Abstract
It is well established that the ferric uptake regulatory protein (Fur) functions as a transcriptional repressor in diverse microorganisms. Recent studies demonstrated that Fur also functions as a transcriptional activator. In this study we defined Fur-mediated activation of gene transcription in the sexually transmitted disease pathogen Neisseria gonorrhoeae. Analysis of 37 genes which were previously determined to be iron induced and which contained putative Fur boxes revealed that only 30 of these genes exhibited reduced transcription in a gonococcal fur mutant strain. Fur-mediated activation was established by examining binding of Fur to the putative promoter regions of 16 Fur-activated genes with variable binding affinities observed. Only ∼50% of the newly identified Fur-regulated genes bound Fur in vitro, suggesting that additional regulatory circuits exist which may function through a Fur-mediated indirect mechanism. The gonococcal Fur-activated genes displayed variable transcription patterns in a fur mutant strain, which correlated with the position of the Fur box in each (promoter) region. These results suggest that Fur-mediated direct transcriptional activation is fulfilled by multiple mechanisms involving either competing with a repressor or recruiting RNA polymerase. Collectively, our studies have established that gonococcal Fur functions as an activator of gene transcription through both direct and indirect mechanisms.
INTRODUCTION
The ferric uptake regulator protein (Fur) is a major transcriptional regulator which responds to iron availability in both Gram-negative and Gram-positive bacteria. Fur and its orthologues share a high degree of sequence conservation, with most Fur proteins being between 15 to 17 kDa in size. Typically, the protein forms a dimer together with ferrous iron, or other divalent cations, and functions as a transcriptional repressor (4, 19). The iron-bound Fur dimer binds to a well-conserved consensus sequence, known as a Fur box, within the promoter region of target genes and functions to block binding of RNA polymerase and thus repress transcription (25). Fur is also able to repress transcription without the presence of a ferrous cofactor in a process termed apo-Fur-mediated regulation (17, 23, 24). In addition to apo-Fur-mediated repression, a subset of genes has been found to be upregulated under iron-replete conditions, suggesting that Fur can also function as a transcriptional activator (10, 14, 23, 30, 32).
Fur-mediated activation was first demonstrated to occur via an indirect mechanism through a small RNA (sRNA). The sRNAs, RyhB of Escherichia coli and Vibrio cholerae (42), PrrF1 and PrrF2 of Pseudomonas aeruginosa (53), FsrA of Bacillus subtilis (29), and nrrF in Neisseria meningitidis (43, 44) were found to be repressed by iron-bound Fur, indicating that the target genes of these small RNAs were indirectly activated by Fur. In Salmonella enterica serovar Typhimurium, Fur represses a repressor protein, the histone-like nucleoid structuring protein (H-NS), which negatively regulates hilA, resulting in indirect activation of hilA by Fur (52). Alternatively, other studies suggest that Fur can activate gene transcription via Fur binding to promoter regions and occluding the binding of a repressor protein, thus derepressing gene transcription (36, 47). In E. coli, Fur-mediated activation of ftnA transcription is due to Fur binding to the ftnA promoter region, which results in competition for H-NS binding (47). However, several genes which are directly activated by Fur have no known counteracting repressor protein, and it is plausible that other mechanisms of Fur activation are at work in these cases.
Neisseria gonorrhoeae is a Gram-negative nonmotile diplococcus which causes the sexually transmitted disease gonorrhea, one of the most common infectious diseases worldwide. Infection of the male urethra leads to inflammatory symptoms including a purulent discharge and an influx of polymorphonuclear leukocytes (PMN) (22). Gonococcal infection of the female genital tract can induce inflammatory cytokines (27) although typically a large percentage of infected females are asymptomatic (34). This can be a serious medical issue as prolonged gonococcal infection can cause more dangerous complications such as pelvic inflammatory disease (PID), a common cause of infertility and ectopic pregnancy (12, 22). We previously demonstrated through in vivo studies that gonococcal genes involved in iron homeostasis are upregulated during infection in men and women. These include genes encoding the transferrin binding proteins and the Fur protein itself (1, 2).
The gonococcal Fur protein is 52% homologous to that of E. coli and 99% identical to that of N. meningitidis. Transcriptome studies of N. meningitidis MC58 wild-type and fur mutant strains have defined the Fur regulon of this organism (15, 18, 32, 51). In N. meningitidis Fur controls at least 116 genes or operons including 32 iron-repressed and 10 iron-activated genes (32). In contrast, the Fur regulon of N. gonorrhoeae is only beginning to be analyzed. Microarray analysis of N. gonorrhoeae FA1090 grown under iron-replete and -depleted conditions reported that 300 genes were repressed by growth under iron-replete conditions while 107 genes were induced (21, 38). For those genes regulated by iron, putative Fur boxes were predicted based on the consensus Fur binding sequences from N. gonorrhoeae, P. aeruginosa, and E. coli (21, 38). Using this analysis 92 gonococcal genes were shown to contain predicted Fur boxes, and of these 28 were shown to bind Fur in a Fur titration assay (FURTA), including 10 iron-activated genes (38). However, the Fur-dependent repression or activation of these genes was not examined. A separate report has demonstrated that in N. gonorrhoeae Fur can function to activate transcription of the norB gene. In this case, gonococcal Fur was demonstrated to bind to the same site in the promoter region of norB as ArsR, resulting in derepression of transcription in a mechanism similar to that found in E. coli with H-NS (36). Despite these recent studies, more global knowledge of Fur-dependent activation of transcription in N. gonorrhoeae is lacking due to the absence of a defined gonococcal fur mutant.
In this study, using a newly constructed gonococcal fur mutant, we have characterized a subset of gonococcal genes which are activated by Fur. Fur-mediated activation was established through binding of Fur to the putative promoter regions of Fur-activated genes. However, this analysis revealed that only ∼50% of such genes which contained predicted Fur boxes bound Fur in vitro, indicating that Fur may activate several genes indirectly. Fur-dependent regulation of these genes was analyzed in a gonococcal fur mutant strain, and variable transcriptional patterns were observed. Furthermore, the transcriptional profiles of Fur-activated genes were associated with different positions of Fur boxes within the promoter regions of these genes. Our results suggest multiple mechanisms by which Fur functions to activate gene transcription in the gonococcus.
MATERIALS AND METHODS
Bacterial strains and growth.
The bacterial strains and plasmids used in this study are listed in Table 1. N. gonorrhoeae F62 (49) strains were grown on gonococcal base (GCB) plates (Invitrogen) or in chemically defined medium (CDM) containing 0.042% Na2CO3 at 37°C. E. coli strains were grown in LB medium. When required, ampicillin was used at 100 μg/ml.
Table 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| N. gonorrhoeae F62 | ||
| Wild type | Isolated from a case of uncomplicated infection; highly competent | 49 |
| fur mutant | fur gene was deleted and replaced by a kanamycin resistance cassette | Daou et al., submitted |
| E. coli | ||
| HBMV119 | fur mutant strain (QC1732) infected with λDE3 to construct an expression strain | 7, 50 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74recA1araD139 Δ(ara-leu)7697galUgalKrpsL (Strr) endA1nupG λ− | Invitrogen |
| Plasmid | ||
| pET15b_NgFur | Gonococcal fur ORF of the strain F62 was cloned into pET15b using NdeI and BamHI sites | This study |
We have recently constructed a gonococcal fur mutant strain (N. Daou, C. Yu, C. Gudino, and C. A. Genco, submitted for publication). Briefly, we used the plasmid pGEMFkoB::Km to construct a fur mutant strain in N. gonorrhoeae F62 since the sequences of fur homologues and the flanking regions are highly identical between N. gonorrhoeae F62 and N. meningitidis MC58. In pGEMFkoB::Km a kanamycin gene was flanked by the sequences upstream and downstream from the fur gene from N. meningitidis MC58 at each side (16). Using pGEMFkoB::Km, the fur gene of N. gonorrhoeae F62 was deleted and replaced by the kanamycin gene. The resulting fur mutant strains were confirmed by PCR on the extracted chromosome DNA, and the absence of fur expression was confirmed by reverse transcription-PCR (RT-PCR) (Daou et al., submitted).
Purification of gonococcal Fur.
The open reading frame (ORF) of the fur gene was amplified from genomic DNA of N. gonorrhoeae F62 using two primers: Fw, 5′-GAATCATATGGAAAAATTCA-3′, and Rv, 5′-CAAAGGATCCGGATTTAACGTT-3′ (restriction sites are underlined). The PCR product was cloned into a pET15b vector (Novagen, San Diego, CA) at the restriction sites NdeI and BamHI. The protein was overexpressed in E. coli HBMV119 (7, 50) with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) overnight at room temperature (RT) and purified using Ni-charged resin according to the manufacturer's protocol (Novagen). The purified Fur was dialyzed against buffer (50 mM Tris-Cl, 500 mM NaCl, 100 μM MnCl2, 10% glycerol, pH 7.9), and the His6 tag was cleaved using biotinylated thrombin (Novagen). The uncleaved protein and the cleaved His6 tag were removed by passing through another Ni-charged resin.
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed as previously described (50). Briefly, the putative promoter regions of each ORF were PCR amplified from the genomic DNA of N. gonorrhoeae F62 wild-type strain with primers engineered with the restriction sites BamHI (Fw) or HindIII (Rv). The primers are listed in Table S1 in the supplemental material. PCR products were digested with both BamHI and HindIII and then radiolabeled with [α-32P]ATP by Klenow treatment (Applied Biosystems, Carlsbad, CA). The radiolabeled probes were purified using a G25 Sephadex QuickSpin column (GE Health Care, Pittsburgh, PA). Probes (0.1 nM) were incubated with purified gonococcal Fur in binding buffer [20 mM Tris-Cl, pH 7.9, 5 mM MgCl2, 40 mM KCl, 0.125 mM MnCl2, 2 mM dithiothreitol (DTT), 10% glycerol, 0.19 μg/μl poly(dI-dC), 3.125 μg/μl bovine serum albumin (BSA)] at RT for 30 min. A cold competition assay was used to determine the specificity of Fur protein binding to the putative promoter regions of each ORF. Unlabeled competitor DNA, as indicated in the legend for Fig. 1 (50- to 1,000-fold), was added to the reaction mixture to compete out the binding of labeled DNA. The reaction mixture was run on a native 6% polyacrylamide gel (acrylamide/bisacrylamide ratio, 37.5:1 [wt/wt]) (Bio-Rad, Hercules, CA) and then dried on a filter paper at 80°C for 1 h. The radiolabeled bands were detected by autoradiography. The integrated density of bound and unbound DNA in the film was quantified using ImageJ software to calculate the percentage of bound DNA in one lane corresponding to a specific Fur concentration. Subsequently, the percentage of bound DNA (y axis) was plotted with the concentration of Fur protein (x axis) for each promoter region using GraphPad Prism, and a KD (equilibrium dissociation constant) value was calculated using the following equation: y = (Bmax × x)/(KD + x), where Bmax is the total number of receptors expressed in the same units as the y values.
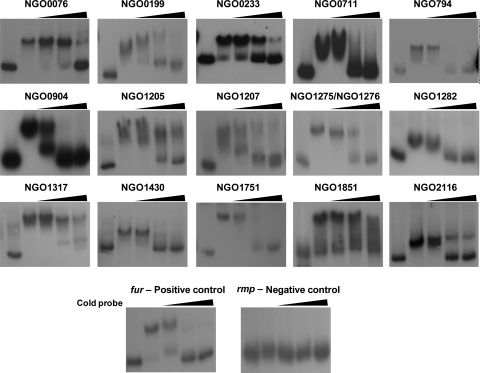
Fig 1.
Cold competition assay for the specificity of Fur binding to the putative promoter regions. 32P-labeled putative promoter DNA was analyzed after incubation with Fur and unlabeled DNA (cold probe). If the unlabeled probes competed for the binding of labeled probes, the binding of Fur to the promoter regions was considered specific. Lane 1, free 32P-labeled DNA; lanes 2 through 5, gonococcal Fur. For the fur, NGO0711, NGO0794, NGO0904, NGO1205, NGO1275/NGO1276, NGO1430, NGO1751, NGO1851, and NGO2116 probes, the Fur protein was added at a concentration of 150 nM, and for the other regions 700 nM Fur protein was added. The fold excess of the cold probes was increased from lane 3 to lane 5, ranging from 50-fold to 1,000-fold (indicated by the triangles). The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
RNA purification and RT-PCR.
N. gonorrhoeae F62 wild-type and fur mutant strains were plated on a GCB plate and grown overnight at 37°C with 5% CO2 and then inoculated into CDM containing 0.042% Na2CO3 and incubated with shaking for 2 h. The culture was diluted into fresh CDM at an optical density at 600 nm (OD600) of 0.1 and incubated for an additional 2 h. Subsequently, 100 μM ferric nitrate or 150 μM Desferal (deferoxamine mesylate) was added to the culture. The bacterial pellets were collected at 1 h and 3 h after the addition of iron or Desferal. RNA was purified from these pellets using an RNeasy kit (Qiagen, Hilden, Germany) and treated with DNase I. The semiquantitative RT-PCR was performed using Superscript III One-Step reagents (Invitrogen, Carlsbad, CA) and visualized on an agarose gel. Quantitative RT-PCR was carried out using a One-Step QuantiTect SYBR green RT-PCR kit (Qiagen) on an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA). A total of 25 ng of RNA was used in each reaction mixture. The relative mRNA levels were evaluated using the comparative cycle threshold (ΔΔCT) method (3). The relative expression level of each gene was normalized to the endogenous rmp gene and is represented as the ratio to that of the wild-type sample under iron-replete (+Fe) conditions. The results are presented as the mean ± the standard deviation of three independent experiments. Statistics were performed using a Student's t test. The mRNA levels observed for the wild type under iron depletion (−Fe), the fur mutant under +Fe conditions, and the fur mutant under −Fe conditions were compared to those of the wild type under +Fe conditions. A P value of <0.05 is considered significant.
DNase I foot printing.
The PCR products of the putative promoter regions of each ORF were cloned into pGEMT-easy vector (Qiagen). One primer, either T7 (5′-TAATACGACTCACTATAGGG-3′) or Sp6 (5′-ATTTAGGTGACACTATAGA-3′), was labeled with 6-carboxyfluorescein ([FAM] Invitrogen). The single-side-labeled probes of the putative promoter regions of each gene were PCR amplified with one labeled primer and one coupled unlabeled primer from the respective pGEMT-easy vectors and purified by ethanol precipitation. The footprinting assay was carried out in 50 μl of buffer (10 mM Tris-Cl, pH 7.9, 100 mM KCl, 1 mM CaCl2, 200 μg/ml BSA, 2 mM MgCl2, 0.5 mM β-mercaptoethanol [β-ME], 8 mM MnCl2, 10% glycerol), containing 1 μg of poly(dI-dC). After a 30-min incubation of the protein and the 2,000-ng probes at RT, 1 μl of DNase I (2 U/μl; Ambion/Applied Biosystems) was added, and DNA digestion was carried out at 37°C for 3 min. Fifty microliters of stop solution (0.1 M EDTA, pH 8.0, 0.6 M Na acetate) was subsequently added to the reaction mixture to terminate DNA digestion. Digested DNA was purified by a QIAquick PCR purification kit (Qiagen) and eluted into 30 μl of H2O. Ten microliters of the digested DNA was mixed with 9.7 μl of HiDi formamide (Applied Biosystems, Foster City, CA) and 0.3 μl of GENEScan-600 LIZ size standards (Applied Biosystems). The samples were analyzed using fragment analysis at the Tufts University Core Facility, and the results were visualized using the software Peakscanner (Applied Biosystems). The experiments were repeated three times, and a gradient of Fur concentrations (50 nM, 250 nM, 500 nM, 750 nM, and 1000 nM) was used for each promoter region.
Alignment of Fur boxes of Fur-activated genes.
The primary Fur boxes of each promoter region of 16 Fur-activated genes were aligned using the software MultAlin (13), and a sequence logo was created by MEME (6).
RESULTS
Identification of Fur-activated genes in N. gonorrhoeae F62.
A gonococcal fur mutant strain was constructed by our group by insertional inactivation of the fur gene. This gonococcal fur mutant strain does not exhibit a growth deficiency compared to the wild-type strain (Daou et al., submitted). To initially identify genes which were activated by Fur, we first screened N. gonorrhoeae F62 fur mutant and wild-type strains for the expression of a subset of genes which were previously reported in N. gonorrhoeae strain FA1090 to be activated in the presence of iron and to contain a predicted Fur box in the promoter region (21, 38). Initial screening was performed in cultures grown under iron-replete and -depleted conditions using semiquantitative RT-PCR (data not shown). This analysis revealed strain-specific differences in the transcription of these genes in response to growth under iron-replete versus iron-depleted conditions. Transcription of NGO1913 and NGO2068 was not detected under either iron-replete or iron-depleted conditions in strain F62, nor did we observe differences in transcription of NGO0711 in response to growth under iron-replete or iron-depleted conditions (Table 2). Consistent with what has been observed in strain FA1090, we observed increased transcription of the remaining 34 genes during growth of N. gonorrhoeae strain F62 under iron-replete conditions (Table 2). Transcription of NGO0233 (nspA) was increased during growth of gonococci under iron-replete conditions in the N. gonorrhoeae strain F62 similar to what has been shown in N. meningitidis (51). Of the iron-upregulated genes we examined, NGO0376, NGO0432, NGO1290, NGO1789, and NGO1957 showed increased transcription in the fur mutant strain (Fur-repressed genes). We propose a mechanism mediated via apo-Fur repression for these genes since they were, in fact, repressed by Fur although they were upregulated under iron-replete conditions. This apo-Fur-mediated regulation was similar to that previously described in Helicobacter pylori (8, 17, 23, 24). Transcription of one operon (NGO1842-NGO1841) was not altered in the N. gonorrhoeae fur mutant strain compared to the level in the wild-type strain (Table 2). Transcription of the remaining 30 genes, including NGO0711, was decreased in the N. gonorrhoeae fur mutant strain (Table 2).
Table 2.
Transcriptional regulation and Fur binding analysis in N. gonorrhoeae F62e
| Gene(s)a | Transcription regulation |
Fur bindingd | |
|---|---|---|---|
| Ironb | Furc | ||
| NGO0037 | + | + | − |
| NGO0043-NGO0042 | + | + | − |
| NGO0076 | + | + | + |
| NGO0199 | + | + | + |
| NGO0233 | + | + | + |
| NGO0376 | + | − | NA |
| NGO0432 | + | − | NA |
| NGO0575 | + | + | − |
| NGO0711 | NC | + | + |
| NGO0775 | + | + | − |
| NGO0794 | + | + | + |
| NGO0904-NGO0906 | + | + | + |
| NGO1205 | + | + | + |
| NGO1207-NGO1209 | + | + | + |
| NGO1275 | + | + | + |
| NGO1276 | + | + | + |
| NGO1282 | + | + | + |
| NGO1290 | + | − | NA |
| NGO1317 | + | + | + |
| NGO1430 | + | + | + |
| NGO1685 | + | + | − |
| NGO1751-NGO1747 | + | + | + |
| NGO1788 | + | + | − |
| NGO1789 | + | − | NA |
| NGO1842-NGO1841 | + | NC | NA |
| NGO1850 | + | + | − |
| NGO1851 | + | + | + |
| NGO1859 | + | + | − |
| NGO1865-NGO1864 | + | + | − |
| NGO1913 | ND | ND | NA |
| NGO1937 | + | + | − |
| NGO1957 | + | − | NA |
| NGO1960 | + | + | − |
| NGO2011 | + | + | − |
| NGO2068 | ND | ND | NA |
| NGO2116 | + | + | + |
| NGO2137 | + | + | − |
| NGO2162 | + | + | − |
For convenience, the gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
The mRNA levels of each gene in the wild-type strain were compared between growth under iron-replete (+Fe) and iron-depleted (−Fe) conditions. +, increased expression under +Fe conditions relative to −Fe condition; −, decreased expression under +Fe condition relative to −Fe condition; NC, expression levels showed no changes under either condition; ND, transcription was not detectable. The cutoff for upregulation (+) and downregulation (−) was 1.5-fold.
The mRNA levels of each gene were compared between the wild-type strain and the fur mutant strain.+, increased expression in the wild-type strain relative to the fur mutant strain; −, decreased expression in the wild-type strain relative to the fur mutant strain; NC, expression levels and regulation patterns responding to iron showed no changes in fur mutant strain compared to levels and patterns in the wild-type strain; ND, transcription was not detectable. The cutoff for upregulation (+) and downregulation (−) was 1.5-fold.
Fur binding to the promoter regions of each gene was determined using EMSAs (see Fig. S2 in the supplemental material). +, Fur bound to the putative promoter region at 1,200 nM; −, Fur did not bind to the putative promoter region at 1,200 nM; NA, EMSA was not performed because the gene was not transcriptionally activated by Fur.
The integrated density of bands in the agarose gel was measured using ImageJ.
To determine if the gonococcal Fur protein bound to the promoter region of the 30 Fur-regulated genes, EMSAs were performed using the promoter regions of these genes. Since Fur boxes may be localized further upstream in the promoter regions of Fur-activated genes (18), we used a long sequence of 400 to 500 bp upstream of the translational start codon ATG. We observed gonococcal Fur binding to the promoter regions of 16 out of 30 genes (Table 2; see also Fig. S1 in the supplemental material). Fur binding to these promoter regions was abolished in the presence of 50 mM EDTA, a chelator of metal ions (see Fig. S1), indicating that metal ions function as cofactors in Fur-mediated binding. We also performed a cold competition assay to further verify specificity of Fur binding. For all 16 genes examined, we observed that the addition of cold probes successfully competed out radiolabeled probes (Fig. 1). Collectively, these results indicate that in N. gonorrhoeae strain F62, 16 genes are activated via a Fur-mediated mechanism which is characterized by Fur binding to the promoter region of these genes (Table 3). Furthermore, 14 additional genes appear to be activated by Fur through a yet to be defined pathway which does not require Fur binding to the promoter regions of these genes (Table 2).
Table 3.
Gonococcal genes activated by Fur via a direct interaction between Fur and the operator regions
| Gene designation in F62a | Gene designation in FA1090 | Function |
|---|---|---|
| * | NGO0076 | Hypothetical protein |
| NGNG_00592 | NGO0199 | Transcription termination factor Rho |
| NGNG_01608 | NGO0233 | nspA, outer membrane protein |
| NGNG_00185 | NGO0711 | Putative alcohol dehydrogenase |
| NGNG_00263 | NGO0794 | bfrA, iron acquisition and storage |
| NGNG_00360-00362 | NGO0904-0906 | Glycolate oxidase; hypothetical protein, Fe-S oxidoreductase |
| NGNG_01845 | NGO1205 | Putative tonB-dependent receptor protein |
| NGNG_01847-01848** | NGO1207-1209 | Excinuclease ABC subunit A, hypothetical protein |
| NGNG_02055 | NGO1275 | norB |
| NGNG_02054 | NGO1276 | aniA, outer membrane protein |
| * | NGO1282 | Hypothetical protein |
| NGNG_01361 | NGO1317 | Transposase |
| NGNG_01242 | NGO1430 | Hypothetical protein |
| NGNG_01666-01662 | NGO1751-1747 | nuo, NADH-quinone oxidoreductase subunit ABCDE |
| NGNG_01772 | NGO1851 | rpoB, DNA direct RNA polymerase subunit β |
| NGNG_00850 | NGO2116 | ABC transporter, ATP binding protein |
A single asterisk (*) indicates that the gene is not present in the genome of N. gonorrhoeae F62; a double asterisk (**) indicates that the operon in F62 is different from that in FA1090.
Differential transcriptional activation in response to Fur.
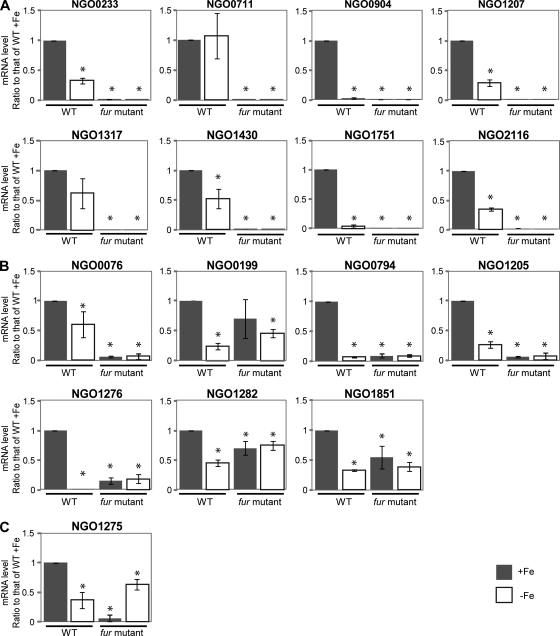
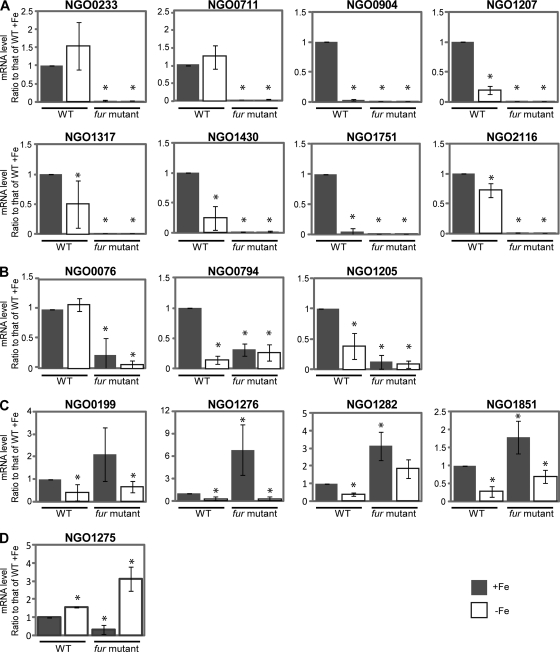
Transcriptional regulation of the newly identified genes which may be directly activated by Fur in response to iron was subsequently studied in more detail using quantitative RT-PCR. We observed increased transcription of 15/16 genes in the wild-type strain grown under iron-replete conditions compared to iron-depleted conditions after 1 h of growth under either condition. Only NGO0711 failed to show any difference. Based on the observed transcriptional regulation, we identified each of these 16 genes with one of three patterns. In one pattern, represented by a set of eight genes, we observed that transcription was completely abolished in the fur mutant strain under either iron-replete or iron-depleted conditions (Fig. 2A). In a second pattern, transcriptional regulation in response to growth under iron-replete and iron-depleted conditions was abolished in the absence of Fur although some basal level of transcription was retained (Fig. 2B). The third pattern of gene transcription was represented by NGO1275, in which transcription was decreased in the gonococcal fur mutant strain, with increased transcription observed during growth under iron-depleted conditions compared to growth under iron-replete conditions in the fur mutant strain (Fig. 2C).
Fig 2.
Transcriptional regulation patterns of Fur-activated genes at 1 h after the addition of iron or Desferal. Transcriptional regulation of the genes which were demonstrated to bind Fur was analyzed using quantitative real-time PCR. The RNA samples were purified from cultures of the wild-type (WT) and fur mutant strains under iron-replete or iron-depleted conditions at 1 h after addition of 100 nM iron or 150 nM Desferal. The transcriptional regulation of these Fur-activated genes in the fur mutant strain displayed different patterns. (A) The genes lose expression completely in the fur mutant strain at the 1-h time point. (B) The genes lose the transcriptional regulation (activation) in response to iron in the fur mutant strain, but basal expression is retained. (C) The gene is repressed under the iron-replete condition in the fur mutant strain. The mRNA levels observed for the three conditions (wild type under −Fe] and fur mutant under +Fe and −Fe conditions) were compared to the value of wild type under +Fe conditions. *, significantly different compared to the mRNA level of the wild type +Fe. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
Examination of gene transcription patterns at later time points (3 h) revealed similar transcriptional control of 15 genes in the wild-type strain in response to growth under iron-replete and -depleted conditions. However, for NGO0233 (nspA), NGO0076, and NGO1275, we observed increased transcription during growth under iron-depleted conditions (Fig. 3). The transcriptional regulation pattern in the fur mutant strain observed for a group of eight genes (designated group A) at 3 h was consistent with that found at 1 h (Fig. 3A). Three genes, NGO0076, NGO0794, and NGO1205 (group B), exhibited similar transcriptional patterns at 3 h and 1 h (Fig. 3B). We observed that the transcriptional regulation patterns of NGO0199, NGO1276, NGO1282, and NGO1851 (group C) were different at 3 h from those at 1 h. Transcription of these four genes was greatly increased during growth under iron-replete conditions in the fur mutant strain at 3 h (Fig. 3C). Although the difference in NGO0199 transcript levels was not significantly different during growth under iron-replete and iron-depleted conditions in the fur mutant strain, the transcriptional patterns were consistent in each independent experiment (data not shown). Therefore, NGO0199 was grouped into this category. The transcription pattern observed for NGO1275 represented a fourth group (group D) showing increased transcription during growth under iron-depleted conditions in both the wild-type and fur mutant strains (Fig. 3D). These results suggest that the variable transcriptional regulation patterns observed for Fur-activated genes may result from complex Fur- and iron-mediated pathways.
Fig 3.
Transcriptional regulation patterns of Fur-activated genes at 3 h after the addition of iron or Desferal. The RNA samples were purified from cultures of the wild-type strain (WT) and fur mutant strain under iron-replete or iron-depleted conditions at 3 h after addition of 100 nM iron or 150 nM Desferal. The transcriptional regulation patterns of the genes directly activated by Fur in the fur mutant strain were further differentiated at the 3-h time point. (A) The genes lose expression completely in the fur mutant strain at the 3-h time point. (B) The genes lose the transcriptional regulation (activation) in the fur mutant strain, but basal expression is retained. (C) The genes are upregulated under the iron-replete condition in the fur mutant strain. (D) The gene is repressed under the iron-replete condition in the fur mutant strain. The mRNA levels observed for the three conditions (wild type −Fe, fur mutant +Fe or fur mutant −Fe) were compared to the value of wild type +Fe. *, significantly different compared to the mRNA level of wild type +Fe. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
Variable Fur binding affinities of the 16 Fur-activated genes.
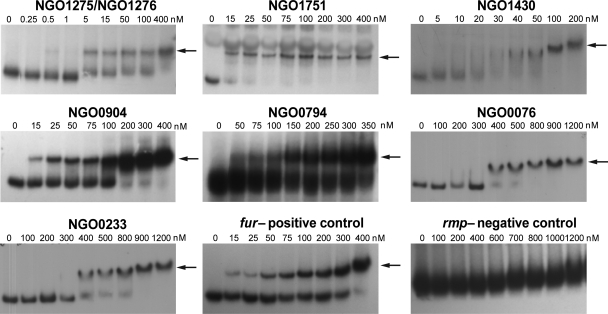
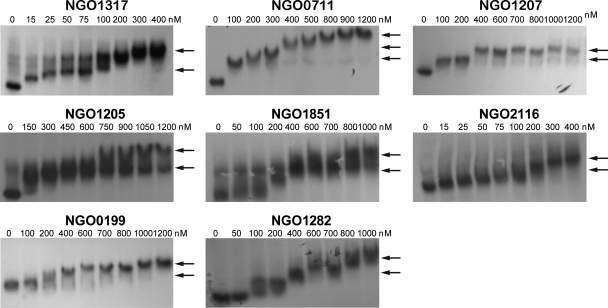
To determine if the transcription patterns observed for the gonococcal Fur-activated genes correlated with varying Fur binding affinities, we further characterized the interaction between Fur and the promoter regions of each of the Fur-activated genes. For this analysis we utilized as a positive control the promoter region of fur gene itself (NGO1779), which exhibits a high binding affinity, with a KD of 26.1 ± 7.0 nM (Fig. 4; Table 4). We observed a wide range of Fur binding affinities for the 16 Fur-activated genes. We observed high binding affinities for NGO1275-NGO1276 and NGO1751, with KDs of 1.2 ± 0.5 nM and 11.6 ± 6.2 nM, respectively (Fig. 4). Lower Fur binding affinities were observed for NGO1430, NGO0904, NGO0794, NGO0076, and NGO0233 (nspA) than for the fur promoter, with KDs of 45.9 ± 15.8 nM, 59.1 ± 13.0 nM, 125.2 ± 24.1 nM, 535.0 ± 201.9 nM, and 724.8 ± 233.0 nM, respectively (Fig. 4; Table 4). Although the putative promoter regions of NGO0199, NGO0711, NGO1207, NGO1282, NGO1851, and NGO2116 showed variable binding affinities to Fur, we were unable to calculate KD values due to a lack of gradation in Fur binding (Fig. 5). Complete shifts for the probes were observed with both low Fur concentrations (≤200 nM; NGO1317, NGO0711, NGO1207, NGO1205, NGO1851, and NGO2116) and relatively high Fur concentrations (≥400 nM; NGO0199 and NGO1282) (Fig. 5). Multiple shifts of the Fur-bound probes correlated with increasing Fur concentrations, as observed by EMSAs of the promoter regions of these six genes (Fig. 5). While we predict that the multiple shifts of Fur-bound DNA observed in EMSA may result from multiple Fur dimers binding to the promoter regions, this was not confirmed (see below). Although we observed a wide range of Fur binding affinities to the 16 Fur-activated genes, this did not appear to correlate with the variable transcriptional regulation patterns of these genes observed in the gonococcal fur mutant strain.
Fig 4.
Variable Fur binding affinities determined by EMSA. Arrows indicate the shift of Fur-bound probes. Since NGO1275 and NGO1276 share the same promoter regions, we used one probe for both genes to demonstrate Fur binding. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
Table 4.
Fur binding affinities
| Groupa | Geneb | KD (nM)c | Primary Fur box | Position of primary Fur box (nt)d |
|---|---|---|---|---|
| fur | 26.1 ± 7.0 | AAAGCGAACGATAATCATACGCTTAAG | −32 to −5 | |
| A | NGO0233 | 724.8 ± 233.0 | AGAAAATTTCAGTATAATACGGCAGGATTCTT | −27 to +5 |
| NGO0711 | ND | TTTATTGTGGTATTGAAATTATTTATCAACAAGCAAGG | +53 to +90 | |
| NGO0904 | 59.1 ± 13.0 | GCATTGTAATGATAATTATTATCGAAAATCATCAGAG | −69 to −105 | |
| NGO1207 | ND | TGCTACAATATCGGTTTTCCTTTATTTT | −18 to +9 | |
| NGO1317 | ND | TAGATTGGCAGATATGTTACCCTCGAAAT | +100 to +128 | |
| NGO1430 | 45.9 ± 15.8 | ATTATTATCATGAGCTAAGAA | +15 to +36 | |
| NGO1751 | 11.6 ± 6.2 | ATCAAATAAGAATCGTTATCATAACATGATTG | −30 to +2 | |
| NGO2116 | ND | TGATAAAGGTTATCATTTGAAAGATAACAT | −24 to +6 | |
| B | NGO0076 | 535.0 ± 201.9 | AAGCTGCAAATCAATGCCGACAACATCTTCAAC | −35 to −3 |
| NGO0794 | 125.2 ± 24.1 | ATTTATTTTGATAATTGTTTGTATTTTAAA | −37 to −8 | |
| NGO1205 | ND | TTGCCTATGCCCTCTGAGTATTCTCAAAATCTTATGATAT | −39 to +1 | |
| C | NGO0199 | ND | GAATTGAACAAATAATCAATTCCCATACCCC | −153 to −112 |
| NGO1276 | 1.2 ± 0.5 | TGATAGTTATTATCATATATGTATTTGTT | −189 to −161 | |
| NGO1282 | ND | ACGATATTCATCCTGATGCGCGCTCCAAAATGAACGTGGTCTT | −168 to −126 | |
| NGO1851 | ND | AGCATTAAGTATTGTTTACATTTATTTGCAT | −10 to +21 | |
| D | NGO1275 | 1.2 ± 0.5 | TGATAGTTATTATCATATATGTATTTGTT | −93 to −65 |
Groups are based on the categories of the transcriptional patterns in the fur mutant strain.
The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
The integrated density of bound and unbound DNA in a film was quantified using ImageJ software to calculate the percentage of bound DNA in one lane corresponding to a specific Fur concentration. Subsequently, the percentages of bound DNA were plotted against the concentration of Fur protein for each promoter region using GraphPad Prism, and a KD value was calculated (see Materials and Methods). ND, the binding was not applicable for the calculation of KD.
The position of the Fur box was counted according to the transcriptional start sites (+1). The −10 and −35 promoter motifs were predicted using BPROM.
Fig 5.
Variable Fur binding affinities and multiple levels of Fur-bound probes determined by EMSA. Arrows indicate the shift of Fur-bound probe. For NGO1317, NGO0711, NGO1205, and NGO1851, multiple shifts were observed per lane. For NGO1207, NGO0199, NGO1282, and NGO2116, different shifts were observed when different concentrations of Fur were used. Multiple shifts may result from multiple Fur dimers binding to the same probe. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
Locations of Fur boxes within the promoter regions.
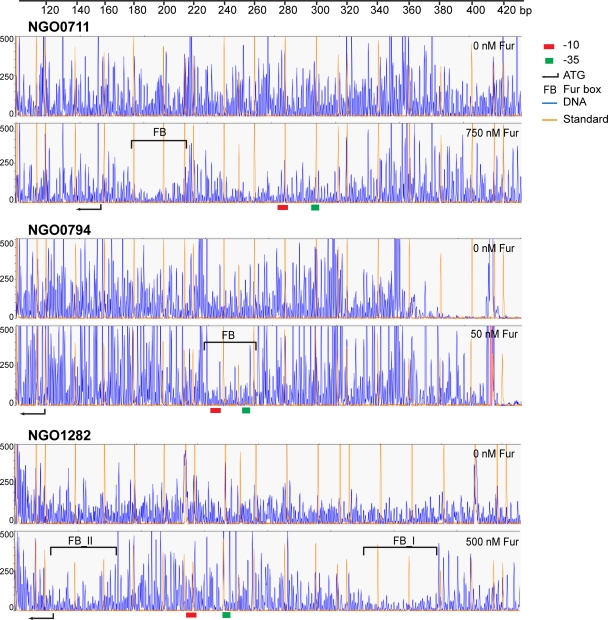
We next reasoned that the sequence specificity and the position of the Fur binding site within the promoter region of the 16 Fur-activated genes might predict possible mechanisms by which gene transcription was initiated. Thus, we performed DNase I footprinting analysis of the 16 Fur-activated genes. The primary Fur binding site within the putative promoter region for each gene is shown in Fig. 6 and Table 4 and in Fig. S2 in the supplemental material. For 5 of the 16 genes (NGO1275, NGO1282, NGO1317, NGO1851, and NGO2116), we observed multiple regions of protection with the addition of increasing concentrations of Fur (data not shown), while the remaining nine genes exhibited only one protected region. The multiple Fur binding sites observed for NGO1282, NGO1317, NGO1851, and NGO2116 correlated with complete shifts of all the probes observed by EMSA. Interestingly, the localization of Fur binding sites relative to the −10 and −35 motifs for each gene was variable and correlated with the transcriptional patterns observed for these genes in the gonococcal fur mutant strain. For the genes with transcriptional patterns belonging to group A (Fig. 3A), the Fur boxes were localized overlapping or downstream of the −10 motif, as represented by NGO0711 (Fig. 6 and Table 4; see also Fig. S2). We observed only one gene (NGO0904) within this group which did not show this pattern. The Fur binding site identified for NGO0904 was localized upstream of the −10 and −35 motifs (see also Fig. S2). For the genes in group B (Fig. 3B), the Fur binding site overlapped with both the −10 and −35 motifs as represented by NGO0794 (Fig. 6 and Table 4). In contrast, the Fur binding sites of the genes in groups C and D (Fig. 3C and D) were localized further upstream from the −35 motif as represented by NGO1282 (Fig. 6 and Table 4; see also Fig. S2). Since NGO1275 and NGO1276 shared the same promoter regions, it is highly likely that they also shared the same Fur binding sites in N. gonorrhoeae strain F62 (see Fig. S2). Although the sequence of the promoter region of NGO1276 is identical to that of the N. meningitidis aniA gene, we did not detect a Fur binding site in NGO1276. In this group, only NGO1851 is an exception in that the Fur box of NGO1851 overlapped with the −10 motif (see Fig. S2). Collectively, these results indicate that Fur can bind to a variety of sequences and positions with the promoter regions of Fur-activated genes, and this may likely relate to the ability of Fur to ultimately activate gene transcription via different mechanisms.
Fig 6.
DNase I footprinting of the promoter regions of representative genes for groups A (NGO0711), B (NGO0794), and C (NGO1282) to Fur. Fur-protected regions are indicated by brackets. The promoter motifs, −10 and −35, are labeled as a red and green box, respectively. The translational start site (ATG) is indicated by an arrow. The gene designations of N. gonorrhoeae F62 were assigned according to their homologues in N. gonorrhoeae FA1090.
DISCUSSION
Previous transcriptome studies of N. gonorrhoeae FA1090 have shown that roughly 20% of the genes or operons in the gonococcal genome are differentially regulated by iron availability. Approximately half of these iron-regulated genes contain predicted Fur boxes in their promoter regions, including 54 iron-repressed and 38 iron-activated genes (21, 38). However, Fur has been demonstrated to bind to the promoter regions of only a small subset of these genes (20, 28, 36, 50). These genes include those encoding iron-scavenging receptors and iron-regulated transporters (fbpA, hemO, tbpA, tbpB, fetB, and tonB) and genes encoding enzymes which require iron as a cofactor such as sodB (20, 28, 36, 50). This small number of genes is far less than the predicted number in N. gonorrhoeae, according to studies mentioned above, and is also less than the 38 iron-repressed and 9 iron-activated genes that have been demonstrated to be directly regulated by Fur in the closely related pathogen N. meningitidis (15, 32). Furthermore, in the absence of a defined gonococcal fur mutant strain, definitive proof of Fur-mediated regulation was not possible.
The studies described here are the first to define Fur-dependent regulation of several genes in N. gonorrhoeae and to characterize Fur-mediated direct activation of gene transcription. Experimental evidence of direct binding of Fur to promoter regions identified 16 genes which were activated by Fur. These 16 genes encode proteins with a broad range of functions including iron storage, cell structure, and energy metabolism. Several of these, including nspA, norB, aniA, and nuoA, have been demonstrated to be directly activated by Fur in N. meningitidis (15, 32, 35, 36, 51). Our analysis identified six genes (NGO0199, NGO0233 [nspA], NGO0794 [bfrA], NGO1207, NGO1282, and NGO1851) which were activated by Fur in N. gonorrhoeae. BfrA is a subunit of the iron storage protein bacterioferritin, which has been proposed to play a role in protection from iron-related oxidative stress (11). NspA is an outer membrane protein, which has been studied extensively in N. meningitidis. The N. meningitidis NspA protein induces antibody protection against N. meningitidis serogroup A, B, and C and binds to the alternative complement pathway inhibitor factor H (40, 41, 46). NGO1207 encodes an exonuclease ABC subunit A that is crucial in DNA damage recognition and may play a role in survival of N. gonorrhoeae under oxidative stress conditions. NGO1282 encodes a hypothetical protein with unknown function. NGO0199 encodes the transcription termination factor Rho, while NGO1851 encodes the RNA polymerase subunit β. It is possible that via regulating these genes Fur may play a broad role in gonococcal gene transcription. Such a wide range of possible functions for Fur-activated genes indicates that Fur-mediated control extends beyond merely regulating genes required for iron metabolism.
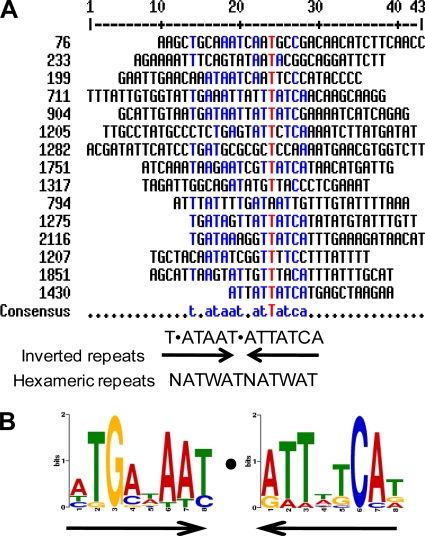
An important observation from our study was that only ∼50% (16/30) of the genes which were activated by Fur and contained predicted Fur boxes were demonstrated to bind Fur in vitro. These results indicate that additional regulatory circuits exist which function via a Fur-mediated indirect mechanism of control. These results also indicate that predictions based on Fur boxes found in the promoters of Fur-repressed genes are not a particularly reliable method to predict Fur boxes within Fur-activated genes. The Fur box within the promoter regions of Fur-repressed genes has been determined to be a highly conserved ∼19-bp sequence among both Gram-negative and Gram-positive bacteria, which is represented as GATAATGATAATCATTATC in E. coli (19). This sequence can be interpreted as three repetitions of NATWAT or 9-1-9 inverted repeats (5, 26, 31). An alignment of the Fur binding sites of the 16 genes which were activated by Fur did not show a high degree of consensus; however, we did identify a consensus comprising a shorter motif of T-ATAAT-ATTATCA (Fig. 7A), and the sequence logo created by MEME software was similar to this consensus sequence (Fig. 7B). This motif is similar to the inverted repeats found in the Fur boxes of Fur-repressed genes. Although the critical nucleotides for Fur binding within this Fur box have not been defined, it is possible that specific sequences contribute to Fur binding affinity. In turn, different Fur binding affinities may determine the relative hierarchy of Fur transcriptional control. A greater degree of deviation from the Fur consensus sequence was observed for those genes with lower Fur binding affinities. Hence, it is plausible that the iron-induced genes that were not predicted to have Fur binding sites are in fact recognized and activated by Fur. We acknowledge that the presence of a Fur binding box may not be the only criterion required for Fur-mediated activation of gene transcription. As discussed below, additional regulatory proteins may facilitate cooperative binding of Fur to sequences which deviate from the consensus binding site.
Fig 7.
Alignment of the primary Fur boxes of 16 Fur-activated genes. A consensus sequence for the Fur box of the Fur-activated genes was generated by aligning the 16 primary Fur boxes using Multalin software (A), and a similar sequence logo was created by aligning the same 16 primary Fur boxes using the software MEME (B). The consensus sequence and the logo could be interpreted as an invert repeat or hexameric repeats, which is similar to the Fur boxes reported for Fur-repressed genes.
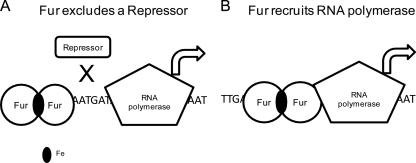
The mechanisms by which Fur functions as a transcriptional repressor are well known. An iron-bound Fur dimer binds to the −10 and −35 motifs in the promoter region, blocks the binding of RNA polymerase, and prevents transcription (25, 26). Fur-mediated activation was initially thought to be an indirect consequence via repression of a negative regulator such as a small RNA. In N. meningitidis we previously reported that a sRNA nrrF is directly repressed by Fur and that its target genes, sdhA and sdhC, are indirectly activated by Fur (43, 44). An nrrF homologue is also present in N. gonorrhoeae (Daou et al., submitted). Studies in N. meningitidis first reported that Fur functions directly as an activator and binds upstream of the −10 and −35 promoter regions of norB, aniA, and nuoA (15). The Fur boxes of NGO1276 (aniA) and NGO1751 (nuoA) of N. gonorrhoeae are different from their homologues in N. meningitidis although their promoter sequences are highly conserved. In addition, the Fur box of nuoA overlaps with the −10 motif. We observed that the Fur boxes of NGO1275 (norB) and NGO1276 (aniA) are upstream of the −10 and −35 motifs of both genes. It has been demonstrated that Fur activates transcription of norB under iron-replete conditions, yet deletion of the Fur box results in activation of norB independent of Fur (36). Subsequently, it was discovered that ArsR, a newly identified repressor, bound to the same site in the promoter region of norB as Fur (36). This mechanism of direct Fur-mediated activation thus occurs via prevention of a repressor binding to the promoter region and occurs when Fur binds to a site upstream of the −10 and −35 motifs (Fig. 8A). We have demonstrated here that for NGO0199, NGO1275 (NGO1276), and NGO1282 (groups C and D), the Fur binding sites are all located upstream of the promoter motifs with only NGO1851 as an exception. These results suggest that Fur activation of these genes occurs via a pathway similar to that of norB regulation. Future studies will focus on the identification of the corresponding repressor proteins. It is also possible that an additional regulator, which also responds to iron availability, may be involved in the regulation of genes in groups C and D. This scenario has been reported for the regulation of the norB (NGO1275) gene that is under the control of the repressor NsrR, which is activated by NO and contains a 2Fe-2S cluster. Under iron-depleted conditions, NsrR is inactivated and results in the derepression of norB (36, 37). aniA is also known to be under the control of several additional regulators including FNR, an activator functioning under anaerobic conditions, and NarP/NarQ, a two-component system which is not well characterized (36). It is possible that these regulators may interact with Fur since they are localized in the same operator regions. The gonococcus possess ∼60 regulators although only a very small number of these have been characterized, including NsrR, FNR, OxyR, RegF, NmlR, PerR, MtrR, and FarR (48). In addition, we recently identified a novel phage repressor, FarP, which is directly activated by Fur (Daou et al., submitted). Binding of FarP to the operator region of the operon NGNG_00460–00463 prevents Fur-mediated direct activation (Daou et al., submitted) of this operon. Collectively, it is plausible that a Fur box localized further upstream of the promoter region allows additional regulators to play a role in transcriptional control. Under these conditions, Fur can participate in cross talk with other regulatory cascades to enable gonococci to adapt to different environmental signals at the same time.
Fig 8.
A schematic of two possible mechanisms by which Fur directly activates gene transcription. (A) Binding of Fur dimers excludes the binding of a repressor, which reverses the repression of the gene transcription. (B) Binding of Fur dimers to the promoter region facilitates the binding of RNA polymerase, which upregulates gene transcription. In both mechanisms the Fur-mediated control may occur independently of iron.
An important observation from our study was that the primary Fur box for the majority of Fur-activated genes (10/16) was not localized upstream of −10 and −35 motifs but was found either overlapping with, or downstream of, the two motifs. This suggests that Fur can directly activate transcription through additional mechanisms. Several well-described transcriptional activators carry out their function by recruiting RNA polymerase to the promoter region to enhance transcription initiation (9). In this case, the binding position of the activator is close to the −10 and −35 motifs. Therefore, Fur may be able to recruit RNA polymerase to activate transcription (Fig. 8B). Fur binding adjacent to the RNA polymerase binding site could, in fact, facilitate the interaction of RNA polymerase with a subunit to enhance transcription. Given the diversity of positions of Fur binding sites relative to the −10 and −35 motifs in the gonococcal genes identified in group A and group B, it is possible that Fur may interact with RNA polymerase in various ways. For example, Fur may interact with different sigma factors in specific promoters. The σ70 family members recognize −35 and −10, while the σ54 family members bind to −24 and −12 motifs upstream of the transcription start site (39). We predict that once transcription is initiated, Fur may dissociate from the promoter, and current studies are aimed at characterizing these interactions in greater detail.
Proper control of the Fur regulon is not only important for iron homeostasis of bacteria but also critical for the virulence of several bacterial pathogens. In a number of bacterial pathogens, fur-deficient strains exhibit reduced virulence in vivo. For example, a V. cholerae fur mutant strain exhibits decreased colonization in the infant mouse gut compared to the wild-type strain (45). A Bacillus cereus fur mutant strain had a higher 50% lethal dose LD50 (4,932 CFU) than the LD50 of the wild-type strain (1,859 CFU) (33). In contrast, an N. gonorrhoeae F62 fur mutant strain exhibited increased adherence to and invasion of human endocervical epithelial cells and increased colonization in a mouse model compared to the levels in the wild type (Daou et al., submitted). These results suggest that Fur functions in a pathogen-specific manner to control genes required for colonization and subsequent pathology. We previously reported that a subset of Fur-repressed genes was expressed in cervical swab specimens from women with uncomplicated gonorrhea and in urethral swab specimens from men with urethral infections (1, 2). In contrast, we did not observe the expression of Fur-activated genes in cervical swab specimens from women with uncomplicated gonorrhea. Collectively, these results suggest that Fur-mediated repression and activation of gene expression may play an important role during mucosal gonococcal infection.
In summary, our studies have established that gonococcal Fur functions as an activator of gene transcription through multiple mechanisms which include Fur binding to the promoter region of Fur activated genes regions. Fur-mediated direct activation may be achieved by a variety of mechanisms acting either alone or in concert with other proteins in the gonococcus. Future work will focus on defining the interactions of Fur with RNA polymerase and in defining the mechanisms utilized by this global regulatory protein to both activate and repress gene transcription.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nadine Daou, Kenneth R. Barth, and Ryan McClure for helpful discussions and critical review of the manuscript.
This work is supported by the NIH/NIAID grant RO1AI048611.
Footnotes
Published ahead of print 27 January 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Agarwal S, et al. 2005. The gonococcal Fur-regulated tbpA and tbpB genes are expressed during natural mucosal gonococcal infection. Infect. Immun. 73: 4281–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agarwal S, Sebastian S, Szmigielski B, Rice PA, Genco CA. 2008. Expression of the gonococcal global regulatory protein Fur and genes encompassing the Fur and iron regulon during in vitro and in vivo infection in women. J. Bacteriol. 190: 3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Applied Biosystems 1997. User bulletin no. 2: relative quantification of gene expression. Applied Biosystems, Foster City, CA [Google Scholar]
- 4. Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26: 5471–5477 [DOI] [PubMed] [Google Scholar]
- 5. Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184: 5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36 [PubMed] [Google Scholar]
- 7. Baumler AJ, et al. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183: 207–213 [DOI] [PubMed] [Google Scholar]
- 8. Bereswill S, et al. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182: 5948–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2: 57–65 [DOI] [PubMed] [Google Scholar]
- 10. Butcher BG, et al. 2011. Characterization of the Fur Regulon in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 193: 4598–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen CY, Berish SA, Morse SA, Mietzner TA. 1993. The ferric iron-binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol. Microbiol. 10: 311–318 [DOI] [PubMed] [Google Scholar]
- 12. Cohen MS, Sparling PF. 1992. Mucosal infection with Neisseria gonorrhoeae. Bacterial adaptation and mucosal defenses. J. Clin. Invest. 89: 1699–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Danielli A, et al. 2006. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J. Bacteriol. 188: 4654–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delany I, Grifantini R, Bartolini E, Rappuoli R, Scarlato V. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 188: 2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delany I, Ieva R, Alaimo C, Rappuoli R, Scarlato V. 2003. The iron-responsive regulator fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 185: 6032–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delany I, Pacheco AB, Spohn G, Rappuoli R, Scarlato V. 2001. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183: 4932–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delany I, Rappuoli R, Scarlato V. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52: 1081–1090 [DOI] [PubMed] [Google Scholar]
- 19. de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169: 2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Desai PJ, Angerer A, Genco CA. 1996. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J. Bacteriol. 178: 5020–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187: 4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edwards JL, Apicella MA. 2004. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin. Microbiol. Rev. 17: 965–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ernst FD, et al. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151: 533–546 [DOI] [PubMed] [Google Scholar]
- 24. Ernst FD, et al. 2005. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J. Bacteriol. 187: 3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Escolar L, de Lorenzo V, Perez-Martin J. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26: 799–808 [DOI] [PubMed] [Google Scholar]
- 26. Escolar L, Perez-Martin J, de Lorenzo V. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283: 537–547 [DOI] [PubMed] [Google Scholar]
- 27. Fichorova RN, Desai PJ, Gibson FC, III, Genco CA. 2001. Distinct proinflammatory host responses to Neisseria gonorrhoeae infection in immortalized human cervical and vaginal epithelial cells. Infect. Immun. 69: 5840–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forng RY, Ekechukwu CR, Subbarao S, Morse SA, Genco CA. 1997. Promoter mapping and transcriptional regulation of the iron-regulated Neisseria gonorrhoeae fbpA gene. J. Bacteriol. 179: 3047–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaballa A, et al. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U. S. A. 105: 11927–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao H, et al. 2008. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190: 3063–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Genco CA, Desai PJ. 1996. Iron acquisition in the pathogenic Neisseria. Trends Microbiol. 4: 179–184 [DOI] [PubMed] [Google Scholar]
- 32. Grifantini R, et al. 2004. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 54: 962–979 [DOI] [PubMed] [Google Scholar]
- 33. Harvie DR, Vilchez S, Steggles JR, Ellar DJ. 2005. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 151: 569–577 [DOI] [PubMed] [Google Scholar]
- 34. Hedges SR, Mayo MS, Mestecky J, Hook EW, III, Russell MW. 1999. Limited local and systemic antibody responses to Neisseria gonorrhoeae during uncomplicated genital infections. Infect. Immun. 67: 3937–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Householder TC, Belli WA, Lissenden S, Cole JA, Clark VL. 1999. cis- And trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Isabella V, et al. 2008. cis- And trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154: 226–239 [DOI] [PubMed] [Google Scholar]
- 37. Isabella VM, Lapek JD, Jr, Kennedy EM, Clark VL. 2009. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol. Microbiol. 71: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jackson LA, et al. 2010. Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J. Bacteriol. 192: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69: 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis LA, et al. 2010. The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog. 6: e1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin D, et al. 2000. Candidate Neisseria meningitidis NspA vaccine. J. Biotechnol. 83: 27–31 [DOI] [PubMed] [Google Scholar]
- 42. Masse E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. 2007. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189: 3686–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mellin JR, et al. 2010. Role of Hfq in iron-dependent and -independent gene regulation in Neisseria meningitidis. Microbiology 156: 2316–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73: 8167–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moe GR, Tan S, Granoff DM. 1999. Differences in surface expression of NspA among Neisseria meningitidis group B strains. Infect. Immun. 67: 5664–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nandal A, et al. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe2+-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75: 637–657 [DOI] [PubMed] [Google Scholar]
- 48. Schielke S, Frosch M, Kurzai O. 2010. Virulence determinants involved in differential host niche adaptation of Neisseria meningitidis and Neisseria gonorrhoeae. Med. Microbiol. Immunol. 199: 185–196 [DOI] [PubMed] [Google Scholar]
- 49. Schneider H, Griffiss JM, Williams GD, Pier GB. 1982. Immunological basis of serum resistance of Neisseria gonorrhoeae. J. Gen. Microbiol. 128: 13–22 [DOI] [PubMed] [Google Scholar]
- 50. Sebastian S, Agarwal S, Murphy JR, Genco CA. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184: 3965–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shaik YB, et al. 2007. Expression of the iron-activated nspA and secY genes in Neisseria meningitidis group B by Fur-dependent and -independent mechanisms. J. Bacteriol. 189: 663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Troxell B, et al. 2011. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilderman PJ, et al. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101: 9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.