Abstract
The rat parvovirus H-1PV is a promising anticancer agent given its oncosuppressive properties and the absence of known side effects in humans. H-1PV replicates preferentially in transformed cells, but the virus can enter both normal and cancer cells. Uptake by normal cells sequesters a significant portion of the administered viral dose away from the tumor target. Hence, targeting H-1PV entry specifically to tumor cells is important to increase the efficacy of parvovirus-based treatments. In this study, we first found that sialic acid plays a key role in H-1PV entry. We then genetically engineered the H-1PV capsid to improve its affinity for human tumor cells. By analogy with the resolved crystal structure of the closely related parvovirus minute virus of mice, we developed an in silico three-dimensional (3D) model of the H-1PV wild-type capsid. Based on this model, we identified putative amino acids involved in cell membrane recognition and virus entry at the level of the 2-fold axis of symmetry of the capsid, within the so-called dimple region. In situ mutagenesis of these residues significantly reduced the binding and entry of H-1PV into permissive cells. We then engineered an entry-deficient viral capsid and inserted a cyclic RGD-4C peptide at the level of its 3-fold axis spike. This peptide binds αvβ3 and αvβ5 integrins, which are overexpressed in cancer cells and growing blood vessels. The insertion of the peptide rescued viral infectivity toward cells overexpressing αvβ5 integrins, resulting in the efficient killing of these cells by the reengineered virus. This work demonstrates that H-1PV can be genetically retargeted through the modification of its capsid, showing great promise for a more efficient use of this virus in cancer therapy.
INTRODUCTION
Parvoviridae are small, nonenveloped, single-stranded DNA viruses that infect a wide variety of animal species, from insects to humans (60). Rodent members of the genus Parvovirus (PV), such as minute virus of mice (MVM) and rat H-1PV, attract high levels of interest as novel anticancer agents, because they can replicate autonomously in oncogene-transformed cells and exert both oncolytic and oncosuppressive activities in various cell culture and animal models while being nonpathogenic for humans (41, 57). The oncoselectivity of PVs is not due to better virus uptake by transformed cells but to a more efficient viral replication and/or toxicity in these cells. This results in part from the fact that PV DNA replication and gene expression are dependent on cellular factors such as E2F, CREB, ATF, cyclin A (57), and others, all of which are known to be upregulated in cancer cells. Moreover, in contrast to normal cells, cancer cells are unable to mount an efficient antiviral defense against PV (22), thus providing more favorable conditions for the viral life cycle. Besides their antineoplastic activities, another advantage of rodent PVs for cancer therapy is the lack of previous exposure of (most) humans to these agents, precluding the rapid elimination of the virus inoculum through preexisting antiviral immunity (11). Taken together, these properties make these viruses very attractive candidates for use as anticancer agents.
This study focuses on rat H-1PV, which infects and kills human tumor cell lines of various origins (e.g., of brain [23], pancreas [4, 14], blood [3], colon [38], cervix [20], and breast [66, 67]) and which is currently under evaluation in a phase I/IIa clinical trial for the treatment of patients with recurrent glioblastoma multiforme (57). H-1PV has the ability to induce different cell death pathways in cancer cells, including necrosis (53), apoptosis (28, 46, 54, 65), and lysosome-dependent cell death (16), while sparing nontransformed cells. Recently, we have reported the capacity of the virus to induce oxidative stress in cancer cells leading to DNA damage, cell cycle arrest, and apoptosis. These effects are mediated by the nonstructural NS1 protein (28).
Although the anticancer potential of H-1PV is supported by a large set of preclinical studies, its efficacy in clinical applications may be limited by the fact that PVs can still enter normal cells. The uptake of the virus by nontumor cells is expected to result in the sequestration of a significant portion of the administered viral dose away from the tumor target. Targeting PV entry specifically to tumor cells thus would increase the efficacy of PV-based treatments and provide additional safety against possible side effects on normal cells.
Several attempts have been made to modify the natural tropism of PVs through the adaptation of the wild-type strains to specific cell types in culture (19) or through in vivo passaging (35, 58). These approaches, however, lack predictability and are limited to initially semipermissive cell lines and preexisting viral tropism. PVs with altered tropism also have been generated by replacing the whole capsid with the one from a related virus (33). However, this pseudotyping strategy is limited by the fact that modifications are not heritable, i.e., progeny viruses do not keep the retargeting ability of the initial pseudotype. An ideal approach to increase the oncotropism of PVs would be to genetically redirect the binding of the virus to cancer cell-specific receptors. This strategy has proven successful in retargeting other nonenveloped viruses for gene therapy or virotherapeutic purposes, for instance, adeno-associated virus (AAV) (40) or adenovirus (39). Attempts to retarget autonomous PVs in this way have not yet been reported and are the subject of the present study.
Genetic tropism modification generally consists of two steps. First, it is essential to abrogate the natural tropism of the virus to prevent it from entering cells that originally were competent for uptake. This is achievable by modifying the capsid residues involved in cell recognition and binding. Second, it is necessary to retarget the virus specifically to cancer cells by grafting into the viral capsid a foreign peptide with high affinity for receptors that are exclusively or preferentially expressed on cancer cells. This requires (i) the identification of a position within the viral capsid tolerating the insertion of the retargeting peptide and (ii) the identification of suitable ligand peptides that, once inserted into the viral capsid, retain the affinity for their receptors. Both steps are very challenging due to the structural constraints imposed by the icosahedral viral capsid. Indeed, changes in the capsid often are incompatible with efficient particle formation and viral fitness. The retargeting steps also imply a precise knowledge of the capsid structural and functional elements and, in particular, of the region(s) involved in binding to one or several specific cell receptors, none of which has been studied so far in the case of H-1PV.
The capsid of members of the Parvoviridae family consists of 60 copies of two to three nested polypeptide sequences assembled in a T=1 icosahedral symmetry (12). In the case of H-1PV, VP1 and VP2 are encoded by alternatively spliced transcripts and share a C-terminal core sequence while having N-terminal extensions of different lengths. The outer architecture of the PV capsids typically shows (i) a spike-like protrusion at the 3-fold axis of symmetry, (ii) a depression at the 2-fold axis, called the dimple, and (iii) a pore connecting the inside of the virion to the exterior of the particle at the 5-fold axis of symmetry (12). A cellular receptor has been identified for some of the members of the PV genus, e.g., for the feline PV (FPV) and its canine-tropic variant (CPV) that both enter through binding to the transferrin receptor (TfR) (48). A number of serotypes of AAV enter cells via binding to heparin sulfate proteoglycans (62), αvß5 integrins (61), fibroblast growth factor receptor (52), or platelet-derived growth factor receptor alpha (15). Globoside (Gb4Cer) (8), Ku80 autoantigen (43), and αvß1 integrin (69) have been shown to represent cell receptors/coreceptors for another member of the Parvoviridae family, the human pathogen B19. However, the cognate cell receptors for the group of rodent parvoviruses remain to be identified (29). It is known that binding to sialic acid is required for cell surface receptor recognition by MVM, since cell membrane attachment and infection both are neuraminidase sensitive (35). The X-ray crystal structure of MVM prototype strain (MVMp) capsids soaked with sialic acid (N-acetyl neuraminic acid) reveals that the sugar is positioned within the dimple recess surrounding the icosahedral 2-fold symmetry axis of the viral capsid immediately adjacent to residues I362 and K368. Point mutations of these residues result in lower affinity for the sialic acid component of the cell receptor (32, 35, 36, 45).
Among the peptides that have been successfully used for the specific tumor targeting of chemicals, diagnostic tools, or viral anticancer agents, one of the most extensively studied is the Arg-Gly-Asp (RGD) peptide (63). The RGD sequence can be found in many extracellular matrix (ECM) proteins, e.g., fibronectin, vitronectin, etc., and is responsible for the binding of these ECM proteins to their cellular receptors (26). The RGD peptide, particularly in its cyclic form CDCRGDCFC (termed RGD-4C), binds strongly to αvß3 and αvß5 integrins (5, 44), which typically are overexpressed in cancer cells and angiogenic blood vessels (7, 71). The RGD-4C–integrin interaction has been successfully exploited for the retargeting of adenoviruses (24, 63) and adeno-associated viruses (21) to cancer cells.
In this study, we first discovered that sialic acid plays an important role in H-1PV infection. We then provided proof of concept that it is possible to genetically reprogram H-1PV entry through the modification of its viral capsid. In the absence of structural data, we modeled the H-1PV capsid based on its homology with the capsid of the closely related MVM, whose crystal structure has been resolved and residues involved in sialic acid binding characterized. Based on this analysis, we identified putative amino acids involved in cell membrane recognition and entry and confirmed that the substitution of these residues abrogated the entry of H-1PV into cells permissive for wild-type virus infection. Furthermore, we identified a site within the viral capsid's 3-fold axis spike tolerating the insertion of a foreign peptide, and the insertion of the RGD-4C peptide in this position rescued the ability of the virus to infect and kill transformed cells, thereby conferring a new level of oncotropism to the engineered H-1PV.
MATERIALS AND METHODS
VP2 homology modeling.
Homology modeling of H-1PV VP2 was performed using the CPHmodels 3.0 server of the Center for Biological Sequence Analysis from the Technical University of Denmark (http://www.cbs.dtu.dk/services/CPHmodels/). The 3.25-Å resolved baculovirus-expressed VP2 virus-like capsids (MVMpb) VP2 1Z14 three-dimensional (3D) crystal structure model of parvovirus MVM, which shares 66.4% homology with the H-1PV VP2 capsomer within the subsequence of positions 38 to 593, was used as a template (41). The model obtained then was verified for stereochemical quality and further refined using PROCHECK (34), WHAT-CHECK (27), ERRAT (10), VERIFY-3D (37), and PROVE (51), all indicating that the overall capsomer was modeled with particularly high confidence.
H-1PV capsid 3D model.
To produce a 3D model of H-1PV capsid, 60 copies of the VP2 model were assembled in PyMol (13) and aligned on the MVMpb 1Z14 model, focusing on the Cα of the backbone. Alignment was achieved using the routine tool that first performs sequence alignment followed by structural alignment and then carries out cycles of refinement to reject structural outliers found during the fit. To obtain a topographic representation of the capsid surface, an application was developed in house (see the supplemental material). This application calculates the location of the center of mass of the virion from the capsid Protein Data Bank (PDB) file and then computes the distance between the center of mass and each of the atoms forming the capsid. Each so-called center of mass-atom distance is loaded into the original PDB file, replacing the b-factor field, and is finally displayed as a colored gradient using the PyMol program.
Generation of H-1PV mutants.
For the construction of H-1PV mutants, a fragment of the viral genome containing the VP transcription unit, obtained by digesting the pSR19 clone (30) with HindIII-HpaI, was first subcloned into the pBSK-HpaI vector, generating the pVPsub construct. pBSK-HpaI is a pBluescript SK+ plasmid (Stratagene) with a modified polylinker constructed by replacing the HindIII-XhoI fragment with an adapter containing an HpaI restriction site (underlined), which was obtained by annealing the 5′-AGC TTA TCG ATA CCG TCG ACG TTA ACC-3′ and 5′-TCG AGG TTA ACG TCG ACG GTA TCG ATA-3′ oligonucleotides. In situ mutagenesis was performed using pVPsub as a template as previously described (59), using the following primers (the mutations introduced are in boldface): for clone pVPsub I367S, 5′-GGT ACC GCT AGA CAG CAC AGC TGG CGA GG-3′ and 5′-CCT CGC CAG CTG TGC TGT CTA GCG GTA CC-3′; for clone pVPsub H373R, 5′-GCT GGC GAG GAC CGT GAT GCA AAC GGA GC-3′ and 5′-GCT CCG TTT GCA TCA CGG TCC TCG CCA GC-3′. The modified VP2s finally were cloned back into their parental pSR19 backbone to generate pH-1PV I367S (abbreviated to I367S) and pH-1PV-H373R (abbreviated to H373R) detargeted mutants, using HindIII and HpaI restriction enzymes. Mutations were introduced in positions I367 and R373 of the VP2 protein, corresponding to I509 and R515 in the VP1 protein, respectively (according to the Swiss-Prot P03136.1 sequence). The H-1PV-H373R-RGD-4C retargeted mutant (abbreviated to RGD-4C) was constructed by inserting the RGD-4C (CDCRGDCFC) peptide (31) into A441 of VP2 capsid protein (corresponding to A583 of the VP1 protein) of the pH-1PV H373R mutant. For this purpose, overlap extension PCR (25) was performed using the following primers: RGD 1, 5′-GCC GCG GAG ACT GTT TCT GCG GCA GAA CTA ACA TGC A-3′; RGD 2, 5′-AAC AGT CTC CGC GGC AGT CAC AAG CTA TGG CGT CTT CTC-3′ (with RGD-4C-related sequences indicated in boldface); External 1, 5′-CGA AGA TTG GGC CAA AC-3′; and External 2, 5′-TTT GTC CCA AAT TTG TCC-3′. The resulting PCR fragment then was cloned into pH-1PV H373R using the MfeI and DraIII restriction enzymes.
Cells.
HEK293T (transformed human embryonic kidney), NB324K (newborn human kidney), SK-MEL-28 (skin melanoma), CHO Pro-5, Lec1, and Lec2 (hamster ovary) cell lines were obtained from the ATCC (LGS Standards GmBH, Wesel, Germany). HeLa (cervical carcinoma) cells were a gift from Angel Alonso (German Cancer Research Center, Heidelberg, Germany). Primary keratinocytes and HEMa-LP (primary human adult melanocyte lightly pigmented) were purchased from PromoCell and Invitrogen, respectively. CS-1 hamster melanoma cells were obtained from the Wistar Institute of Anatomy and Biology (Philadelphia, PA). CS-1/β3 and CS-1/β5, which are derived from CS-1 and constitutively express β3 and β5 integrins, were a gift from David Cheresh (University of California, La Jolla, CA). 293T, HeLa, and SK-MEL-28 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS; Gibco, Invitrogen, Germany). NB324K cells were grown in minimum essential medium supplemented with 5% FBS. CHO Pro-5, Lec1, and Lec2 cells were cultured in alpha minimum essential medium with ribonucleosides, deoxyribonucleosides, and the addition of 10% FBS. Primary melanocytes were grown in medium 254 supplemented with human melanocyte growth supplement (HMGS; Invitrogen). Primary keratinocytes were grown in EpiLife-based medium containing human keratinocyte supplement (HKGS; CAScade Biologics). CS-1, CS-1/β3, and CS-1/β5 were grown in RPMI–10% FBS. All media, except the ones of primary melanocytes and keratinocytes, contained 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were kept at 37°C in 5% CO2 and 92% humidity.
Cell transfection and virus production.
HEK293T cells (6 × 106 cells) were plated in 175-cm2 flasks and transiently transfected with 15 μg of viral plasmid/flask according to Reed et al. (55). After 3 days, cells were harvested and viral particles released from cell suspension through three freeze-thaw cycles. Crude cell extracts were digested with 50 U/ml Benzonase nuclease (ultrapure grade) (Sigma) at 37°C for 30 min. Viruses were purified using an iodixanol discontinuous gradient according to Zolotukhin et al. (72). Produced viral particles were quantified by quantitative real-time PCR (qPCR) (see below) and represented as encapsidated viral genomes (Vg). The further amplification of the viral stocks was conducted via the infection of the NB324K cell line at a multiplicity of infection (MOI) of 100 Vg/cell. Cells were harvested 5 to 7 days postinfection and treated as described above. Recombinant H-1-green fluorescent protein (recPV-GFP) (harboring the enhanced green fluorescent protein [EGFP]-encoding gene) was produced according to reference 17.
Short interfering RNAs (siRNAs) against β3 (catalogue name Hs_ITGB3_1) and β5 (catalogue name Hs_ITGB5_5) integrins and nonspecific AllStars negative control were purchased from Qiagen. siRNA transfections (10 nM) in SK-MEL-28 cells were performed with Lipofectamine RNAimax (Invitrogen) according to the manufacturer's instructions.
Virus titration.
qPCR and plaque assays using NB324K indicator cells were performed as previously described (18).
Infection unit (IU) assays were carried out in 96-well plates seeded with 7.5 × 103 cells/well (NB324K or HEK293T) or 5 × 103 cells/well (HeLa). One day after seeding, cells were infected with 10-fold serial dilutions of the virus stocks and incubated for 72 h at 37°C in 5% CO2. After the alkaline lysis (0.75 M NaOH) of infected cells, DNA was transferred to a nylon membrane, cross-linked, and hybridized to a 32P-radiolabeled NS-1-specific probe corresponding to the EcoRV (nucleotide [nt] 385)-EcoRI (nt 1084) fragment of pMVM plasmid. Blots were exposed to X-ray film for autoradiography. Titration experiments were always performed at least in duplicate. recPV-GFP virus was titrated in HEK-293T cells, and viral titers were expressed as transduction units (TU)/ml.
Electron microscopy.
Carbon-coated 300-mesh copper grids were placed face down onto 5-μl aliquots of virus suspension for 2 min, stained with 2% uranylacetate for 30 s, and dried for approximately 1 min. Micrographs were taken at a magnification of 38,000-fold with a Zeiss 10A electron microscope (Zeiss, Oberkochen, Germany) using an acceleration voltage of 80 kV. The magnification indicator was routinely controlled by comparison with a grating replica.
Virus uptake assays.
Cells (5 × 104 cells/well) were seeded in a 24-well plate and grown in 1 ml of complete medium. After 21 h, cells were grown for a further 3 h at 37°C with or without 0.1 U/ml neuraminidase (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). The culture medium then was removed and replaced with 0.5 ml serum-free medium containing H-1PV wild-type or mutant virus at an MOI of 500 Vg/cell. Two different assays were carried out to measure virions bound to the cell surface (binding assay) or both bound and internalized (binding/entry assay) viral particles. For the binding assay, infection was carried out in triplicate for 1 or 2 h at 4°C. For the binding/entry assay, infection was carried out for 2 h (NB324K, normal keratinocytes, and HeLa) or 6 h (Pro-5, Lec1, Lec2, normal melanocytes, and SK-MEL-28) at 37°C. Infected cells were washed with 500 μl of phosphate-buffered saline (PBS), harvested by treatment with trypsin (Gibco, Invitrogen, Germany), and resuspended in 200 μl of complete medium without further washing. In the case of binding/entry assays (37°C), the whole-cell suspension contained cell surface-bound particles (released by the trypsin treatment) as well as the internalized virions. Cells then were subjected to three snap-freeze-thaw cycles to release the viral particles. Viral DNA was purified from the original virus inoculum and cell lysates using the QiaAmp MinElute virus kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and was quantified using the parvovirus-specific qPCR described above. For the knockdown experiment in SK-MEL-28 cells, 2 × 104 cells/well were seeded in a poly-l-lysine-coated 24-well plate and grown in 0.5 ml of complete medium without antibiotics. Twenty-four h later, cells were transfected with appropriate siRNAs in serum-free medium. After 24 h the medium was changed and cells were further grown for an additional 24 h. The culture medium then was removed and replaced with 0.2 ml serum-free medium containing H-1RGD mutant parvovirus at an MOI of 500 Vg/cell. Infection was carried out in triplicate for 6 h at 37°C to allow cell surface binding and the internalization of viral particles. Cells then were treated as described above.
Determination of integrin expression by flow cytometry.
Cells were harvested and centrifuged at 1,500 rpm for 5 min at room temperature. Pellets were resuspended in fresh medium, cooled on ice, and then washed in fluorescence-activated cell sorter (FACS) buffer (PBS supplemented with 10% fetal calf serum [FCS] and 0.01% NaN3) at 4°C. Cells (2 × 105) were incubated in a total volume of 100 μl for 90 min on ice with one of the following antibodies (all from Millipore, Temecula, CA): (i) 1:50 dilution of mouse IgG1-fluorescein isothiocyanate (FITC) isotype control, clone Ci4; (ii) 4 μg/ml mouse αvβ3-FITC, clone LM609; and (iii) 4 μg/ml mouse αvβ5-FITC, clone P1F6. Cells were washed twice with 1 ml FACS buffer and finally resuspended in 700 μl FACS buffer. Cells were analyzed by flow cytometry (FACSort; Becton Dickinson, Franklin Lakes, NJ), and data were evaluated using FCS Express version 3 (De Novo Software, Los Angeles, CA).
Protein extraction and Western blot analysis.
Cells were harvested by trypsinization and washed with PBS. Pellets were dissolved in 5 volumes of lysis buffer (50 mM Tris, pH 8, 200 mM NaCl, 0.5% NP-40, 1 mM dithiothreitol [DTT]) containing protease inhibitors (complete EDTA-free; Roche, Mannheim, Germany) and incubated on ice for 30 min. After centrifugation (10,000 rpm for 5 min at 4°C) to remove cellular debris, total extracted proteins were measured by Bradford assay. Western blot analysis was performed on 20 μg of total extract using 10% SDS-PAGE. The following antibodies were used for immunoblotting: anti-β-tubulin (clone TUB 2.1; Sigma-Aldrich, St. Louis, MO), polyclonal anti-NS1 SP8 antiserum (6), and polyclonal anti-VP2 antiserum (30).
Cell viability assay.
Virus toxicity for primary melanocytes and SK-MEL-28 cells was determined using the CellTiter-Glo assay (Promega, Mannheim, Germany), which measures the culture's ATP content as an indicator of metabolically active viable cells. Briefly, cells were seeded in an opaque-walled 96-well plate at a density of 2,000 cells/well in 50 μl of medium. After incubation for 24 h, cells were infected with purified virus (200,000 Vg for primary melanocytes and 500,000 Vg for SK-MEL-28) diluted in 50 μl of medium. At day 5 after infection, 80 μl of CellTiter-Glo reagent was directly added to the cell medium. After orbital shaking for 2 min, cultures were further incubated at room temperature for 10 min. The cellular glowing signal then was measured using a Fluoroskan plate-reading luminometer (Ascent FL; Thermo Labsystems, Dreieich, Germany). Wells containing only medium were used for background evaluation. Percentages of cell viability were calculated from the ratios of the luminescent values of virus-infected versus mock-treated cultures after background subtraction.
RESULTS
In silico modeling of the H-1PV capsid.
As the crystal structure of the H-1PV capsid has not yet been resolved, we constructed an in silico 3D model of the H-1PV capsid by homology with the resolved crystal structure of the closely related MVM parvovirus. We first modeled the structure of the H-1PV VP2 capsid protein. As shown in Fig. S1 in the supplemental material, the topology of the modeled H-1PV VP2 is similar to that of other parvovirus capsid proteins. An eight-stranded β-barrel motif (βB to βI) forms the core contiguous capsid decorated by loop insertions between the β-strands. Other smaller β-strand secondary structures also can be found inside the loops, like in CPV or MVM, but in different places. According to our model, the H-1PV VP2 protein contains a small α-helix (αA) domain spanning residues 126 to 137, a sequence that is conserved in all the parvovirus structures determined so far (9) (see Fig. S1 in the supplemental material).
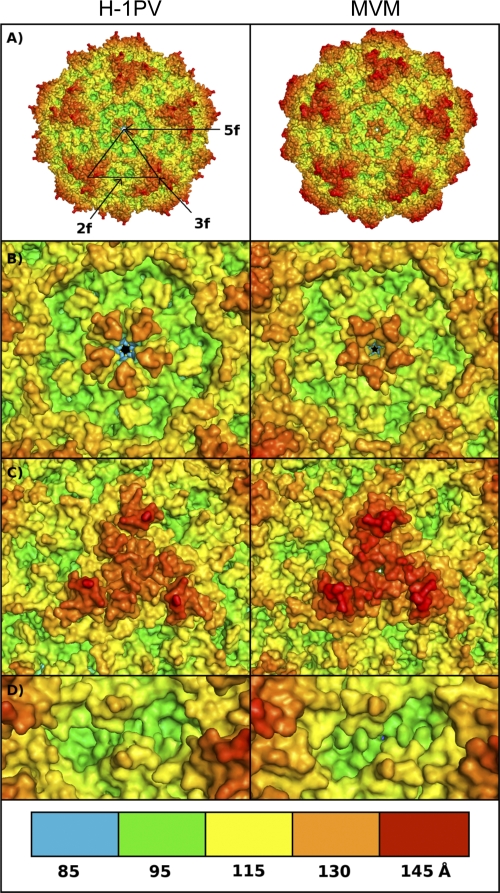
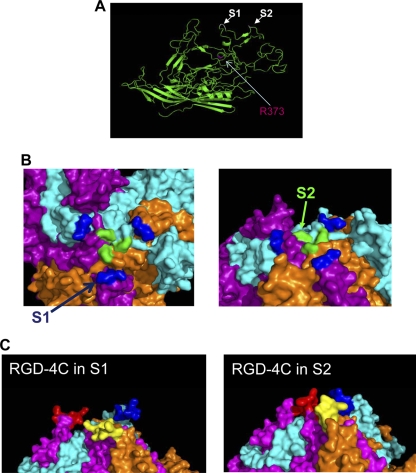
The VP2 protein represents more than 90% of the 60 H-1PV VP units (49). In the case of MVM, it has been described that virus-like particles only made of VP2 were not significantly different from models obtained from full wild-type virions (43). Therefore, we decided to model the H-1PV capsid based on VP2 only. To form the complete H-1PV capsid 3D model, 60 copies of the VP2 model were cloned in PyMol and aligned with the MVMpb crystal structure model (see Materials and Methods for a detailed description of the parameters adopted for the modeling). The resulting T=1 icosahedral capsid model displays all the main structural features of a characteristic mammalian autonomous parvovirus capsid, such as (i) a 5-fold axis pore surrounded by a canyon formed by the clustering of five symmetry-related ribbons; (ii) a 3-fold axis spike resulting from the clustering of six large surface loops, two from each 3-fold-symmetry-related VP2 subunit; and (iii) a 2-fold axis dimple (Fig. 1A). H-1PV and MVM capsids are very similar in this respect, and they have comparable geometries. In both capsids, the 5-fold axis pore structure is approximately 130 Å away from the hypothetical center of the capsid (Fig. 1B), while the 3-fold axis spike (Fig. 1C) and the 2-fold axis dimple (Fig. 1D) are 145 and 100 Å away from the center, respectively. The center of the 3-fold axis spikes seems slightly less protruding in H-1PV than in MVM (∼130 versus ∼135 Å) and is surrounded by three off-centered apexes due to the exposed side chain of Glu 233.
Fig 1.
In silico model of H-1PV and MVM parvovirus capsids. (A) The capsid of H-1PV (left) was modeled using the available crystal structure model of the MVM (right) capsid as a template. The approximate positions of icosahedral 5-fold (5f), 3-fold (3f), and 2-fold (2f) axes of symmetry are shown for a viral asymmetric unit. (B to D) Close-up views of the H-1PV (left) and MVM (right) capsid surfaces at the 5-fold (B), 3-fold (C), and 2-fold (D) icosahedral axes. The panel at the bottom depicts the color range (in Å) for the depth-cued distances from the viral center of the particles. The color coding of the particles was generated by an in-house-developed algorithm, and images were generated with PyMol software as indicated in Materials and Methods.
Sialic acid is involved in H-1PV cell membrane recognition and entry.
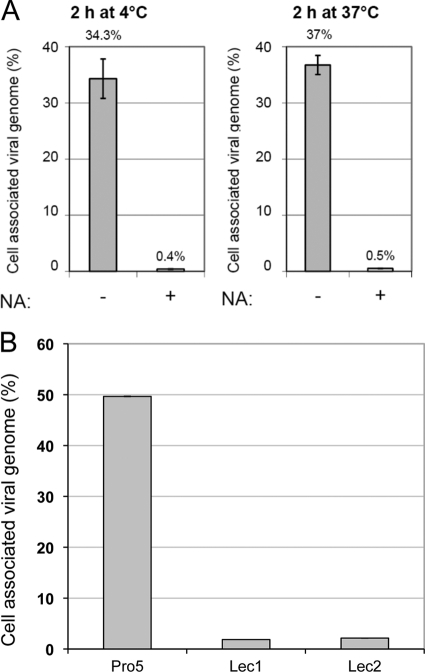
The analysis of the model suggests that the 2-fold axis dimple is conserved in MVM and H-1PV (Fig. 1D). In MVM, this region binds to sialic acid, which appears to be important for cellular entry (35). Treatment with neuraminidase, which cleaves sialic acid from surface proteins, was shown to greatly impair MVM cellular entry. Thus, we asked whether sialic acid is also involved in the binding of the H-1PV capsid to the cell membrane. For this purpose, two highly H-1PV-permissive cell lines, HeLa and NB324K, were grown in the presence or absence of neuraminidase before H-1PV infection. Pretreatment with neuraminidase dramatically decreased (by approximately 95%) the capacity of both HeLa and NB324K cells to take up the virus, suggesting that, similarly to MVM, H-1PV interacts with sialic acid and that this binding is important for viral infection (Fig. 2A and data not shown). To confirm these results, we have repeated the binding/entry assay using the CHO Pro-5 cells, which express sialic acid on their surface, and CHO Lec1 and Lec2 isogenic mutants, which lack surface-exposed sialic acid. H-1PV was able to associate exclusively with the surface of the CHO Pro-5 parental cell line but not with one of the sialic acid-defective cell lines, confirming the importance of the sugar in viral cell surface recognition. (Fig. 2B).
Fig 2.
Involvement of sialic acid in H-1PV cell surface recognition and entry. (A) HeLa cells were treated with 0.1 U/ml neuraminidase (NA) (+) or left untreated (−) for 3 h at 37°C. Cultures were then infected with wild-type H-1PV, incubated for 2 h at either 4 or 37°C, and processed for the measurement of cell-associated viral genomes by quantitative real-time PCR as described in Materials and Methods. The results are presented as the percentage of viral particles taken up by the cells relative to input virus. Numbers on top of columns and bars indicate average values and standard deviations from triplicate measurements, respectively. (B) The Pro-5 parental cell line (expressing sialic acid) or the sialic acid-deficient isogenic mutant Lec1 and Lec2 cells were seeded in 24-well plates (1 × 105 cells/well) and infected with H-1PV (2,000 Vg/cell). After 6 h of incubation at 37°C, cells were collected and processed for the measurement of cell-associated viral genomes.
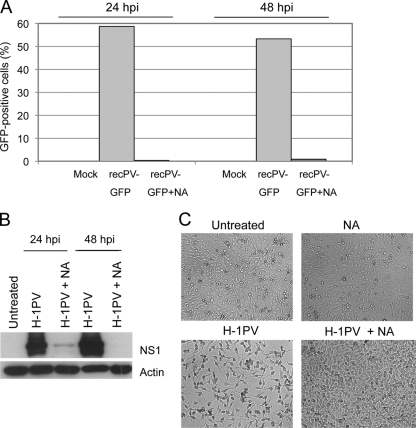
We next tested the effect of long-term neuraminidase treatment on virus infection. HeLa cells were grown in the presence or absence of neuraminidase and then infected with a recombinant H-1PV carrying the EGFP-encoding gene. In agreement with the hypothesis that sialic acid plays a key role in H-1PV cellular entry, EGFP transduction was observed only in cells grown in a neuraminidase-free medium (Fig. 3A). Consistently, similar experiments performed with wild-type H-1PV showed that the neuraminidase treatment prevented the expression of parvovirus nonstructural NS1 protein and protected the cells from viral cytotoxicity (Fig. 3B and C). Taken together, these results indicate that sialic acid is required for H-1PV infection.
Fig 3.
Sialic acid is required for efficient viral infection. (A) Virus transduction. HeLa cells (2 × 105 cells/well) were seeded in a 6-well plate and infected with recPV-GFP at a concentration of 1 transduction unit (TU)/cell in the presence or absence of neuraminidase (0.1 U/ml). At 24 and 48 h postinfection, cells were collected and GFP-positive cells quantified by FACS analysis. (B) Virus infectivity. HeLa cells were infected with H-1PV at 500 Vg/cell and grown with or without neuraminidase (NA) for 24 and 48 h. Twenty μg of total protein extracts from these cultures were analyzed for the presence of parvovirus NS1 protein or actin (used as a loading control). (C) Viral cytotoxicity. Representative images of the cells showing that neuraminidase treatment (NA) prevented H-1PV-induced cytotoxicity.
H-1PV detargeting by in situ mutagenesis.
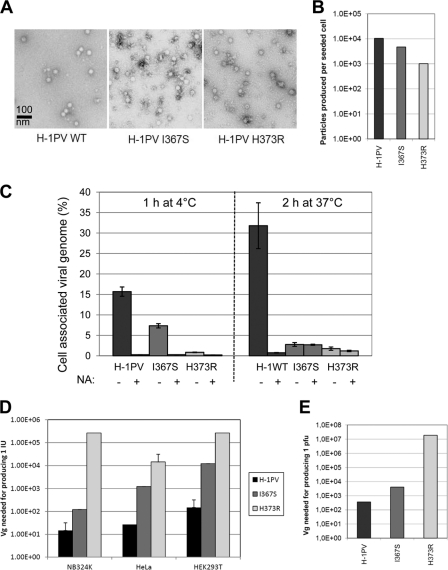
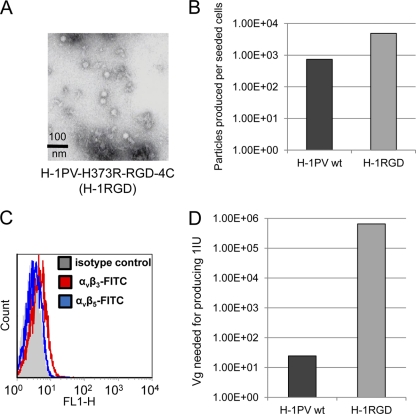
To redirect viral tropism at the level of cell surface receptor recognition, it is first important to ablate the binding of the virus to its natural receptor(s) (detargeting). As sialic acid mediates the cell surface recognition of both MVM and H-1PV, we hypothesized that, similarly to MVM, the dimple region of H-1PV also is involved in the interaction with the sugar. In MVM, both I362 and K368 are involved in the binding to sialic acid (35). These residues were predicted to correspond to I367 and H373 in H-1PV, as revealed by the alignment of the 3D models of H-1PV and MVM VP2s (see Fig. S2 in the supplemental material). Based on mass charge properties, our in silico modeling, and the fact that analogous substitutions in MVM are compatible with proper particle assembly (36), we decided to replace I367 and H373 with S and R, respectively, thus generating the H-1PV I367S and H-1PV H373R mutants. After viral DNA transfection in HEK293T cells, viral particles with apparently normal morphology were formed (Fig. 4A). Although mutants viruses were produced at titers lower than those of wild-type H-1PV (Fig. 4B), no differences in terms of stability were observed between the different viral batches (data not shown). However, the mutations did strongly affect viral binding and entry into both NB324K and HeLa cells (Fig. 4C and data not shown), although some differences were observed between the two mutants. Indeed, the binding to the cell surface was almost completely abolished for the H373R mutant at both 4 and 37°C (more than 90% reduction), while the I367S mutant maintained some capacity to bind to the cell surface in a neuraminidase-sensitive manner at 4°C. At 37°C, the I367S mutant behaved similarly to the H373R mutant and was not taken up by the cells. Viral DNA hybridization assays performed in NB324K, HeLa, and HEK293T cells confirmed that as a result of the introduced substitutions the number of mutant particles being able to enter the cells to initiate their replication cycle was strongly reduced (Fig. 4D). In agreement with previous results, inhibition was much more evident for the H373R than for the I367S mutant. Consistently, both mutations also impaired the capacity of the virus to form plaques in NB324K with a strong effect observed for the H373R substitution (Fig. 4E). It is important to point out that while entry was reduced by approximately 12-fold for the H373R mutant, the ability of the virus to cause productive infection was decreased at least 100 times more, suggesting that the mutation affects not only cell entry but also the capacity of the virus to reach the nucleus and propagate. The two mutant viruses also displayed a reduced ability to lyse cells and halt their proliferation, as measured by lactate dehydrogenase (LDH) and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays, respectively (data not shown).
Fig 4.
Detargeting H-1PV by I367S and H373R substitution. (A) Morphological appearance. Capsids made of wild-type and mutant VP proteins were analyzed by electron microscopy. (B) Virus production. HEK293T cells were transfected with the indicated viral plasmids and grown for a total of 5 days. After benzonase treatment for digesting cellular DNA and nonencapsidated viral DNA, crude cell extracts were analyzed for the presence of full virions. Viral particles were determined by the PCR quantification of encapsidated viral genomes. Columns represent the numbers of produced particles divided by the number of cells seeded prior to DNA transfection. (C) Binding/entry. HeLa cells were infected with wild-type H-1PV (H-1PV) or H-1PV mutants (I367S and H373R) for the time and at the temperature indicated. The cell-associated viral genome was determined as described in Materials and Methods and in the legend to Fig. 2. (D) Infectivity. Wild-type and mutant H-1PV were tested for their capacity to form infectious centers in NB324K, HeLa, and HEK293T indicator cells as described in Materials and Methods. IU, infectious units. Values indicate the number of purified particles (expressed as encapsidated Vg) that need to be inoculated per cell to produce 1 IU. (E) Plaque formation. Wild-type and mutant H-1PV were tested for their capacity to form plaques in NB324K indicator cells. Values represent the amounts of purified particles (expressed as encapsidated Vg per cell) needed to produce 1 plaque.
Retargeting of entry-defective H-1PV H373R mutant to αvβ5 integrins by insertion of a cyclic RGD peptide into its capsid.
Our results indicated that the H373R substitution within the VP proteins dramatically reduced both virus cell surface recognition and entry while maintaining the capacity of the proteins to properly assemble and form the capsid. The second step in the genetic reprogramming of H-1PV was to further modify this entry-defective mutant (here called H373R) by inserting cancer-specific retargeting peptides into its capsid. As a first proof-of-principle test, we decided to insert a cyclic RGD-containing peptide (amino acid sequence CDCRGDCFC), known as RGD-4C, that displays high affinity for αvβ3 and αvβ5 integrins (5, 44). We started by identifying a position within the viral capsid that would ensure the exposure of the peptide at the outer surface of the capsid while preserving its retargeting characteristics. We relied on our in silico capsid model to screen for potential insertion sites, paying special attention to two aspects of the capsid surface: (i) the level of protrusion and (ii) the hydrophilic nature increasing the chances of the peptide being exposed to the outside. There were two VP residues (depicted as S1 and S2 sites in Fig. 5) that met these criteria in particular. They correspond to Gly 234 and Ala 441 in the VP2 sequence and are located in two loops present at the spikes of the 3-fold axis of symmetry (Fig. 5A and B). We predicted that the RGD-4C peptide inserted in these positions would be well exposed at the outer surface (Fig. 5C) and inserted the RGD-4C next to a nitro-carboxy group of Ala 441 of the VP2 protein sequence (S2 site) to generate the H-1PV H373R-RGD-4C mutant (abbreviated to H-1RGD). After the transfection of the viral DNA, virions were formed in similar amounts and had the same morphological appearance as wild-type virions, indicating that the insertion at this position did not impair virus assembly or packaging (Fig. 6A and B and data not shown). We then tested the capacity of H-1RGD to infect NB324K compared to that of the wild-type virus. The expression of αvβ3 and αvβ5 integrins was under our detection limit in these cells (Fig. 6C). Consistently with the lack of αvβ3 and αvβ5 integrins on the surface of NB324K cells, H-1RGD behaved similarly to the H373R detargeted virus and did not efficiently infect these cells (Fig. 6D).
Fig 5.
Model-based search for candidate sites tolerating the insertion of retargeting peptides in the H-1PV capsid. (A) The VP2 structural model reveals the presence of loops exposed at the surface of the viral capsid. The screening of these loops for hydrophilic regions better suited for peptide insertion reveals two candidate sites, S1 and S2 (indicated in white), corresponding to amino acids Gly 234 and Ala 441, respectively, of the VP2 protein sequence. The amino acid R373, which was substituted for H373 to detarget H-1PV, is shown in red. (B) 3D model of three capsomers of the viral capsid showing the position of the S1 (blue) and S2 (green) sites that were selected as the most favorable positions for the insertion of the retargeting peptide. (C) Simulation of the insertion of the RGD-4C peptide (CDCRGDCFC) into the S1 and S2 positions. The three capsomers are represented with different colors, magenta, cyan, and orange, with the inserted sequence shown in red, blue, and yellow, respectively. According to the model, the RGD-4C ligand remains well exposed at the outer surface.
Fig 6.
Genetic insertion of the RGD-4C peptide into the capsid of the detargeted H-1PV-H373R mutant. (A) Morphology. The capacity of the H-1PV-H373R RGD-4C mutant for capsid assembly was tested by electron microscopy visualization of purified virus preparation. (B) Production. Viruses were produced in HEK293T cells through DNA transfection as shown in Fig. 3. The H-1PV-H373R RGD-4C mutant is abbreviated to H-1RGD. (C) FACS analysis showing the protein levels of αvβ3 and αvβ5 integrins on the surface of NB324K. (D) Infectivity. NB324K cells were infected with purified wild-type or RGD-4C-containing (RGD-4C) H-1PV. Cells were collected 7 days postinfection (pi) and processed for viral DNA hybridization assay to assess the capacity of the viral particles (expressed as encapsidated Vg) to form infection units.
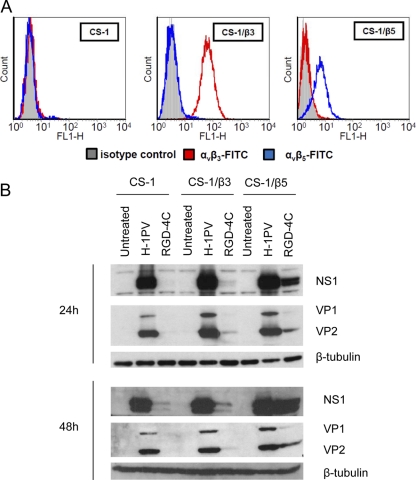
To verify whether the insertion of the RGD-4C peptide into the viral capsid would provide the virus with a novel tropism specific for cells presenting αvβ3 and αvβ5 integrins on their surface, we investigated the capacity of H-1RGD to infect hamster melanoma CS-1 cells lacking both αvβ3 and αvβ5 integrins (64) or the isogenic subclones CS-1/β3 and CS-1/β5 expressing αvβ3 and αvβ5 integrins, respectively (Fig. 7A) (70). Cells were infected with either wild-type or H-1RGD viruses, and infection efficiency was analyzed by checking the production levels of NS1 and VPs at 24 and 48 h postinfection. As shown in Fig. 7B, wild-type H-1PV was able to infect all three cell lines tested with similar efficiency irrespective of their integrin content. On the contrary, differences in viral protein production were observed for the RGD-4C-containing H-1PV mutant. In agreement with the lack of αvβ3 and αvβ5 integrin expression, H-1RGD infected CS-1 cells very poorly compared to the wild-type virus. A slightly better infection rate was found in CS-1/β3 cells, while viral proteins were efficiently produced in CS-1/β5 cells. These results suggest that the primary cognate receptor involved in the internalization of the RGD-4C-containing H-1PV mainly consists of αvβ5 rather than αvβ3 integrins (Fig. 7B).
Fig 7.
Specific infection of cells overexpressing αvβ5 integrins with the RGD-4C-containing H-1PV-H373R mutant. (A) CS-1 parental cells and derivative CS-1/β3 and CS-1/β5 cell lines were assessed for αvβ3 and αvβ5 protein expression by flow-cytometric analysis as described in Materials and Methods. (B) CS-1, CS-1/β3, and CS-1/β5 cell lines were grown in the absence (untreated) or presence of the wild type (H-1PV) or the RGD-4C-containing H-1PV mutant (H-1RGD) for 24 or 48 h before they were processed for Western blot analysis and the detection of NS1, VP1, and VP2 viral protein levels. β-Tubulin was used as a loading control.
H-1RGD specificity for melanoma cells expressing αvβ5 integrins.
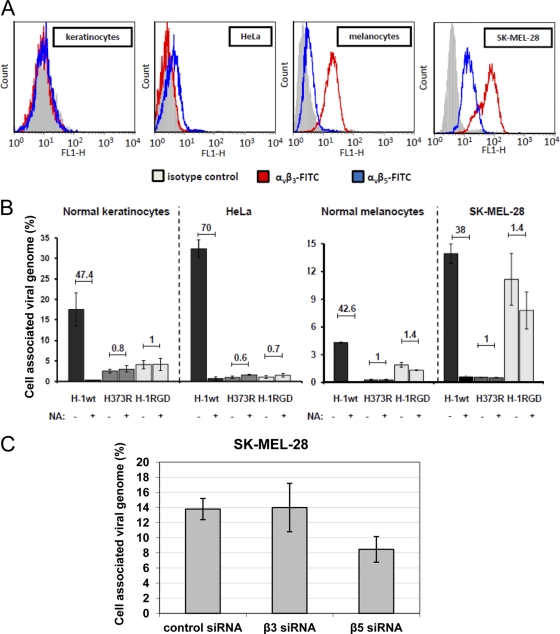
We then tested whether the RGD-4C insertion led to virus retargeting to human cancer cells versus normal human primary cells. Internalization assays were performed using SK-MEL-28 skin melanoma cells, which overexpress αvβ3 and αvβ5 integrins (2, 68). For comparison, we also tested HeLa cells together with primary cultures of nontransformed human melanocytes and keratinocytes. Flow-cytometric analysis confirmed the high expression of αvβ3 and αvβ5 integrins in SK-MEL-28 cells. We also found that primary melanocytes preferentially expressed αvβ3 integrins, while almost no expression of both αvβ3 and αvβ5 integrins was detected in primary keratinocytes and HeLa cells (Fig. 8A). As shown in Fig. 8B, the wild-type virus was taken up by all of these different cultures in a sialic acid-dependent way, as viral entry was dramatically reduced by pretreating cultures with neuraminidase. These results further confirmed the previous data obtained from HeLa and NB324K cells, showing that wild-type virus uptake is dependent on surface sialic acid. As expected, the H373R detargeted virus was unable to bind to the membrane and enter HeLa cells. Similar inefficiency was found in SK-MEL-28 and normal cell cultures. Remarkably, the insertion of the RGD-4C peptide into the detargeted virus rescued its entry into the αvβ3+/αvβ5+ melanoma cells to a great extent. The binding and entry of H-1RGD was only marginally sensitive to neuraminidase treatment, indicating that the H-1RGD virus infected the cells through an alternative route independent of sialic acid (Fig. 8B, right). Conversely, siRNA against β5 integrins reduced H-1RGD binding/entry, confirming the involvement of the integrins in these events (Fig. 8C). The RGD peptide displayed on H-1RGD virus also correlated with some rescue of αvβ3+ normal melanocytes but at a much lower level compared to that of melanoma cells. In contrast, RGD insertion-dependent rescue was not significant in integrin-negative HeLa cells and normal keratinocytes. Taken together, these data were consistent with the results obtained using the CS-1 cellular system (Fig. 7) (i.e., the presence of αvβ3 and αvβ5 integrins at the cell surface, which allowed H-1RGD uptake with low and high efficiency, respectively) (Fig. 8A and B).
Fig 8.
Preferential H-1RGD infection of SK-MEL-28 cells displaying αvβ5 integrins on their surfaces. (A) Integrin content. Cervical carcinoma-derived HeLa cells, SK-MEL-28 melanoma cells, and their normal counterparts (primary normal keratinocytes and melanocytes) were grown for 48 h and analyzed by FACS for their surface expression of αvβ3 and αvβ5 integrins as described in Materials and Methods. (B) These cells were treated with 0.1 U/ml neuraminidase (NA) (+) or left untreated (−) for 3 h at 37°C, infected with the indicated viruses at an MOI of 500 Vg/cell, and processed for the measurement of cell-associated virions (including both cell surface-bound and internalized viruses) as described in the legend to Fig. 2. Columns represent the percentage of input virions that were taken up. Numbers on top of columns give the ratios of cell-associated virions in untreated versus neuraminidase-treated cells. (C) SK-MEL-28 melanoma cells were transfected with specific siRNAs targeted against αvβ3 and αvβ5 integrins or nonspecific AllStars negative controls and were grown for 48 h before being infected with the H-1RGD virus. The binding/entry assay was carried out for 6 h, and then cells were processed for the measurement of internalized virus particles as described in Materials and Methods. The results are presented as the percentage of cell-associated virus particles relative to input virus. The columns represent average values with relative standard deviations.
We next investigated whether the RGD-mediated retargeting of H-1RGD onto integrin-positive melanoma (SK-MEL-28) cells was reflected in an enhanced oncolytic capacity of this virus compared to that of the detargeted H373R mutant. As shown in Fig. 9A (left), normal melanocytes resisted infection with both modified viruses as well as wild-type H-1PV, in keeping with the tumor specificity of parvovirus cytotoxicity. In contrast, the H-1RGD and parental viruses could be distinguished from the H373R mutant by their capacity to effectively kill melanoma cells (Fig. 9A, right). Thus, the RGD insertion into the H373R mutant rescued not only the entry but also the cytopathic effect of this virus in melanoma cells, suggesting that after internalization, H-1RGD was competent for the expression of viral toxic proteins. Indeed, the production of NS1, known to be the main effector of parvovirus-induced cell disturbances, could be detected at high levels in melanoma cells infected with H-1RGD (and wild-type) virus but not the H373R mutant (Fig. 9B). In conclusion, these experiments demonstrate that the H-1PV capsid can be genetically modified to direct this virus at the surface of distinct cancer cells while preserving the viral oncolytic potential.
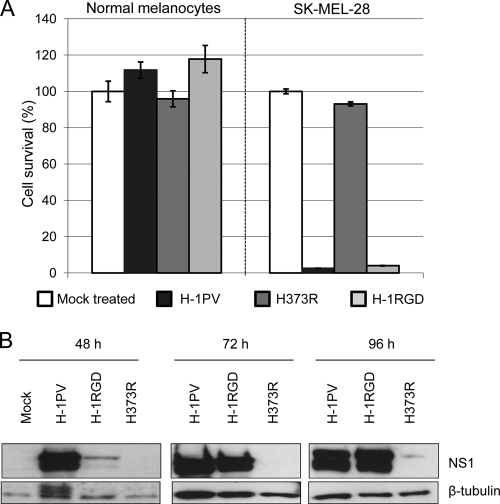
Fig 9.
Toxicity of H-1RGD for melanoma cells. (A) Cell survival. The toxicity of the H-1PV mutants and parental viruses was evaluated in melanoma SK-MEL-28 cells and normal primary melanocytes. The survival of infected cells was determined as described in Materials and Methods and expressed as the percentage relative to values for mock-treated cultures. Columns show means of triplicate measurements with standard deviation bars from one typical experiment performed in triplicate. (B) Expression of the viral cytotoxic NS1 protein. SK-MEL-28 cells were infected with the indicated viruses (5,000 Vg/cell) and harvested at 48, 72, and 96 h postinfection. Total protein extracts were prepared and analyzed by SDS-PAGE for their NS1 content. β-Tubulin was used as a loading control.
DISCUSSION
H-1PV replicates preferentially in cancer cells, but it enters most normal cells with a similar efficiency. The uptake by normal cells will likely dilute the therapeutic dose of the virus available to target cancer cells, reducing its antineoplastic potential and raising some safety concerns. The major objective of this study was to explore the possibility of genetically modifying the H-1PV capsid so as to increase its specificity for cancer cells at the level of viral entry. To this end, two equally important steps were successfully attained. First, we largely ablated the natural tropism of the virus, reducing its uptake by originally permissive cells. Second, we reprogrammed viral cell binding and entry by genetically inserting an RGD-4C retargeting peptide into its capsid. By using this peptide as a model ligand, we showed for the first time that redirecting the cell binding and entry of H-1PV by genetic peptide insertion is indeed a feasible approach.
To achieve this, we heavily relied on our in silico 3D model of the H-1PV capsid that was developed based on the homology with the already-resolved crystal structure of the closely related MVM (1). The analysis of this model suggested a high degree of homology between the 2-fold axis of symmetry of MVM and H-1PV. In MVM, this region was described to take part in the binding to sialic acid (SA) (35). The treatment of permissive cells with neuraminidase, which removes SA moieties from cell surfaces, abolished MVM infection, suggesting that the primary receptor(s) for MVM contains SA residues. We found that SA is also an important determinant of H-1PV infectivity. SA also has been described to serve as a cellular attachment molecule for other parvoviruses, including Aleutian mink disease virus (AMDV), bovine parvovirus (BPV), canine parvovirus, feline parvovirus, and adeno-associated virus types 4, 5, and 6 (49). Therefore, our results further support the concept that SA represents a common mediator of parvovirus cell attachment and most likely is an important component of the receptor(s) engaged by all of these viruses.
In MVM, the two I362 and K368 residues (corresponding to I367 and H373 in the H-1PV capsid), located on the wall of the dimple region at the 2-fold axis of symmetry, have been described to be involved in the interaction with SA (32, 36, 45). Modifications of these two residues (single or double substitution of I362 with S and K368 with R) made the binding of MVM particles more sensitive to cell treatment with neuraminidase, implying that the mutated viruses had a reduced affinity to SA (36). We showed, in the present study, that analogous substitutions introduced into the H-1PV capsid resulted in an even more dramatic phenotype. In particular, the replacement of H373 with R greatly impaired the infectivity of the virus toward originally permissive NB324K, HEK293T, HeLa, and SK-MEL-28 cells as well as that of normal cells, such as primary keratinocyte and melanocytes. As a result of this reduced infectivity, the efficient production of the mutated virus required cell transfection with viral DNA and could not be amplified through secondary rounds of infection. The conversion of I367 to S also affected the capacity of the virus to associate with the cells, but it had a milder effect than that of H373R substitution. Interestingly, we also noted that the H373R mutation strongly inhibited viral entry and had an even more dramatic effect on virus infectivity, reducing the ability of the virus to replicate its genome as well as to form progeny viral particles. We hypothesize that the mutation affects not only the interaction with sialic acid but also the affinity of the capsid for other cellular components involved in mediating viral postentry events. Taken together, these results demonstrate that, similarly to MVM, the 2-fold axis of symmetry plays a crucial role in dictating H-1PV infectivity. We presume that other residues present in this region, in addition to I367 and H373, also contribute to the interaction with the cell membrane and participate in the early steps of the infection of H-1PV.
A second important outcome of this study was the identification of a position within the viral capsid tolerating the insertion of a foreign peptide without preventing the assembly and packaging of the virus. As the H373R substitution dramatically impaired the binding/entry of H-1PV into originally permissive cells, we decided to use this modified virus as the backbone for the insertion of an RGD-4C peptide. This peptide has been extensively studied and was previously employed to specifically deliver therapeutics and imaging agents to αvβ3 and αvβ5 integrin-expressing cells. These integrins are often overexpressed in cancer cells and angiogenic blood vessels (7, 71). In particular, the RGD-4C peptide (as well as other RGD-containing peptides) has been successfully engrafted on the surface of viral vectors, for example, of adenovirus (63), AAV vectors (21), and measles viruses (47), increasing their transduction efficiency and antitumor activities.
Based on our in silico model analysis, we selected the residue A441 of the VP2 protein (corresponding to A583 in VP1), located in one of the most protruding loops of the 3-fold spike, as a putative position to which we could graft the RGD-C peptide. This residue is surrounded by hydrophilic residues that we reasoned would favor the extrusion of the peptide, allowing better recognition of αvβ3 and αvβ5 integrins. In silico modeling also predicted that the peptide would be well exposed on the exterior surface of the viral capsid. This idea was further supported by previous studies performed in AAV and AAV-based vectors, where the 3-fold spike was exploited for the insertion of RGD-containing peptides or other sequences (42). AAV capsids modified in this way were found to keep the capacity to properly assemble and package the viral DNA while acquiring a new tropism, although a reduction of viral transduction efficiency was observed in some cases (49). In agreement with our prediction, the insertion of the RGD-4C peptide into the 3-fold spike of the H-1PV capsid did not impair particle assembly, viral DNA replication, and virus production in cells transfected with the corresponding molecular clone. Furthermore, the RGD-containing H-1 virions were infectious and displayed a novel tropism which appeared to be specific for αvβ5 integrins, as shown by our analysis of the melanoma CS-1 cellular system, in which the expression of αvβ5 but not αvβ3 integrins on the cell surface was required for efficient viral infection. Consistently, H-1PV was taken up more efficiently by SK-MEL-28 cells expressing high levels of αvβ5 integrins than by normal melanocytes, NB324K cells, HeLa cells, and normal keratinocytes, in which αvβ5 integrin expression is low or absent. Moreover, the siRNA-mediated silencing of αvβ5 integrins in SK-MEL-28 cells strongly reduced H-1RGD cell surface binding and cellular entry, providing further evidence of the involvement of these molecules in these events. Surprisingly, the H-1RGD virus had a much lower affinity for αvβ3 integrins. This was not expected, since the RGD-4C peptide is also known to target αvβ3-expressing cells with high affinity (63). The high selectivity of H-1RGD for αvβ5-expressing cells is quite unique, and to the best of our knowledge there are no other examples in the literature showing this degree of specificity. For example, the RGD-4C peptide previously has been used to redirect the tropism of adenovirus toward HeLa cells which express αvβ3 integrins (56), whereas these cells were quite refractory to infection with H-1RGD under our experimental conditions. It is clear that the retargeting ability of a peptide not only depends on its specific sequence but also heavily relies on the chemical properties of the site at which the peptide is inserted. We assume that the RGD-4C peptide inserted at the VP2 residue 441 may adopt, in the context of the viral capsid, a particular conformation that favors binding to αvβ5 instead of αvβ3 integrins. Alternatively, the neighboring VP sequences/structures may affect the affinity of the inserted peptide for integrins. When we modeled the RGD insertion in another loop of the viral capsid (position S1 in Fig. 4), the RGD-4C peptide appeared to have a more open conformation. Therefore, the insertion of the same peptide into other positions, if compatible with proper particle assembly, may extend the viral tropism to αvβ3 integrin-expressing cells. It is also possible that the insertion of slightly different RGD-containing sequences would redirect the virus tropism to αvβ3 integrins.
Our incentive to carry out the present work was to test the possibility of engineering the parvoviral capsid to minimize virus uptake and sequestration by normal cells while maintaining viral entry into tumor cells. The retargeting strategy adopted allowed the H-RGD virus to enter and kill melanoma cells while keeping the impairment of the intermediate (H373R) detargeted mutant in entering normal cells. It should be stated, however, that while it gained in specificity, the H-1RGD virus lost some effectiveness in infecting (as deduced by a delayed expression of NS1 cytotoxic protein) and killing SK-MEL-28 cells compared to the effectiveness of the wild-type virus (Fig. 9 and data not shown). This is probably because the two viruses use alternative routes of entry. Hence, the melanoma cell line could express critical factors involved in the early steps of the life cycle of the respective viruses at different levels (both at binding or postbinding levels, e.g., receptors and coreceptors and/or proteins involved in viral internalization and trafficking to and into the nucleus), accounting for the diversity observed. Attempts at improving the oncolytic activity of the retargeted virus by means of other exogenous peptides inserted in the same or other sites of the viral capsid go beyond the scope of the present proof-of-principle study but are certainly worth pursuing. The successful ablation of the natural viral tropism together with the identification of a site tolerating the insertion of foreign peptides into the viral capsid pave the way for new lines of research toward the in vivo cancer retargeting of H-1PV. One may consider grafting peptidic ligands of known human specific receptors, such as the epidermal growth factor receptor that has been successfully used to retarget adenovirus to gliomas (50). Another attractive strategy would be to graft a peptide library into the insertion site identified in this study, thus generating a pool of H-1PV virions that display a different peptide on the capsid. Such a library could be used to select retargeted H-1PVs specifically infecting certain types of cancer cells through in vitro or in vivo screenings. A similar approach has been successfully applied for the retargeting of AAV (42). While additional work is still required to explore, validate, and optimize the anticancer potential of retargeted H-1PV, the present study gives evidence of the possibility of genetically reprogramming parvovirus entry, adding a new attractive element to the oncolytic parvovirus toolbox.
Supplementary Material
ACKNOWLEDGMENTS
We thank the team of the DKFZ Virus Production and Development Unit, in particular Markus Müller, Silvia Münstermann, and Barbara Liebertrau, for their valuable help with virus production and characterization. We are also grateful to Barbara Hub for electron microscopy analyses, Nathalie Salomé, Michele Vogel, and Christiane Dinsart for the anti-NS1 and anti-VP antibodies, and David Cheresh and the Wistar Institute of Anatomy and Biology (Philadelphia, PA) for the CS-1 cell model.
N.E.A. was the recipient of an EMBO long-term fellowship. J.K.K. is the recipient of a fellowship from the Helmholtz International Graduate School for Cancer Research. D.M.N. is supported by a Helmholtz-University Group grant. This study has been partly supported by grants from the Federal Ministry of Education and Research (BMBF) and the Helmholtz Association in the framework of the Deutsches Krebsforschungszentrum/Cancéropôle du Grand-Est Joint Programme in Applied Tumor Virology.
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369–1381 [DOI] [PubMed] [Google Scholar]
- 2. Alonso MM, et al. 2008. Delta-24-RGD in combination with RAD001 induces enhanced anti-glioma effect via autophagic cell death. Mol. Ther. 16:487–493 [DOI] [PubMed] [Google Scholar]
- 3. Angelova AL, et al. 2009. Oncolytic rat parvovirus H-1PV, a candidate for the treatment of human lymphoma: in vitro and in vivo studies. Mol. Ther. 17:1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angelova AL, et al. 2009. Improvement of gemcitabine-based therapy of pancreatic carcinoma by means of oncolytic parvovirus H-1PV. Clin. Cancer Res. 15:511–519 [DOI] [PubMed] [Google Scholar]
- 5. Assa-Munt N, Jia X, Laakkonen P, Ruoslahti E. 2001. Solution structures and integrin binding activities of an RGD peptide with two isomers. Biochemistry 40:2373–2378 [DOI] [PubMed] [Google Scholar]
- 6. Bodendorf U, Cziepluch C, Jauniaux JC, Rommelaere J, Salome N. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brooks PC, Clark RA, Cheresh DA. 1994. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science 264:569–571 [DOI] [PubMed] [Google Scholar]
- 8. Brown KE, Anderson SM, Young NS. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114–117 [DOI] [PubMed] [Google Scholar]
- 9. Chapman MS, Agbandje-McKenna M. 2006. Atomic structure of viral particles. Hodder Arnold, London, United Kingdom [Google Scholar]
- 10. Colovos C, Yeates TO. 1993. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2:1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cornelis JJ, Deleu L, Kock U, Rommelaere J. 2006. Parvovirus oncosuppression. Edward Arnold Publishers Ltd., London, United Kingdom [Google Scholar]
- 12. Cotmore SF, Tattersall P. 2007. Parvoviral host range and cell entry mechanisms. Adv. Virus Res. 70:183–232 [DOI] [PubMed] [Google Scholar]
- 13. DeLano WL. The PyMOL molecular graphics system. DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 14. Dempe S, Stroh-Dege AY, Schwarz E, Rommelaere J, Dinsart C. 2010. SMAD4: a predictive marker of PDAC cell permissiveness for oncolytic infection with parvovirus H-1PV. Int. J. Cancer 126:2914–2927 [DOI] [PubMed] [Google Scholar]
- 15. Di Pasquale G, et al. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 9:1306–1312 [DOI] [PubMed] [Google Scholar]
- 16. Di Piazza M, et al. 2007. Cytosolic activation of cathepsins mediates parvovirus H-1-induced killing of cisplatin and TRAIL-resistant glioma cells. J. Virol. 81:4186–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Andaloussi N, Barbara L, Serena B, Rommelaere J, Marchini A. Efficient recombinant parvovirus production with the help of adenovirus-derived systems. J. Vis. Exp., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El-Andaloussi N, et al. 2011. Novel adenovirus-based helper system to support production of recombinant parvovirus. Cancer Gene Ther. 18:240–249 [DOI] [PubMed] [Google Scholar]
- 19. Etingov I, et al. 2008. An extension of the minute virus of mice tissue tropism. Virology 379:245–255 [DOI] [PubMed] [Google Scholar]
- 20. Faisst S, et al. 1998. Dose-dependent regression of HeLa cell-derived tumours in SCID mice after parvovirus H-1 infection. Int. J. Cancer 75:584–589 [DOI] [PubMed] [Google Scholar]
- 21. Girod A, et al. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1052–1056 [DOI] [PubMed] [Google Scholar]
- 22. Grekova S, et al. 2010. Activation of an antiviral response in normal but not transformed mouse cells: a new determinant of minute virus of mice oncotropism. J. Virol. 84:516–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herrero Y, Calle M, et al. 2004. Parvovirus H-1 infection of human glioma cells leads to complete viral replication and efficient cell killing. Int. J. Cancer 109:76–84 [DOI] [PubMed] [Google Scholar]
- 24. Hesse A, Kosmides D, Kontermann RE, Nettelbeck DM. 2007. Tropism modification of adenovirus vectors by peptide ligand insertion into various positions of the adenovirus serotype 41 short-fiber knob domain. J. Virol. 81:2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 26. Hodivala-Dilke KM, Reynolds AR, Reynolds LE. 2003. Integrins in angiogenesis: multitalented molecules in a balancing act. Cell Tissue Res. 314:131–144 [DOI] [PubMed] [Google Scholar]
- 27. Hooft RW, Vriend G, Sander C, Abola EE. 1996. Errors in protein structures. Nature 381:272. [DOI] [PubMed] [Google Scholar]
- 28. Hristov G, et al. 2010. Through its nonstructural protein NS1, parvovirus H-1 induces apoptosis via accumulation of reactive oxygen species. J. Virol. 84:5909–5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hueffer K, Parrish CR. 2003. Parvovirus host range, cell tropism and evolution. Curr. Opin. Microbiol. 6:392–398 [DOI] [PubMed] [Google Scholar]
- 30. Kestler J, et al. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619–1632 [DOI] [PubMed] [Google Scholar]
- 31. Koivunen E, Wang B, Ruoslahti E. 1995. Phage libraries displaying cyclic peptides with different ring sizes: ligand specificities of the RGD-directed integrins. Biotechnology 13:265–270 [DOI] [PubMed] [Google Scholar]
- 32. Kontou M, et al. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 79:10931–10943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kruger L, et al. 2008. Augmented transgene expression in transformed cells using a parvoviral hybrid vector. Cancer Gene Ther. 15:252–267 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067 [DOI] [PubMed] [Google Scholar]
- 35. Lopez-Bueno A, et al. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80:1563–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopez-Bueno A, et al. 2008. Evolution to pathogenicity of the parvovirus minute virus of mice in immunodeficient mice involves genetic heterogeneity at the capsid domain that determines tropism. J. Virol. 82:1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luthy R, Bowie JU, Eisenberg D. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83–85 [DOI] [PubMed] [Google Scholar]
- 38. Malerba M, Daeffler L, Rommelaere J, Iggo RD. 2003. Replicating parvoviruses that target colon cancer cells. J. Virol. 77:6683–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mathis JM, Stoff-Khalili MA, Curiel DT. 2005. Oncolytic adenoviruses-selective retargeting to tumor cells. Oncogene 24:7775–7791 [DOI] [PubMed] [Google Scholar]
- 40. Michelfelder S, et al. 2009. Successful expansion but not complete restriction of tropism of adeno-associated virus by in vivo biopanning of random virus display peptide libraries. PLoS One 4:e5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mousset S, Rommelaere J. 1982. Minute virus of mice inhibits cell transformation by simian virus 40. Nature 300:537–539 [DOI] [PubMed] [Google Scholar]
- 42. Muller OJ, et al. 2003. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 21:1040–1046 [DOI] [PubMed] [Google Scholar]
- 43. Munakata Y, et al. 2005. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood 106:3449–3456 [DOI] [PubMed] [Google Scholar]
- 44. Nagel H, et al. 2003. The alphavbeta5 integrin of hematopoietic and nonhematopoietic cells is a transduction receptor of RGD-4C fiber-modified adenoviruses. Gene Ther. 10:1643–1653 [DOI] [PubMed] [Google Scholar]
- 45. Nam HJ, et al. 2006. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 281:25670–25677 [DOI] [PubMed] [Google Scholar]
- 46. Ohshima T, et al. 1998. Induction of apoptosis in vitro and in vivo by H-1 parvovirus infection. J. Gen. Virol. 79:3067–3071 [DOI] [PubMed] [Google Scholar]
- 47. Ong HT, et al. 2009. Intravascularly administered RGD-displaying measles viruses bind to and infect neovessel endothelial cells in vivo. Mol. Ther. 17:1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parker JS, Murphy WJ, Wang D, O'Brien SJ, Parrish CR. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parrish CR. 2010. Structures and functions of parvovirus capsids and the process of cell infection. Curr. Top. Microbiol. Immunol. 343:149–176 [DOI] [PubMed] [Google Scholar]
- 50. Piao Y, et al. 2009. Oncolytic adenovirus retargeted to delta-EGFR induces selective antiglioma activity. Cancer Gene Ther. 16:256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pontius J, Richelle J, Wodak SJ. 1996. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 264:121–136 [DOI] [PubMed] [Google Scholar]
- 52. Qing K, et al. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71–77 [DOI] [PubMed] [Google Scholar]
- 53. Ran Z, Rayet B, Rommelaere J, Faisst S. 1999. Parvovirus H-1-induced cell death: influence of intracellular NAD consumption on the regulation of necrosis and apoptosis. Virus Res. 65:161–174 [DOI] [PubMed] [Google Scholar]
- 54. Rayet B, Lopez-Guerrero JA, Rommelaere J, Dinsart C. 1998. Induction of programmed cell death by parvovirus H-1 in U937 cells: connection with the tumor necrosis factor alpha signalling pathway. J. Virol. 72:8893–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 138:85–98 [DOI] [PubMed] [Google Scholar]
- 56. Rein DT, et al. 2004. Gene transfer to cervical cancer with fiber-modified adenoviruses. Int. J. Cancer 111:698–704 [DOI] [PubMed] [Google Scholar]
- 57. Rommelaere J, et al. 2010. Oncolytic parvoviruses as cancer therapeutics. Cytokine Growth Factor Rev. 21:185–195 [DOI] [PubMed] [Google Scholar]
- 58. Rubio MP, Lopez-Bueno A, Almendral JM. 2005. Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J. Virol. 79:11280–11290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sambrook J, Russell DW. 2001. In vitro mutagenesis using double-stranded DNA templates: selection of mutants with DpnI. In Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 60. Siegl G. 1983. Biology and pathogenicity of autonomous parvoviruses, p. 297–362 In Berns KI. (ed), The parvoviruses. Plenum Press, Inc., New York, NY [Google Scholar]
- 61. Summerford C, Bartlett JS, Samulski RJ. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78–82 [DOI] [PubMed] [Google Scholar]
- 62. Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Temming K, Schiffelers RM, Molema G, Kok RJ. 2005. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resist. Updat. 8:381–402 [DOI] [PubMed] [Google Scholar]
- 64. Thomas L, Chan PW, Chang S, Damsky C. 1993. 5-Bromo-2-deoxyuridine regulates invasiveness and expression of integrins and matrix-degrading proteinases in a differentiated hamster melanoma cell. J. Cell Sci. 105:191–201 [DOI] [PubMed] [Google Scholar]
- 65. Ueno Y, et al. 2001. Propagation of rat parvovirus in thymic lymphoma cell line C58(NT)d and subsequent appearance of a resistant cell clone after lytic infection. J. Virol. 75:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Pachterbeke C, et al. 1997. Varying sensitivity of human mammary carcinoma cells to the toxic effect of parvovirus H-1. Eur. J. Cancer 33:1648–1653 [DOI] [PubMed] [Google Scholar]
- 67. Van Pachterbeke C, et al. 1993. Parvovirus H-1 inhibits growth of short-term tumor-derived but not normal mammary tissue cultures. Int. J. Cancer 55:672–677 [DOI] [PubMed] [Google Scholar]
- 68. Volk AL, et al. 2003. Enhanced adenovirus infection of melanoma cells by fiber-modification: incorporation of RGD peptide or Ad5/3 chimerism. Cancer Biol. Ther. 2:511–515 [DOI] [PubMed] [Google Scholar]
- 69. Weigel-Kelley KA, Yoder MC, Srivastava A. 2003. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood 102:3927–3933 [DOI] [PubMed] [Google Scholar]
- 70. Wickham TJ, Filardo EJ, Cheresh DA, Nemerow GR. 1994. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zitzmann S, Ehemann V, Schwab M. 2002. Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 62:5139–5143 [PubMed] [Google Scholar]
- 72. Zolotukhin S, et al. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973–985 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.