Abstract
The largest tegument protein of herpes simplex virus type 1 (HSV1), pUL36, is a multivalent cross-linker between the viral capsids and the tegument and associated membrane proteins during assembly that upon subsequent cell entry releases the incoming capsids from the outer tegument and viral envelope. Here we show that pUL36 was recruited to cytosolic progeny capsids that later colocalized with membrane proteins of herpes simplex virus type 1 (HSV1) and the trans-Golgi network. During cell entry, pUL36 dissociated from viral membrane proteins but remained associated with cytosolic capsids until arrival at the nucleus. HSV1 UL36 mutants lacking C-terminal portions of increasing size expressed truncated pUL36 but could not form plaques. Cytosolic capsids of mutants lacking the C-terminal 735 of the 3,164 amino acid residues accumulated in the cytosol but did not recruit pUL36 or associate with membranes. In contrast, pUL36 lacking only the 167 C-terminal residues bound to cytosolic capsids and subsequently colocalized with viral and host membrane proteins. Progeny virions fused with neighboring cells, but incoming capsids did not retain pUL36, nor could they target the nucleus or initiate HSV1 gene expression. Our data suggest that residues 2430 to 2893 of HSV1 pUL36, containing one binding site for the capsid protein pUL25, are sufficient to recruit pUL36 onto cytosolic capsids during assembly for secondary envelopment, whereas the 167 residues of the very C terminus with the second pUL25 binding site are crucial to maintain pUL36 on incoming capsids during cell entry. Capsids lacking pUL36 are targeted neither to membranes for virus assembly nor to nuclear pores for genome uncoating.
INTRODUCTION
Infections with herpes simplex virus type 1 (HSV1; human alphaherpesvirus 1) cause the common herpes labialis, herpes keratitis, and keratoconjunctivitis, as well as life-threatening neonatal infections, herpes encephalitis in patients with primary immune deficiencies, and eczema herpeticum in patients with atopic dermatitis (46, 54, 101, 102). The virions contain the DNA genomes of 152 kb encased in icosahedral capsids that interact with the surrounding tegument; this protein layer consists of a partially icosahedrally ordered inner portion and a less organized outer portion that connects to the viral lipid envelope (42, 88, 101, 118). HSV1 packages up to 26 different tegument proteins that have been grouped into inner and outer tegument on the basis of their preferred association with capsids or membranes during assembly and entry as well as their fractionation behavior during virion lysis (40, 60, 62, 68, 75, 96, 116).
Herpesvirus morphogenesis commences in the nucleus, where preassembled capsids package newly synthesized viral genomes (12, 33, 47, 75). According to the most widely accepted secondary reenvelopment model, nuclear capsids traverse the nuclear membranes by primary envelopment at the inner nuclear membrane and primary fusion with the membranes of the endoplasmic reticulum to enter the cytosol. Inner tegument proteins may bind to nuclear or cytosolic capsids, while outer tegument proteins can associate with the cytosolic tails of viral membrane proteins that are targeted to the sites of secondary envelopment containing marker proteins of the trans-Golgi network (TGN) as well as of early or late endosomes (9, 11, 20, 34, 39, 43, 86, 93, 96, 97, 109, 112, 115). Partially tegumented capsids may then travel to these cytoplasmic membranes, and interactions between inner and outer teguments could mediate secondary envelopment, giving rise to enveloped virions enclosed by a host membrane. In contrast, the luminal single-envelopment model proposes that all tegument proteins bind to nuclear capsids prior to their final envelopment at the inner nuclear membrane (12, 33, 48, 58). Common to both scenarios is the formation of large secretory vesicles that contain fully assembled virions and that move to the cell periphery, where their fusion with the plasma membrane releases virions from infected cells (14, 39, 58, 79, 87, 97, 99).
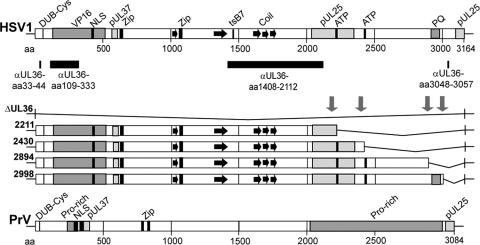
The HSV1 open reading frame UL36 encodes pUL36, an essential inner tegument protein of 3,164 amino acid (aa) residues (Fig. 1) that is evolutionarily conserved among all herpesviruses (71, 72). With its N-terminal binding sites for the tegument proteins VP16 and pUL37 and its C-terminal binding sites for pUL25, it serves as a reversible, multivalent cross-linker between the capsid and the tegument during assembly (16, 52, 53, 74, 98, 114). pUL25 is a minor structural protein located on the capsid surface that is required for the stable retention of progeny viral genomes within the capsids, nuclear egress, and the release of incoming genomes at the nuclear pores (17, 70, 92, 95, 96, 100, 108, 111). In cells infected with HSV1 ΔUL36 or a mutant encoding only the first 361 aa of pUL36, capsids reach the cytosol, but there is no secondary envelopment, no cell egress, and no plaque formation (25, 98). The same phenotype has been reported for the UL36 homolog of pseudorabiesvirus (PrV) (Fig. 1), an alphaherpesvirus of swine (36). However, PrV ΔUL36 with an additional green fluorescent protein (GFP) tag on the small capsid protein VP26 is already impaired in nuclear egress (63). Thus, the combination of a tag on VP26 with a deletion of pUL36 reduces, at least in PrV, nuclear capsid egress (63), whereas deletion of only pUL36 does not seem to influence capsid translocation into the cytoplasm (36). PrV pUL36 versions lacking only 144 aa of the very C terminus or containing insertional transposon mutations in the center also do not replicate, indicating the existence of functional domains between aa 1540 and 1987, while the large internal portion of aa 2087 to 2795 of PrV pUL36 is dispensable, at least in cell culture (8, 56, 80).
Fig 1.
Primary sequence of HSV1 pUL36 and HSV1 UL36 mutants. The N-terminal third of HSV1 pUL36 harbors deubiquitinase and deneddylase activity with a conserved Cys at aa residue 65 (DUB-Cys), an NLS (aa 367 to 498), and binding sites for the major tegument protein VP16 and the inner tegument protein pUL37 (2, 37, 51, 53, 77, 114). The tsB7 mutation that prevents DNA release from incoming capsids at the nuclear pores has been mapped to aa 1453 in the middle of pUL36 (1). Furthermore, two potential leucine zipper motifs (Zip) at aa 632 to 653 and aa 1070 to 1091 and several coiled coils (black arrows) have been identified by in silico analysis (6, 61, 71, 103). Within the C-terminal third of HSV1 pUL36, there are two potential ATP binding motifs between aa 2211 to 2219 and 2430 to 2443, a prominent repeat of 35 PQ between aa 2911 to 2980, and two binding sites for pUL25 between aa 2037 to 2352 and 3102 to 3164 (16, 71, 92). The pUL36 regions against which polyclonal antibodies have been raised are indicated by black bars: a peptide of aa 33 to 44 at the N terminus (Nterm, pAb R28), recombinant protein fragments of aa 109 to 333 (pAb VP1-2 N) or aa 1408 to 2112 (middle, pAb 146 and pAb 147), and a peptide of aa 3048 to 3057 at the C terminus (Cterm, pAb R29) (49, 71, 116). Using the BAC pHSV1(17+)blueLox as a parental wild-type background (86), we have constructed HSV1(17+)blueLox-ΔUL36 lacking the entire UL36 open reading frame or inserted stop codons together with a kanamycin resistance cassette (downward-pointing arrows) upstream of the two potential ATP binding sites or up- and downstream of the prominent PQ repeat to generate the mutants HSV1(17+)blueLox-UL36codon2211stop, -2430stop, -2894stop, and -2998stop that express C-terminally truncated versions of pUL36. Similarly, PrV pUL36 also has a DUB-Cys at position 26, two NLSs, and a PrV pUL37 binding region, predicted leucine zippers at aa 779 to 800 and 827 to 848, several Pro-rich regions between aa 226 to 299 and 2026 to 3970, and a PrV pUL25 binding region at the very C terminus (8, 16, 51, 52, 56, 57, 81).
Extracellular mature alphaherpesvirus virions contain pUL36, hence, its other names viral protein 1/2 and 1-3 (52, 107), but it is unclear when and how pUL36 associates with viral particles during assembly. So far, the subcellular localization of pUL36 has been determined only using strains in which either pUL36 or the small capsid protein VP26 had been tagged with a fluorescent protein. Furthermore, infected cells contain several forms of pUL36 of lower molecular weight (36, 49, 51, 52) that may be targeted to distinct viral and host structures. Antibodies generated against full-length HSV1 pUL36 or its N-terminal aa 1 to 287 label the nucleus as well as the cytoplasm of infected cells (2, 72), whereas antibodies generated against recombinant fragments derived from the entire PrV pUL36, including the N terminus, do not yield nuclear signals but prominently yield cytoplasmic signals (52, 81). Upon transient expression, HSV1 pUL36 and PrV pUL36 are detected in the nucleus, and eliminating their nuclear localization signals (NLSs) abrogates this nuclear targeting (2, 56, 81).
HSV1 virions enter cells by fusion of their envelope with a host membrane, thereby exposing capsids and tegument proteins to the cytosol (38, 45, 68, 78, 89, 106). The motor protein dynein then propels the cytosolic capsids along microtubules toward the nucleus, where they dock with one capsid corner facing the nuclear pore and release the viral genome into the nucleoplasm for viral transcription and replication (27, 40, 41, 64, 65, 67, 90, 100, 101, 106). At least some HSV1 and PrV pUL36 remains associated with incoming capsids until arrival at the nucleus (4, 18, 40, 62, 81, 83). Inner HSV1 tegument proteins such as pUL36 or pUL37 are required for recruiting microtubule motors onto viral capsids, for capsid transport along microtubules in vitro, and for binding to nuclear pores in vitro (90, 96, 104, 116). HSV1(HFEM)-tsB7 and HSV1(17+)-ts1213 with temperature-sensitive mutations in pUL36 still dock at the nuclear pores but do not release their genomes at the nonpermissive temperature, and HSV1 gene expression requires proteolysis of the incoming pUL36 (1, 5, 49, 94). Furthermore, antibodies against pUL36 that have been introduced into cells reduce the number of incoming capsids targeted to the nucleus, and capsids lacking pUL36 introduced into uninfected cells cannot initiate an infection (18, 98).
Here we show that the authentic, untagged HSV1 pUL36 was present on cytosolic capsids after their nuclear egress but prior to secondary envelopment and remained capsid associated upon cell entry and virus fusion until arrival at the nuclear pore. New HSV1 UL36 mutant strains revealed that even the very C-terminal 167 aa of the large tegument protein pUL36 were required for virus propagation and plaque formation. Our data suggest that the residues 2430 to 2893 of HSV1 pUL36, containing one binding site for the capsid protein pUL25 (92), are sufficient to recruit pUL36 onto cytosolic capsids for secondary envelopment. In contrast, the very C terminus with the second pUL25 binding site (16, 92) is crucial to stabilize pUL36 on incoming capsids. Thus, several C-terminal domains are required to maintain pUL36 on incoming capsids during cell entry, whereas the C-terminal 271 aa are dispensable for the association of pUL36 with cytosolic progeny capsids. Our results show that capsids lacking pUL36 are targeted neither to membranes for virus assembly nor to the nuclear pores for genome uncoating.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells (ATCC CCL-10) were grown in minimal essential medium (MEM) containing 10% (vol/vol) fetal calf serum (FCS; Life Technologies Gibco, Darmstadt, Germany), Vero cells (ATCC CCL-81) in MEM containing 7.5% FCS, and the Vero cell-derived pUL36-complementing cell line HS30 (25) in MEM containing 7.5% FCS; and in every fifth passage, 500 μg/ml G418 was added (PAA Laboratories GmbH, Pasching, Austria). HSV1 strain 17+ (from John Subak-Sharpe, MRC Virology Unit, Glasgow, United Kingdom) and HSV1 strain KOS (ATCC VR-1493; from Pat Spear, Northwestern University, Chicago, IL) were propagated in BHK-21 cells and prepared as described previously (106). For assembly experiments, we used extracellular virus pelleted from the medium of infected cells (86), while for cell entry experiments, we further purified the sedimented virions on linear Nycodenz gradients (26).
Virus stocks were plaque titrated in three independent cycles (27, 106). To determine the ratio of HSV1 UL36 potential mutant to potential revertant virions, the virus stocks of the HSV1 UL36 mutants were titrated on Vero-HS30 cells, which complemented UL36 in trans (25), and on Vero cells. Wild-type (wt) HSV1 inocula had genome-to-PFU ratios of less than 40, indicative of a low concentration of defective viral particles (26). The bacterial artificial chromosome (BAC)-derived medium pellets that were used to inoculate cells had genome-to-PFU ratios of about 200 and thus were comparable to each other.
Antibodies.
To detect pUL36, we used the rabbit polyclonal antibodies (pAbs) R28 (Nterm) raised against the N-terminal peptide aa 33 to 44 (71), pAb VP1-2 N raised against a recombinant fragment of aa 109 to 333 (49), pAb 146 and pAb 147 raised against a recombinant fragment of aa 1408 to 2112 (116), and pAb R29 (Cterm) raised against the C-terminal fragment from aa 3048 to 3057 (71). To detect VP5, we used the rabbit pAb NC-1 (15) and the mouse monoclonal antibody (MAb) H1.4 (Meridian Life Science, Saco, ME) or 5C10 (110). VP19c and VP26 were labeled by rabbit pAb NC2 (15) and a pAb against aa 95 to 112 of VP26 (24), respectively, VP16 was labeled by mouse MAb LP1 (69) or by rabbit pAb 230 (83), and VP22 was labeled by rabbit pAb AGV30 (32). Glycoprotein B (gB) and gD were detected using rabbit pAb R68 and mouse MAb DL6, respectively (29, 30). Actin was labeled using the mouse MAb 1501 (Millipore, Billerica, MA) or tetramethyl rhodamine isocyanate (TRITC)-phalloidin (1 μg/ml in phosphate-buffered saline [PBS]; Sigma-Aldrich, Schnelldorf, Germany). We also used mouse MAb 414 (Covance, Princeton, NJ) to detect nuclear pores as well as MAb α-p230 and MAb EEA1 to label specific host membranes (BD Transduction Laboratories, Lexington, KY). The goat-derived secondary antibodies for fluorescence microscopy were conjugated with lissamine-rhodamine B sulfonyl chloride (LRSC) or with fluorescein isothiocyanate (FITC) and highly preabsorbed against cross-reactivity to species other than the intended one (rabbit or mouse; Dianova, Hamburg, Germany), or for immunoblotting, they were coupled with alkaline phosphatase (Thermo Scientific, Bonn, Germany).
Construction of HSV1 UL36 mutants.
The parental BAC pHSV1(17+)blueLox contains the entire HSV1 genome, together with a β-galactosidase expression cassette integrated into the thymidine kinase locus, as well as the genes required for low-copy replication in Escherichia coli flanked by loxP sites and a eukaryotic cre recombinase expression cassette (86). We amplified a kanamycin resistance cassette flanked by FLP recombination target (FRT) sites by PCR from the plasmid pGP704-Kan (7). The primers contained a homology to the pGP704-Kan plasmid as well as a homology to UL36 that defined the insertion site of the fragment. For the pUL36 truncation mutants, one primer also encoded the stop codon TAG (see Table S1 in the supplemental material). The resulting PCR products were transformed into E. coli DH10B harboring pHSV1(17+)blueLox and pKD46 (22) and which expressed the RED recombination enzymes upon induction (19, 117). We used seven different restriction digests and in-house sequencing of the mutated regions to analyze kanamycin resistant recombinants.
To reconstitute viruses, BAC DNA was prepared using a NucleoBond BAC 100 kit (Macherey & Nagel, Düren, Germany). Subconfluent Vero or HS30 cells were transfected with 10 μg per 60-mm dish BAC DNA (MBS mammalian transfection kit; Stratagene, La Jolla, CA) and grown until cytopathic effects developed. Cells and medium were combined, and virus was released by three cycles of freeze-thawing. About a tenth of such a passage 1 (P1) was used to infect one 175-cm2 flask of subconfluent Vero or HS30 cells until 60 to 70% of the cells had rounded up. Virions from this P2 were pelleted from the culture medium, resuspended in buffer (100 mM NaCl, 30 mM morpholineethanesulfonic acid, 20 mM Tris-HCl, pH 7.4), and stored in aliquots at −80°C as described previously (106). HSV1 UL36 mutants, transcomplemented by growth in HS30 cells but not in the parental Vero cells, contained the full-length pUL36 in their teguments but harbored the different pUL36 mutations in their genomes.
Immunoblotting.
Subconfluent Vero cells in 60-mm dishes either were infected at a multiplicity of infection (MOI) of 10 by adding virus-containing medium (see Fig. 2A) or were precooled for 20 min on ice and inoculated for 2 h on ice in CO2-independent medium at an MOI of 10 (see Fig. 2B and 7) or 200 (see Fig. 5) PFU/cell. After removing unbound virus, the cells were transferred to regular growth medium at 37°C in 5% CO2. At the indicated times, the cells, including those that had already detached from the substrate, were washed with PBS and collected. They were lysed in hot sample buffer (50 mM Tris-HCl, pH 6.8, 1% [wt/vol] SDS, 1% [vol/vol] β-mercaptoethanol, 5% [vol/vol] glycerol, 0.001% [wt/vol] bromphenol blue) with protease inhibitors aprotinin, E-64, and leupeptin (AEL; Sigma-Aldrich) in H2O, antipain, bestatin, and pepstatin (ABP; Sigma-Aldrich) in methanol, and phenylmethylsulfonyl fluoride (PMSF; Roth) in isopropanol. After SDS-PAGE in linear gradient gels (5 to 15% polyacrylamide, 8-cm length, 0.75-mm thickness or 7.5 to 18% polyacrylamide, 16-cm length, 1.5-mm thickness), the proteins were transferred onto nitrocellulose membranes and incubated with PBS containing 0.1% (vol/vol) Tween 20 (PBS-T). After blocking in 5% (wt/vol) low-fat milk in PBS-T, the membranes were probed with primary antibodies, followed by secondary goat antirabbit or antimouse antibodies conjugated to alkaline phosphatase. The membranes were stained with 0.2 mM nitroblue tetrazolium chloride and 0.8 mM 5-bromo-4-chloro-indolyl-3-phosphate in 100 mM NaCl, 5 mM MgCl2, 100 mM Tris-HCl, pH 9.5, and subsequently scanned with a ScanJet 6300C scanner (Hewlett Packard, Wilmington, DE).
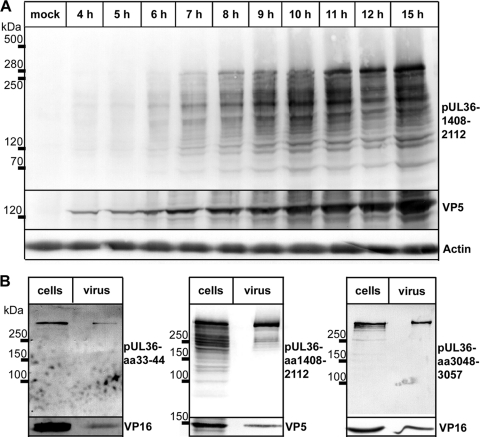
Fig 2.
Packaging of HSV1 pUL36 proteins into virions. (A) Vero cells were infected with HSV1(17+) at an MOI of 10 PFU/cell or mock treated, harvested at the indicated time points, and separated on a linear 7.5 to 18% SDS-polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and probed with antibodies against pUL36 aa 1408 to 2112 (middle, pAb 146), VP5 (pAb NC-1), or actin (MAb 1501). (B) Vero cells infected with HSV1(17+) at an MOI of 10 PFU/cell for 12 h (cells) or gradient purified HSV1(17+) secreted from infected BHK cells and purified on a linear Nycodenz gradient (virus) were lysed; and the proteins were separated on linear 5 to 15% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with antibodies directed against pUL36 aa 33 to 44 (Nterm, pAb R28), pUL36 aa 1408 to 2112 (middle, pAb 146), pUL36 aa 3048 to 3057 (Cterm, pAb R29), VP16 (MAb LP1), or VP5 (MAb H1.4).
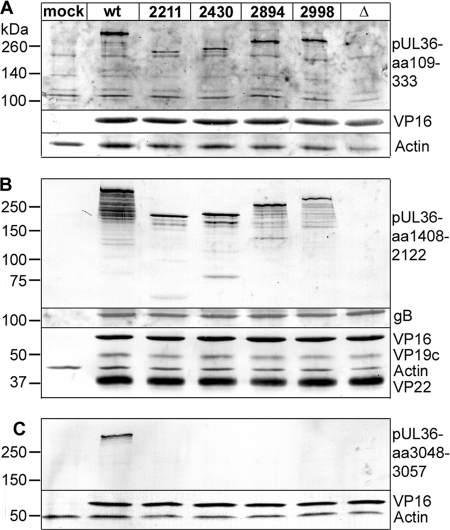
Fig 7.
Synthesis of truncated HSV1 pUL36 proteins. Vero cells were mock treated (mock) or infected at an MOI of 10 PFU/cell with HSV1(17+)blueLox (wt) or with mutant HSV1(17+)blueLox-UL36codon2211stop, -UL36codon2430stop, -UL36codon2894stop, or -UL36codon2998stop or HSV1(17+)blueLox-ΔUL36 (Δ), transcomplemented with pUL36 by amplification on Vero-HS30 cells, and harvested at 10 h p.i. The proteins were separated by linear 5 to 15% gradient SDS-PAGE, transferred to nitrocellulose membranes, and probed with antibodies directed against pUL36 aa 109 to 333 (A; pAb VP1-2 N), pUL36 aa 1408 to 2112 (B; middle, pAb 146), and pUL36 aa 3048 to 3057 (C; Cterm, pAb R29) and, as loading controls, antibodies against VP16 (A to C; MAb LP1), gB (B; pAb R68), VP19c (B; pAb NC2), VP22 (B; pAb AGV30), or actin (A to C; MAb 1501).
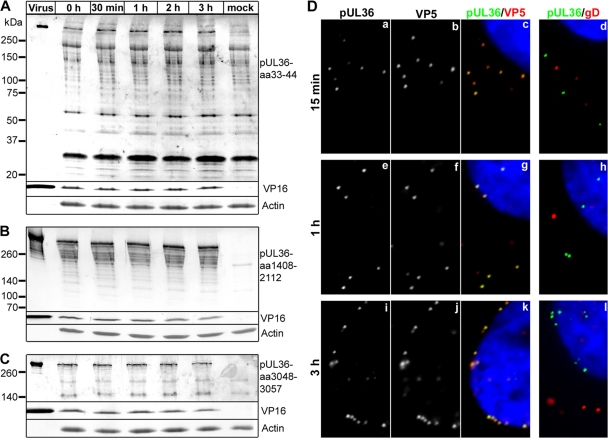
Fig 5.
Full-length pUL36 remains associated with parental, incoming HSV1 capsids. (A to C) Vero cells were either infected with HSV1(17+) at an MOI of 200 PFU/cell in the presence of cycloheximide and harvested at the indicated time points or mock treated for 3 h (mock). Additionally, gradient-purified HSV1(17+) secreted from infected BHK cells and purified on a linear Nycodenz gradient (virus) was loaded. Proteins were separated on linear 5 to 15% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with antibodies directed against pUL36 aa 33 to 44 (A; Nterm, pAb R28), pUL36 aa 1408 to 2112 (B; middle, pAb 146,), pUL36 aa 3048-3057 (C; Cterm, pAb R29), VP16 (MAb LP1), or actin (MAb 1501). (D) Vero cells were infected with HSV1(17+) at an MOI of 50 PFU/cell in the presence of cycloheximide and fixed at the indicated time points with 3% paraformaldehyde, followed by TX-100 permeabilization. The cells were labeled with antibodies directed against pUL36 aa 1408 to 2112 (green and a, e, and i; middle, pAb 147), the major capsid protein VP5 (b, f, and j and red in c, g, and k; MAb 5C10), or the viral membrane protein gD (red in d, h, and l; MAb DL-6) and the nuclei (Hoechst dye; blue in c, d, g, h, k, and l) and analyzed by epifluorescence microscopy.
Light microscopy.
Subconfluent Vero cells grown on coverslips in 24-well dishes were precooled for 20 min on ice and inoculated in 0.2 ml CO2-independent medium containing 0.1% (wt/vol) bovine serum albumin (BSA) with HSV1 for 2 h on ice (26, 103). After removing unbound virus, we transferred the cells to regular growth medium containing FCS at 37°C and 5% CO2. After 1 h, any residual inoculum was inactivated by low-pH treatment (40 mM citrate, pH 3, 135 mM NaCl, 10 mM KCl) for 3 min, followed by incubation in fresh FCS containing growth medium at 37°C (86). For cell entry experiments, 0.5 mM cycloheximide prevented the synthesis of progeny viral proteins (106). In the viral spread experiments, the medium contained 10 μg/ml pooled, human IgGs that contained sufficient amounts of HSV1-neutralizing antibodies (Sigma-Aldrich). The cells were either fixed and permeabilized by incubating them for 4 min in water-free methanol at −20°C (data not shown) or fixed with 3% (wt/vol) paraformaldehyde in PBS for 20 min and subsequently permeabilized with 0.1% Triton X-100 for 5 min (100, 106). The specimens were labeled with antibodies as described before (27, 86, 100, 106). To block unspecific antibody interactions with the abundantly expressed HSV1 Fc receptor (28) and thus to reduce antigen-independent antibody labeling, we used 10% (vol/vol) human serum of a healthy HSV1-seronegative volunteer and 0.5% (wt/vol) BSA in PBS as blocking reagent (26). The DNA was stained using Hoechst 33258 (Invitrogen) or TO-PRO-3 (Invitrogen).
For assembly and spread experiments, we recorded optical confocal sections using an LSM 510 Meta microscope with a Plan-Apochromat ×63 objective and argon (488 nm) and helium-neon (543 nm, 633 nm) lasers controlled by the LSM 510 software (Zeiss, Göttingen, Germany). Cell entry experiments were analyzed with an inverted AxioObserver Z1 microscope equipped with a Plan-Apochromat ×63 objective, a mercury lamp, appropriate fluorescence filter sets, and an Axiocam HRm camera controlled by the Axiovision software (version 4.6.3.0 SP1; Zeiss). Digital images were processed with the LSM Image Browser (version 3.5.0.376, 2005; Zeiss), ImageJ (version 1.35j; Wayne Rasband, NIH [http://rsb.info.nih.gov/ij/]), and Adobe Photoshop CS (version 7.0; Adobe Systems, San Jose, CA) programs.
To quantify any colocalizations, we used the CellProfiler software (13). For VP5 and pUL36 colocalization (see Fig. 4), we analyzed 5 to 14 cells per time point. All pixel intensities were rescaled to 0 to 1. Using the Otsu Global thresholding method (91) in the TO-PRO-3 channel, the nuclear area was defined and the apparent nuclear VP5 signals were erased digitally. Capsids were then identified using the Kapur thresholding method (50). pUL36 fluorescence intensities were measured in those areas overlapping with the capsid signal. To determine the amount of pUL36 that was not recruited to capsids, the pUL36 fluorescence intensities were measured in manually defined cytoplasmic areas after subtracting the areas occupied by capsids (diffuse cytoplasmic signal). Capsids with a pUL36 fluorescence larger than the average diffuse cytoplasmic signal were scored as pUL36 positive. For VP26 and p230 colocalizations (see Fig. 9C), we randomly documented 60 cells derived from 2 independent experiments. Prior to object segmentation, each image was rescaled to 100 by 100 mm at 300 pixels/inch. To segment the viral capsids from the background staining of the anti-VP26 antibody, we converted the gray values from 0, 1.00, and 255 to 60, 1.00, and 255, and to segment the p230 signal, we converted the gray values from 0, 1.00, and 255 to 60, 1.00, and 170 using the “Levels” function of Adobe Photoshop. After defining the nuclear areas using differential interference contrast, the apparent nuclear capsid signals were erased digitally. After overlaying these processed images, we determined the number of pixels per cell labeled for both VP26 and p230 to obtain an estimate of the degree of membrane association of the cytoplasmic capsids of the different HSV1 mutant strains in comparison to wild type. Standard errors of the means and statistically significant differences were determined using Mann-Whitney tests (version 5.02; GraphPad Software, Inc.)
Fig 4.
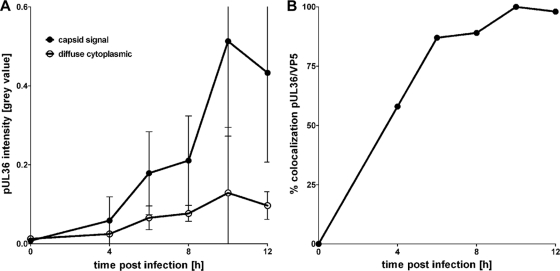
Colocalization of pUL36 with cytoplasmic capsids. Vero cells infected with HSV1(17+) at an MOI of 10 PFU/cell were fixed at 4, 6, 8, 10, or 12 h postinfection and prepared for immunofluorescence using antibodies directed against pUL36 aa 1408 to 2112 (middle, pAb 147) and the major capsid protein VP5 (MAb 5C10) and TO-PRO-3, as for Fig. 3A. (A) For each time point postinfection, the pUL36 fluorescence intensity (gray value) was measured on cytoplasmic capsids (capsid signal) and the diffuse cytoplasmic signal (diffuse cytoplasmic) was determined. The error bars indicate the standard deviation. (B) These data were used to calculate the percentage of cytoplasmic capsids positive for pUL36.
Fig 9.
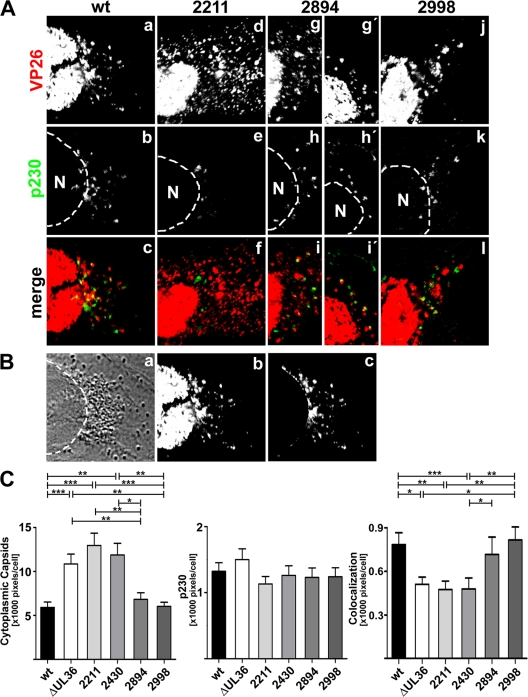
Amino acids 2430 to 2893 of HSV1 pUL36 are required for capsid association with the trans-Golgi network. (A) Vero cells were infected at an MOI of 0.2 PFU/cell with pHSV1(17+)blueLox (wt; a to c) or the mutant HSV1(17+)blueLox-UL36codon2211stop (2211; d to f), -UL36codon2894stop (2894; g to i and g′ to i′), or -UL36codon2998stop (2998; j to l), transcomplemented with pUL36 by amplification on Vero-HS30 cells, and fixed at 15 h p.i. with 3% paraformaldehyde, followed by TX-100 permeabilization. The specimens were labeled with antibodies directed against VP26 (red and a, d, g, g′, and j; pAb) and a TGN marker protein (green and b, e, h, h′, and k; MAb p230) and analyzed by confocal fluorescence microscopy. (B) For quantification, the position of the nuclei was determined by differential interference contrast (a), and the nuclear but not the cytoplasmic VP26 signal (b) was digitally eliminated (c). (C) After infection with pHSV1(17+)blueLox (wt) or the mutants, the average number of cytoplasmic VP26-positive or p230-positive pixels and the degree of apparent colocalization were measured for each virus in 60 cells derived from two independent experiments. The error bars indicate the standard error of the mean. Significant differences according to a Mann-Whitney test are indicated: ***, P < 0.001; **, P = 0.001 to 0.01; *, P = 0.01 to 0.05.
Electron microscopy.
Vero cells were infected synchronously at an MOI of 10 as described for light microscopy. After 12 or 14 h, the cells were fixed for 1 h at room temperature with 2% (wt/vol) glutaraldehyde in 130 mM cacodylate buffer at pH 7.4 containing 2 mM CaCl2 and 10 mM MgCl2. Then, the cells were washed and postfixed with 1% (wt/vol) OsO4 in 165 mM cacodylate buffer at pH 7.4 containing 1.5% (wt/vol) K3Fe(III)CN6 for 1 h, followed by incubation in 0.5% (wt/vol) uranyl acetate in 50% (vol/vol) ethanol overnight. The cells were flat embedded in Epon, and 50-nm sections were cut parallel to the substrate. Images were taken with an FEI Tecnai G2 T20 electron microscope.
RESULTS
pUL36 wild type is recruited onto cytosolic capsids during assembly.
We first analyzed the expression of pUL36 in Vero cells infected with HSV1 at an MOI of 10 PFU/cell by immunoblotting using antibodies raised against the middle aa 1408 to 2112 of HSV1 pUL36 (116). The expression of pUL36 commenced at about 6 h postinfection (p.i.; Fig. 2A). In addition to a major band of more than 250 kDa, infected cells contained several bands of lower apparent molecular mass. The trace amounts detected at 4 h most likely represented pUL36 already present in the inoculum. Antibodies raised against the N-terminal aa 33 to 44 or the C-terminal aa 3048 to 3057 (71) detected the major pUL36 band of more than 250 kDa in gradient-purified virions that therefore represented the full-length pUL36 forms (Fig. 2B). In addition, in particular the antibodies against the middle region aa 1408 to 2112 detected several smaller pUL36 forms in infected cells. In contrast, virions predominantly contained the full-length and two smaller rather prominent bands that displayed middle but no N- or C-terminal pUL36 epitopes.
Next, we analyzed the subcellular localization of pUL36 after synchronous infection for 4 to 12 h with HSV1 strain 17+ at an MOI of 10 PFU/cell after fixation with formaldehyde followed by detergent permeabilization (Fig. 3). As early as 6 h and 8 h p.i., the antibodies raised against the pUL36 middle region showed several flecks that had accumulated in close proximity to the nucleus (Fig. 3Ab and f) and were later also distributed throughout the cytoplasm (data not shown). Double labeling for hexons, using an antibody directed against mature VP5, the major capsid protein, revealed that the cell nuclei contained many capsids but that only a few of them had traversed the nuclear envelope this early in infection (Fig. 3Ac and g). In contrast to nuclear capsids (Fig. 3A, red arrowhead), the majority of the cytoplasmic capsids possessed pUL36 (Fig. 3A, yellow arrowheads). At 8 h p.i., capsids with pUL36 had accumulated in a perinuclear region (Fig. 3Ah, yellow). Using the software CellProfiler (12), we segmented the cytoplasmic capsids based on the anti-VP5 labeling and determined the average pUL36 intensity on the capsids and in cytoplasmic regions excluding the capsids (Fig. 4). The amount of pUL36 present on cytoplasmic capsids continuously increased from 4 to 10 h postinfection (Fig. 4A), and already at 4 h postinfection, more than 50% of the cytoplasmic capsids contained a larger amount of pUL36 than that present in the cytoplasm (Fig. 4B).
Fig 3.
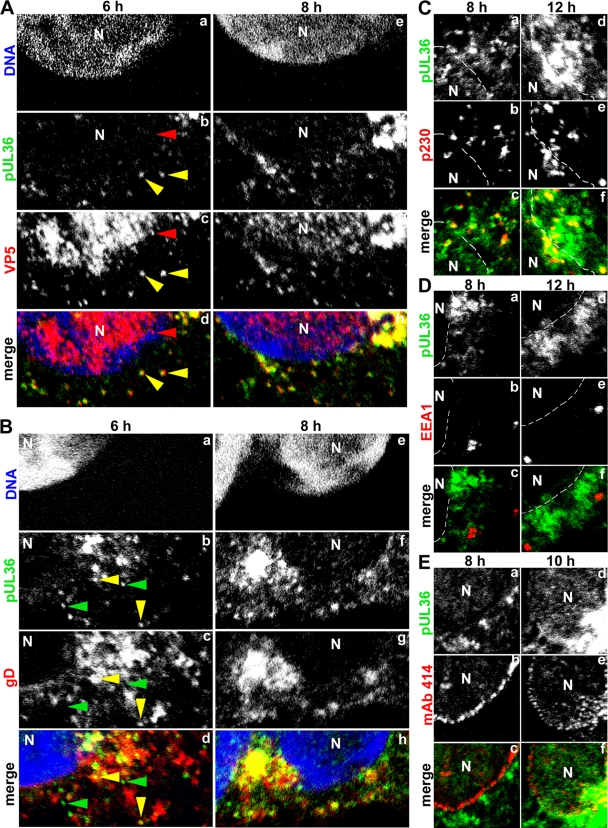
HSV1 pUL36 binds to progeny, cytosolic capsids prior to secondary envelopment. (A) Vero cells were infected with HSV1(17+) at an MOI of 10 PFU/cell and fixed with 3% paraformaldehyde at the indicated time points, followed by TX-100 permeabilization. The specimens were labeled with antibodies directed against pUL36 aa 1408 to 2112 (green; middle, pAb 147), the major capsid protein VP5 (A; red; MAb 5C10), the viral membrane protein gD (B; red; MAb DL-6), a marker for the trans-Golgi network (C; red; MAb α-p230), a marker for early endosomes (D; red; MAb α-EEA1), or the nuclear pores (E; red; MAb 414) and analyzed by confocal fluorescence microscopy. The nuclei (N) were labeled with the DNA stain TO-PRO-3 (blue in panels A and B) and are indicated by dashed lines (C to E).
At 6 h p.i., the HSV1 envelope gD was mainly located in a perinuclear region along with smaller patches in the cytoplasm. The majority of the pUL36 flecks did not colocalize with gD and thus most likely represent unenveloped, cytosolic capsids prior to secondary envelopment and membrane acquisition (Fig. 3B, green arrowheads). However, as the infection progressed to 8 h and further (data not shown), more and more pUL36-positive capsids had acquired gD. Some of these pUL36 particles colocalizing with gD (Fig. 3B, yellow) most likely represent capsids in the process of or after secondary envelopment. These perinuclear pUL36-containing capsids also colocalized to some extent with the host protein p230, a marker of the trans-Golgi network (Fig. 3C, yellow), but pUL36 did not associate with all types of membranes, as indicated by its lack of colocalization with the early endosome marker EEA1 both in the perinuclear region (Fig. 3D, green) (84) and in the cell periphery (data not shown).
Labeling of the chromosomes using TO-PRO-3 (Fig. 3Aa and e and Ba and e) or of the nuclear pores using MAb 414 (Fig. 3Eb and e), which detects nuclear pore proteins with phenylalanine-glycine repeats (23), enabled a distinction between nuclear and cytoplasmic pUL36 labeling. As the infection progressed, pUL36 was also increasingly located in regions enclosed by the nuclear envelope but typically in smaller amounts than in the cytoplasm. It did not accumulate on nuclear capsids but appeared to be mainly diffuse. Since nuclear envelopes have indentations (66), confocal images often contained tangential cross sections of such indentations. Therefore, the apparent nuclear lumen occasionally appeared to contain membrane markers such as gD (Fig. 3B), p230 (Fig. 3C), or nuclear pores (Fig. 3E), but these were located in the cytoplasm above or below the nucleus within the same optical confocal section. Infection experiments using HSV1 strain KOS (data not shown) or strain 17+ and fixation with methanol (data not shown) or formaldehyde resulted in similar labeling patterns.
In summary, these results indicate that the inner tegument protein HSV1 pUL36 was localized on progeny cytosolic capsids lacking membrane proteins. HSV1 pUL36 remained attached to capsids during their transport to membranes that contained gD and host markers of the TGN and where secondary envelopment presumably occurred. As the HSV1 infection progressed, a minor fraction of pUL36 was located in the cell nuclei but not on nuclear capsids.
Wild-type pUL36 remains capsid associated during cell entry.
To determine the subcellular fate of HSV1 pUL36 during cell entry, we inoculated Vero cells at an MOI of 200 PFU/cell and in the presence of cycloheximide, an inhibitor of protein synthesis, to limit our analysis to parental, incoming viral proteins (106). Antibodies directed against the N-terminal aa 33 to 44 (Fig. 5A) or C-terminal aa 3048 to 3057 (Fig. 5C) mainly detected full-length pUL36 of more than 250 kDa, besides several unspecific bands that were also present in uninfected cells (mock treated). In contrast, the antibodies generated against the middle aa 1408 to 2112 detected full-length pUL36 and several smaller pUL36 forms that were also present in the inoculum (Fig. 5B; cf. Fig. 2B). This pattern did not change during the first 3 h of inoculation. To analyze the subcellular localization of incoming pUL36, Vero cells were inoculated in the presence of cycloheximide with an MOI of 50 PFU/cell (Fig. 5D). After 15 min, the parental capsids were randomly distributed over the entire cytoplasm as described before (26, 27, 65, 100, 106), and pUL36 colocalized with capsids (Fig. 5Dc, yellow) but not with gD (Fig. 5Dd, green). While capsids targeted to the nucleus contained pUL36 (Fig. 5Dg and k, yellow), gD remained in more peripheral regions of the cytoplasm (Fig. 5Dh and l, red), as reported before (26, 27, 92). Over time, more capsids arrived at the nucleus, and nearly all of them colocalized with pUL36. Together, these data indicate that during internalization into cells, pUL36 and the viral envelope separated and that pUL36, most likely in its full-length form, remained attached to incoming HSV1 capsids until their arrival at the nuclear pores.
Generation of HSV1 UL36 mutant strains.
To construct HSV1 mutants lacking specific regions of pUL36, we used the BAC pHSV1(17+)blueLox and RED-mediated recombination in E. coli (19, 85, 86, 117). We inserted stop codons and did not delete parts of the UL36 gene in order to minimize modifications in genomic regions potentially involved in other functions. Since we aimed to characterize regions of pUL36 required for its recruitment onto cytosolic capsids, we focused on the C-terminal third of pUL36 that contains binding domains for the capsid protein pUL25 between aa 2037 to 2352 and 3104 to 3164 (16, 92). In silico sequence analysis of pUL36 also indicates potential ATP binding sites, one in the middle and one just downstream of the pUL25 binding region from aa 2037 to 2352, and a striking proline-glutamine repeat located between the two pUL25 binding sites (cf. Fig. 1) (71). We inserted a stop codon together with a resistance cassette flanked by FRT sites upstream of each potential ATP binding site or up- or downstream of the PQ repeat and generated HSV1(17+)blueLox-UL36codon2211stop (2211stop), -UL36codon2430stop (2430stop), -UL36codon2894stop (2894stop), and -UL36codon2998stop (2998stop) (Fig. 1). As an additional control to the parental wild-type HSV1(17+)blueLox, we also replaced the UL36 gene with the resistance cassette and generated HSV1(17+)blueLox-ΔUL36.
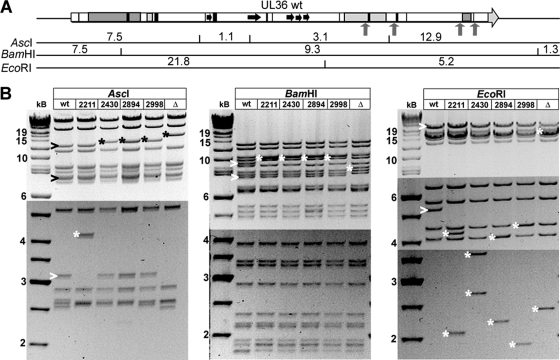
Besides the desired mutations, homologous recombination can change DNA regions rich in repeats, as they are present in many herpesviral genomes (3, 21). We therefore characterized each mutant BAC using seven restriction enzymes diagnostic for the entire HSV1 genome (86). The expected restriction fragment sizes (Fig. 6A) were based on the HSV1(17+) sequence (GenBank accession no. NC_001806). All shifted and other fragments of the mutants were as expected (Fig. 6B). These restriction digests as well as sequencing of PCR fragments spanning the mutated region (data not shown) proved that the resistance cassettes had been correctly integrated into UL36 without any major undesired recombination events elsewhere in the pHSV1(17+)blueLox backbone.
Fig 6.
Generation and characterization of pHSV1(17+)-UL36 mutants. (A) Using the BAC pHSV1(17+)blueLox as parental wild type (86), we have constructed pHSV1(17+)blueLox-ΔUL36 lacking the entire UL36 open reading frame. As further mutants, we have inserted stop codons together with a kanamycin resistance cassette (upward-pointing arrows) upstream of the two potential ATP binding sites or up- and downstream of the prominent PQ repeat and generated the BAC plasmids pHSV1(17+)blueLox-UL36codon2211stop, -UL36codon2430stop, -UL36codon2894stop, and -UL36codon2998stop to generate HSV1 strains expressing C-terminally truncated versions of pUL36. The expected fragment sizes after restriction digestions of the UL36 region with AscI, BamHI, or EcoRI were calculated according to the published HSV1(17+) sequence (GenBank accession no. NC_001806). (B) Agarose gels of restriction endonuclease digests of the parental pHSV1(17+)blueLox (wt); pHSV1(17+)blueLox-UL36codon2211stop, -UL36codon2430stop, -UL36codon2894stop, or -UL36codon2998stop; or pHSV1(17+)blueLox-ΔUL36 (Δ) with AscI, BamHI, or EcoRI. The sizes of the restriction fragments and molecular mass markers are indicated in kilobases. Altered fragments in the parental wt (>) and bands of the mutants shifted due to the insertion of the kanamycin resistance cassette with the stop codon (*) are indicated. The bands of various sizes of about 2.6 kb after AscI digestion most likely reflected the repetitive a-sequences that occur in various numbers in HSV1 genomes (86, 113).
Characterization of HSV1 mutants with C-terminal truncated pUL36 forms.
Transfection of Vero cells with the mutated pHSV1(17+)blueLox did not result in plaque formation, even after 6 days, although the parental pHSV1(17+)blueLox generated plaques within 3 days (data not shown). In contrast, transfection of the complementing cell line Vero-HS30, in which full-length pUL36 is provided in trans upon HSV1 infection (25), resulted in plaque formation (Table 1). Thus, the Vero-HS30 cells synthesized pUL36 forms that rescued the BAC mutant genomes. This indicates that the new HSV1 strains had been mutated in UL36 and that no other regions of the HSV1 genome required for replication had been altered. Therefore, even the most C-terminal 167 aa (2998stop) of pUL36 were essential for HSV1 replication.
Table 1.
Revertant formation of HSV1(17+) UL36 mutantsa
| HSV1 strain titrated | Titer (PFU/ml [103]) |
Ratio of mutant to revertant plaques | |
|---|---|---|---|
| Vero-HS30 | Verob | ||
| HSV1(17+)blueLox | NDc | 400,000 | ND |
| HSV1(17+)blueLox ΔUL36 | 12,000 | <1 | >12,000:1 |
| HSV1(17+)blueLox-UL36codon2211stop | 33,000 | 14 | 2,357:1 |
| HSV1(17+)blueLox-UL36codon2430stop | 29,000 | 9 | 3,222:1 |
| HSV1(17+)blueLox-UL36codon2894stop | 19,000 | <1 | >19,000:1 |
| HSV1(17+)blueLox-UL36codon2998stop | 29,000 | <1 | >29,000:1 |
Preparations of HSV1 UL36 mutants were titrated in triplicate on the complementing cell line Vero-HS30 and also on the noncomplementing parental Vero cells to determine the amount of revertants in parallel. For comparison, the parental HSV1(17+)blueLox strain from a parallel preparation was also titrated.
The input virus caused severe cytopathic effects at higher concentrations.
ND, not determined.
To characterize the phenotypes of the new UL36 mutants in detail, we prepared virus preparations in Vero-HS30 cells, in which capsids could recruit pUL36 wild type for assembly, while their genomes would contain the desired UL36 mutations. To reduce the occurrence of revertants, we harvested the UL36 mutant viruses prior to the development of full cytopathic effects. Revertants could arise by homologous recombination between the HSV1(17+)blueLox genomes and the homologous regions of the HSV1 UL36 gene or its flanking 2,100-bp upstream and 474-bp downstream regions present in the Vero-HS30 cells, thus repairing the mutations introduced into the BACs in E. coli. To determine the ratio of mutated to reverted genomes, we titrated all stocks in parallel on complementing Vero-HS30 cells and on noncomplementing Vero cells. This ratio was always at least greater than 2,000 (Table 1), indicating that such infection experiments reflected the biology of the mutated and not the pUL36 wild-type forms. Furthermore, the ratio of the number of DNase-protected viral genomes to the number of PFU (26, 96) was about 200, and thus, the specific infectivity of these preparations was similar to that of the parental strain prepared in parallel. However, the pUL36 levels in the complementing cell lines did not restore growth of the mutants to wild-type titers, as has also been reported before for another HSV1 ΔUL36 strain (98).
Next, we performed immunoblot analysis of Vero cells infected with transcomplemented HSV1 UL36 virions which had packaged functional wild-type pUL36 forms from the Vero-HS30 cells (Fig. 7). The HSV1 structural proteins gB, VP16, VP19c, and VP22 were expressed in comparable amounts after infection with HSV1 wild type or UL36 mutants. In cells infected with HSV1 ΔUL36, none of the antibodies directed against the N terminus, the middle region, or the C terminus of pUL36 detected any bands, indicating the absence of the entire protein. In contrast, infection with HSV1(17+)blueLox yielded full-length pUL36 containing N-terminal, middle, and C-terminal epitopes. Cells inoculated with transcomplemented UL36 mutants synthesized truncated versions of pUL36 containing N-terminal and middle but not C-terminal epitopes. Thus, the stop codons that had been inserted into HSV1 UL36 were functional.
The internal HSV1 pUL36 amino acid residues 2430 to 2893 but not the C-terminal amino acid residues 2894 to 3164 are required for targeting capsids to cytoplasmic membranes.
Further immunofluorescence microscopy studies revealed that nuclear capsid egress was unimpaired since newly synthesized capsids were located in the nucleus and in the cytoplasm, irrespective of the HSV1 UL36 mutant analyzed (Fig. 8, red). At 15 h p.i., the majority of wild-type HSV1 cytoplasmic capsids were concentrated in cytoplasmic perinuclear patches (Fig. 8a and b). In contrast, after infection with mutants lacking the entire protein (ΔUL36; Fig. 8e and f), the C-terminal 954 aa (2211stop; data not shown), or just 735 aa (2430stop; Fig. 8i and j), individual capsids were randomly dispersed over the entire cytoplasm. Capsids of the mutant lacking only the C-terminal 167 aa behaved similar to wild-type capsids and were also concentrated in perinuclear patches (2998stop; Fig. 8q and r). Capsids of the mutant lacking the C-terminal 271 aa (2894stop) showed an intermediate phenotype; the capsids were dispersed over the entire cytoplasm (Fig. 8m and n) or concentrated in perinuclear patches (Fig. 8m′ and n′).
Fig 8.
Subcellular localization of truncated HSV1 pUL36 proteins. Vero cells were infected at an MOI of 0.2 PFU/cell with pHSV1(17+)blueLox (wt; a to d), with the mutant HSV1(17+)blueLox-ΔUL36 (ΔUL36; e to h), or with HSV1(17+)blueLox-UL36codon2430stop (2430; i to l), -UL36codon2894stop (2894; m to p and m′ to p′), or -UL36codon2998stop (2998; q to t), transcomplemented with pUL36 by amplification on Vero-HS30 cells, and fixed at 15 h p.i. with 3% paraformaldehyde, followed by TX-100 permeabilization. The specimens were labeled with antibodies directed against VP5 (red and b, f, j, n, n′, and r; MAb 5C10) or pUL36 aa 1408 to 2112 (green and c, g, k, o, o′, and s; middle, pAb 147) and analyzed by confocal fluorescence microscopy. Parts of the overviews (a, e, i, m, m′, and q) are shown at higher magnification in the horizontally neighboring panels.
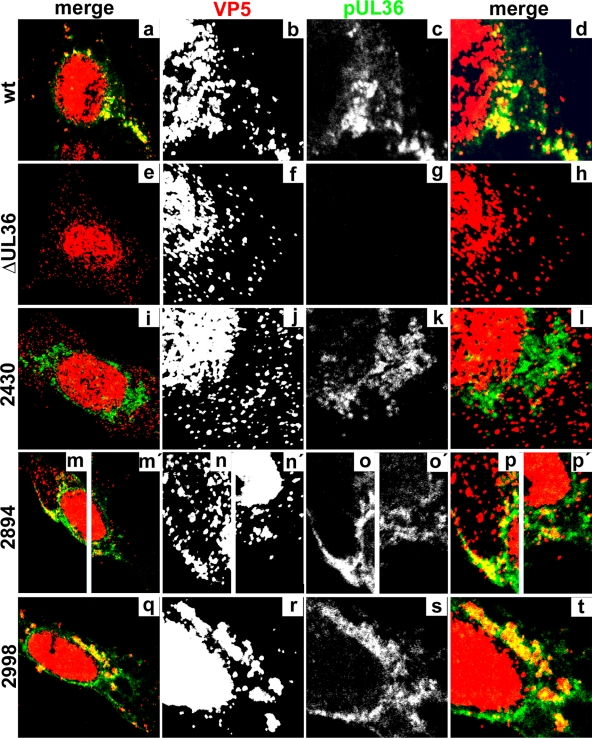
pUL36 was mainly localized in the perinuclear patches with wild-type HSV1 (Fig. 8c, green), and inoculation with HSV1 ΔUL36 revealed no background signal (Fig. 8g). The capsids in the perinuclear patches of wild-type HSV1 (Fig. 8b), the mutant lacking the C-terminal 167 aa (2998stop; Fig. 8r), and most of 2894stop (Fig. 8n and n′) colocalized with pUL36 (Fig. 8d, p, p′, and t, yellow). In contrast, the dispersed capsids of the other UL36 truncation mutants (shown only for 2430stop; Fig. 8j) colocalized only rarely with cytoplasmic pUL36 (Fig. 8l, yellow). To examine whether capsids associated with cytoplasmic membranes, infected cells were double labeled with antibodies against capsids and gD (data not shown) or against capsids and p230, the TGN marker (Fig. 9A). The perinuclear capsid patches of wild-type HSV1 (Fig. 9Aa), the mutant lacking 271 aa (2894stop; Fig. 9Ag and g′), and the mutant lacking 167 aa (2998stop; Fig. 9Aj) colocalized with gD (data not shown) and p230 (Fig. 9Ac, i, i′, and l, yellow). In contrast, the dispersed cytoplasmic capsids (Fig. 9Ad) rarely colocalized with gD (not shown) or p230 after infection with HSV1 ΔUL36 (data not shown) or with the mutants lacking 954 aa (2211stop; Fig. 9Af) or 735 aa (2430stop; data not shown), indicating that they were impaired in association with cytoplasmic membranes and thus most likely in secondary envelopment.
To estimate the amount of cytoplasmic capsids, we determined the position of the nuclei based on transmission images (Fig. 9Ba) and digitally erased the nuclear but not the cytoplasmic capsid signal (compare Fig. 9Bb with Bc). Using the software CellProfiler (13), we determined the number of pixels occupied by cytoplasmic capsids, by p230, and by both as an indicator of the degree of colocalization of capsids with membranes containing the TGN marker (Fig. 9C). In cells infected with wild-type HSV1, cytoplasmic capsids occupied about 5,900 pixels per cell. This amount almost doubled after infection with HSV1 ΔUL36 or mutants lacking the C-terminal 954 aa (2211stop) or 735 aa (2430stop). These data suggest that the capsids of these mutants did not leave the cells but, rather, accumulated in the cytoplasm. After infection with the mutant lacking the C-terminal 271 aa (2894stop), the cytoplasmic capsids covered about 6,800 pixels per cell, and after infection with the mutant lacking only 167 aa (2998stop), the cytoplasmic capsids covered about 6,000 pixels per cell, similar to wild type. There was no significant difference in the number of cytoplasmic p230 signals among the HSV1 strains. In contrast, the area labeled for both p230 and the capsid protein VP26 revealed a significantly lower association of HSV1 capsids with the TGN after infection with HSV1 ΔUL36 or the mutants lacking the C-terminal 954 aa (2211stop) or 735 aa (2430stop). On the other hand, infection with wild-type HSV1 and the mutants lacking 271 aa (2894stop) or 167 aa (2998stop) displayed about 800 pixels per cell occupied by both capsids and p230. These differences between the mutants were particularly significant in relation to the higher concentration of cytoplasmic capsids for HSV1 ΔUL36, 2211stop, and 2430stop, which nevertheless did not result in a higher degree of colocalization with the TGN marker p230.
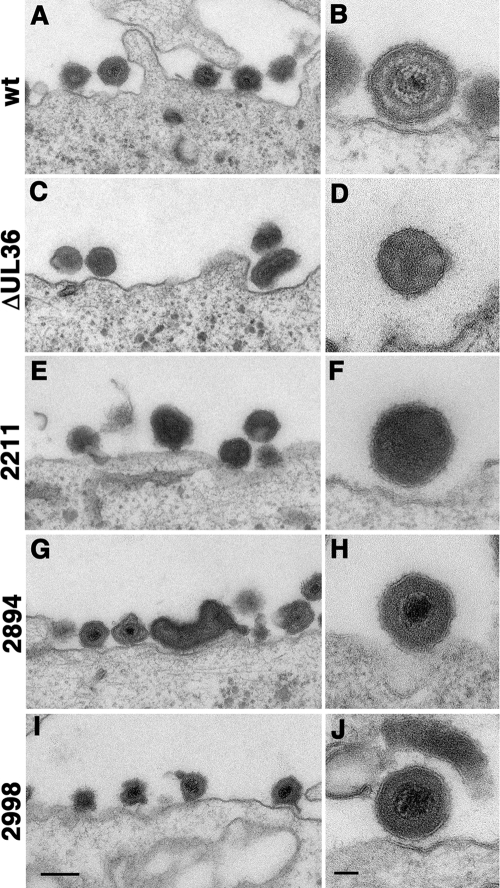
To determine whether the pUL36 mutants underwent secondary envelopment and formed virions, we infected Vero cells at an MOI of 10 and analyzed them by electron microscopy (Fig. 10). Whereas after infection with HSV1 wild type many virions containing capsids were attached to the plasma membrane of infected cells at 12 or 14 h postinfection, infection with HSV1 ΔUL36 resulted in the formation only of L particles lacking viral capsids. The mutant HSV1 2211stop produced only L particles, similar to the phenotype of the complete deletion of UL36 (Fig. 10F). However, the short deletions of HSV1 2894stop (Fig. 10H) and HSV1 2998stop (Fig. 10J) still secreted complete virions. Our data indicate that we have generated two different types of dysfunctional HSV1 UL36 mutants. First, in the absence of pUL36 or a large C-terminal portion of pUL36, capsids accumulated in the cytosol, no longer recruited pUL36, did not associate with cytoplasmic membranes, and did not form virions. Second, those mutants lacking only a smaller portion of the C terminus recruited the truncated pUL36 forms onto cytosolic capsids, still associated with membranes, and could assemble virions but nevertheless did not form plaques and thus were not infectious.
Fig 10.
The C-terminal 270 amino acids of HSV1 pUL36 are dispensable for secondary envelopment and virion assembly. Vero cells were infected at an MOI of 10 PFU/cell with pHSV1(17+)blueLox (wt; A and B), the mutant HSV1(17+)blueLox-ΔUL36 (ΔUL36; C and D), or HSV1(17+)blueLox-UL36codon2211stop (2211; E and F), -UL36codon2894stop (2894; G and H), or -UL36codon2998stop (2998; I and J) and prepared for electron microscopy after 14 h or 12 h (E and F, respectively). Bars = 200 nm (A, C, E, G, and I) and 50 nm (B, D, F, H, and J).
The C-terminal 167 amino acid residues of HSV1 pUL36 are required for targeting incoming capsids to the nucleus but not for virus assembly and virus internalization.
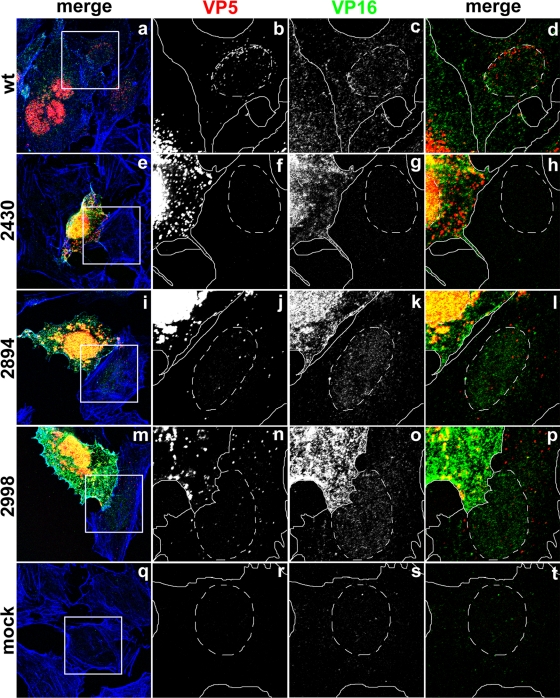
HSV1 mutants lacking the very C terminus of pUL36 still formed virions but did not generate plaques, possibly because subsequent cell entry was perturbed. We therefore inoculated Vero cells at an MOI of 0.2 PFU/cell with UL36 mutants transcomplemented with wild-type pUL36 and labeled the filamentous actin to indicate the cell boundaries as well as the capsids and the transcriptional activator VP16. Single seeder cells expressed large amounts of VP5 and VP16 in the nucleus (Fig. 11, yellow). Cells infected with wild-type HSV1 formed plaques (Fig. 11a), and the primary seeder cells were surrounded by neighboring cells containing several capsids that had been targeted to the nucleus (Fig. 11b; red in panels a and d). Inoculation with HSV1 ΔUL36 (data not shown) or mutants lacking C-terminal 954 aa (2211stop; data not shown) or 735 aa (2430stop; Fig. 11e) resulted in isolated seeder cells with nuclear and cytoplasmic capsids, but no capsids were detected in neighboring cells (Fig. 11f).
Fig 11.
The C-terminal 167 amino acid residues of HSV1 pUL36 are required for targeting incoming capsids to the nucleus, while the C-terminal 271 amino acid residues are dispensable for egress and internalization. Vero cells were infected at an MOI of 0.2 PFU/cell with pHSV1(17+)blueLox (wt; a to d) or mutant HSV1(17+)blueLox-UL36codon2430stop (2430; e to h), -UL36codon2894stop (2894; i to l), or -UL36codon2998stop (2998; m to p), transcomplemented with pUL36 by amplification on Vero-HS30 cells or not infected (mock; q to t), and fixed at 15 h p.i. with 3% paraformaldehyde, followed by TX-100 permeabilization. The specimens were labeled with antibodies directed against VP5 (red and b, f, j, n, and r; MAb 5C10) or VP16 (green and c, g, k, o, and s; pAb R230) and analyzed by confocal fluorescence microscopy. The insets (white squares) are shown at higher magnification in the horizontally neighboring panels. The location of the cell borders was estimated based on staining with TRITC-phalloidin for filamentous actin (blue in a, e, i, m, and q; continuous white lines), and the position of the cell nuclei was determined by differential interference contrast (dashed white lines).
In seeder cells inoculated with mutants lacking only the C-terminal 271 aa (2894stop; Fig. 11i) or 167 aa (2998stop; Fig. 11m), there were nuclear and cytoplasmic capsids. Furthermore, the neighboring cells surrounding those seeder cells contained several cytoplasmic capsids, but only a minor fraction of those had been targeted to the nucleus (Fig. 11j and n; Fig. 11l and p, red). To assess whether such capsids represented intact virions or unenveloped capsids that had been released into the cytosol upon viral fusion with host membranes, we determined the subcellular localization of VP16. As for the capsids, VP16 is released into the cytosol only upon HSV1 fusion and then imported into the nucleus, where it promotes early HSV1 transcription (5, 55). Cells infected with wild-type HSV1 (Fig. 11c) or with the mutants lacking 271 aa (2894stop; Fig. 11k) or 167 aa (2998stop; Fig. 11o) contained VP16 in the nucleus, demonstrating that fusion of virions had indeed occurred. In contrast, the cells in the immediate neighborhood of the seeder cells inoculated with the mutant that lacks the C-terminal 735 aa (2430stop; Fig. 11g) did not reveal any VP16 signal, supporting the observation that these cells did not contain any viral particles. Control experiments demonstrated a low background signal in uninfected cells (Fig. 11q to t).
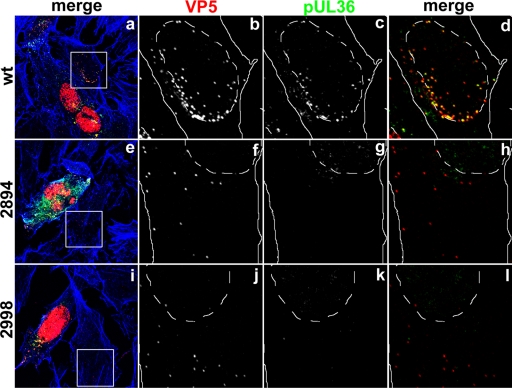
Next, we addressed whether pUL36 was associated with the incoming capsids in the neighboring cells. To this end, we labeled cells inoculated with 0.2 PFU/cell with antibodies against the capsid protein VP5 and pUL36 as well as for filamentous actin (Fig. 12). Incoming wild-type capsids targeted to the nucleus in neighboring cells of the primary seeder cells and contained pUL36 (Fig. 12d, yellow), whereas neighboring cells of those inoculated with the mutants lacking 954 aa (2211stop) or 735 aa (2430stop) did not contain either capsids or pUL36 (data not shown). In contrast, neighboring cells of those inoculated with the mutants lacking only 271 aa (2894stop) or 167 aa (2998stop) contained capsids (Fig. 12f and j), but pUL36 could not be detected on those (Fig. 12g and k). Thus, any truncated pUL36 forms potentially packaged into virions upon secondary envelopment had been lost from incoming capsids during cell entry and were most likely undetectable by immunofluorescence microscopy due to their free diffusion within the cytosol.
Fig 12.
The C-terminal 167 amino acid residues of HSV1 pUL36 are required to maintain capsid association during cell entry. Vero cells were infected at an MOI of 0.2 PFU/cell with pHSV1(17+)blueLox (wt; a to d) or mutant HSV1(17+)blueLox-UL36codon2894stop (2894; e to h) or -UL36codon2998stop (2998; i to l), transcomplemented with pUL36 by amplification on Vero-HS30 cells, and fixed at 15 h p.i. with 3% paraformaldehyde, followed by TX-100 permeabilization. The specimens were labeled with antibodies directed against VP5 (red and b, f, and j; MAb 5C10) or pUL36 (green and c, g, and k; middle, pAb 147) and analyzed by confocal fluorescence microscopy. The insets (white squares) are shown at higher magnification in the horizontally neighboring panels. The location of the cell borders was estimated based on staining with TRITC-phalloidin for filamentous actin (blue in a, e, and i; continuous white lines), and the position of the cell nuclei was determined by differential interference contrast (dashed white lines).
In summary, our data suggest that HSV1 pUL36 mutants lacking only the very 271 or 167 aa of the C terminus were able to assemble virions that left infected seeder cells and spread into neighboring cells, where they released capsids into the cytosol upon HSV1 fusion. However, such cytosolic capsids were not efficiently targeted to the nucleus, and the truncated pUL36 forms apparently dissociated from these incoming capsids after viral fusion.
DISCUSSION
In this study, we investigated the functions of the HSV1 essential large tegument protein pUL36 during assembly and cell entry. Using different antibodies, we show that various pUL36 forms were synthesized during infection but that HSV1 virions predominantly packaged the full-length form, as also reported for PrV pUL36 (76). Our subcellular localization studies indicate that HSV1 pUL36 wt was associated with cytosolic capsids after nuclear egress but prior to secondary envelopment and remained capsid associated during virus egress and upon cell entry until arrival at the nuclear pores. By deleting the entire gene or inserting a functional stop at codon position 2211, 2430, 2894, or 2998 in the BAC pHSV1(17+)blueLox (cf. Fig. 1), we have generated two functionally distinct sets of mutants (summarized in Table 2). Progeny capsids of the mutants lacking pUL36 entirely or with larger C-terminal deletions were still able to egress from the nucleus but could not recruit pUL36, nor could they associate with membranes for secondary envelopment or assemble virions. In contrast, the mutants with shorter C-terminal deletions recruited pUL36 onto capsids and underwent secondary envelopment, virion formation, cell egress, and entry into neighboring cells. However, the incoming cytosolic capsids lost pUL36, were not targeted to the nucleus, and were unable to induce HSV1 gene expression.
Table 2.
Characterization of HSV1 UL36 mutantsa
| HSV1 strain | Result by the following assay: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nuclear egress and assembly |
Spread and cell entry |
||||||||
| Nuclear capsids | Capsids in cytoplasm | Capsids with pUl36 | Capsids at TGN and gD | Virion assembly | Spread to neighbor cells | Fusion, nuclear VP16 | Capsids with pUL36 | Plaque formation | |
| wt | + | + | + | + | + | + | + | + | + |
| ΔUL36 | + | + | − | − | − | − | − | − | − |
| 2211stop | + | + | − | − | ND | − | − | − | − |
| 2430stop | + | + | − | − | − | − | − | − | − |
| 2894stop | + | + | + | + | + | + | + | − | − |
| 2998stop | + | + | + | + | + | + | + | − | − |
The HSV1 mutants of UL36 characterized in this study can be classified into two groups. In the absence of pUL36 (ΔUL36) or large C-terminal portions (2211stop, 2430 stop), capsids left the nucleus and accumulated in the cytosol, but they no longer recruited pUL36 and did not associate with cytoplasmic membranes. Those mutants lacking only a smaller portion of UL36 (2894stop, 2998stop) recruited the truncated pUL36 forms onto cytosolic capsids, still associated with cytoplasmic membranes, and formed virions that entered neighboring cells. However, the incoming capsids did not contain pUL36, did not bind to the nucleus, and did not initiate viral gene expression. ND, not determined.
pUL36 capsid association during assembly.
pUL36 was detected as early as 4 h p.i. on unenveloped, cytosolic capsids but not on nuclear capsids. As the infection progressed, pUL36 remained capsid associated and accumulated in cytoplasmic patches that also contained viral and host membrane proteins and thus most likely represented the sites of secondary envelopment. Such a transient appearance of unenveloped cytosolic capsids is consistent with our studies using dual-color HSV1(17+) strains, in which the small capsid protein VP26 and gD had been tagged with different autofluorescent protein domains (86).
Only later in infection, a mainly diffuse signal for HSV1 pUL36 appeared in the nucleus, while the homologous PrV pUL36 was also detected in the cytoplasm but not in the nucleus (52, 81). Such a subcellular localization of pUL36 supports the secondary reenvelopment model of herpesvirus assembly in which the inner tegument protein pUL36 is acquired by cytosolic capsids prior to secondary envelopment but is harder to reconcile with the luminal single-envelopment model that postulates that all tegument proteins are recruited onto nuclear capsids prior to the envelopment at the inner nuclear membrane.
However, our experiments do not exclude the possibility that there may be other pUL36 forms, particularly those harboring the NLS or pUL25 binding domains and possibly occurring later in infection, that may associate with nuclear capsids in the nucleus (2, 10, 16, 56, 92) but that were not detected by the antibodies directed against epitopes of the middle region. Smaller C-terminal fragments of PrV pUL36 or HSV1 pUL36 are redirected from the cytosol to nuclear capsid assemblons upon virus infection (16, 56). Furthermore, if pUL36 binding to nuclear capsids would trigger efficient nuclear egress, any steady-state concentration of nuclear capsids decorated with pUL36 would be low. Consistent with our morphology data, biochemical studies also reveal significant amounts of pUL36 C-terminal and middle epitopes on capsids isolated from intact virions but only very few C-terminal and no middle region epitopes on similar amounts of capsids isolated from the nuclei of infected cells (96, 111, 116). Moreover, PrV pUL36 has been detected by immunoelectron microscopy on cytosolic capsids and cytoplasmic enveloped virions but not on primary virions within the nuclear envelope after primary envelopment (52, 81). These apparent discrepancies between HSV1 and PrV may be due to differences between tagged and untagged forms of pUL36 or indicate different subcellular localizations of different pUL36 forms (2, 16, 52, 56, 72, 81).
The truncated pUL36 of the mutant 2211stop no longer bound to capsids but was nevertheless not impaired in nuclear egress. It lacks the very C-terminal pUL25 binding site between aa 3104 and 3164 and the stop codon has most likely destroyed the second pUL25 binding site between aa 2037 to 2353, but it still contains a potential capsid binding region within the N-terminal 161 aa (16, 92, 98). Surprisingly, pUL36 of 2430stop, which contains the second pUL25 binding site, also no longer associated with cytosolic capsids. This suggests either that regions downstream of aa 2429 contribute to the folding of the upstream pUL25 binding site or that they comprise further binding domains that are required for stable capsid association. The pUL36 of 2894stop was also reduced on capsids. The colocalization of pUL36 with capsids was delayed, even though this pUL36 was truncated about 540 aa downstream of the region containing the pUL25 binding site, but the deleted region may also either affect the folding of the upstream pUL25 binding site or contain further unknown capsid binding motifs. The pUL36 of 2998stop colocalized with capsids as efficiently as the full-length pUL36. Thus, the very C-terminal 167 aa containing the second C-terminal pUL25 binding site were not essential to recruit pUL36 onto cytosolic capsids during assembly.
Furthermore, the pUL36 of the mutants 2998stop and, to a slightly lesser extent, 2894stop also colocalized with cytoplasmic membranes. Their cytosolic capsids recruited the C-terminal truncated pUL36 forms that most likely bound via their N-terminal third to the inner tegument protein pUL37 and to the major tegument protein VP16 (cf. Fig. 1). Besides the two capsid proteins VP19c and VP26, pUL37 could also bind to the outer tegument protein VP11/12 (pUL46); furthermore, VP16 could strengthen the association with the outer tegument via its binding to VP22 (pUL49), vhs (pUL41), and VP11/12 and with the viral envelope via binding to the viral glycoproteins gB, gD, and gH (31, 35, 53, 56, 96, 105, 114). Thus, pUL36 mutants that associated with cytosolic capsids and still contained the binding motifs for tegument proteins were able to form virions that were secreted from inoculated cells. In the absence of pUL36, the other inner tegument protein, pUL37, and also the recently characterized interactions between pUL21-pUL16-pUL11 (44, 59, 114) apparently did not suffice for cross-linkage between the capsid and the outer tegument. This notion is also supported by the accumulation of cytoplasmic capsids that lack pUL36 after inoculation with HSV1(17+)blueLox-ΔUL36, -2211stop, or -2430stop. In contrast, cytoplasmic capsids of 2894stop or 2998stop contained pUL36, and their concentration was similar to that after infection with HSV1(17+)blueLox (Fig. 9C).
pUL36 capsid association during cell entry.
HSV1 pUL36 remained capsid associated until arrival at the nuclear envelope, as reported using fluorescent tags or other antibodies (4, 18, 40, 62, 82) and consistent with its proposed functions in binding to microtubule motors and nuclear pores (5, 63, 96, 98, 116). When only the most C-terminal pUL25 binding site was lacking (2894stop, 2998stop), the producer cells still formed virions that upon viral fusion delivered capsids into the cytosol. However, those incoming capsids no longer maintained pUL36 on their surface and were not efficiently targeted to the nucleus, either because they could no longer recruit microtubule motors or because they could not bind to nuclear pores. Similarly, newly assembled cytosolic capsids of an HSV1 mutant lacking UL36 entirely are unable to target to other nuclei, after inoculated cells have been artificially fused with neighboring uninfected cells into syncytia to overcome the two plasma membrane barriers (98). However, the history of these incoming capsids is different from that of the pUL36 mutants 2894stop and 2998stop. Mutants 2894stop and 2998stop had been able to recruit C-terminally truncated versions of pUL36 during assembly, presumably due to the pUL25 binding site located between aa 2037 and 2353 (92) and the potential capsid binding site within the 300 N-terminal amino acids of pUL36 (98). It seems that in the absence of the second C-terminal pUL25 binding site between aa 3104 and 3164 (16, 56, 92), the association of pUL36 with capsids was sufficient for assembly but weakened after viral fusion with the neighboring cells. Apparently, in addition to aa 2037 to 2353 (92), the region between aa 2430 and 2893 of pUL36 also contributed to its affinity for capsids and was required to maintain pUL36 on incoming capsids.
Interestingly, the amount of incoming wild-type capsids in the neighboring cells (Fig. 12b) was similar to that after synchronous infection (Fig. 5Dj), demonstrating that cell entry experiments using MOIs of 50, as we had reported previously (26, 27, 65, 100, 106), represent cell entry conditions similar to those that occur in a plaque assay.
Our results support the notion that different domains of pUL36 form an assembly cross-linker between the capsid surface and the other tegument proteins. The very C-terminal domain binding to pUL25 is essential during cell entry, when the cytosolic concentration of capsids and of pUL36 is low, whereas during assembly, this binding site seems dispensable, since both capsids and pUL36 are present in high concentrations. Furthermore, the association between pUL36 and the capsids becomes irreversible upon secondary envelopment, or the middle pUL25 binding site on pUL36 may be active during assembly but specifically reversed during cell entry.
On the other end of pUL36, the N-terminal binding to VP16, as well as the bonds between pUL37 and other tegument proteins, must be untied upon cell entry to permit transport of cytosolic capsids from the host membranes toward the nucleus. It is tempting to speculate that binding of the viral glycoproteins to host receptors such as heparan sulfate proteoglycans, αVβIII integrin, or nectin-1 (45) may transmit structural signals across the viral membrane into the tegument, as shown, for example, in a change in the binding properties of the tegument protein pUL16 (73). Furthermore, the tegument kinases pUL13 and pUS3 might become activated during cell entry, and the cytosol of cells late in infection may comprise protein kinase and phosphatase activities different from those in the naïve cytosol that incoming capsids face upon entering uninfected neighboring cells. Indeed, the major tegument proteins VP13/14, VP16, and VP22 are phosphorylated upon cell entry (83), and Jovasevic et al. (2008) have shown that pUL36 is cleaved into smaller forms upon cell entry (49). Our studies indicate that small interfering RNAs that perturb the synthesis of pUL36 or small chemical compounds that compete with the multiple interactions of pUL36 with capsids would effectively block viral infection, plaque formation, and pathogenesis. The evolutionarily conserved pUL36 may therefore provide a potential target structure for the development of new drugs against infections with herpesviruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Malte Sandbaumhüter for his support with the electron microscopy experiment and particularly K. Radtke (Department of Pathology, University of Montreal) and Katinka Döhner, as well as Fenja Anderson, Anna Buch, and Deepika Devadas (Institute of Virology, Hannover Medical School), for fruitful discussions and critical feedback on the manuscript. We also acknowledge the MHH Confocal Light Microscopy Facility for continuous support. We are grateful to P. Desai (Johns Hopkins University, Baltimore, MD), J. Subak-Sharpe (Glasgow, United Kingdom), P. G. Spear (Chicago, IL), and E. Borst and M. Messerle (Institute of Virology, Hannover Medical School) for providing cells, viruses, and plasmids. H. Browne and A. Minson (University of Cambridge), R. Courtney (Pennsylvania State University), G. H. Cohen and R. J. Eisenberg (University of Pennsylvania), P. Desai (Johns Hopkins University, Baltimore, MD), D. M. Meredith (University of Leeds), G. Elliott and P. O'Hare (Imperial College, London, United Kingdom), as well as W. W. Newcomb and J. C. Brown (University of Virginia) generously donated antibodies.
This study was supported by the Volkswagen-Stiftung (Niedersächsisches Vorab; Deutsche Technion-Gesellschaft e.V.) and the Deutsche Forschungsgemeinschaft (German Research Council; program project grant SPP1175 with So 403/3; Excellence Cluster EXC 62/1, REBIRTH—From Regenerative Biology to Reconstructive Therapy to B.S.). J.S. was a Ph.D. student of the Hannover Biomedical Research School (HBRS; Hannover Medical School, Centre of Infection Biology [ZIB]).
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abaitua F, Daikoku T, Crump CM, Bolstad M, O'Hare P. 2011. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1-2 protein of herpes simplex virus. J. Virol. 85:2024–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abaitua F, O'Hare P. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batterson W, Furlong D, Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berger B, et al. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. U. S. A. 92:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borst EM, Benkartek C, Messerle M. 2007. Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Curr. Protoc. Immunol. Chapter 10:Unit 10.32 [DOI] [PubMed] [Google Scholar]
- 8. Böttcher S, et al. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910–9915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunetti CR, Dingwell KS, Wale C, Graham FL, Johnson DC. 1998. Herpes simplex virus gD and virions accumulate in endosomes by mannose 6-phosphate-dependent and -independent mechanisms. J. Virol. 72:3330–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calistri A, et al. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81:11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campadelli Fiume G. 2007. The egress of alphaherpesviruses from the cell. In Arvin A, et al. (ed), Human herpesviruses, biology, therapy and immunoprophylaxis. Cambridge University Press, New York, NY: [PubMed] [Google Scholar]
- 13. Carpenter AE, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng SB, Ferland P, Webster P, Bearer EL. 2011. Herpes simplex virus dances with amyloid precursor protein while exiting the cell. PLoS One 6:e17966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen GH, et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alpha herpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conway JF, et al. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Copeland AM, Newcomb WW, Brown JC. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Court DL, Sawitzke JA, Thomason LC. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36:361–388 [DOI] [PubMed] [Google Scholar]
- 20. Crump CM, Yates C, Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dambach MJ, Trecki J, Martin N, Markovitz NS. 2006. Oncolytic viruses derived from the gamma34.5-deleted herpes simplex virus recombinant R3616 encode a truncated UL3 protein. Mol. Ther. 13:891–898 [DOI] [PubMed] [Google Scholar]
- 22. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davis LI, Blobel G. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. U. S. A. 84:7552–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Desai P, DeLuca NA, Person S. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115–124 [DOI] [PubMed] [Google Scholar]
- 25. Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Döhner K, Radtke K, Schmidt S, Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Döhner K, et al. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dubin G, Frank I, Friedman HM. 1990. Herpes simplex virus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG) and IgG complexes. J. Virol. 64:2725–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eisenberg RJ, et al. 1985. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J. Virol. 53:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eisenberg RJ, et al. 1987. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb. Pathog. 3:423–435 [DOI] [PubMed] [Google Scholar]
- 31. Elliott G, Mouzakitis G, O'Hare P. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elliott G, O'Hare P. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223–233 [DOI] [PubMed] [Google Scholar]
- 33. Enquist LW, Husak PJ, Banfield BW, Smith GA. 1998. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237–347 [DOI] [PubMed] [Google Scholar]
- 34. Farnsworth A, Johnson DC. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fossum E, et al. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gastadello S, et al. 2010. A deneddylase encoded by EBV promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat. Cell Biol. 12:351–361 [DOI] [PubMed] [Google Scholar]
- 38. Gianni T, Campadelli-Fiume G, Menotti L. 2004. Entry of herpes simplex virus mediated by chimeric forms of nectin1 retargeted to endosomes or to lipid rafts occurs through acidic endosomes. J. Virol. 78:12268–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Granzow H, et al. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Granzow H, Klupp BG, Mettenleiter TC. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Granzow H, et al. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grünewald K, et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398 [DOI] [PubMed] [Google Scholar]
- 43. Harley CA, Dasgupta A, Wilson DW. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harper AL, et al. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84:2963–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. James SH, Kimberlin DW, Whitley RJ. 2009. Antiviral therapy for herpesvirus central nervous system infections: neonatal herpes simplex virus infection, herpes simplex encephalitis, and congenital cytomegalovirus infection. Antiviral Res. 83:207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 48. Johnson DC, Spear PG. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jovasevic V, Liang L, Roizman B. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kapur JN, Sahoo PK, Wong AKC. 1985. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vision Graph. Image Processing 29:273–285 [Google Scholar]
- 51. Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547–557 [DOI] [PubMed] [Google Scholar]
- 52. Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koelle DM, Corey L. 2008. Herpes simplex: insights on pathogenesis and possible vaccines. Annu. Rev. Med. 59:381–395 [DOI] [PubMed] [Google Scholar]
- 55. La Boissiere S, Hughes T, O'Hare P. 1999. HCF-dependent nuclear import of VP16. EMBO J. 18:480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee JI, Luxton GW, Smith GA. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee JI, et al. 2009. A herpesvirus encoded deubiquitinase is a novel neuroinvasive determinant. PLoS Pathog. 5:e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leuzinger H, et al. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 79:13047–13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Loomis JS, Courtney RJ, Wills JW. 2006. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 80:10534–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 62. Luxton GW, et al. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lycke E, et al. 1988. Herpes simplex virus infection of the human sensory neuron. An electron microscopy study. Arch. Virol. 101:87–104 [DOI] [PubMed] [Google Scholar]
- 65. Mabit H, et al. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 76:9962–9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maeshima K, et al. 2006. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J. Cell Sci. 119:4442–4451 [DOI] [PubMed] [Google Scholar]
- 67. Marozin S, Prank U, Sodeik B. 2004. Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell-cell contact sites. J. Gen. Virol. 85:775–786 [DOI] [PubMed] [Google Scholar]
- 68. Maurer UE, Sodeik B, Grunewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. U. S. A. 105:10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McLean C, et al. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63:297–305 [DOI] [PubMed] [Google Scholar]
- 70. McNab AR, et al. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McNabb DS, Courtney RJ. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 66:7581–7584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McNabb DS, Courtney RJ. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221–232 [DOI] [PubMed] [Google Scholar]
- 73. Meckes DG, Jr, Wills JW. 2008. Structural rearrangement within an enveloped virus upon binding to the host cell. J. Virol. 82:10429–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 76. Michael K, Klupp BG, Mettenleiter TC, Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mijatov B, Cunningham AL, Diefenbach RJ. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31 [DOI] [PubMed] [Google Scholar]
- 78. Milne RS, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Miranda-Saksena M, et al. 2009. Herpes simplex virus utilizes the large secretory vesicle pathway for anterograde transport of tegument and envelope proteins and for viral exocytosis from growth cones of human fetal axons. J. Virol. 83:3187–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Möhl BS, et al. 2010. Random transposon-mediated mutagenesis of the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 84:8153–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Möhl BS, et al. 2009. Intracellular localization of the pseudorabies virus large tegument protein pUL36. J. Virol. 83:9641–9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Morrison EE, Stevenson AJ, Wang YF, Meredith DM. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517–2528 [DOI] [PubMed] [Google Scholar]
- 83. Morrison EE, Wang YF, Meredith DM. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mu FT, et al. 1995. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 270:13503–13511 [DOI] [PubMed] [Google Scholar]
- 85. Muyrers JP, Zhang Y, Testa G, Stewart AF. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nagel CH, et al. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J. Virol. 82:3109–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Negatsch A, et al. 2010. Ultrastructural analysis of virion formation and intraaxonal transport of herpes simplex virus type 1 in primary rat neurons. J. Virol. 84:13031–13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Newcomb WW, Brown JC. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84:9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nicola AV, Hou J, Major EO, Straus SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 79:7609–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Otsu N. 1979. A threshold selection method from gray-level histograms. IEEE Trans. Sys. Man. Cyber. 9:62–66 [Google Scholar]
- 92. Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pawliczek T, Crump CM. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Preston VG. 1990. Herpes simplex virus activates expression of a cellular gene by specific binding to the cell surface. Virology 176:474–482 [DOI] [PubMed] [Google Scholar]
- 95. Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 82:6654–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Radtke K, et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Remillard-Labrosse G, Mihai C, Duron J, Guay G, Lippe R. 2009. Protein kinase D-dependent trafficking of the large herpes simplex virus type 1 capsids from the TGN to plasma membrane. Traffic 10:1074–1083 [DOI] [PubMed] [Google Scholar]
- 98. Roberts AP, et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Roberts KL, Baines JD. 2010. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J. Virol. 84:9889–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rode K, et al. 2011. Uncoupling uncoating of herpes simplex virus genomes from their nuclear import and gene expression. J. Virol. 85:4271–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex viruses, p 2502–2601 In Knipe DM, Howley PM. (ed), Fundamental virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 102. Sancho-Shimizu V, et al. 2007. Genetic susceptibility to herpes simplex virus 1 encephalitis in mice and humans. Curr. Opin. Allergy Clin. Immunol. 7:495–505 [DOI] [PubMed] [Google Scholar]
- 103. Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 82:7388–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Smibert CA, Popova B, Xiao P, Capone JP, Smiley JR. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Spear PG, Roizman B. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Stow ND. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755–10765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sugimoto K, et al. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Trus BL, Newcomb WW, Booy FP, Brown JC, Steven AC. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. U. S. A. 89:11508–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Trus BL, et al. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Umene K, Sakaoka H. 1991. Homogeneity and diversity of genome polymorphism in a set of herpes simplex virus type 1 strains classified as the same genotypic group. Arch. Virol. 119:53–65 [DOI] [PubMed] [Google Scholar]
- 114. Vittone V, et al. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wisner TW, Johnson DC. 2004. Redistribution of cellular and herpes simplex virus proteins from the trans-Golgi network to cell junctions without enveloped capsids. J. Virol. 78:11519–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wolfstein A, et al. 2006. The inner tegument promotes herpes simplex virus transport along microtubules in vitro. Traffic 7:1–11 [DOI] [PubMed] [Google Scholar]
- 117. Zhang Y, Buchholz F, Muyrers JP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123–128 [DOI] [PubMed] [Google Scholar]
- 118. Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.