Abstract
Although highly active antiretroviral therapy (HAART) has converted HIV into a chronic disease, a reservoir of HIV latently infected resting T cells prevents the eradication of the virus from patients. To achieve eradication, HAART must be combined with drugs that reactivate the dormant viruses. We examined this problem in an established model of HIV postintegration latency by screening a library of small molecules. Initially, we identified eight molecules that reactivated latent HIV. Using them as templates, additional hits were identified by means of similarity-based virtual screening. One of those hits, 8-methoxy-6-methylquinolin-4-ol (MMQO), proved to be useful to reactivate HIV-1 in different cellular models, especially in combination with other known reactivating agents, without causing T-cell activation and with lower toxicity than that of the initial hits. Interestingly, we have established that MMQO produces Jun N-terminal protein kinase (JNK) activation and enhances the T-cell receptor (TCR)/CD3 stimulation of HIV-1 reactivation from latency but inhibits CD3-induced interleukin-2 (IL-2) and tumor necrosis factor alpha (TNF-α) gene transcription. Moreover, MMQO prevents TCR-induced cell cycle progression and proliferation in primary T cells. The present study documents that the combination of biological screening in a cellular model of viral latency with virtual screening is useful for the identification of novel agents able to reactivate HIV-1. Moreover, we set the bases for a hypothetical therapy to reactivate latent HIV by combining MMQO with physiological or pharmacological TCR/CD3 stimulation.
INTRODUCTION
HIV-infected individuals successfully treated with antiretroviral therapy (ART) control viremia down to undetectable levels. Nevertheless, viremia reemerges rapidly after the interruption of treatment, which is consistent with the existence of a latent viral reservoir. This reservoir consists mainly of long-lived latently infected resting memory CD4+ T cells (16, 28). The absence of viral protein expression shields latently infected cells from the immune system. Latently infected cells can be maintained for years by cellular quiescence. Antigen stimulation or cytokine induction reactivates the latent provirus and leads to viral replication and reinfection. This reservoir prevents the total eradication of the virus. As a result, the infection is converted into a chronic disease. To achieve HIV eradication from infected patients, ART might have to be combined with drugs that reactivate the dormant viruses (72).
Latently infected cells contain replication-competent provirus blocked at the transcriptional level by effective and reversible silencing (molecular mechanisms of HIV latency are reviewed in references 3, 17, 18, and 45). HIV transcription depends on cellular factors (Sp1, NF-κB, NFAT, and others) in addition to the viral Tat transactivator, and consequently the activity of the HIV promoter is tightly linked to the level of activation of its host cell. A suboptimal cellular environment for HIV expression at the transcriptional or posttranscriptional level collaborates to maintain the latent state.
The transcription factor Tat binds an RNA element (TAR) at the 5′ end of the viral transcript. Its main role is to recruit the P-TEFb complex to the long terminal repeat (LTR) to promote efficient elongation (40, 58). Tat is absent from infected cells until inefficient transcription from the HIV promoter (depending on host cell factors) allows some synthesis of this viral protein. After Tat is produced, HIV transcription enters a second, more efficient phase. In latently infected cells, HIV transcription is blocked at the initial phase, Tat is absent, and chromatin imposes a barrier to the clearance of the preinitiation complex and polymerase progression. Chromatin plays an essential role in the transcriptional regulation of HIV (61, 64). Key regulatory nucleosomes are positioned around the transcription start site of HIV when the promoter is inactive and are acetylated or remodeled upon activation (50, 75). Previous models of HIV latency, cell lines ACH2 and U1, harbored proviruses with mutations in their Tat-TAR transcriptional axis, strengthening the concept that transcription inhibition is key to establishing and maintaining HIV latency (26, 27). Thus, the lack of specific host factors, a defective Tat-TAR axis, and a repressive chromatin environment could contribute to postintegration latency.
HIV transcription is determined by the site of integration in the human genome (39). The survival of the infected cell to allow conversion to the resting state may require integration at a genomic site unfavorable for HIV transcription. Rare latent infection has been associated with provirus integration at regions of constitutive heterochromatin, such as centromeric alphoid repeats, gene deserts, or introns of very highly expressed genes (35, 38, 49). The silenced HIV promoter has been associated with repressive epigenetic marks, histone deacetylases and methyltransferases, heterochromatin proteins, and CpG methylation (5, 24, 30, 37, 41, 80). Transcriptional interference has been proposed as the cause of the silencing of HIV integrated at active genes (33, 48, 49). Chromatin reassembly factors traveling with the machinery transcribing a host gene where a latent HIV is integrated participate in this repressing phenomenon (31).
Several agents or chemical compounds stimulate transcription from the HIV-1 promoter and, consequently, reactivate latently integrated viruses (reviewed in reference 18). Treatment with single cytokines, such as interleukin-2 (IL-2) or IL-7, or in different combinations, reactivate latent HIV-1 in resting CD4+ T cells obtained from ART-treated patients or in a chronically infected cell line (8, 15, 47, 60, 78). Tumor necrosis factor alpha (TNF-α) stimulates HIV-1 reactivation in U1 cells but not in resting CD4+ T cells unless combined with IL-6 and IL-2 (14, 75). The drawback of treatments with cytokines is that they promote the global activation of resting T cells, decreasing the immunological memory of the organism. Mitogenic phorbol esters such as phorbol-12-myristate-13-acetate (PMA), as well as the non-tumor-promoting phorbol ester prostratin, also activate the dormant HIV-1 promoter through the activation of protein kinase C (PKC) and, ultimately, NF-κB pathways (7, 8, 43, 75, 79). In addition to PKC, the stimulation of the nuclear factor of activated T cells (NF-AT) is also a key pathway leading to HIV-1 activation.
Histone deacetylase (HDAC) inhibitors, such as trichostatin A (TSA) or butyric acid, were described earlier as activators of the dormant HIV-1 promoter in chronically infected cell lines by promoting chromatin remodeling at the proviral promoter (74). Later, valproic acid was extensively used to investigate the potential of HDAC inhibitors for reactivating HIV-1 in many different model systems or even in volunteer patients, with contradictory results (46, 65, 69, 84). HDAC inhibitors do not globally activate resting T cells but may have general effects on transcription. Recently, new HDAC inhibitors, more potent and selective, were developed and tested on HIV-1 reactivation, including SAHA (suberoylanilide hydroxamic acid) (1, 12, 20, 53, 83). Combining nonmitogenic phorbol esters, such as prostratin, with HDAC inhibitors has a synergistic effect on HIV-1 reactivation and could be the basis of future therapies to eradicate the virus from patients (62). Additionally, inhibitors of other chromatin modifiers involved in HIV-1 repression could enter into play, such as inhibitors of histone or DNA methylation (30, 37, 41).
Using a model of HIV postintegration latency consisting of the infection of Jurkat cells with a green fluorescent protein (GFP)-encoding HIV minigenome and selecting for reversible silent integrations (38), we searched for new chemical agents that reactivate latent HIV-1. Initially, eight molecules were identified from a library of 6,000 small molecules, and other derivative molecules were proposed by applying virtual screening with computational tools and were validated in the latency cellular model. One particular drug, 8-methoxy-6-methylquinolin-4-ol (MMQO), has been useful to reactivate HIV in different cellular models, especially in combination with TNF-α or other known reactivating agents. Additionally, we explored the pathway that could be involved in the activation of the silenced HIV promoter by MMQO. MMQO produced Jun N-terminal protein kinase (JNK) activation and T-cell receptor (TCR)/CD3 costimulation to activate the NF-κB and the extracellular signal-regulated kinase (ERK) pathways. Interestingly, MMQO prevents TCR-induced cell proliferation in primary T cells.
MATERIALS AND METHODS
Cell lines, culturing conditions, and cell treatments.
We used an established model of HIV-1 latency based on the infection of Jurkat T cells at a low multiplicity of infection (MOI) with HIV-derived particles containing the minigenome carried by plasmid pEV731 (LTR-Tat-internal ribosome entry site [IRES]-GFP-LTR) (38). Briefly, GFP-negative cells were treated with TNF-α, and resulting GFP-positive, latently infected cells were isolated (latent pool), and clones were generated from single cells. Clones J-Lat H2, A1 and A2 (38), E27 (31), and Jurkat-LAT-GFP clone 8 (52) have been described elsewhere. Additionally, clones J-Lat 6.3 and 15.4, generated similarly with a full-length GFP-encoding HIV-1 vector (HIV-R7/E−/GFP), were described already (38).
Jurkat-derived clones, ACH2 and U1 cells, were grown in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C under a 5% CO2 atmosphere. HEK293T and HeLa cells were grown under the same conditions as those of Jurkat cells in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin.
When indicated, cells were treated with 10 nM PMA (Sigma-Aldrich), 400 nM TSA (Sigma-Aldrich), 10 ng/ml TNF-α (Sigma-Aldrich), 2 μM prostratin (LC Laboratories), 5 μM 5-aza-2′deoxycitidin (5-azadC; Sigma-Aldrich), or 10 mM hexamethylene bisacetamide (19) (HMBA; Sigma-Aldrich), unless other concentrations are indicated in the figure legends. Similarly, cells were stimulated with immobilized α-CD3 monoclonal antibody (MAb) (1 μg/ml; clone OKT3) or soluble α-CD28 MAb (0.5 μg/ml; clone 15E8). 5-azadC was added 24 h before other treatments (preincubation). Details of treatment with other drugs are in particular figure legends.
Culturing of PBMCs from patients.
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats from aviremic, highly active antiretroviral therapy (HAART)-treated HIV-1 patients (Hospital Clínic, Barcelona, Spain) by centrifugation through a Ficoll-Hypaque gradient (Sigma Chemical Co., St. Louis, MO). Primary cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C under a 5% CO2 atmosphere. These cell cultures were treated as soon as they were seeded with MMQO or other drugs, as indicated in each experiment, without phytohemagglutinin addition. All of the patients gave informed written consent, and this study was reviewed and approved by the Institutional Ethical Committee board of Hospital Clínic (Barcelona, Spain).
Drug screening.
For screening a library of 6,000 small molecules (Chembridge, San Diego, CA), each compound was added at 40 μM to 5 × 104 J-Lat A2 cells in 200 μl of medium in 96-well plates. After 24 h of incubation, the percentage of GFP-expressing cells was determined by flow cytometry. Each plate contained wells with PMA (10 nM) or TSA (400 nM) as a positive control for HIV-1 reactivation.
Flow cytometry analysis and sorting.
GFP fluorescence was measured in a Cytomics FC500 MPL cytometer (Beckman Coulter, Fullerton, CA). A two-parameter analysis was used to distinguish GFP-derived fluorescence from the background. Fluorescence was represented in a logarithmic scale. Cell sorting was carried out with the Moflo high-speed sorter (Dako-Cytomatin, Fort Collins, CO).
Cell cycle analysis.
Cells were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS), resuspended in PBS, and fixed in 70% ethanol at −20°C overnight. Fixed cells were washed three times with PBS, treated with RNase A (1 mg/ml) at 37°C for 1 h, and stained with propidium iodide (0.1 mg/ml). DNA content was determined on a Coulter Epics XL cytometer (Beckman Coulter Inc., Fullerton, CA) and analyzed with MultiCycle AV (Phoenix Flow Systems) for peak definitions corresponding to the different cell cycle phases.
Apoptosis monitoring.
Cultured cells were labeled with annexin V conjugated to the red-sensitive dye CF647 and 7-aminoactinomycin (7-AAD), which were included in the FlowCellect annexin red kit (Millipore), by following the manufacturer's instructions and were analyzed in a Gallios cytometer (Beckman Coulter Inc., Fullerton, CA).
Human T-cell proliferation assays.
PBMCs from healthy adult volunteer donors were isolated by the centrifugation of venous blood on Lymphoprep density gradients (Axis-Shield PoC, Oslo, Norway). Cells (105) were cultured in triplicate in 96-well round-bottom microtiter plates in 200 μl of complete medium and stimulated with staphylococcal enterotoxin B (SEB) (1 μg/ml) (Sigma Chemical Co., St. Louis, MO) in the presence or absence of MQQO as indicated. The cultures were carried out for 3 days and pulsed with 0.5 μCi [3H]thymidine (TdR)/well (MP Biomedicals, Irvine, CA) for the last 12 h of culture. Radioactivity incorporated into DNA was measured by liquid scintillation counting.
Infectivity assay.
Viral infectivity from treated ACH2 cell culture supernatant fluids was measured using TZM-bl indicator cells harboring an integrated LTR-luciferase reporter system as described elsewhere (54). Viruses were quantified by determining the concentration of p24 in the supernatant with an antigen (Ag) capture assay (enzyme-linked immunosorbent assay [ELISA]; Innogenetics NV, Ghent, Belgium).
RNA extraction, reverse transcription-PCR (RT-PCR), and quantitative PCR (qPCR) for expression analysis.
Total RNA was extracted using High Pure RNA isolation (Roche) according to the manufacturer's instructions. cDNA was obtained from 100 ng of total RNA using SuperScript VILO cDNA synthesis (Invitrogen). The indicated gene products were analyzed by PCR with specific oligonucleotides, followed by visualization in agarose gels. Where indicated, the quantification of gene products was performed by real-time PCR using LightCycler 480 SYBR green I master mix (Roche, Indianapolis, IN). Each value was corrected by human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and expressed as relative units. Sequences of oligonucleotides used for real-time PCR were the following: 5′LTR (R-gag) forward (AGTAGTGTGTGCCCGTCTGT) and reverse (TCGCTTTCAGGTCCCTGTTCG); Tat forward (CAAAAGCCTTAGGCATCTCCT) and reverse (CCACCTTCTTCTTCGATTCCT); GFP forward (GAAGCAGCACGACTTCTTCAA) and reverse (GCTTGTCGGCCATGATATAGA); and 3′LTR (U3) forward (ATCCACTGACCTTTGGATGG) and reverse (GTACTCCGGATGCAGCTCTC) (31).
Transient transfections and luciferase assays.
Jurkat cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations and treated as indicated. For the luciferase assays, the cells were washed twice in PBS and lysed in 25 mM Tris-phosphate, pH 7.8, 8 mM MgCl2, 1 mM dithiothreitol (DTT), 1% Triton X-100, and 7% glycerol during 15 min at room temperature in a horizontal shaker. After centrifugation, the supernatant was used to measure luciferase activity using an Autolumat LB 9510 (Berthold) by following the instructions of the luciferase assay kit (Promega, Madison, WI).
Protein extraction, gel electrophoresis, and immunoblotting.
Jurkat cells (106 cells/ml) were stimulated with α-CD3 (1 μg/ml) together with 1 μg protein A from Staphylococcus aureus for cross-linking and MMQO during 15 min. Cells were then washed with PBS and proteins extracted in 50 μl of lysis buffer (20 mM HEPES, pH 8.0, 10 mM KCl, 0.15 mM EGTA, 0.15 mM EDTA, 0.5 mM Na3VO4, 5 mM NaF, 1 mM DTT, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin, 0.5 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 0.5% NP-40. Protein concentration was determined by the Bradford assay (Bio-Rad, Richmond, CA), and 30 μg of protein was boiled in Laemmli buffer and electrophoresed in 10% SDS-polyacrylamide gels. Separated proteins were transferred to nitrocellulose membranes (0.5 A at 100 V; 4°C) for 1 h. Blots were blocked in Tris-buffered saline (TBS) solution containing 0.1% Tween 20 and 5% nonfat dry milk overnight at 4°C, and the immunodetection of specific proteins was carried out with primary antibodies using an ECL system (GE Healthcare). The antibodies anti-phospho-IκBα (ser32/ser36), anti-phospho-p65 (ser536), and phospho-ERK1+2 (Tyr204) were from Santa Cruz Biotechnology (Santa Cruz, CA), and antibodies against anti-phospho-JNK1+2 (Thr183/Tyr185) and anti-phospho-p38 (Thr180/Tyr182) were from Cell Signaling (Danvers, MA). The MAb anti-β-tubulin was purchased from Sigma Co. (St. Louis, MO).
Virtual screening.
Similarity-based virtual screening is based on encoding chemical structures using some sort of mathematical descriptors. In this work, three types of two-dimensional descriptors were used, namely, pharmacophoric fragments (PHRAG), feature-pair distributions (FPD), and Shannon entropy distributions (SHED), each one of them characterizing chemical structures with a different degree of fuzziness and thus complementing each other in terms of structural congenericity and hopping abilities. PHRAG are all possible fixed-length segments of five atom features that can be extracted from the topology of a molecule. In contrast, FPD capture the overall spreading of pairs of atom-centered features at different predefined bond lengths. Finally, SHED quantifies the variability within all possible FPD using the concept of Shannon entropy. Detailed definitions can be found elsewhere (34, 77).
When using PHRAG and FPD, the similarity between two molecules corresponds to the overlapping fraction of their respective profiles, whereas with SHED, Euclidean distances are calculated instead (77). The biological relevance of the similarity and distance values obtained with these descriptors was assessed on their ability to discriminate active from random compounds for all targets chemically represented in publicly available sources. As a result of this validation analysis, compounds with similarity values of greater than 0.76 and 0.87 for PHRAG and FPD, respectively, and below a distance value of 0.52 for SHED were considered to be within the applicability domain (OAD) of these descriptors (77).
RESULTS
Screening molecules that reactivate HIV-1 from latency in an in vitro cellular model.
Using a cell line containing a latent HIV-based retroviral vector (clone J-Lat A2 [38]), we screened a library of 6,000 small molecules (purchased from Chembridge, San Diego, CA). Viral expression was monitored by GFP expression in a flow cytometer. Eight compounds were identified that reproducibly lead to an induction of HIV expression as measured by increased GFP expression (data not shown). Six of the hits were grouped in two classes according to chemical similarities. Class 1 contains three molecules with a common quinolin-8-ol structure. Class 2 molecules share an acyl hydrazone functionality. Hits 4 and 1 were chosen for further analysis as representatives of class 1 and 2 compounds, respectively (Fig. 1A; see Fig. S1A in the supplemental material). A dose-response experiment on a heterogenous population of latently infected cells (J-Lat pool) showed that maximal activation was achieved with 40 to 80 μM both compounds (Fig. 1B). This result indicated that the ability of these agents to promote HIV-1 reactivation involved different latent integration events and was not restricted to one particular cell clone. This was confirmed by testing the reactivation of J-Lat clones H2, A1, and E27, which were previously described (31, 38) to harbor the provirus in different genome environments: centromeric alphoid repeats, gene deserts, and gene introns, respectively (data not shown). Nonetheless, these agents were deleterious to cell viability, as analyzed by flow cytometry (Fig. 1B and C). Hit 1 was less toxic than hit 4 and produced a better reactivation of the latent pool.
Fig 1.
Screening of molecules that reactivate HIV from latency in an in vitro cellular model. (A) Chemical structures of active compounds identified in a screening for HIV activation. Upon screening a library of 6,000 small molecules using a cell line containing a latent HIV-based retroviral vector (J-Lat clone A2), eight compounds were identified that reproducibly lead to an induction of HIV expression. Six of these compounds were grouped into two families according to their structural similarity, classes 1 and 2. Hits 4 and 1 are representative of these classes 1 and 2, respectively. (B) Dose response of hits 1 and 4. Cells from the J-Lat heterogeneous population were incubated with increasing concentrations of drug for 24 h and analyzed by flow cytometry. HIV-GFP reactivation is reported as the percentage of GFP-expressing cells (%GFP). Cell viability was measured by flow cytometry gating on the live population at the forward scatter (FSC) and side scatter (SSC) plot. An example is shown in panel C.
Because of the deleterious effect of the agents obtained so far, we next used different methods to search for derivatives with increased reactivating activity and/or lower cell toxicity. We tested 49 compounds (Chembridge) with various permutations of the R1 to R5 groups of class 1 hits for their ability to reactivate latent HIV in J-Lat cells (data not shown). This analysis identified nine new active compounds closely related to hits 4, 7, and 8 (see Fig. S1B in the supplemental material). This analysis demonstrated that both R3 and R5 could be replaced while conserving biological activity and cell toxicity.
Virtual screening identifies MMQO as a novel compound that reactivates HIV-1 from latency with no cell toxicity.
An efficient, cost-effective strategy to access a significantly larger portion of chemical space is to use computational tools to virtually screen the commercial catalogues of chemical vendors, from which selected numbers of molecules are purchased for experimental testing. In cases where target information is missing but bioactive molecules are known, ligand-based approaches to virtual screening are the best option (25). Accordingly, a collection of 7.5 million unique compounds available commercially was rank ordered by similarity against the eight hits identified above (2). The list of top-ranking molecules was further filtered using criteria based on cost and availability to optimize resources and vendors. A priority list of 43 compounds was finally selected for purchase from just three different chemical providers (Chembridge, Enamine, and Specs). Upon reception, testing for the reactivation of HIV-1 in the J-Lat pool led to the identification of 3 additional hits (percentage of GFP-expressing cells, >4). One of those, 8-methoxy-6-methylquinolin-4-ol (MMQO), was particularly active (Fig. 2A). MMQO resembles class 1 initial hits, but it contains an OH group at position 4 instead of 8. On a dose-response curve, MMQO was active at concentrations of up to 320 μM, with a calculated 50% effective concentration (EC50) close to 80 μM (Fig. 2B). The maximal reactivation potential at 24 h in the J-Lat pool was higher than that with hits 1 and 4, and more importantly, the toxicity was greatly reduced. No effect on cell viability was observed at concentrations of up to 80 μM, the concentration chosen for further analysis.
Fig 2.
Virtual screening identifies MMQO as a novel compound that reactivates HIV from latency with no cell toxicity. (A) A library of 43 small molecules (A2 to A44) obtained by virtual screening for compounds pharmacophorically related to the initial hits was purchased and tested for HIV reactivation in a J-Lat heterogeneous cell population. Drugs were added to cells at 40 μM for 48 h, and the percentage of GFP-expressing cells (%GFP) was measured by flow cytometry. The chemical structure of the positive-hit 8-methoxy-6-methylquinolin-4-ol (MMQO) is indicated. (B) Dose response of MMQO. Cells from the J-Lat heterogeneous population were incubated with increasing concentrations of MMQO for 24 h and analyzed by flow cytometry. HIV-GFP reactivation is reported as a percentage of GFP-expressing cells (%GFP) and mean fluorescence intensity (MFI). Cell viability was measured on the FSC-SSC plot as described for Fig. 1C. Data are represented as means ± standard errors of the means (SEM) (n = 3). (C and D) Cell cycle profile of cells treated with reactivating hits. Jurkat cells were treated for 36 h with MMQO, hit 1, or hit 4 at 40 μM or were maintained untreated as a mock control, followed by propidium iodide staining and the flow cytometry analysis of the proportion of cells in each cell cycle phase. The percentage of cells in sub-G1, G1, S, and G2/M phases is shown in panel D. Data are represented as means ± SEM (n = 3). (E) Hits 1 and 4, but not MMQO, produce cell death. HeLa cells were treated for 16 h with MMQO, hit 1, or hit 4 at 80 μM or were maintained untreated as a mock control, followed by annexin V-CF647 and 7-AAD staining and the flow cytometry analysis of the proportion of cells in each staining gate. The percentage of cells in early (annexin-positive) or late (annexin-positive plus 7-AAD-positive) apoptosis is shown. Data are represented as means ± SEM (n = 3).
The effect of MMQO on cell proliferation was further studied and compared to the initial hits by the propidium iodide staining of cells treated for 36 h and followed by cell cycle analysis by flow cytometry. MMQO produced a slight increase in the proportion of cells in the G1 phase of the cycle (Fig. 2C and D). In contrast, the cell cycle profile was clearly altered in cells treated with hit 1 or 4, including the accumulation of cells in sub-G1, which is indicative of cell death. To further compare consequences of these compounds on cell viability, treated cells were stained with annexin V and 7-AAD. The hits produced an increased proportion of cells in early or late apoptosis (Fig. 2E).
Similarly to the initial hits, MMQO reactivated latent HIV-1 in different genomic contexts, including active genes (J-Lat E27 and A2) and centromeric alphoid repeats (J-Lat H2), with no cell toxicity (Fig. 3A).
Fig 3.
MMQO activates HIV at the transcriptional level independently of genome context. (A) MMQO activates latent HIV in different genomic contexts. Cells of latently infected clones J-Lat E27, A2, and H2 were incubated with increasing concentrations of MMQO for 24 h and analyzed by flow cytometry for GFP-expressing cells and cell viability. (B) MMQO activates a transfected HIV-1 LTR-luciferase reporter. 293T cells were transfected with an HIV-1 LTR-luciferase reporter plasmid. One day after transfection, cells were treated with MMQO (80 μM), EGF (50 ng/ml), PMA (10 nM), TSA (400 nM), or TNF-α (10 ng/ml) for 36 h, followed by cell lysis, protein normalization (5 μg), and luciferase activity assay. Data are expressed as relative light units (RLU). (C) Time course response of HIV-1 transcription to MMQO analyzed by RT-qPCR. J-Lat E27 cells were treated with 80 μM MMQO for the time indicated, and RNA was extracted. PMA (10 nM) and TNF-α (10 ng/ml) treatments for 12 h were added as controls. HIV transcription was measured by RT-qPCR using primers corresponding to the HIV 5′LTR (R-gag) and normalizing with GAPDH. The fold change (FC) between treated and untreated cells is shown. (D) MMQO stimulates HIV transcription initiation. J-Lat E27 cells were untreated or were treated with 80 μM MMQO for 24 h, RNA was extracted, and HIV reactivation was measured by RT-qPCR. Amplicons corresponding to the 5′LTR (R-gag), Tat, GFP, and 3′LTR (U3) were used. GAPDH expression was measured for normalization. To compare between different amplicons, qPCR was performed in parallel from genomic DNA (gDNA). Data are expressed as relative units (RU) (cDNA amplification/GAPDH)/gDNA amplification in a log scale. A scheme of the HIV minigenome with positions of qPCR primers is shown below. Throughout the figure data are represented as means ± SEM (n = 3).
We next analyzed whether MMQO induced HIV-1 expression at the transcriptional level. To determine if MMQO was acting through the HIV-1 promoter, we analyzed its effect on an HIV-1 promoter reporter assay upon transient transfection into 293T cells. MMQO activated the LTR-luciferase reporter as efficiently as other HIV inducers, such as TNF-α, PMA, or TSA (Fig. 3B). Further, we performed quantitative PCR on synthesized cDNA generated from J-Lat E27 cells left untreated or treated with MMQO. In a time course experiment, the increase in HIV-1 mRNA was detected as early as 2 h after MMQO addition. At 12 h, HIV-1 expression was increased ca. 12-fold, which is lower than the induction that was observed with control treatments with PMA or TNF-α (Fig. 3C). To explore whether MMQO induced initiation or enhanced elongation, four amplicons covering the HIV-1 minigenome transcript from the 5′LTR to 3′LTR were used: 5′LTR (R-gag), Tat, GFP, and 3′LTR (U3). Any of these primer pairs distinguished between spliced and unspliced transcripts due to the nature of the minimal HIV-1 minigenome in J-Lat cells (Fig. 3D). To compare different amplicons, the amplification of cDNA was corrected by the amplification of genomic DNA with the same sets of primers. MMQO treatment enhanced the expression of all HIV-1 regions by approximately one order of magnitude (Fig. 3D). A noted decrease of transcription along the HIV minigenome at the basal and induced conditions was observed not only in J-Lat E27 but also in other clones and in the latent pool (data not shown), and this has been described before (31). This suggests that latency in J-Lat cells is partially produced by a block in the elongation of HIV transcripts. MMQO seems to stimulate transcription initiation instead of being able to specifically overcome the elongation block. This is similar to the effect of stimulation with TNF-α reported before (31).
Synergistic activation of HIV-1 expression by combination of MMQO with small molecules that reactivate latent virus.
We next compared the abilities of MMQO, other agents, and combinations of them to reactivate latent HIV-1 in J-Lat E27 cells. Because the percentage of GFP-positive cells approaches 100% upon strong reactivation by combined agents, data also were reported as mean fluorescence intensity (MFI). MMQO was less active than PMA, TNF-α, prostratin, or HMBA, and its activity was similar to that of TSA (Fig. 4A and B). When combined, MMQO showed synergy with PMA, TNF-α, and prostratin and only additive effects with HMBA and TSA. MMQO could further stimulate the reactivating ability of combinations such as PMA with TNF or PMA with TSA. The synergistic effect of MMQO and other agents on the reactivation of latent HIV-1 was observed in all J-Lat clones tested, including latent integrations at centromeric alphoid repeats (Fig. 4C, J-Lat H2).
Fig 4.
Synergistic activation of HIV expression by combination of MMQO with small molecules known to reactivate latent HIV. (A) MMQO synergizes with TNF-α and PMA on the activation of a latent HIV-1 minigenome. Cells from clone J-Lat E27 were incubated for 36 h with combinations of MMQO (80 μM), TSA (400 nM), PMA (10 nM), TNF-α (10 ng/ml), and HMBA (10 mM) and analyzed by flow cytometry. HIV-GFP reactivation is reported as MFI or as the percentage of GFP-expressing cells for a selection of the treatments together with the percentage of cell viability. (B) MMQO synergizes with prostratin. J-Lat E27 cells were incubated for 36 h with combinations of MMQO (80 μM) and prostratin (2 μM) and analyzed as described for panel A. Data are represented as means ± SEM (n = 3). (C) Combined activation of clone J-Lat H2. Cells from clone J-Lat H2 were treated for 36 h with PMA (10 nM), TSA (400 nM), TNF-α (10 ng/ml), HMBA (10 mM), or 5-azadC (5 μM) in the absence or presence of MMQO (80 μM). HIV-GFP reactivation is reported as MFI. (D) Activation of HIV-1 with low doses of MMQO and PMA. J-Lat E27 cells were incubated for 16 h with combinations of suboptimal doses of MMQO and PMA and analyzed as described for panel A. Data are represented as means ± SEM (n = 3). (E) J-Lat E27 cells were treated with increasing concentrations of TNF-α (100 to 1,000 ng/ml) or PMA (10 to 160 nM) in the absence or presence (50 or 100 μM) of MMQO. HIV-GFP reactivation is reported as MFI. (F) J-Lat E27 cells were treated with increasing concentrations of MMQO (50 to 150 nM) in the absence or presence of HMBA(10 or 20 mM) or TSA (0.4 or 1 μM). HIV-GFP reactivation is reported as MFI. (G) Combinatorial treatment with MMQO and hit 1 or 4. J-Lat E27 cells were treated with increasing concentrations of hit 1 or 4 (5 to 80 μM) in the absence or presence (50 or 100 μM) of MMQO. HIV-GFP reactivation is reported as MFI. Cell viability extracted from the FSC-SSC plots is shown in the lower panels. (H) Combinatorial treatment with hits 1 and 4 and MMQO. J-Lat E27 cells were treated with increasing concentrations of hit 1 or 4 (10 to 40 μM) in the absence or presence (80 μM) of MMQO. HIV-GFP reactivation is reported as MFI.
Interestingly, the synergism between PMA and MMQO also was observed at suboptimal doses of both drugs, as low as 15 to 40 μM MMQO and 0.3 to 1 mM PMA (Fig. 4D). Synergistic activation suggests that the two agents use different pathways unless they are used at suboptimal concentrations. To explore whether MMQO's effect on HIV-1 reactivation was equivalent to the effect of PMA, TNF-α, HMBA, or TSA, we performed experiments of saturation with one of the agents to see if the other was still active. In a reactivation experiment with clone J-Lat E27, when the concentrations of TNF-α or PMA were increased until saturation, MMQO still showed a clear synergistic effect that further increased HIV-1 reactivation (Fig. 4E). Despite the high concentrations of agents used, cell viability was not greatly compromised (data not shown). Nonetheless, high concentrations of HMBA or TSA (more than 10 or 400 mM, respectively) were highly toxic, and these experiments were not conclusive. Instead, we used increasing concentrations of MMQO up to 150 μM, close to saturation, and observed that the addition of suboptimal concentrations of HMBA or TSA was still stimulatory (Fig. 4F) despite the associated cell toxicity (data not shown). Taken together, our results indicate that MMQO synergizes with other known activating agents on the reactivation of latent HIV-1 in the J-Lat model and also suggest that its cellular target is different from the known targets for these compounds, i.e., protein kinase C (PMA), TNF receptor signaling (TNF-α), histone deacetylases (TSA), and P-TEFb release (HMBA).
MMQO, which was obtained by virtual screening from the initial hits, was chemically closer to class 1 than to class 2 hits. To explore whether MMQO and hits 1 and 4 synergize, we performed a dose response curve with increasing concentrations of these hits in the absence or presence of MMQO (Fig. 4G). MMQO synergized with both compounds, more notably with hit 1 (class 2). This suggested that MMQO and class 2 hits have different cellular targets to promote HIV-1 reactivation. In a different experiment, hits 1 and 4 enhanced each other at low doses, but when the concentration of one of them was increased it blocked the response to the other. When MMQO was added, there was synergy with both compounds, but when the dose of hit 1 or 4 was increased in the presence of the other one, it became inhibitory (Fig. 4H). Mainly, hit 4 (class 1) blocks the synergy between hit 1 and MMQO, suggesting that MMQO and class 1 hits act on the same target.
HIV-1 reactivation in alternative latency cellular models.
Cell clones latently infected with a full-length HIV vector (HIV-R7/E−/GFP) (38) also were tested for reactivation. The effect of MMQO alone or in combination with other agents (TNF-α, TSA, and 5-azadC) was analyzed in clone J-Lat 15.4 (Fig. 5A). Single treatments were almost inactive. The reactivation of such clones required combinations including the demethylating drug 5-azadC, as previously reported (41). MMQO synergized with treatments such as TNF-α plus 5-azadC. In the presence of 5-azadC, the response to increasing concentrations of MMQO was linear for up to 7% of GFP-positive cells at 120 μM this drug (data not shown).
Fig 5.
MMQO reactivates latent HIV from different HIV latency models containing a full-length provirus. (A) MMQO synergizes with treatments that activate full-length HIV-1 J-Lat cells. Cells from clone J-Lat 15.4 were incubated for 36 h with combinations of TNF-α (10 ng/ml), TSA (400 nM), and 5-azadC (5 μM) in the absence or presence of MMQO (80 μM) and analyzed by flow cytometry. 5-azadC was added 24 h before other treatments (preincubation). HIV-GFP reactivation is reported as a percentage of GFP-expressing cells. (B) HIV reactivation from latently infected cell lines U1 and ACH2. U1 and ACH2 cells were incubated for 6 h with MMQO (80 μM), PMA (10 nM), TSA (400 nM), TNF-α (10 ng/ml), HMBA (10 mM), or 5-azadC (5 μM) as indicated, and RNA was extracted to assess HIV expression by RT-qPCR using primers corresponding to HIV 5′LTR (R-gag) and normalizing with GAPDH. Data are represented relative to the maximal activation for each cell line. (C) MMQO enhances HIV reactivation by TNF-α and 5-azadC in cell lines U1 and ACH2. U1 and ACH2 cells were incubated for 6 h with TNF-α (10 ng/ml) and 5-azadC (5 μM) or left untreated and were in the absence or presence of MMQO (80 μM), and RNA was extracted to assess HIV expression by RT-qPCR using primers corresponding to the HIV 5′LTR (R-gag) and normalizing with GAPDH. Data are represented as fold increases compared to the expression of untreated cells. (D) MMQO reactivation of HIV-1 from patients. Twenty to 30 million PBMC were obtained from several HAART-treated patients (named A to I) and incubated with 80 μM MMQO for 36 h or left untreated. RNA was extracted and HIV expression measured by RT-qPCR using primers corresponding to the HIV 5′LTR (R-gag) and normalizing with GAPDH. Data are expressed as relative units (RU) of HIV/GAPDH expression. (E) MMQO induces viral particle formation in ACH2 cells. ACH2 cells were incubated for 24 h with MMQO (80 μM), PMA, ionomycin (IO), or prostratin. Cell culture supernatant media were collected 24 and 72 h after the initial addition of drugs and assayed for the Gag-derived p24 HIV protein using an ELISA to measure the production of viral particles. (F) Viral particles induced in MMQO-treated ACH2 cells are infectious. Human PBMC or ACH2 cells were incubated for 24 h with MMQO (80 μM) or prostratin (2 μM), supernatant media were collected, and viral infectivity was measured using TZM-bl indicator cells harboring an LTR-luciferase reporter system. After 72 h of incubation, cell-associated luciferase activity was determined. Data are expressed as the fold increase of treated versus untreated cells in relative luciferase units (RLU). Throughout the figure data are represented as means ± SEM (n = 3).
Further, we explored the ability of MMQO to reactivate latent HIV from two additional widely used cellular models, the human monocytic and T-lymphoid cell lines U1 and ACH-2, respectively, persistently infected with HIV-1 (Fig. 5B and C). The reactivation of HIV by MMQO, as analyzed by RT-qPCR, was very limited in U1 cells but was still similar to that of PHA or TSA. In these cells, MMQO greatly synergized with an effective treatment such as TNF-α plus 5-azadC. In ACH-2 cells, MMQO upregulated HIV expression almost 10-fold and was additive to the activation by TNF-α plus 5-azadC.
The reactivation of HIV by MMQO was analyzed in cultured PBMCs obtained from aviremic, HAART-treated patients. Seeded PBMCs were treated with MMQO or left untreated for 36 h without prior PHA stimulation, followed by RNA extraction and the analysis of HIV expression by RT-qPCR. In at least two out of nine tested patients, HIV reactivation was clearly detected in MMQO-treated but not mock-treated PBMCs at the RNA level (Fig. 5D). Attempts to detect viral particles (Gag-derived p24 antigen) in media of treated PBMCs were unsuccessful. Instead, infective viral particles were detected in media of MMQO-treated ACH2 cells, comparably to treatments with PMA or prostratin, either measuring p24 antigen (Fig. 5E) or using TZM-bl indicator cells harboring an LTR-luciferase reporter system (Fig. 5F).
MMQO enhances TCR/CD3 stimulation of HIV-1 reactivation from latency and prevents TCR-induced proliferation in primary T cells.
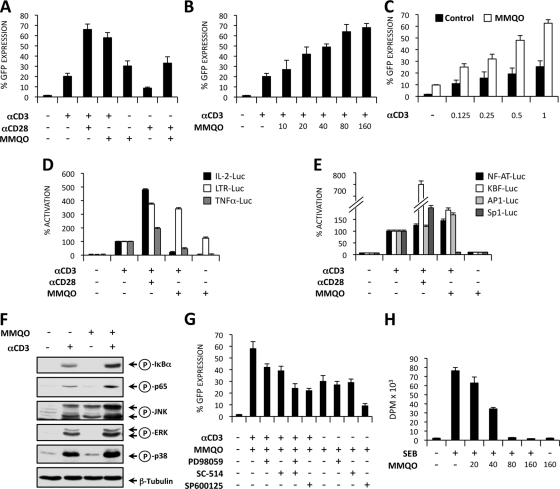
It is thought that in latently infected T cells HIV-1 can be reactivated in vivo through the stimulation of the T-cell receptor. The activation of the TCR induces multiple signal transduction pathways, leading to the activation of the HIV transcription initiation factors NF-κB, NFAT, AP-1, and Sp1, as well as the regulation of HIV transcriptional elongation. To study the effects of MMQO on TCR-induced HIV-1 reactivation, Jurkat-LAT-GFP cells were stimulated with the agonist MAbs α-CD3 and α-CD28 in the absence or the presence of MMQO. In Fig. 6A it is shown that CD3 (1 μg/ml) and MMQO activation antagonized HIV-1 latency, and an additive effect was observed when both stimuli were provided together. This effect was comparable to the one observed after stimulation with a pair of α-CD3 and α-CD28 MAbs which mimic full TCR activation. The effect of MMQO on CD3-induced HIV-1 reactivation was dose dependent, reaching a plateau at a concentration of 80 μM (Fig. 6B). At concentrations lower than 80 μM, MMQO alone did not antagonize HIV-1 latency in Jurkat-LAT-GFP cells (data not shown). Moreover, MMQO clearly synergizes with lower concentrations of MAb α-CD3 to reactivate HIV-1 from latency in this cell model (Fig. 6C).
Fig 6.
MMQO enhances TCR/CD3 stimulation of HIV-1 reactivation from latency and prevents TCR-induced proliferation in primary T cells. (A and B) MMQO synergizes with CD3 to reactivate HIV-1 from latency. Jurkat-LAT-GFP cells were stimulated for 18 h on 96-well plates with coated anti-CD3 (1 μg/ml), soluble anti-CD28 (0.5 μg/ml), and MMQO (160 μM) separately or in combination. (B) Jurkat-LAT-GFP cells were stimulated with coated anti-CD3 and increasing concentrations of MMQO as indicated for 18 h. GFP expression was measured by flow cytometry, and results indicate the percentage of GFP+ cells. (C) Jurkat-LAT-GFP cells were stimulated with increasing concentrations of coated anti-CD3 and MMQO at 80 μM for 18 h. (D) Differential effects of MMQO on IL-2 and TNF-α promoters and HIV-LTR transactivation. Jurkat cells were transiently transfected with the plasmids IL-2-Luc, TNF-α-Luc, and LTR-Luc, and 24 h later they were stimulated with coated anti-CD3 (1 μg/ml) in the absence or the presence of either soluble anti-CD28 (0.5 μg/ml) or MMQO (160 μM) or with MMQO alone for 24 h. The results are expressed as the percentage of activation considering CD3-induced transactivation as 100% activation. (E) Activity of MMQO on NF-κB-, NFAT-, AP-1-, and Sp1-responsive minimal promoters. Jurkat cells were transfected with NF-κB, NFAT, AP-1, and Sp1 luciferase reporter plasmid DNA and stimulated as described for panel C. Luciferase activity was measured 24 h later after protein normalization. The results are expressed as the percentage of activation considering CD3-induced transactivation as 100% activation. (F) Effect of MMQO on α-CD3-induced NF-κB and MAPK activation. Jurkat cells were stimulated with anti-CD3 and MMQO (160 μM) separately or in combination for 15 min, and the levels of phospho-IκBα, phosho-p65 (Ser536), phospho-JNK, phospho-ERK, and phospho-p38 were assessed by immunoblotting with specific antibodies and anti-tubulin MAb as a loading control. (G) MMQO antagonizes HIV-1 latency through the JNK pathway. Jurkat-LAT-GFP cells were treated with the ERK inhibitor PD98059 (25 μM), IKK2 inhibitor (25 μM), and JNK inhibitor SP600125 (1 μM) or left untreated, and 30 min later they were stimulated with anti-CD3 plus MMQO (80 μM) or with MMQO alone for 18 h. HIV-GFP reactivation was assessed by flow cytometry, and results indicate the percentage of GFP+ cells. (H) Effects of MMQO on T-cell proliferation. Human PBMCs were stimulated with SEB (1 μg/ml) in the presence or the absence of increasing concentrations of MMQO for 72 h. [3H]thymidine incorporation was measured by liquid scintillation counting and is represented as disintegrations per minute (DPM). Throughout the figure data are represented as means ± SEM (n = 3).
We next investigated the effect of MMQO on the transcriptional activity of IL-2, TNF-α, and LTR promoters in a Tat-independent manner. We found that in Jurkat cells α-CD3 treatment induced the transcriptional activity of the three promoters. In contrast, MMQO induced HIV-LTR transactivation but failed to stimulate IL-2 and TNF-α gene promoters. Interestingly, MMQO that enhanced α-CD3-induced HIV-LTR transcription was able to repress CD3-induced IL-2 and TNF-α promoter activation (Fig. 6D). These results prompted us to investigate the effects of MMQO on the transcriptional activities of minimal promoters containing binding sites for the transcription factors NF-κB, NFAT, AP-1, and Sp1, which are involved in the transcriptional regulation of IL-2, TNF-α, and HIV-LTR gene promoters. In Fig. 6E we showed that MMQO alone did not induce the transcriptional activity of any of these minimal promoters, enhanced CD3-induced NFAT, NF-κB, and AP-1 activation, and repressed CD3-induced Sp1 transactivation. Similarly, MMQO alone showed no activity on these reporters when transfected into HeLa cells, but it synergized strongly with treatments that optimally activated them, namely, TNF-α or PMA plus ionomycin (data not shown).
To identify the biochemical pathways activated by MMQO, Jurkat cells were stimulated with the MAb α-CD3 and MMQO separately or in combination for 15 min, and the phosphorylation of the NF-κB inhibitor IκBα and the NF-κB subunit p65, as well as the phosphorylation (activation) of the mitogen-activated protein kinases (MAPKs) JNK1+2, ERK1+2, and p38, were investigated by Western blotting using specific MAbs. CD3 stimulation induced phosphorylation of all proteins analyzed that was clearly enhanced by costimulation with MMQO, which by itself activated JKN1+2 but not the other pathways (Fig. 6F). Moreover, MMQO alone was unable to induce IκB degradation (data not shown).
To further assess the NF-κB and MAPK dependence of CD3 and MMQO-mediated antagonism of HIV latency, we pretreated Jurkat-LAT-GFP cells with the chemical inhibitors SC-514 (IKK2 inhibitor), PD98059 (MEK1 inhibitor), and SP600125 (JNK inhibitor). As depicted in Fig. 6G, JNK inhibition, but not IKK2 and ERK inhibition, greatly prevented both MMQO- and CD3/MMQO-induced HIV reactivation, further suggesting that JNK activation is a central event in MMQO action on HIV reactivation. Combined IKK2 and ERK inhibition also inhibited CD3/MMQO-induced GFP expression (Fig. 6G). The further use of chemical inhibitors ruled out the participation of calcium signaling/NFAT, PKC, phosphatidylinositol 3-kinase (PI3K), or p38, among others, on MMQO-induced HIV reactivation from J-Lat cells (data not shown).
We next studied the effects of MMQO on several T-cell activation events. The SEB activation model was used, since B cells and macrophages present this superantigen and activate T cells through the TCR and costimulators. In Fig. 6H it is shown that DNA synthesis measured by [3H]TdR uptake in SEB-stimulated T cells was markedly inhibited by MMQO in a concentration-dependent manner. Cell cycle analysis in unstimulated T cells showed that after 4 days in culture most of the cells remained largely in the G0/G1 phase of the cell cycle (64%), and a significant percentage (30%) of cells undergo apoptosis that is reflected in the sub-G0/G1 fraction. Five days following activation with SEB, T cells were fully cycling and progressed through the S, G2, and M phases of the cell cycle (34% of the cells), while pretreatment with MMQO (80 and 160 μM) almost completely prevented the entry of the cells in the S phase of the cell cycle. Moreover, MMQO did not induce a significant increase in the percentage of hypodiploid cells (sub-G0/G1), suggesting that at the used doses MMQO did not induce cytotoxicity or apoptosis in primary T cells (data not shown).
DISCUSSION
In this study, we used a simple model of HIV-1 latency to screen small-molecule libraries for new agents that reactivate the dormant virus. Of the compounds identified here, MMQO has been the most effective and least toxic for Jurkat cells. Although its reactivating potential is less than that of other known agents, it may represent a new class of latency-reversing therapy that can be combined with others to obtain a synergistic effect. Additionally, although the precise target of MMQO is unknown, it may eventually reveal a new plausible intervention into a pathway involved in the regulation of HIV-1 expression.
The J-Lat model for HIV-1 latency, containing either a minimal HIV-1 minigenome or a recombinant full-length virus (38), is useful for the study of the determinants of postintegration latency associated with distinct integration events (4, 5, 22–24, 41, 48, 49, 59, 80) or to investigate the reactivating potential of several agents (21, 52, 57, 62, 66, 68, 79, 81, 83). Latency is established due to transcriptional silencing (i.e., the repression of the integrated HIV-1 promoter). In vivo, other factors also may be involved, but once latency has been established, flushing out the HIV promoter with external agents is useful to purge the latently infected cells. Many agents proven to reactivate latent virus in J-Lat cells also activate HIV-1 in other latency models or in PBMCs from infected patients, such as PMA, prostratin, PHA, HMBA, or HDAC inhibitors (7, 43, 82). TNF-α is an exception, as it reactivates HIV-1 in many cell lines but not in other systems based on primary cells (reviewed in reference 11). In latently infected, resting CD4+ T cells derived from HIV-infected individuals under ART, TNF-α activates viral replication in conjunction with cytokines IL-2 and IL-6 (14). In this system, NF-κB activation by TNF-α and binding to the LTR is not sufficient for HIV-1 activation. TNF-α may need to be combined with inhibitors of DNA methylation for the efficient reactivation of full-length HIV-1 in J-Lat cells (41).
Additional, more sophisticated, in vitro models for HIV-1 latency in primary CD4+ T cells have been described (6, 9, 10, 51, 71, 73, 82). Some of them are not appropriate for high-throughput screening for activators of latent HIV-1, either because of the short life span of resting T cells in vitro or because cells are maintained in the presence of IL-2 or IL-7, which may activate latent HIV-1 and alter the quiescent state. In the primary cell HIV-1 latency model described by Yang and coworkers, cells were maintained in vitro in the absence of activating cytokines and were used to screen drug libraries, obtaining a new compound that reactivated latent HIV-1, 5-hydroxynaphthalene-1,4-dione (5HN) (82). 5HN activated HIV-1 by forming reactive oxygen species (ROS) that indirectly activated NF-κB without activating NFAT or PKC signaling and, consequently, without inducing global T-cell activation. 5HN resembles class 1 hits and MMQO, as all contain a double aromatic ring, but they are chemically divergent. 5HN is a quinone that can be enzymatically reduced to a semiquinone radical that, under aerobic conditions, generates ROS. The role of ROS as intermediates in activating NF-κB and, eventually, expressing HIV-1 has been described (32, 67). The compounds described here are unable to support this chemistry and form ROS, so they might act through a different mechanism. In this line, MMQO does not promote IκB phosphorylation/degradation, activate an NF-κB reporter by itself, or induce IκB gene expression (Fig. 6 and data not shown). Moreover, pretreatment with agents that remove oxygen radicals, such as N-acetyl-l-cysteine (NAC), did not block HIV reactivation by MMQO, as has been reported for 5HN-mediated reactivation (data not shown). In conclusion, MMQO is unable to induce the NF-κB pathway by itself, but it induces some signaling pathway that cross-talks with it, as well as with pathways affecting AP-1 and NFAT reporter promoters.
A recently reported high-throughput screening using SupT1 cells latently infected with a GFP and secretable alkaline phosphatase encoding HIV identified a new activator (AV6) of HIV-1 which required NFAT and synergized with the HDAC inhibitor valproic acid (55). Our data have shown that MMQO activity on HIV is independent of NFAT activation, as it was not affected by cyclosporine or FK506 (data not shown).
Our data suggest that MMQO promotes HIV-1 transcription initiation and rapid increase in mRNA synthesis, presumably by stimulating a signaling pathway that directs a transcription factor to the viral LTR. Because MMQO has synergy with agents such as PMA or TNF-α even at saturating concentrations, we ruled out the idea that it acts by activating PKC or TNF receptor signaling. In addition, the absence of the blockage of MMQO effects with inhibitors of PKC, PI3K, or ERK, among others, eliminated the participation of these pathways. Another hypothesis we explored is that MMQO damages DNA and promotes the activation of cytokines or the ATM/ATR response that, through NF-κB, could stimulate the HIV-1 promoter. It has been previously described that damaging agents such as UV light may induce the HIV-1 promoter (70, 76). We ruled out the participation of ATM/ATR, as an inhibitor of this pathway does not block the stimulating effect of MMQO. Additionally, as already mentioned, the direct activation of NF-κB by MMQO has not been observed in our experiments. The participation of p38 MAPK, which is involved in reactivation by UV light or cytokines (44), also has been ruled out by using a specific inhibitor (data not shown).
In some primary models of HIV-1 latency, most common NF-κB inducers did not reactivate latent virus in primary T cells, but TCR stimulation was very effective at antagonizing HIV-1 latency (6). Therefore, we investigated the activity of MMQO in the context of TCR activation. Although MMQO is not very potent by itself to reactivate latent HIV-1, this drug efficiently cooperates with TCR activation even at the lower concentration tested. This is of particular relevance because MMQO could be developed as an antilatency therapy specific for T cells at nontoxic concentrations that could be reached at the anatomic sites where memory T cells recognize antigen recall. Moreover, we found that the antagonizing activity of MMQO on HIV-1 latency was also enhanced in the presence of low concentrations of OKT3. This MAb (Muromonab) is used in clinical practice to prevent the rejection of organ allografts, but it is also a potent mitogen for T lymphocytes, therefore it has been investigated for its ability to purge the HIV-1 reservoir. In this clinical trial, high concentrations of OKT3 (5 mg/day for four consecutive days) in combination with recombinant IL-2 were used, and it was concluded that high-level stimulation might result in a negative outcome because of the depletion of noninfected T cells (29). However, lower concentrations of OKT3 are well tolerated in humans and can be immunostimulatory in cancer patients (63). Thus, a possible combination therapy based on low concentrations of humanized OKT3 and MMQO may be of interest for the viral eradication strategies. In this sense, a major goal for the development of HIV-1 reactivation therapies is to induce HIV-1 replication from latent reservoirs without inducing polyclonal T-cell activation. Our results fit well with this principle, as we show that MMQO inhibits antigen-induced T-cell proliferation and the TCR/CD3 induction of IL-2 and TNF-α gene transcription (Fig. 6).
The identification of the exact mechanism of action of MMQO requires further research, but our findings indicate that this compound activates signal pathways that differentially regulate IL-2 and HIV-LTR gene transcription. While the mechanisms for IL-2 repression are unknown at the moment, the enhancement of CD3-induced HIV-1 reactivation can be explained by the coactivation of the NF-κB and MAPK pathways. Recently, it has been shown that T-cell receptor signaling is sufficient to activate transcription elongation in the absence of Tat by a mechanism that involves the ERK-dependent activation of P-TEFb (42). Thus, it is likely that MMQO favors transcription initiation by activating NF-κB and transcription elongation through the ERK pathway, overriding the requirement of high levels of HIV-1 Tat protein that probably are not achievable in latently infected primary memory cells.
Interestingly, MMQO alone produced JNK phosphorylation in Jurkat cells. Because MMQO did not show stimulating activity on an AP1-Luc reporter, it is possible that substrates other than c-jun are involved in MMQO-induced HIV-1 reactivation. Because MMQO-activated signals are similar to those induced through FAS/CD95, such as JNK activation and TCR/CD3 costimulation to activate the NF-κB and ERK pathways (36, 56), we analyzed the potential involvement of the CD95 adaptor protein FADD on the mechanism of action of MMQO. We generated a pool of cells containing the HIV-1-GFP minigenome latently integrated in Jurkat cells stably expressing a dominant-negative mutant of FADD (Jurkat-FADDDN) (36), and we analyzed reactivation by MMQO and other agonists (data not shown). We found in this model that FADD was not required for the MMQO activation of latent HIV, arguing against this hypothesis.
MMQO synergizes with the class 1 and 2 hits initially identified in our screening. On the other hand, class 1 and 2 hits have additive effects at low doses but interfere with each other at higher concentrations. When the three compounds are combined, hit 4 (class 1) is a potent inhibitor of the synergism between MMQO and hit 1 (class 2). Our data suggest that MMQO and class 1 hits act through the same target, but class 2 compounds function on a different pathway or on a different component of the same pathway. Alternatively, all compounds could act on the same pathway, but its overstimulation could render the pathway inactive and leave the HIV-1 promoter repressed.
To improve the reactivating activity of MMQO-derived molecules, we performed a second virtual screening to identify compounds pharmacophorically similar to MMQO. From the list of suggested molecules, we purchased and tested a new set of 15 compounds (Chembridge). A new compound able to reactivate HIV-1 to levels similar to that of MMQO was obtained, 6-methoxy-2-methylquinolin-4-amine (MMQA) (see Fig. S2 in the supplemental material). Its structure was a new variation, different from the class 1 quinolin-8-ol and the MMQO quinolin-4-ol, and it partially resembles the AV6 compound recently reported elsewhere (55). Due to the structural coincidence between MMQA and the quinolinamine moiety of AV6, we cannot rule out the possibility that MMQA acts on a different pathway than that of MMQO. As MMQA showed significant cell toxicity, it was ruled out as a possibility in this study, and we further focused on MMQO.
HIV-1 latency has been associated with rare proviral integration at genome regions unfavorable for promoter accessibility and transcription initiation, such as centromeric heterochromatin, gene deserts, or introns of very highly expressed genes (35, 38, 49). Interestingly, MMQO and the initial hits could reactivate latent HIV-1 in different J-Lat clones representing these diverse genome environments. This indicates that the targeted pathway is generally involved in stimulating transcription initiation from the HIV-1 promoter, not affecting particular targets that may differ between integration sites, such as repressive chromatin components associated with centromeric heterochromatin, or with the interfering transcriptional machinery traveling over the proviral promoter while reading a host gene. Other chromatin-related enzymes or components may be repressive in most integration sites, such as HDACs. Nonetheless, our data suggest that MMQO is not acting as an HDAC inhibitor. J-Lat clones generated with a full-length recombinant HIV-1 vector contain proviruses heavily repressed by DNA methylation (5, 41). Reactivation by agents such as TNF-α or MMQO required cotreatment with DNA methylation inhibitors such as 5-azadC.
HIV-1 reactivation by MMQO also occurred to a different extent in the U1 and ACH2 models of HIV latency, harboring proviruses with mutations in their Tat-TAR transcriptional axis, and MMQO showed an additive or synergistic effect with inducing treatments, such as TNF-α plus 5-azadC. Finally, MMQO was also able to induce HIV-1 expression in blood cells obtained from some of the infected patients analyzed. Considering the extremely low proportion of latently infected cells in PBMC (estimated to be ≤10−6 in lymphocytes) (13), the lack of response in some of the patients may be due to stochastic effects. Taken together, these findings show that MMQO is able to reactivate latent HIV-1 from diverse environments and in different models obtained with minimal or complete viral genomes.
The discovery of MMQO as a new inducer expands the repertoire of available strategies to be combined with standard ART to reduce the reservoir of latent HIV-1. Further studies are required to identify the precise target of the agents described here, and their capacity to reactivate HIV-1 may be improved by combining computational and chemical approaches. After additional cellular models of HIV latency have been tested, preclinical trials could be undertaken to further assess the viability of a therapy based on MMQO combined with physiological or pharmacological TCR/CD3 stimulation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Fundación para la Investigación y Prevención del SIDA en España (FIPSE; 36602/06 and 360946/10), Fundación Eugenio Rodríguez Pascual, Instituto de Salud Carlos III-Fondo de Investigación Sanitaria (PI05/1831), Ministerio de Ciencia e Innovación (MICINN) and FEDER (BFU2008-00359/BMC), and the Generalitat de Catalunya (2009-SGR-1222) to A.J.; grant BIO2008-02329 to J.M.; and grants SAF2010-19292, FIPSE 36710/08, and ISCIII-RETIC RD06/006 to E.M. E.G. was the recipient of a fellowship from the Generalitat de Catalunya and was also supported by intramural resources from the Centre de Regulació Genòmica and Institut de Biologia Molecular de Barcelona-CSIC. J.-M.T. was the recipient of a fellowship from the Fondation pour la Recherche Medicale and a JAE-Doc contract from CSIC-MICINN.
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Archin NM, et al. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retrovir. 25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Areias FM, et al. 2010. In silico directed chemical probing of the adenosine receptor family. Bioorg. Med. Chem. 18:3043–3052 [DOI] [PubMed] [Google Scholar]
- 3. Bisgrove D, Lewinski M, Bushman F, Verdin E. 2005. Molecular mechanisms of HIV-1 proviral latency. Expert Rev. Anti Infect. Ther. 3:805–814 [DOI] [PubMed] [Google Scholar]
- 4. Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U. S. A. 104:13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blazkova J, et al. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brooks DG, Arlen PA, Gao L, Kitchen CM, Zack JA. 2003. Identification of T cell-signaling pathways that stimulate latent HIV in primary cells. Proc. Natl. Acad. Sci. U. S. A. 100:12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brooks DG, et al. 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19:413–423 [DOI] [PubMed] [Google Scholar]
- 9. Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7:459–464 [DOI] [PubMed] [Google Scholar]
- 10. Burke B, et al. 2007. Primary cell model for activation-inducible human immunodeficiency virus. J. Virol. 81:7424–7434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan JK, Greene WC. 2011. NF-kappaB/Rel: agonist and antagonist roles in HIV-1 latency. Curr. Opin. HIV AIDS 6:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi BS, et al. 2010. Novel histone deacetylase inhibitors CG05 and CG06 effectively reactivate latently infected HIV-1. AIDS 24:609–611 [DOI] [PubMed] [Google Scholar]
- 13. Chun TW, et al. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188 [DOI] [PubMed] [Google Scholar]
- 14. Chun TW, Engel D, Mizell SB, Ehler LA, Fauci AS. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chun TW, et al. 1999. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 5:651–655 [DOI] [PubMed] [Google Scholar]
- 16. Chun TW, Fauci AS. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl. Acad. Sci. U. S. A. 96:10958–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7:798–812 [DOI] [PubMed] [Google Scholar]
- 18. Colin L, Van Lint C. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Contreras X, Barboric M, Lenasi T, Peterlin BM. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3:1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Contreras X, et al. 2009. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Daoubi M, et al. 2007. Isolation of new phenylacetylingol derivatives that reactivate HIV-1 latency and a novel spirotriterpenoid from Euphorbia officinarum latex. Bioorg. Med. Chem. 15:4577–4584 [DOI] [PubMed] [Google Scholar]
- 22. De Marco A, et al. 2008. Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 5:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dieudonne M, et al. 2009. Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery. EMBO J. 28:2231–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. du Chene I, et al. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26:424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ekins S, Mestres J, Testa B. 2007. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br. J. Pharmacol. 152:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emiliani S, et al. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emiliani S, et al. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. U. S. A. 93:6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finzi D, et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517 [DOI] [PubMed] [Google Scholar]
- 29. Fraser C, et al. 2000. Reduction of the HIV-1-infected T-cell reservoir by immune activation treatment is dose-dependent and restricted by the potency of antiretroviral drugs. AIDS 14:659–669 [DOI] [PubMed] [Google Scholar]
- 30. Friedman J, et al. 2011. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85:9078–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gallastegui E, Millan-Zambrano G, Terme JM, Chavez S, Jordan A. 2011. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 85:3187–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gloire G, Legrand-Poels S, Piette J. 2006. NF-kappaB activation by reactive oxygen species: fifteen years later. Biochem. Pharmacol. 72:1493–1505 [DOI] [PubMed] [Google Scholar]
- 33. Greger IH, Demarchi F, Giacca M, Proudfoot NJ. 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 26:1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gregori-Puigjane E, Mestres J. 2006. SHED: Shannon entropy descriptors from topological feature distributions. J. Chem. Infect. Model. 46:1615–1622 [DOI] [PubMed] [Google Scholar]
- 35. Han Y, et al. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hofmann TG, et al. 2001. CD95-induced JNK activation signals are transmitted by the death-inducing signaling complex (DISC), but not by Daxx. Int. J. Cancer 93:185–191 [DOI] [PubMed] [Google Scholar]
- 37. Imai K, Togami H, Okamoto T. 2010. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 285:16538–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jordan A, Defechereux P, Verdin E. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karn J. 1999. Tackling Tat. J. Mol. Biol. 293:235–254 [DOI] [PubMed] [Google Scholar]
- 41. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410:896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kulkosky J, et al. 2004. Expression of latent HAART-persistent HIV type 1 induced by novel cellular activating agents. AIDS Res. Hum. Retrovir. 20:497–505 [DOI] [PubMed] [Google Scholar]
- 44. Kumar S, et al. 1996. Activation of the HIV-1 long terminal repeat by cytokines and environmental stress requires an active CSBP/p38 MAP kinase. J. Biol. Chem. 271:30864–30869 [DOI] [PubMed] [Google Scholar]
- 45. Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10:525–531 [DOI] [PubMed] [Google Scholar]
- 46. Lehrman G, et al. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lehrman G, Ylisastigui L, Bosch RJ, Margolis DM. 2004. Interleukin-7 induces HIV type 1 outgrowth from peripheral resting CD4+ T cells. J. Acquir. Immune Defic. Syndr. 36:1103–1104 [DOI] [PubMed] [Google Scholar]
- 48. Lenasi T, Contreras X, Peterlin BM. 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lewinski MK, et al. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lusic M, Marcello A, Cereseto A, Giacca M. 2003. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 22:6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marini A, Harper JM, Romerio F. 2008. An in vitro system to model the establishment and reactivation of HIV-1 latency. J. Immunol. 181:7713–7720 [DOI] [PubMed] [Google Scholar]
- 52. Marquez N, et al. 2008. Differential effects of phorbol-13-monoesters on human immunodeficiency virus reactivation. Biochem. Pharmacol. 75:1370–1380 [DOI] [PubMed] [Google Scholar]
- 53. Matalon S, et al. 2010. The histone deacetylase inhibitor ITF2357 decreases surface CXCR4 and CCR5 expression on CD4(+) T-cells and monocytes and is superior to valproic acid for latent HIV-1 expression in vitro. J. Acquir. Immune Defic. Syndr. 54:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Medina-Ramirez M, et al. 2011. Broadly cross-neutralizing antibodies in HIV-1 patients with undetectable viremia. J. Virol. 85:5804–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Micheva-Viteva S, et al. 2011. High-throughput screening uncovers a compound that activates latent HIV-1 and acts cooperatively with a histone deacetylase (HDAC) inhibitor. J. Biol. Chem. 286:21083–21091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paulsen M, et al. 2011. Modulation of CD4+ T-cell activation by CD95 co-stimulation. Cell Death Differ. 18:619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Perez M, et al. 2010. Bryostatin-1 synergizes with histone deacetylase inhibitors to reactivate HIV-1 from latency. Curr. HIV. Res. 8:418–429 [DOI] [PubMed] [Google Scholar]
- 58. Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297–305 [DOI] [PubMed] [Google Scholar]
- 59. Pion M, et al. 2003. Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. J. Virol. 77:4025–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Poli G, Kinter AL, Fauci AS. 1994. Interleukin 1 induces expression of the human immunodeficiency virus alone and in synergy with interleukin 6 in chronically infected U1 cells: inhibition of inductive effects by the interleukin 1 receptor antagonist. Proc. Natl. Acad. Sci. U. S. A. 91:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Quivy V, De Walque S, Van Lint C. 2007. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell. Biochem. 41:371–396 [PubMed] [Google Scholar]
- 62. Reuse S, et al. 2009. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One 4:e6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Richards J, et al. 1999. Phase I evaluation of humanized OKT3: toxicity and immunomodulatory effects of hOKT3gamma4. Cancer Res. 59:2096–2101 [PubMed] [Google Scholar]
- 64. Sadowski I, Lourenco P, Malcolm T. 2008. Factors controlling chromatin organization and nucleosome positioning for establishment and maintenance of HIV latency. Curr. HIV Res. 6:286–295 [DOI] [PubMed] [Google Scholar]
- 65. Sagot-Lerolle N, et al. 2008. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS 22:1125–1129 [DOI] [PubMed] [Google Scholar]
- 66. Savarino A, et al. 2009. “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology 6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schreck R, Rieber P, Baeuerle PA. 1991. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 10:2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shehu-Xhilaga M, et al. 2009. The novel histone deacetylase inhibitors metacept-1 and metacept-3 potently increase HIV-1 transcription in latently infected cells. AIDS 23:2047–2050 [DOI] [PubMed] [Google Scholar]
- 69. Siliciano JD, et al. 2007. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J. Infect. Dis. 195:833–836 [DOI] [PubMed] [Google Scholar]
- 70. Stanley SK, Folks TM, Fauci AS. 1989. Induction of expression of human immunodeficiency virus in a chronically infected promonocytic cell line by ultraviolet irradiation. AIDS Res. Hum. Retrovir. 5:375–384 [DOI] [PubMed] [Google Scholar]
- 71. Swiggard WJ, et al. 2005. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J. Virol. 79:14179–14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Trono D, et al. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174–180 [DOI] [PubMed] [Google Scholar]
- 73. Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Van Lint C, Emiliani S, Ott M, Verdin E. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 75. Verdin E, Paras P, Jr., Van Lint C. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vicenzi E, Poli G. 1994. Ultraviolet irradiation and cytokines as regulators of HIV latency and expression. Chem. Biol. Interact. 91:101–109 [DOI] [PubMed] [Google Scholar]
- 77. Vidal D, Garcia-Serna R, Mestres J. 2011. Ligand-based approaches to in silico pharmacology. Methods Mol. Biol. 672:489–502 [DOI] [PubMed] [Google Scholar]
- 78. Wang FX, et al. 2005. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J. Clin. Investig. 115:128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Williams SA, et al. 2004. Prostratin antagonizes HIV latency by activating NF-kappaB. J. Biol. Chem. 279:42008–42017 [DOI] [PubMed] [Google Scholar]
- 80. Williams SA, et al. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Williams SA, Kwon H, Chen LF, Greene WC. 2007. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 81:6043–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang HC, et al. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Investig. 119:3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ying H, Zhang Y, Lin S, Han Y, Zhu HZ. 2010. Histone deacetylase inhibitor Scriptaid reactivates latent HIV-1 promoter by inducing histone modification in in vitro latency cell lines. Int. J. Mol. Med. 26:265–272 [DOI] [PubMed] [Google Scholar]
- 84. Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:1101–1108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.