of different experiments, the “wild-type” strain (PB198/pYtvA, with all four genes of the rsbR family present) was always included as a control. The GSR activation was then normalized on the level of activation in the “wild-type” strain in the light. Thus, a value of 1 corresponds to the activity of the control strain in the light. To create the bar graphs in Fig. 1 and 2, the time point at 120 min after IPTG induction was chosen because it was previously determined to be most suitable (5).
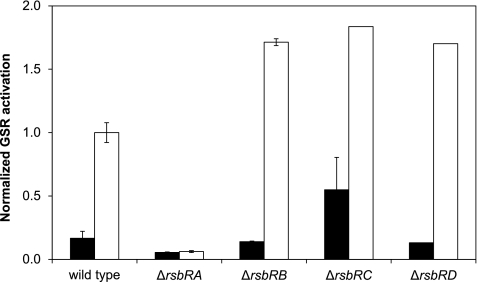
Fig 1.
Effects of single null mutations of rsbR genes on the YtvA-dependent light activation of the GSR. All strains contained a copy of lacZ, controlled by the σB-dependent ctc promoter, and pYtvA, a plasmid for overexpression of ytvA. The β-galactosidase activity was determined 2 h after IPTG induction of YtvA overexpression. Absolute activation expressed in Miller units was normalized on the activation of the “wild-type” strain PB198/pYtvA in the light, which was set to 1. Filled bars correspond to dark conditions; open bars correspond to light conditions. The other strains used were PB491/pYtvA (ΔrsbRA), PB528/pYtvA (ΔrsbRB), PB574/pYtvA (ΔrsbRC), and PB520/pYtvA (ΔrsbRD).
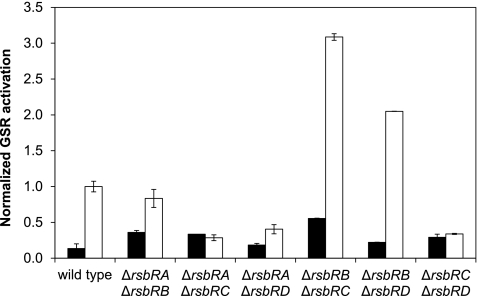
Fig 2.
Effects of double null mutations of rsbR genes on the YtvA-dependent light activation of the GSR. The GSR activation was determined and normalized as described for Fig. 1. Filled bars correspond to dark conditions; open bars correspond to light conditions. The strains used were MA4/pYtvA (ΔrsbRA ΔrsbRB), MA1/pYtvA (ΔrsbRA ΔrsbRC), MA2/pYtvA (ΔrsbRA ΔrsbRD), MA5/pYtvA (ΔrsbRB ΔrsbRC), MA6/pYtvA (ΔrsbRB ΔrsbRD), and MA3/pYtvA (ΔrsbRC ΔrsbRD).
A mutant that lacks RsbRA has no YtvA-dependent light activation of the GSR.
Figure 1 shows the relative σB activation of each strain that contains a single null mutation in one of the rsbR genes (PB491/pYtvA, PB528/pYtvA, PB574/pYtvA, and PB520/pYtvA), compared to the control strain (PB198/pYtvA), grown both in the absence or presence of light. The absence of RsbRA abolishes the YtvA-dependent light activation of the GSR, whereas the absence of any of the other three RsbR proteins increases the level of the activation of the GSR by light by ca. 75%. In the case of the rsbRC-null mutation the GSR activation in the dark is also increased, which could imply a small suppressive role for RsbRC even in unstressed cells.
RsbRB is a strong negative regulator of the YtvA-dependent light activation of the GSR.
When comparing the GSR activation in strains that contain null mutations in two of the four rsbR genes (Fig. 2), it is clear that the presence of rsbRB is of key importance. When an rsbRA-null mutation is combined with the rsbRB mutation (MA4/pYtvA) a small light effect is restored. Combining an rsbRB deletion with an rsbRD (MA6/pYtvA) or rsbRC (MA5/pYtvA) deletion increases the response to light, compared to the single mutants (Fig. 1). Similarly, the presence of RsbRB is the common factor of the other three strains (MA1/pYtvA, MA2/pYtvA and MA3/pYtvA), in all of which the light effect is significantly decreased or completely abolished compared to the single mutants (Fig. 1).
RsbRA, in contrast, appears to have a positive effect on the light activation of the GSR. This is especially clear when comparing null mutants in both rsbRA and either rsbRC (MA1/pYtvA) or rsbRD (MA2/pYtvA) with the single null mutants of rsbRC and rsbRD (Fig. 1): all of the light enhancement of the GSR in those single null mutants is lost in the double-deletion strains. In strains in which RsbRA is present but RsbRB is absent (MA5/pYtvA and MA6/pYtvA) the activity of the GSR is further enhanced, but in the strain with RsbRA and RsbRB as the only RsbR proteins (MA3/pYtvA) the light activation is lost, indicating that RsbRB is able to suppress the activating effect of RsbRA.
RsbRA is sufficient for YtvA-dependent light activation of the GSR.
Based on the results presented thus far, it is difficult to distinguish between direct effects caused by the activity of the RsbR protein itself and indirect effects caused by displacement of other RsbR proteins by YtvA. Therefore, the four strains with three of the four rsbR genes knocked out were assayed next. Figure 3A shows that the strain with RsbRA as the only remaining RsbR protein is capable of transmitting the light signal to activate the GSR, even when YtvA overproduction is not induced (time zero, open triangles). The strain with only RsbRB, in contrast, shows no detectable activation of the GSR by light (Fig. 3B), which is consistent with the results discussed above.
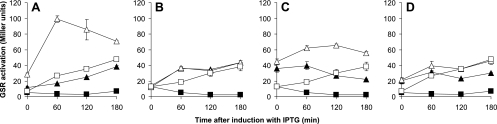
Fig 3.
Effects of triple null mutations of rsbR genes on the YtvA-dependent light activation of the GSR. All strains contained a copy of lacZ, controlled by the σB-dependent ctc promoter, and pYtvA, a plasmid for overexpression of ytvA. The β-galactosidase activity was determined at various time points after IPTG induction of YtvA overexpression and expressed in Miller units. Open symbols represent cultures grown in the light; closed symbols represent cultures grown in the dark. The “wild type”-like strain PB198/pYtvA is represented by squares in all panels, while each one of the triple null mutant strains is represented by triangles. (A) The triple null mutant of rsbRB, rsbRC, and rsbRD with pYtvA (MA7/pYtvA, with RsbRA as the only RsbR protein). (B) The triple null mutant of rsbRA, rsbRC and rsbRD with pYtvA (MA8/pYtvA, with only RsbRB). Note that the lines for this mutant in the light and the dark overlay each other. (C) The triple null mutant of rsbRA, rsbRB, and rsbRD with pYtvA (MA9/pYtvA, with only RsbRC). (D) The triple null mutant of rsbRA, rsbRB, and rsbRC with pYtvA (MA10/pYtvA, with only RsbRD).
The other two triple null mutants show less pronounced, but nevertheless clear, effects. The strain with only RsbRC is able to transmit the light signal and activate the GSR (Fig. 3C), whereas the strain with only RsbRD shows a light effect that is very small (Fig. 3D).
We observed an increase of the activity of the GSR after induction of YtvA overproduction in the dark in the strain with only RsbRA (Fig. 3A). This observation may point toward an effect of overproduced YtvA, even in the dark. The strains with only RsbRC or RsbRD show a high background activation that remains approximately constant throughout the experiment (Fig. 3C and D), as has been observed before (1, 23).
There is no light effect in the absence of all four RsbR proteins.
It has previously been shown that a strain with null mutations in all four rsbR genes has a high, constitutive activation of the GSR in unstressed conditions (1, 23). This suggests that YtvA is unable to form stressosomes without the presence of at least one of the RsbR proteins. However, wild-type expression levels of ytvA might be insufficient to sequester RsbT, and the light conditions of these previous experiments were not controlled.
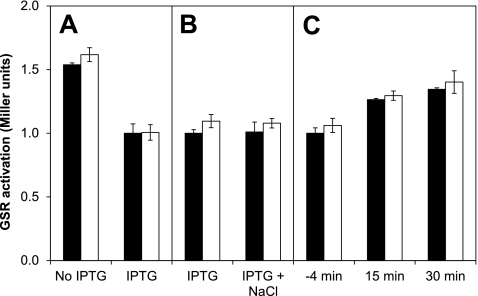
Therefore, we transformed strain MA11 (which lacks all four rsbR genes) with pYtvA. When overproduction of YtvA was induced the level of GSR activation decreased (Fig. 4A), but it is unclear whether this is related to a general effect of overproduction or to a partial ability of an excess of YtvA to capture RsbT and lower the stress response. However, there was no additional induction of the GSR in the light, suggesting that YtvA is nonfunctional in this strain with respect to transmission of a light signal (Fig. 4A). The addition of 300 mM NaCl also did not further activate the GSR, regardless of the light conditions (Fig. 4B), which further suggests that if any stressosome-like structures with only YtvA and RsbT are formed at all, they are nonfunctional. There was also no transient induction of the GSR in response to light (Fig. 4C). In all experiments with this strain the level of GSR activation rose as growth progressed from early to late exponential growth phase (e.g., Fig. 4C), which might be related to various levels of overproduction of YtvA.
Fig 4.
Effect of overproduction of YtvA in a strain lacking all four rsbR genes (MA11/pYtvA). Filled bars correspond to dark conditions; open bars correspond to light conditions. The data from each panel were normalized to the activity of the IPTG-induced strain in the dark for easy comparison. (A) With or without IPTG induction of overproduction of YtvA after continuous exposure to light (1.75 h). (B) 30 min after addition of 300 mM NaCl in cultures kept in the dark or in the light. (C) Before and after turning on the light (at 0 min).
Taken together, these data show that YtvA is unable to form active stressosomes (capable of sequestering RsbT and transmitting a stress signal) without the presence of at least one RsbR protein, independent of the light conditions.
The observed negative effect of RsbRB is not due to a defect in the environmental stress pathway.
We wondered if the striking negative effect of RsbRB on signals generated by YtvA was in fact a general effect on the activity of the entire environmental branch of the GSR. First, we probed the four strains with a single RsbR protein on a Western blot with polyclonal anti-N-terminal RsbRA (Fig. 5) and observed a band that was present in all triple knockout mutants but not in the quadruple mutant that lacks all RsbR proteins. This suggests that the antibody recognizes an epitope common to all RsbR proteins. The intensities of the bands for all mutants were similar, hinting at similar protein levels and an expression-level-control mechanism for the total amount of RsbR protein in the cell.
Fig 5.
Western blot with anti-N-terminal RsbRA in a strain lacking all RsbR proteins (MA11) (lane 1), a strain with RsbRA as the only RsbR protein (MA7) (lane 2), a strain with RsbRB as the only RsbR protein (MA8) (lane 3), a strain with RsbRC as the only RsbR protein (MA9) (lane 4), a strain with RsbRD as the only RsbR protein (MA10) (lane 5), and a strain with all four RsbR proteins (PB198) (lane 6).
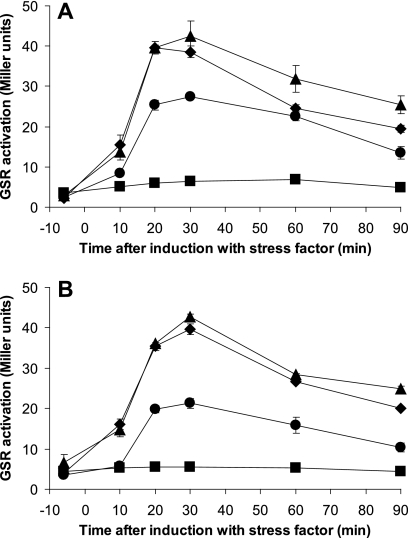
We then stressed strains with either RsbRA (MA7) or RsbRB (MA8) as the only RsbR protein with 4% (685 mM) and 2% (342 mM) ethanol and 300 mM NaCl and with water as a control. The entire experiment was performed in strict darkness, to prevent light-activation of YtvA. The results (Fig. 6) show that both strains are very similar in both the dynamics and the absolute level of GSR activation. This observation confirms that both RsbRA and RsbRB are able to mediate a fully functional environmental stress response under our experimental conditions and that the results presented in Fig. 3 are specific for the YtvA-mediated light-induced GSR.
Fig 6.
Response of strains containing only a single RsbR protein to environmental stresses other than light. At time point zero, the cultures were induced with 4% ethanol (▲), 2% ethanol (♦), 300 mM NaCl (●), or water as a control (■). The experiments were carried out in strict darkness to prevent interference by the wild-type copy of ytvA. The data shown were acquired in the same experiment but split over two panels for clarity. (A) Strain MA7, containing RsbRA as the only RsbR protein (ΔrsbRB ΔrsbRC ΔrsbRD mutants). (B) Strain MA8, containing RsbRB as the only RsbR protein (ΔrsbRA ΔrsbRC ΔrsbRD mutants).
An rsbRB-null mutation is sufficient to detect light-activation of the GSR without overproduction of YtvA.
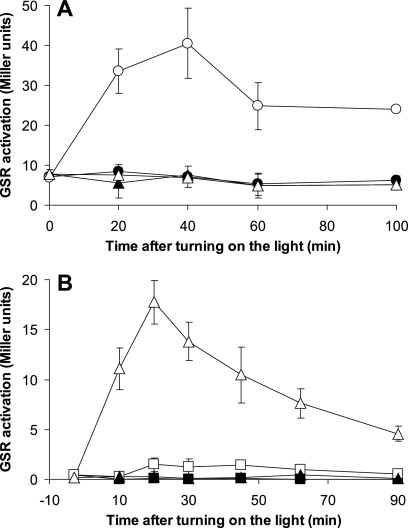
The experiments with the triple null mutant with RsbRA as the only remaining RsbR protein produced an unexpected additional result: without overproduction of YtvA there was a highly reproducible difference between light and dark cultures (time point zero, Fig. 3A). Previously, we and others had been unable to show a light-dependent activation of the GSR at wild-type expression levels of ytvA using the lacZ reporter, except when light stress was combined with an additional environmental stress, such as NaCl shock (3, 36). To exclude the possibility that the observed activation was caused by a leaky promoter in the pYtvA plasmid, the triple null mutant strains, with only RsbRA (MA7) or only RsbRB (MA8) and without the pYtvA plasmid, were grown in the dark and exposed to continuous illumination from time point zero onward. Upon illumination only MA7, and not MA8, showed an increase in GSR activation (Fig. 7A), confirming that a strain with RsbRA as the only RsbR protein has a light-sensitive GSR without overproduction of YtvA.
Fig 7.
Light activation of the GSR without overproduction of YtvA. The β-galactosidase activity, a measure for GSR activation, was determined at various time points and is expressed in Miller units. Cultures were kept strictly in the dark until time point zero, at which time the light was switched on. Filled symbols represent cultures kept in the dark; open symbols represent illuminated cultures. (A) Comparison of strains containing RsbRA (MA7, ΔrsbRB ΔrsbRC ΔrsbRD mutants; circles) or RsbRB (MA8, ΔrsbRA ΔrsbRC ΔrsbRD mutants; triangles) as the only RsbR proteins. (B) Comparison of the single null mutant of rsbRB (PB528, triangles) and the “wild type” control strain (PB198, squares).
The double-deletion mutant of rsbRB and rsbRC (MA5) and the single-deletion mutant of rsbRB (PB528) were also compared to the “wild type” strain (PB198) in a similar assay without YtvA overproduction. The cultures were carefully kept in the dark during the entire experiment (including all precultures) to prevent any unwanted preliminary activation of YtvA. Switching on the light at time point zero resulted in a clear and transient light-dependent activation of the GSR in both MA5 (data not shown) and PB528 (Fig. 7B). To further confirm that the observed light-dependent induction was really due to YtvA and not due to an effect of the rsbRB mutation, the experiment was repeated with a strain with null mutations in both rsbRB and ytvA (JBS10011). In this strain, the observed light-dependent activation of the GSR was completely abolished, confirming the dependence of this effect on YtvA (0.0 ± 0.0 Miller units for JBS10011, compared to 16.8 ± 2.5 for PB528, 15 min after turning on the light). Thus, these results clearly show that the loss of rsbRB is sufficient to make the light sensitivity of the GSR observable in an otherwise wild-type background in exponentially growing cells.
DISCUSSION
Differentiation of function among RsbR proteins with respect to light-induced activation of the stress response.
It has generally been held that the four RsbR proteins are functionally very similar, albeit not identical (23), because early experiments already showed limited differences between them (1). In hindsight, the interpretation of these studies has become difficult because they were performed before YtvA was known to function as a photoreceptor in the GSR (3). Thus, illumination conditions were not controlled.
We have now presented clear evidence of a differentiation of function among the RsbR proteins with respect to YtvA-dependent light-induced stress. Using genetic studies (Fig. 1 to 3), we have established that the presence of RsbRA has a positive effect on the transmission of the light signal, whereas RsbRB has a clear negative effect. When RsbRB is the only RsbR protein present, no light effect is detectable at all (Fig. 3B).
Our results and data available in literature show that the effect of RsbRB in a strain with none of the other RsbR proteins present is not due to differences in protein levels or due to a nonfunctional signaling pathway. First, no increased basal activation of the GSR is observed in this strain (see, for example, Fig. 6B and references 1 and 23), which suggests that there is sufficient protein to form enough stressosomes to capture all RsbT and prevent activation of the GSR. Second, the response to salt and ethanol in strains with either RsbRA or RsbRB as the only RsbR protein is similar (Fig. 6) (23), again indicating normal functionality of the stressosomes. Third, a recent study found the expression levels of rsbRA and rsbRB to be similar (35). The availability of sufficient protein is also suggested by the Western blot shown in Fig. 5. Taken together, these data show that RsbRB cannot simply be regarded as a general inhibitor of the environmental GSR, and its inhibitory effect appears to be exclusive to YtvA.
The roles of RsbRC and RsbRD are less distinct, but RsbRC clearly allows the transmission of the light signal (Fig. 3C), while the results obtained for RsbRD are inconclusive (Fig. 3D). The latter observation might be explained by a lower expression level of rsbRD, as has recently been suggested (35), although our Western blot does not suggest large differences in protein concentration. However, despite the minor roles of RsbRC and RsbRD, the additional presence of either one allows RsbRA to overcome the negative effect of RsbRB (Fig. 1). Thus, the exact roles and interactions of the individual RsbR proteins are likely very complex.
In addition to the well-established light-dependent induction of the GSR by YtvA, we observed a small YtvA-dependent induction of the GSR in the dark in the strain with only RsbRA (Fig. 3A). This may be caused by a lowered capacity to inactivate RsbT when stressosomes incorporate YtvA as an additional Rsb protein (see below), partly replacing RsbRA.
Relevance of having four RsbR proteins.
The four RsbR proteins have been assumed to be at least partially redundant (1, 23), which we have also found for at least RsbRA and RsbRB in the response to NaCl and ethanol shock (Fig. 6). The two main hypotheses proposed for their presence are that (i) one senses a different environmental stress or that (ii) one senses a common signal(s) with a different sensitivity or threshold (23). The latter explanation has been favored because of the redundancy of the RsbR proteins and the fact that their transcriptional regulation is different. The results presented here show that an individual RsbR protein can modulate the response to a specific environmental stress (i.e., RsbRB modulating light stress), which suggests that the biological rationale for the existence of four RsbR proteins may be a mix of both aforementioned hypotheses.
If the total level of RsbR proteins in the cell is indeed approximately constant, as suggested by Fig. 5, this does not necessarily diminish the relevance of the differential regulation of the rsbR genes. Regulation of the pool size of the RsbR proteins may be posttranscriptional, and the upregulation of one RsbR protein may result in a smaller contribution of other RsbR proteins to this pool. However, more extensive experiments are required before such interpretations can be confirmed.
While the present study was in preparation, Martinez et al. (29) reported that triple null mutants containing RsbRC or RsbRD as the sole RsbR protein were able to respond to energy stress, a stress which is normally not sensed by the stressosome in B. subtilis. These researchers could not exclude the possibility that the higher background activation observed in these strains (see also Fig. 3C and D), in combination with a decreased ATP concentration during energy stress, might indirectly result in the inability to keep σB fully inactivated. In addition, the light conditions in that study were not specified. However, in light of our results, it appears probable that the absence of certain RsbR proteins renders the GSR sensitive to stresses that were not previously thought to activate the environmental GSR, which may well include starvation signals.
The loss of RsbRB is sufficient for a light-sensitive GSR.
The loss of rsbRB is sufficient for a clearly observable light-dependent GSR in otherwise wild-type cells, i.e., even without the overproduction of YtvA (Fig. 7B). Previously, no light-dependent effect of YtvA was observed without either overproduction or the imposition of an additional environmental stress (3, 36), which made it less obvious to conclude that YtvA has a significant biological role as a photoreceptor. However, our results now show that downregulation of rsbRB is enough to make the GSR of B. subtilis strongly light sensitive. It will, therefore, be very interesting to uncover the (transcriptional) regulation of rsbRB, because such experiments may reveal conditions in which light is perceived as a strong stress factor for the organism. Intriguingly, the light intensities needed for activation of the GSR through YtvA are far below ambient intensities at moderate latitude at midday.
Mechanism of inhibition of the YtvA-dependent light-induced GSR by RsbRB.
Although interaction between YtvA and the stressosome has been shown previously (1, 15), it is far from clear how this interaction takes place. The most intuitive explanation would be that the C-terminal STAS domain of YtvA occupies a position similar to the STAS domains of the other RsbR proteins (which form the core of the stressosome [27]). However, direct evidence to support this notion is lacking, and it is not known whether the STAS domain of YtvA is equivalent in function to the STAS domains of the RsbR proteins. Also, the quadruple mutant, which lacks all rsbR genes but has YtvA (MA11/pYtvA), has a high basal level of GSR activation, which suggests that YtvA alone is unable to form stressosomes that can inactivate RsbT (Fig. 4). Based on these results the following hypothesis can be proposed: YtvA is part of the stressosome as an RsbR protein but needs at least one of the other RsbR proteins to either capture RsbT or maintain stressosome integrity. This hypothesis also offers a possible explanation for the observed negative role of RsbRB: stressosomes with (mostly) RsbRB might expel or exclude YtvA, thereby inhibiting the ability of YtvA to transmit the light signal. However, more work needs to be done to confirm these hypotheses.
Indirect support of YtvA's presence in the stressosome was obtained in an experiment in which we swapped the N-terminal domain of RsbRA for the LOV-domain of YtvA. Cells containing this fusion protein as the only RsbR paralog display light dependence of their salt-stress response, which shows that a LOV domain is capable of activating the stressosome (J. B. van der Steen et al., unpublished results).
Light as a functional probe.
The lack of knowledge on the nature of the direct input signals for the stressosome has hampered progress in research into the molecular mechanism by which the stressosome operates, in particular with respect to signal integration. We have shown here that the use of YtvA, a photoreceptor with a well-defined input signal, allows one to overcome such limitations. With the new observation that loss of rsbRB is enough to create a light-sensitive GSR, probing the parameters of stressosomes becomes much easier, in vivo as well as in vitro if stressosomes containing YtvA can be isolated or constructed. This is not only true for B. subtilis; the important pathogen L. monocytogenes, for example, also has a stressosome and a YtvA-homolog (25, 32), and a blue-light effect dependent on this homolog has been demonstrated (31), which shows that the approach presented here can be extended to other organisms as well.
ACKNOWLEDGMENTS
We thank Chester Price (University of California at Davis) for generously providing strains and Jon Marles-Wright (Newcastle University) for providing the anti-N-RsbRA antibody.
M.A.-P. was supported by a grant from the Earth and Life Sciences Division of the Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Akbar S, et al. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akbar S, Kang CM, Gaidenko TA, Price CW. 1997. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567–578 [DOI] [PubMed] [Google Scholar]
- 3. Ávila-Pérez M, Hellingwerf KJ, Kort R. 2006. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188:6411–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ávila-Pérez M, van der Steen JB, Kort R, Hellingwerf KJ. 2010. Red light activates the σB-mediated general stress response of Bacillus subtilis via the energy branch of the upstream signaling cascade. J. Bacteriol. 192:755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ávila-Pérez M, et al. 2009. In vivo mutational analysis of YtvA from Bacillus subtilis: mechanism of light activation of the general stress response. J. Biol. Chem. 284:24958–24964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benson AK, Haldenwang WG. 1993. The σB-dependent promoter of the Bacillus subtilis Sigb operon is induced by heat shock. J. Bacteriol. 175:1929–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boylan SA, Redfield AR, Brody MS, Price CW. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931–7937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boylan SA, Rutherford A, Thomas SM, Price CW. 1992. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brody MS, Vijay K, Price CW. 2001. Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen CC, Lewis RJ, Harris R, Yudkin MD, Delumeau O. 2003. A supramolecular complex in the environmental stress signaling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657–1669 [DOI] [PubMed] [Google Scholar]
- 11. Chen CC, Yudkin MD, Delumeau O. 2004. Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J. Bacteriol. 186:6830–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delumeau O, Chen CC, Murray JW, Yudkin MD, Lewis RJ. 2006. High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 188:7885–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delumeau O, et al. 2004. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 279:40927–40937 [DOI] [PubMed] [Google Scholar]
- 14. Eichenberger P, et al. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945–972 [DOI] [PubMed] [Google Scholar]
- 15. Gaidenko TA, Kim TJ, Weigel AL, Brody MS, Price CW. 2006. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188:6387–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaidenko TA, Yang X, Lee YM, Price CW. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J. Mol. Biol. 288:29–39 [DOI] [PubMed] [Google Scholar]
- 17. Hecker M, Pane-Farre J, Volker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236 [DOI] [PubMed] [Google Scholar]
- 18. Helmann JD, et al. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joseph P, Fantino JR, Herbaud ML, Denizot F. 2001. Rapid orientated cloning in a shuttle vector allowing modulated gene expression in Bacillus subtilis. FEMS Microbiol. Lett. 205:91–97 [DOI] [PubMed] [Google Scholar]
- 20. Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenney TJ, Moran CP., Jr 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim TJ, Gaidenko TA, Price CW. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 186:6124–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim TJ, Gaidenko TA, Price CW. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135–150 [DOI] [PubMed] [Google Scholar]
- 24. Kunst F, Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Losi A. 2004. The bacterial counterparts of plant phototropins. Photochem. Photobiol. Sci. 3:566–574 [DOI] [PubMed] [Google Scholar]
- 26. Losi A, Polverini E, Quest B, Gartner W. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marles-Wright J, et al. 2008. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92–96 [DOI] [PubMed] [Google Scholar]
- 28. Marles-Wright J, Lewis RJ. 2008. The Bacillus subtilis stressosome: a signal integration and transduction hub. Commun. Integr. Biol. 1:182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez L, Reeves A, Haldenwang W. 2010. Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow σB activation following exposure to either physical or nutritional stress. J. Bacteriol. 192:6279–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nguyen VD, et al. 2007. Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis. Proteomics 7:1391–1408 [DOI] [PubMed] [Google Scholar]
- 31. Ondrusch N, Kreft J. 2011. Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS One 6:e16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pane-Farre J, Lewis RJ, Stulke J. 2005. The RsbRST stress module in bacteria: a signaling system that may interact with different output modules. J. Mol. Microbiol. Biotechnol. 9:65–76 [DOI] [PubMed] [Google Scholar]
- 33. Petersohn A, et al. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Price CW, et al. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757–774 [DOI] [PubMed] [Google Scholar]
- 35. Reeves A, Martinez L, Haldenwang W. 2010. Expression of, and in vivo stressosome formation by, single members of the RsbR protein family in Bacillus subtilis. Microbiology 156:990–998 [DOI] [PubMed] [Google Scholar]
- 36. Suzuki N, Takaya N, Hoshino T, Nakamura A. 2007. Enhancement of a σB-dependent stress response in Bacillus subtilis by light via YtvA photoreceptor. J. Gen. Appl. Microbiol. 53:81–88 [DOI] [PubMed] [Google Scholar]
- 37. Vijay K, Brody MS, Fredlund E, Price CW. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180–188 [DOI] [PubMed] [Google Scholar]
- 38. Voelker U, et al. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xue G-P, Johnson JS, Dalrymple BP. 1999. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J. Microbiol. Methods 34:183–191 [Google Scholar]
- 40. Yang X, Kang CM, Brody MS, Price CW. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265–2275 [DOI] [PubMed] [Google Scholar]
- 41. Zhang S, Haldenwang WG. 2005. Contributions of ATP, GTP, and redox state to nutritional stress activation of the Bacillus subtilis σB transcription factor. J. Bacteriol. 187:7554–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]