Abstract
Recently, we reported the discovery of three novel coronaviruses, bulbul coronavirus HKU11, thrush coronavirus HKU12, and munia coronavirus HKU13, which were identified as representatives of a novel genus, Deltacoronavirus, in the subfamily Coronavirinae. In this territory-wide molecular epidemiology study involving 3,137 mammals and 3,298 birds, we discovered seven additional novel deltacoronaviruses in pigs and birds, which we named porcine coronavirus HKU15, white-eye coronavirus HKU16, sparrow coronavirus HKU17, magpie robin coronavirus HKU18, night heron coronavirus HKU19, wigeon coronavirus HKU20, and common moorhen coronavirus HKU21. Complete genome sequencing and comparative genome analysis showed that the avian and mammalian deltacoronaviruses have similar genome characteristics and structures. They all have relatively small genomes (25.421 to 26.674 kb), the smallest among all coronaviruses. They all have a single papain-like protease domain in the nsp3 gene; an accessory gene, NS6 open reading frame (ORF), located between the M and N genes; and a variable number of accessory genes (up to four) downstream of the N gene. Moreover, they all have the same putative transcription regulatory sequence of ACACCA. Molecular clock analysis showed that the most recent common ancestor of all coronaviruses was estimated at approximately 8100 BC, and those of Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus were at approximately 2400 BC, 3300 BC, 2800 BC, and 3000 BC, respectively. From our studies, it appears that bats and birds, the warm blooded flying vertebrates, are ideal hosts for the coronavirus gene source, bats for Alphacoronavirus and Betacoronavirus and birds for Gammacoronavirus and Deltacoronavirus, to fuel coronavirus evolution and dissemination.
INTRODUCTION
Coronaviruses (CoVs) are found in a wide variety of animals, in which they can cause respiratory, enteric, hepatic, and neurological diseases of varying severity. Based on genotypic and serological characterization, CoVs were traditionally divided into three distinct groups (3, 22, 54). Recently, the Coronavirus Study Group of the International Committee for Taxonomy of Viruses has proposed three genera, Alphacoronavirus, Betacoronavirus, and Gammacoronavirus, to replace the traditional CoV groups 1, 2, and 3. As a result of the unique mechanism of viral replication, CoVs have a high frequency of recombination (22). Their tendency for recombination and the inherently high mutation rates in RNA virus may allow them to adapt to new hosts and ecological niches (18, 47).
The recent severe acute respiratory syndrome (SARS) epidemic, the discovery of SARS coronavirus (SARS-CoV), and the identification of SARS-CoV-like viruses from Himalayan palm civets and a raccoon dog from wild live markets in China have boosted interest in the discovery of novel CoVs in both humans and animals (5, 16, 33, 36, 39, 40, 46). A novel human CoV (HCoV) of the genus Alphacoronavirus, human coronavirus NL63 (HCoV-NL63), was reported independently by two groups in 2004 (12, 44). In 2005, we also described the discovery, complete genome sequence, clinical features, and molecular epidemiology of another novel HCoV, human coronavirus HKU1 (HCoV-HKU1), in the genus Betacoronavirus (24, 48, 50). As for animal CoVs, we and others have described the discovery of SARS-CoV-like viruses in horseshoe bats in Hong Kong Special Administrative Region (HKSAR) and other provinces of China (25, 30). Based on these findings, we conducted molecular surveillance studies to examine the diversity of CoVs in bats of our locality as well as of the Guangdong province of southern China, where the SARS epidemic originated and wet markets and game food restaurants serving bat dishes are commonly found. In these studies, at least nine other novel CoVs were discovered, including two novel subgroups in Betacoronavirus, subgroups C and D (26, 37, 45, 51). Other groups have also conducted molecular surveillance studies in bats and other animals, and additional novel CoVs were discovered and complete genomes sequenced (4, 6, 7, 9, 10, 13–15, 17, 21, 31, 32, 34, 43, 53).
Birds are the reservoir of major emerging viruses, most notably, avian influenza viruses (29). Due to their flocking behavior and abilities to fly over long distances, birds have the potential to disseminate these emerging viruses efficiently among themselves and to other animals and humans. As for CoVs, the number of known CoVs in birds is relatively small compared to that in bats. Recently, we described the discovery of three novel CoVs in three families of birds, named bulbul coronavirus HKU11 (BuCoV HKU11), thrush coronavirus HKU12 (ThCoV HKU12), and munia coronavirus HKU13 (MunCoV HKU13) (49). These three CoVs formed a unique group of CoV, which probably represented a novel genus of CoV, Deltacoronavirus (8). We hypothesize that there are other previously unrecognized CoVs in this novel genus from mammals and other families of birds. To test this hypothesis, we carried out a territory-wide molecular epidemiology study in 3,137 mammals and 3,519 birds in HKSAR. Based on the results of comparative genome and phylogenetic analysis in the present study, we propose seven novel CoVs in Deltacoronavirus. Our model of bats and birds as the gene source of the four genera of coronaviruses is also discussed.
MATERIALS AND METHODS
Animal surveillance and sample collection.
All specimens of bats, cats, dogs, wild rodents, monkeys, and birds were collected with the assistance of the Department of Agriculture, Fisheries and Conservation, HKSAR, and those of pigs, cattle, chickens, and street rodents were collected with the assistance of the Department of Food, Environmental and Hygiene, HKSAR, from various locations in HKSAR over a 53-month period (February 2007 to June 2011). All specimens of Asian leopard cats were collected in the Guangdong province of southern China over an 8-month period (August 2010 to March 2011). Tracheal, rectal, and cloacal swabs were collected using procedures described previously (47, 49). Nasopharyngeal aspirates from humans were collected from patients in Queen Mary Hospital over a 13-month period (February 2010 to February 2011) (24, 47, 50). A total of 7,140 samples from 11 species of bats, 169 pigs, 230 cats, 231 dogs, 47 cattle, 221 chickens, 389 rodents, 235 monkeys, 1,397 humans, 15 Asian leopard cats, and 3,298 dead wild birds of 134 different species in 38 families had been tested.
RNA extraction.
Viral RNA was extracted from the tracheal, rectal, and cloacal swabs and nasopharyngeal aspirates using RNeasy Mini Spin column (Qiagen, Hilden, Germany) (27, 45, 47, 50). The RNA was eluted in 50 μl of RNase-free water and was used as the template for reverse transcription-PCR (RT-PCR).
RT-PCR of RdRp gene of CoVs using Deltacoronavirus conserved primers and DNA sequencing.
Initial CoV screening was performed by amplifying a 440-bp fragment of the RNA-dependent RNA polymerase (RdRp) gene of CoVs using Deltacoronavirus conserved primers (5′-GTGGVTGTMTTAATGCACAGTC-3′ and 5′-TACTGYCTGTTRGTCATRGTG-3′) designed by multiple alignments of the nucleotide sequences of available RdRp genes of BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13 (49). Reverse transcription was performed using the SuperScript III kit (Invitrogen, San Diego, CA). The PCR mixture (25 μl) contained cDNA, PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 3 mM MgCl2, and 0.01% gelatin), 200 μM each deoxynucleoside triphosphate (dNTP), and 1.0 U Taq polymerase (Applied Biosystems, Foster City, CA). The mixtures were amplified with 60 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystems, Foster City, CA). Standard precautions were taken to avoid PCR contamination, and no false positive was observed in negative controls.
The PCR products were gel purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA), using the two PCR primers. The sequences of the PCR products were compared with known sequences of the RdRp genes of CoVs in the GenBank database.
Complete genome sequencing.
Two complete genomes of porcine coronavirus HKU15 (PorCoV HKU15) and one complete genome each of white-eye coronavirus HKU16 (WECoV HKU16), sparrow coronavirus HKU17 (SpCoV HKU17), magpie robin coronavirus HKU18 (MRCoV HKU18), night heron coronavirus HKU19 (NHCoV HKU19), wigeon coronavirus HKU20 (WiCoV HKU20), and common moorhen coronavirus HKU21 (CMCoV HKU21) were amplified and sequenced using the RNA extracted from the original swab specimens as templates. The RNA was converted to cDNA by a combined random-priming and oligo(dT)-priming strategy. The cDNA was amplified by degenerate primers designed by multiple alignments of the genomes of other CoVs with complete genomes available, using strategies described in our previous publications (28, 45, 48, 49) and the CoV database CoVDB (20) for sequence retrieval. Additional primers were designed from the results of the first and subsequent rounds of sequencing. These primer sequences are available on request. The 5′ ends of the viral genomes were confirmed by rapid amplification of cDNA ends (RACE) using the 5′/3′ RACE kit (Roche, Germany). Sequences were assembled and manually edited to produce final sequences of the viral genomes.
Genome analysis.
The nucleotide sequences of the genomes and the deduced amino acid sequences of the open reading frames (ORFs) were compared to those of other CoVs using EMBOSS needle (http://www.ebi.ac.uk). Phylogenetic tree construction was performed using the neighbor joining method with ClustalX 1.83. Protein family analysis was performed using PFAM and InterProScan (1, 2). Prediction of transmembrane domains was performed using TMpred and TMHMM (19, 41).
Estimation of divergence dates.
Divergence times for the four genera of CoVs, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus, were calculated using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in BEAST (Version 1.6.1) as described previously (11, 23, 27, 47). One parametric model (Constant Size) and one nonparametric model (Bayesian Skyline) tree priors were used for the inference. Analyses were performed under the GTR+I+G substitution model for RdRp gene sequence data and using both a strict and an unrelaxed log-normal-distributed (Ucld) relaxed molecular clock. The MCMC run was 5 × 107 steps long, with sampling every 1,000 steps. Convergence was assessed on the basis of the effective sampling size after a 10% burn-in using Tracer software version 1.5 (11). The mean time of the most recent common ancestor (tMRCA) and the highest posterior density regions at 95% (HPD) (i.e., a credible set that contains 95% of the sampled values) were calculated, and the best-fitting model was selected by a Bayes factor, using marginal likelihoods implemented in Tracer (see Table S1 in the supplemental material) (42). Bayesian Skyline under a relaxed-clock model with Ucld was adopted for making inferences, as Bayes factor analysis indicated that this model fitted the data better than other models tested (see Table S1). The trees were summarized in a target tree by the Tree Annotator program included in the BEAST package by choosing the tree with the maximum sum of posterior probabilities (maximum clade credibility) after a 10% burn-in.
Nucleotide sequence accession numbers.
The nucleotide sequences of the eight genomes of PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21 have been lodged within the GenBank sequence database under accession no. JQ065042 to JQ065049.
RESULTS
Animal surveillance and identification of seven novel mammalian and avian CoVs.
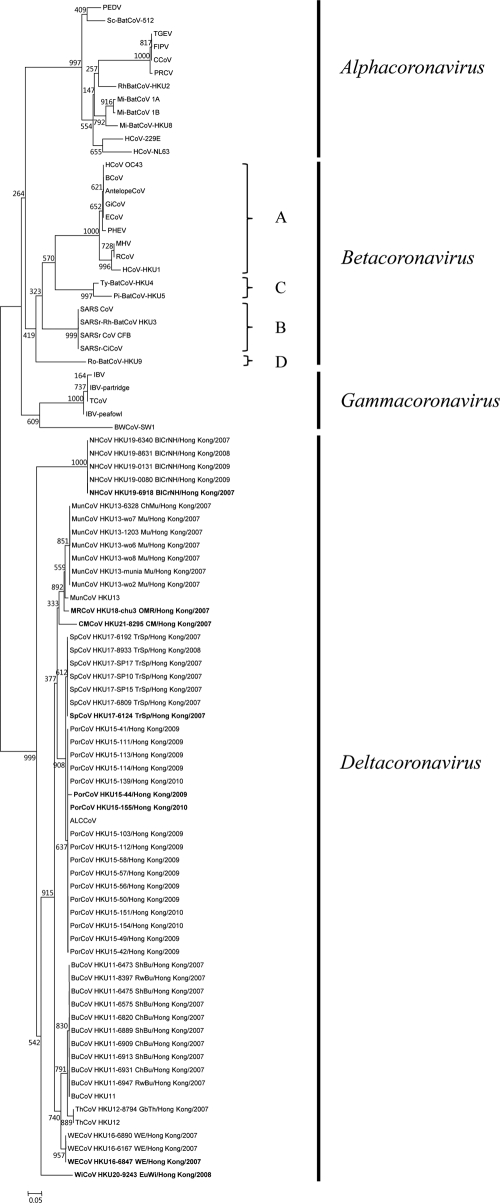
A total of 7,140 respiratory and alimentary specimens from 3,298 dead wild birds, 221 chickens, and 3,137 mammals were obtained (Table 1). RT-PCR for a 440-bp fragment in the RdRp genes of CoVs was positive in specimens from 17 pigs and 35 dead wild birds. Sequencing results suggested the presence of seven novel CoVs (Fig. 1 and Table 1). These seven novel CoVs were most closely related to our recently described BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13, sharing <66% nucleotide identity with all other known CoVs (Fig. 1). No positive results were obtained from any of the 15 Asian leopard cats, 434 bats, 230 cats, 47 cattle, 221 chickens, 231 dogs, 1,387 humans, 235 monkeys, and 389 rodents tested (Table 1).
Table 1.
Animals screened and associated CoVs in the present surveillance study
| Animal | Sample type | No. of specimens tested | No. (%) of specimens positive for CoV | CoV |
|---|---|---|---|---|
| Asian leopard cat | Rectal swab and tracheal swab | 30 | 0 | |
| Bat | Rectal swab | 434 | 0 | |
| Birda | Rectal swab | 3,306 | 35 (1.1%) | WECoV HKU16 (n = 3), SpCoV HKU17 (n = 7), MRCoV HKU18 (n = 1), NHCoV HKU19 (n = 5), WiCoV HKU20 (n = 1), CMCoV HKU21 (n = 1), BuCoV HKU11 (n = 10), ThCoV HKU12 (n = 1), MunCoV HKU13 (n = 6) |
| Cat | Rectal swab and tracheal swab | 460 | 0 | |
| Cattle | Rectal swab | 47 | 0 | |
| Chicken | Cloacal swab | 221 | 0 | |
| Dog | Rectal swab and tracheal swab | 462 | 0 | |
| Human | NPAb | 1,387 | 0 | |
| Monkey | Rectal swab | 235 | 0 | |
| Pig | Rectal swab | 169 | 17 (10.1%) | PorCoV HKU15 |
| Rodent | Rectal swab | 389 | 0 |
No. of birds tested for individual species and their associated CoVs are listed in Table S2 in the supplemental material.
NPA, nasopharyngeal aspirate.
Fig 1.
Phylogenetic analysis of amino acid sequences of the 228-bp fragment (excluding primer sequences) of RNA-dependent RNA polymerase (RdRp) of CoVs identified from dead wild birds and pigs in the present study. The tree was constructed by the neighbor joining method using Kimura correction and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 20 amino acids. The eight genomes completely sequenced are shown in bold. PEDV, porcine epidemic diarrhea virus (NC_003436); Sc-BatCoV-512, Scotophilus bat coronavirus 512 (NC_009657); TGEV, transmissible gastroenteritis virus (NC_002306); FIPV, feline infectious peritonitis virus (AY994055); CCoV, canine coronavirus (GQ477367); PRCV, porcine respiratory coronavirus (DQ811787); Rh-BatCoV-HKU2, Rhinolophus bat coronavirus HKU2 (EF203064); Mi-BatCoV 1A, Miniopterus bat coronavirus 1A (NC_010437); Mi-BatCoV 1B, Miniopterus bat coronavirus 1B (NC_010436); Mi-BatCoV-HKU8, Miniopterus bat coronavirus HKU8 (NC_010438); HCoV-229E, human coronavirus 229E (NC_002645); HCoV-NL63, human coronavirus NL63 (NC_005831); HCoV OC43, human coronavirus OC43 (NC_005147); BCoV, bovine coronavirus (NC_003045); AntelopeCoV, sable antelope CoV (EF424621); GiCoV, giraffe coronavirus (EF424622); ECoV, equine coronavirus (NC_010327); PHEV, porcine hemagglutinating encephalomyelitis virus (NC_007732); MHV, murine hepatitis virus (NC_001846); RCoV, rat coronavirus (NC_012936); HCoV-HKU1, human coronaivurs HKU1 (NC_006577); Ty-BatCoV-HKU4, Tylonycteris bat coronavirus HKU4 (NC_009019); Pi-BatCoV-HKU5, Pipistrellus bat coronavirus HKU5 (NC_009020); SARS CoV, SARS-related human coronavirus (NC_004718); SARSr-Rh-BatCoV HKU3, SARS-related Rhinolophus bat coronavirus HKU3 (DQ022305); SARSr CoV CFB, SARS-related Chinese ferret badger coronavirus (AY545919); SARSr-CiCoV, SARS-related palm civet coronavirus (AY304488); Ro-BatCoV-HKU9, Rousettus bat coronavirus HKU9 (NC_009021); IBV, infectious bronchitis virus (NC_001451); IBV-partridge, partridge coronavirus (AY646283); TCoV, turkey coronavirus (NC_010800); IBV-peafowl, peafowl coronavirus (AY641576); BWCoV-SW1, beluga whale coronavirus SW1 (NC_010646); ALCCoV, Asian leopard cat coronavirus (EF584908); BuCoV HKU11, bulbul coronavirus HKU11(FJ376619); ThCoV HKU12, thrush coronavirus HKU12 (FJ376621); MunCoV HKU13, munia coronavirus HKU13 (FJ376622); PorCoV HKU15, porcine coronavirus HKU15; WECoV HKU16, white-eye coronavirus HKU16; SpCoV HKU17 (TrSp, tree sparrow), sparrow coronavirus HKU17; MRCoV HKU18 (OMR, oriental magpie robin), magpie robin coronavirus HKU18; NHCoV HKU19 (BlCrNH, black-crowned night heron), night heron coronavirus HKU19; WiCoV HKU20 (EuWi, Eurasian wigeon), wigeon coronavirus HKU20; CMCoV HKU21, common moorhen (CM) coronavirus HKU21. Mu, munia; ChMu, chestnut munia; GbTh, gray-backed thrush; ShBu, sooty-headed bulbul; RwBu, red-whiskered bulbul; ChBu, chestnut bulbul.
Genome organization and coding potential of the seven novel mammalian and avian CoVs.
Complete genome sequence data of two strains of PorCoV HKU15 and one complete genome each of WECoV HKU16, SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21 were obtained by assembly of the sequences of the RT-PCR products from the RNA extracted from the corresponding individual specimens.
The size of the genomes of the seven novel CoVs ranged from 25,416 bases (PorCoV HKU15) to 26,674 (MRCoV HKU18) and their G+C contents ranged from 35% (CMCoV HKU21) to 47% (MRCoV HKU18) (Table 2). Their genome organizations are similar to those of other CoVs, with the characteristic gene order 5′-replicase ORF1ab, spike (S), envelope (E), membrane (M), nucleocapsid (N)-3′ (Fig. 2 and Table 3). Both 5′ and 3′ ends contain short untranslated regions. The replicase ORF1ab occupies 18.620 to 18.887 kb of the genomes (Table 3). This ORF encodes a number of putative proteins, including nsp3 [which contains the putative papain-like protease (PLpro)], nsp5 [putative chymotrypsin-like protease (3CLpro)], nsp12 (putative RdRp), nsp13 (putative helicase), and other proteins of unknown functions. Notably, the amino acids upstream to the putative cleavage sites at nsp2/nsp3, nsp3/nsp4, and nsp4/nsp5 are all AG, AG, and LQ for PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, and CMCoV HKU21; however, those at nsp2/nsp3 are VG and DG, those at nsp3/nsp4 are TG and GG, and those at nsp4/nsp5 are VQ for NHCoV HKU19 and WiCoV HKU20 (see Table S3 in the supplemental material).
Table 2.
Comparison of genomic features and amino acid identities among CoVs with complete genome sequences availablea
| CoV | Genome features |
Pairwise amino acid identity (%) |
Pairwise amino acid identity (%) |
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bases) | G+C content | PorCoV HKU15 |
WECoV HKU16 |
SpCoV HKU17 |
MRCoV HKU18 |
NHCoV HKU19 |
WiCoV HKU20 |
CMCoV HKU21 |
|||||||||||||||||||||||||||||
| 3CLpro | RdRp | Hel | S | N | 3CLpro | RdRp | Hel | S | N | 3CLpro | RdRp | Hel | S | N | 3CLpro | RdRp | Hel | S | N | 3CLpro | RdRp | Hel | S | N | 3CLpro | RdRp | Hel | S | N | 3CLpro | RdRp | Hel | S | N | |||
| Alphacoronavirus | |||||||||||||||||||||||||||||||||||||
| PEDV | 28,033 | 0.42 | 35.8 | 48.7 | 49.3 | 38.0 | 23.4 | 37.4 | 49.3 | 47.8 | 38.7 | 22.4 | 36.5 | 48.9 | 48.9 | 39.2 | 24.1 | 36.8 | 48.9 | 49.1 | 40.8 | 22.3 | 37.2 | 48.8 | 47.9 | 36.3 | 20.6 | 39.7 | 50.0 | 48.5 | 37.9 | 21.1 | 38.1 | 49.6 | 46.7 | 39.4 | 21.7 |
| TGEV | 28,586 | 0.38 | 34.9 | 49.6 | 51.6 | 35.5 | 23.2 | 34.5 | 49.4 | 49.6 | 36.1 | 24.5 | 35.3 | 49.8 | 51.4 | 39.4 | 23.5 | 34.6 | 49.6 | 50.7 | 39.9 | 23.7 | 32.9 | 50.8 | 50.8 | 37.1 | 22.3 | 37.3 | 50.2 | 50.2 | 36.7 | 23.7 | 33.7 | 49.3 | 49.4 | 36.4 | 22.9 |
| FIPV | 29,355 | 0.38 | 35.7 | 49.7 | 51.2 | 35.1 | 24.5 | 35.5 | 49.6 | 49.3 | 36.3 | 25.1 | 35.7 | 49.9 | 51.1 | 38.5 | 25.0 | 35.5 | 50.1 | 50.4 | 38.9 | 22.2 | 32.4 | 51.2 | 50.7 | 36.9 | 20.8 | 37.9 | 50.1 | 50.0 | 36.5 | 22.5 | 34.5 | 49.7 | 49.1 | 36.0 | 22.1 |
| CCoV | 29,363 | 0.38 | 35.6 | 49.7 | 51.6 | 34.9 | 23.3 | 35.2 | 49.4 | 49.6 | 35.6 | 24.0 | 35.9 | 49.8 | 51.4 | 38.8 | 23.3 | 35.3 | 49.6 | 50.7 | 39.3 | 23.0 | 32.9 | 50.8 | 50.7 | 36.4 | 23.6 | 37.3 | 49.9 | 50.7 | 36.3 | 23.7 | 33.3 | 49.1 | 49.4 | 35.2 | 23.4 |
| PRCV | 27,550 | 0.37 | 34.9 | 49.5 | 51.6 | 40.3 | 23.2 | 34.5 | 49.3 | 49.6 | 40.5 | 23.2 | 35.3 | 49.7 | 51.4 | 44.8 | 23.5 | 34.6 | 49.5 | 50.7 | 44.4 | 22.8 | 32.9 | 50.7 | 50.8 | 41.4 | 22.0 | 37.3 | 50.3 | 50.2 | 41.9 | 23.7 | 33.7 | 49.1 | 49.4 | 40.1 | 21.8 |
| HCoV-229E | 27,317 | 0.38 | 34.4 | 49.3 | 50.6 | 42.5 | 21.6 | 35.4 | 49.0 | 48.3 | 42.4 | 22.5 | 34.2 | 49.5 | 50.2 | 45.5 | 23.0 | 34.8 | 49.3 | 49.8 | 44.0 | 22.5 | 34.6 | 49.2 | 49.0 | 39.9 | 20.4 | 36.3 | 49.6 | 49.6 | 44.1 | 21.5 | 34.8 | 48.6 | 47.5 | 43.2 | 22.8 |
| HCoV-NL63 | 27,553 | 0.34 | 35.9 | 48.8 | 49.9 | 38.2 | 22.1 | 38.1 | 49.2 | 48.1 | 40.1 | 23.0 | 35.6 | 49.2 | 49.6 | 39.3 | 22.6 | 36.9 | 49.2 | 49.5 | 39.3 | 24.6 | 34.8 | 48.9 | 49.0 | 36.2 | 21.5 | 39.1 | 50.6 | 48.6 | 38.8 | 23.5 | 36.9 | 49.5 | 47.2 | 38.8 | 20.6 |
| Rh-BatCoV-HKU2 | 27,165 | 0.39 | 34.4 | 50.1 | 51.4 | 25.0 | 20.8 | 34.3 | 50.0 | 49.1 | 25.2 | 22.3 | 34.4 | 50.2 | 51.1 | 26.2 | 20.9 | 34.4 | 49.8 | 50.3 | 25.9 | 21.7 | 34.3 | 50.5 | 49.7 | 25.1 | 20.9 | 35.5 | 50.6 | 49.1 | 26.3 | 25.2 | 34.1 | 50.1 | 49.1 | 27.3 | 22.5 |
| Mi-BatCoV 1A | 28,326 | 0.38 | 33.5 | 49.0 | 51.4 | 35.8 | 24.4 | 35.0 | 49.4 | 50.1 | 35.7 | 23.3 | 34.2 | 49.4 | 51.1 | 39.4 | 25.2 | 34.2 | 49.0 | 51.4 | 38.2 | 24.5 | 32.6 | 50.5 | 48.7 | 35.7 | 23.8 | 34.7 | 49.9 | 48.9 | 36.4 | 22.5 | 33.4 | 47.9 | 49.1 | 38.6 | 22.4 |
| Mi-BatCoV 1B | 28,476 | 0.39 | 34.2 | 48.5 | 51.1 | 35.6 | 24.6 | 35.4 | 48.8 | 49.4 | 36.1 | 22.1 | 34.8 | 48.8 | 50.7 | 39.1 | 24.9 | 33.5 | 48.8 | 51.1 | 38.2 | 23.8 | 31.9 | 49.8 | 48.2 | 35.7 | 22.3 | 35.7 | 49.6 | 48.9 | 35.7 | 24.1 | 32.8 | 47.7 | 48.3 | 38.2 | 22.8 |
| Mi-BatCoV-HKU8 | 28,773 | 0.42 | 33.1 | 49.3 | 49.8 | 35.9 | 19.4 | 36.0 | 49.9 | 47.5 | 36.0 | 18.8 | 33.4 | 49.6 | 49.3 | 38.9 | 20.4 | 34.4 | 49.3 | 49.3 | 40.4 | 19.8 | 34.6 | 50.1 | 48.1 | 37.2 | 22.6 | 36.7 | 50.5 | 48.8 | 37.6 | 22.3 | 36.0 | 48.5 | 47.4 | 37.0 | 21.6 |
| Sc-BatCoV-512 | 28,179 | 0.40 | 33.8 | 48.6 | 49.1 | 39.0 | 24.8 | 36.0 | 49.2 | 47.5 | 38.7 | 23.7 | 34.1 | 48.7 | 48.8 | 41.3 | 25.2 | 35.4 | 48.3 | 48.8 | 41.1 | 23.4 | 34.9 | 49.0 | 47.8 | 36.8 | 22.3 | 37.3 | 48.9 | 49.1 | 38.1 | 23.1 | 34.7 | 48.7 | 47.8 | 39.6 | 24.3 |
| Betacoronavirus | |||||||||||||||||||||||||||||||||||||
| Subgroup A | |||||||||||||||||||||||||||||||||||||
| HCoV-OC43 | 30,738 | 0.37 | 38.1 | 51.6 | 48.3 | 26.0 | 22.2 | 38.9 | 51.5 | 48.6 | 25.9 | 23.2 | 37.8 | 51.8 | 48.3 | 26.9 | 22.4 | 37.5 | 51.3 | 48.3 | 26.4 | 21.0 | 34.1 | 54.5 | 48.4 | 25.9 | 22.5 | 38.7 | 51.8 | 48.4 | 25.7 | 20.4 | 37.8 | 51.5 | 49.0 | 25.6 | 24.2 |
| BCoV | 31,028 | 0.37 | 38.5 | 51.8 | 48.4 | 25.7 | 22.9 | 38.8 | 51.7 | 48.6 | 25.8 | 21.7 | 38.5 | 51.8 | 48.4 | 26.7 | 22.8 | 37.8 | 51.5 | 48.4 | 26.9 | 24.0 | 34.4 | 54.5 | 48.5 | 25.6 | 23.4 | 38.3 | 51.7 | 48.5 | 26.0 | 21.5 | 38.2 | 51.6 | 49.0 | 25.7 | 23.4 |
| PHEV | 30,480 | 0.37 | 38.5 | 51.7 | 48.3 | 26.9 | 22.1 | 38.1 | 51.6 | 48.6 | 26.1 | 23.1 | 38.5 | 51.6 | 48.3 | 27.2 | 22.3 | 37.8 | 51.4 | 48.3 | 26.9 | 22.2 | 34.4 | 54.5 | 48.5 | 26.1 | 22.7 | 38.7 | 51.7 | 48.5 | 27.1 | 21.6 | 38.2 | 51.6 | 49.0 | 25.4 | 24.1 |
| AntelopeCoV | 30,995 | 0.37 | 38.5 | 51.8 | 48.4 | 25.8 | 22.9 | 38.8 | 51.7 | 48.5 | 25.6 | 21.7 | 38.5 | 51.8 | 48.4 | 27.0 | 22.1 | 37.8 | 51.4 | 48.4 | 27.0 | 24.0 | 34.4 | 54.4 | 48.5 | 26.1 | 23.9 | 38.3 | 51.7 | 48.5 | 26.2 | 21.5 | 38.2 | 51.6 | 49.2 | 25.8 | 23.4 |
| GiCoV | 30,979 | 0.37 | 38.8 | 51.8 | 48.4 | 25.9 | 22.9 | 38.8 | 51.7 | 48.5 | 25.7 | 21.7 | 38.8 | 51.8 | 48.4 | 27.2 | 22.1 | 37.8 | 51.4 | 48.4 | 27.0 | 24.0 | 34.4 | 54.4 | 48.5 | 25.7 | 23.9 | 38.3 | 51.7 | 48.5 | 26.5 | 21.5 | 38.5 | 51.6 | 49.2 | 25.9 | 23.4 |
| ECoV | 30,992 | 0.37 | 38.5 | 51.7 | 49.8 | 26.0 | 23.9 | 38.8 | 51.6 | 49.0 | 26.4 | 22.6 | 38.5 | 51.7 | 49.9 | 26.5 | 24.0 | 37.8 | 51.4 | 49.8 | 27.5 | 23.5 | 34.4 | 54.6 | 48.5 | 25.3 | 24.6 | 38.3 | 51.5 | 48.5 | 26.9 | 22.0 | 38.2 | 51.6 | 49.7 | 25.6 | 24.9 |
| MHV | 31,357 | 0.42 | 38.3 | 51.9 | 48.1 | 26.3 | 24.3 | 39.0 | 51.3 | 48.5 | 26.1 | 24.0 | 38.3 | 51.8 | 48.3 | 26.5 | 25.3 | 37.6 | 51.9 | 48.3 | 26.3 | 24.2 | 35.0 | 53.6 | 47.5 | 25.3 | 24.6 | 39.6 | 50.8 | 47.9 | 27.1 | 24.0 | 39.2 | 51.2 | 48.6 | 26.0 | 24.6 |
| HCoV-HKU1 | 29,926 | 0.32 | 38.1 | 51.2 | 49.3 | 26.1 | 25.2 | 38.0 | 51.4 | 48.2 | 26.4 | 24.8 | 37.9 | 51.3 | 49.4 | 25.7 | 26.0 | 36.4 | 51.4 | 48.8 | 26.4 | 26.0 | 36.3 | 54.4 | 47.4 | 25.4 | 24.7 | 38.1 | 50.9 | 48.5 | 25.8 | 22.7 | 38.3 | 51.2 | 47.8 | 25.0 | 25.4 |
| RCoV | 31,250 | 0.41 | 38.7 | 51.8 | 47.9 | 27.2 | 24.5 | 39.5 | 51.4 | 48.3 | 27.0 | 24.3 | 38.5 | 51.7 | 48.1 | 25.5 | 25.1 | 38.2 | 51.8 | 48.2 | 25.8 | 25.0 | 35.0 | 53.6 | 47.4 | 24.3 | 25.2 | 39.9 | 50.7 | 47.7 | 27.4 | 24.1 | 38.3 | 51.0 | 48.4 | 26.4 | 23.5 |
| Subgroup B | |||||||||||||||||||||||||||||||||||||
| SARS CoV | 29,751 | 0.41 | 34.5 | 50.7 | 51.4 | 26.1 | 26.5 | 36.1 | 50.3 | 50.6 | 27.9 | 24.7 | 34.2 | 51.1 | 51.6 | 25.3 | 25.6 | 34.2 | 50.8 | 51.4 | 25.4 | 26.2 | 32.1 | 50.5 | 50.2 | 26.3 | 22.7 | 34.8 | 49.8 | 50.3 | 26.9 | 24.3 | 32.9 | 50.8 | 51.0 | 27.3 | 24.8 |
| SARSr-CiCoV | 29,728 | 0.41 | 34.5 | 50.7 | 51.4 | 26.2 | 26.5 | 36.1 | 50.3 | 50.6 | 28.0 | 24.7 | 34.2 | 51.1 | 51.6 | 25.2 | 25.6 | 34.2 | 50.8 | 51.4 | 25.5 | 26.2 | 32.1 | 50.5 | 50.2 | 26.2 | 22.7 | 34.8 | 49.8 | 50.3 | 27.0 | 24.3 | 32.9 | 50.8 | 51.0 | 27.1 | 24.8 |
| SARSr-Rh-BatCoV HKU3 | 29,704 | 0.41 | 34.2 | 50.5 | 51.4 | 26.4 | 25.2 | 35.8 | 50.3 | 50.8 | 26.2 | 24.3 | 33.9 | 51.1 | 51.6 | 25.6 | 24.9 | 33.9 | 50.9 | 51.4 | 26.0 | 25.7 | 32.1 | 50.4 | 50.6 | 25.6 | 23.0 | 34.8 | 49.7 | 50.5 | 26.0 | 23.5 | 32.6 | 50.6 | 51.3 | 27.2 | 24.1 |
| SARSr CoV CFB | 29,734 | 0.41 | 34.5 | 50.6 | 51.4 | 26.1 | 26.5 | 36.1 | 50.2 | 50.6 | 28.0 | 24.7 | 34.2 | 51.0 | 51.6 | 25.5 | 25.6 | 34.2 | 50.7 | 51.4 | 25.2 | 26.2 | 32.1 | 50.4 | 50.2 | 25.9 | 22.7 | 34.8 | 49.9 | 50.3 | 26.8 | 24.3 | 32.9 | 50.7 | 51.0 | 27.2 | 24.8 |
| Subgroup C | |||||||||||||||||||||||||||||||||||||
| Ty-BatCoV-HKU4 | 30,286 | 0.38 | 36.9 | 51.2 | 49.8 | 26.6 | 25.1 | 36.6 | 51.0 | 49.4 | 26.1 | 24.7 | 36.9 | 51.5 | 49.7 | 27.0 | 26.2 | 35.7 | 51.0 | 49.7 | 27.3 | 25.9 | 32.7 | 51.3 | 49.9 | 27.3 | 24.4 | 35.8 | 50.9 | 49.9 | 26.4 | 24.4 | 35.6 | 51.9 | 48.9 | 26.8 | 25.1 |
| Pi-BatCoV-HKU5 | 30,488 | 0.43 | 35.7 | 51.1 | 50.0 | 26.0 | 25.6 | 37.8 | 50.3 | 49.0 | 25.5 | 25.7 | 35.4 | 51.4 | 49.8 | 27.2 | 25.3 | 35.0 | 51.1 | 49.7 | 26.3 | 26.2 | 33.7 | 50.9 | 49.6 | 26.2 | 25.6 | 34.6 | 51.2 | 49.8 | 25.3 | 24.7 | 36.0 | 50.9 | 49.0 | 25.7 | 26.1 |
| Subgroup D | |||||||||||||||||||||||||||||||||||||
| Ro-Bat-CoV HKU9 | 29,114 | 0.41 | 36.4 | 51.6 | 51.2 | 28.4 | 25.1 | 39.2 | 52.6 | 50.1 | 26.5 | 23.1 | 36.4 | 51.7 | 50.9 | 26.6 | 24.9 | 35.8 | 51.9 | 50.9 | 27.7 | 25.0 | 33.8 | 52.2 | 50.9 | 27.0 | 22.8 | 35.0 | 51.4 | 49.2 | 27.2 | 22.9 | 36.9 | 52.3 | 51.4 | 27.8 | 23.3 |
| Gammacoronavirus | |||||||||||||||||||||||||||||||||||||
| IBV | 27,608 | 0.38 | 43.9 | 54.8 | 56.6 | 30.3 | 30.0 | 42.6 | 54.6 | 54.5 | 29.9 | 30.8 | 44.2 | 54.9 | 56.6 | 27.6 | 28.9 | 43.3 | 54.3 | 56.2 | 28.4 | 30.3 | 43.6 | 53.6 | 54.8 | 28.7 | 29.7 | 47.1 | 52.4 | 55.4 | 29.4 | 29.2 | 41.7 | 53.9 | 55.1 | 29.4 | 28.1 |
| TCoV | 27,657 | 0.38 | 43.6 | 54.9 | 57.1 | 30.1 | 29.2 | 43.3 | 54.5 | 55.3 | 30.3 | 29.6 | 43.9 | 55.0 | 57.1 | 29.5 | 29.5 | 42.3 | 54.4 | 57.1 | 30.3 | 29.2 | 44.3 | 53.8 | 55.3 | 30.3 | 30.0 | 46.2 | 52.8 | 55.4 | 30.3 | 29.9 | 41.3 | 53.6 | 55.8 | 30.6 | 28.2 |
| BWCoV-SW1 | 31,686 | 0.39 | 38.8 | 52.9 | 52.8 | 27.1 | 32.1 | 39.5 | 52.9 | 51.6 | 28.3 | 31.1 | 38.8 | 52.9 | 52.8 | 28.5 | 31.9 | 37.2 | 52.3 | 52.2 | 27.3 | 31.6 | 41.1 | 52.8 | 54.2 | 27.4 | 30.2 | 42.0 | 52.2 | 51.1 | 27.2 | 30.0 | 38.8 | 52.1 | 52.7 | 28.2 | 31.5 |
| Deltacoronavirus | |||||||||||||||||||||||||||||||||||||
| BuCoV HKU11 | 26,476 | 0.39 | 81.1 | 88.2 | 89.4 | 69.8 | 74.8 | 82.4 | 90.9 | 96.0 | 62.5 | 73.2 | 80.8 | 88.2 | 89.6 | 43.5 | 75.1 | 79.5 | 88.3 | 90.4 | 44.5 | 71.9 | 57.0 | 72.3 | 76.8 | 41.1 | 50.6 | 58.3 | 70.8 | 75.4 | 43.3 | 51.4 | 77.5 | 84.8 | 91.0 | 51.8 | 60.7 |
| ThCoV HKU12 | 26,396 | 0.38 | 82.1 | 88.2 | 89.7 | 47.9 | 79.7 | 83.1 | 89.5 | 94.7 | 47.8 | 81.0 | 81.8 | 88.2 | 89.9 | 46.7 | 79.4 | 81.4 | 86.8 | 89.9 | 45.8 | 76.7 | 57.3 | 71.9 | 76.4 | 43.6 | 49.4 | 57.7 | 71.3 | 74.5 | 43.6 | 49.6 | 78.2 | 84.4 | 90.5 | 46.2 | 63.3 |
| MunCoV HKU13 | 26,552 | 0.43 | 82.7 | 90.1 | 95.8 | 71.2 | 76.8 | 76.5 | 87.9 | 89.1 | 61.3 | 74.6 | 83.4 | 90.1 | 96.0 | 43.8 | 78.8 | 94.5 | 94.6 | 98.0 | 46.1 | 87.5 | 53.1 | 72.9 | 78.0 | 41.4 | 53.4 | 55.4 | 71.7 | 75.7 | 44.0 | 53.2 | 72.0 | 84.7 | 85.4 | 52.2 | 64.4 |
| PorCoV HKU15 | 25,421 | 0.43 | 76.9 | 88.1 | 88.4 | 61.9 | 75.8 | 97.0 | 97.8 | 99.2 | 44.8 | 96.8 | 84.3 | 90.6 | 96.1 | 44.4 | 77.9 | 54.0 | 72.5 | 78.2 | 41.8 | 52.2 | 57.7 | 71.0 | 74.9 | 43.8 | 52.4 | 73.6 | 84.5 | 84.6 | 50.1 | 62.0 | |||||
| WECoV HKU16 | 26,027 | 0.40 | 76.9 | 88.1 | 88.4 | 61.9 | 75.8 | 77.2 | 88.3 | 88.6 | 46.4 | 76.4 | 77.2 | 87.3 | 89.1 | 45.5 | 75.4 | 55.0 | 71.7 | 76.6 | 42.1 | 49.6 | 59.6 | 71.5 | 74.3 | 43.4 | 50.6 | 76.9 | 84.8 | 90.5 | 51.5 | 64.3 | |||||
| SpCoV HKU17 | 26,067 | 0.45 | 97.0 | 97.8 | 99.2 | 44.8 | 96.8 | 77.2 | 88.3 | 88.6 | 46.4 | 76.4 | 84.9 | 91.0 | 96.3 | 68.1 | 79.0 | 52.8 | 72.3 | 78.4 | 47.2 | 51.9 | 58.0 | 71.0 | 75.1 | 45.8 | 53.2 | 73.0 | 84.7 | 84.9 | 46.0 | 63.5 | |||||
| MRCoV HKU18 | 26,674 | 0.47 | 84.3 | 90.6 | 96.1 | 44.4 | 77.9 | 77.2 | 87.3 | 89.1 | 45.5 | 75.4 | 84.9 | 91.0 | 96.3 | 68.1 | 79.0 | 54.0 | 72.5 | 77.7 | 46.4 | 53.8 | 56.4 | 71.2 | 75.1 | 46.3 | 53.1 | 73.3 | 85.1 | 84.8 | 45.7 | 63.9 | |||||
| NHCoV HKU19 | 26,064 | 0.38 | 54.0 | 72.5 | 78.2 | 41.8 | 52.2 | 55.0 | 71.7 | 76.6 | 42.1 | 49.6 | 52.8 | 72.3 | 78.4 | 47.2 | 51.9 | 54.0 | 72.5 | 77.7 | 46.4 | 53.8 | 58.3 | 69.3 | 75.4 | 41.0 | 54.5 | 55.5 | 71.9 | 77.6 | 43.6 | 54.5 | |||||
| WiCoV HKU20 | 26,211 | 0.39 | 57.7 | 71.0 | 74.9 | 43.8 | 52.4 | 59.6 | 71.5 | 74.3 | 43.4 | 50.6 | 58.0 | 71.0 | 75.1 | 45.8 | 53.2 | 56.4 | 71.2 | 75.1 | 46.3 | 53.1 | 58.3 | 69.3 | 75.4 | 41.0 | 54.5 | 58.3 | 70.8 | 76.4 | 44.1 | 57.0 | |||||
| CMCoV HKU21 | 26,216 | 0.35 | 73.6 | 84.5 | 84.6 | 50.1 | 62.0 | 76.9 | 84.8 | 90.5 | 51.5 | 64.3 | 73.0 | 84.7 | 84.9 | 46.0 | 63.5 | 73.3 | 85.1 | 84.8 | 45.7 | 63.9 | 55.5 | 71.9 | 77.6 | 43.6 | 54.5 | 58.3 | 70.8 | 76.4 | 44.1 | 57.0 | |||||
Comparison of genomic features of PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21 and other CoVs with complete genome sequences available and of amino acid identities between the predicted 3CLpro, RNA-dependent RNA (RdRp), helicase (Hel), S, and N proteins of PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21 and the corresponding proteins of other CoVs. PEDV, porcine epidemic diarrhea virus; TGEV, porcine transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; CCoV, canine coronavirus; PRCV, porcine respiratory coronavirus; HCoV-229E, human coronavirus 229E; HCoV-NL63, human coronavirus NL63; Rh-BatCoV-HKU2, Rhinolophus bat coronavirus HKU2; Mi-BatCoV 1A, Miniopterus bat coronavirus 1A; Mi-BatCoV 1B, Miniopterus bat coronavirus 1B; Mi-BatCoV-HKU8, Miniopterus bat coronavirus HKU8; Sc-BatCoV-512, Scotophilus bat coronavirus 512; HCoV OC43, human coronavirus OC43; BCoV, bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; AntelopeCoV, sable antelope coronavirus; GiCoV, giraffe coronavirus; ECoV, equine coronavirus; MHV, murine hepatitis virus; HCoV-HKU1, human coronavirus HKU1; RCoV, rat coronavirus; SARS CoV, SARS-related human coronavirus; SARSr-CiCoV, SARS-related palm civet coronavirus; SARSr-Rh-BatCoV HKU3, SARS-related Rhinolophus bat coronavirus HKU3; SARSr CoV CFB, SARS-related Chinese ferret badger coronavirus; Ty-BatCoV-HKU4, Tylonycteris bat coronavirus HKU4; Pi-BatCoV-HKU5, Pipistrellus bat coronavirus HKU5; Ro-BatCoV-HKU9, Rousettus bat coronavirus HKU9; IBV, infectious bronchitis virus; TCoV, turkey coronavirus; BWCoV-SW1, Beluga whale coronavirus SW1; BuCoV HKU11, bulbul coronavirus HKU11; ThCoV HKU12, thrush coronavirus HKU12; MunCoV HKU13, munia coronavirus HKU13.
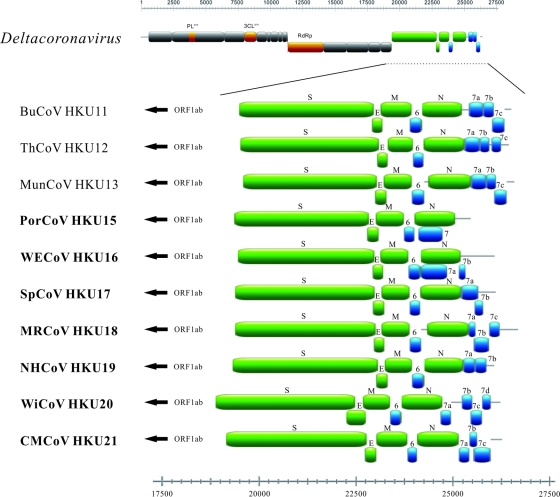
Fig 2.
Genome organization of members in Deltacoronavirus. ORFs downstream of S gene are magnified to show the differences among the genomes of the 10 CoVs. Papain-like protease (PLpro), chymotrypsin-like protease (3CLpro), and RNA-dependent RNA polymerase (RdRp) are represented by orange boxes. Spike (S), envelope (E), membrane (M), and nucleocapsid (N) are represented by green boxes. Putative accessory proteins are represented by blue boxes. The seven CoVs discovered in this study are shown in bold.
Table 3.
Coding potential and putative transcription regulatory sequences of CoV genomesa
| CoV | ORF | Location (nt) | Length (nt) | Length (aa) | Frame | Putative TRS |
|
|---|---|---|---|---|---|---|---|
| TRS location (nt) | TRS sequence(s) (distance in bases to AUG)b | ||||||
| PorCoV HKU15 | 1ab | 540–19342 | 18,803 | 6,268 | +3, +2 | 75 | ACACCA(459)AUG |
| S | 19324–22806 | 3,483 | 1,161 | +1 | 19178 | ACACCA(145)AUG | |
| E | 22800–23051 | 252 | 84 | +3 | 22777 | ACACCG(17)AUG | |
| M | 23044–23697 | 654 | 218 | +1 | 23018 | ACACCA(20)AUG | |
| NS6 | 23697–23981 | 285 | 95 | +3 | 23645 | ACACCA(46)AUG | |
| N | 24002–25030 | 1,029 | 343 | +2 | 23989 | ACACCA(7)AUG | |
| NS7 | 24096–24698 | 603 | 201 | +3 | 24008 | GCACCA(82)AUG | |
| WECoV HKU16 | 1ab | 511–19397 | 18,887 | 6,296 | +1, +3 | 66 | ACACCA(439)AUG |
| S | 19379–22918 | 3,540 | 1,180 | +2 | 19233 | ACACCA(140)AUG | |
| E | 22912–23160 | 249 | 83 | +1 | 22886 | ACACCA(20)AUG | |
| M | 23153–23809 | 657 | 219 | +2 | 23130 | ACACCA(17)AUG | |
| NS6 | 23809–24090 | 282 | 94 | +1 | 23768 | ACAUCA(35)AUG | |
| N | 24115–25158 | 1,044 | 348 | +1 | 24101 | ACACCA(8)AUG | |
| NS7a | 24143–24811 | 669 | 223 | +2 | 24101 | ACACCA(36)AUG | |
| NS7b | 25139–25270 | 132 | 44 | +2 | 25039 | AAACCA(94)AUG | |
| SpCoV HKU17 | 1ab | 520–19352 | 18,833 | 6,278 | +1, +3 | 57 | ACACCA(452)AUG |
| S | 19334–22954 | 3,621 | 1,207 | +2 | 19188 | ACACCA(140)AUG | |
| E | 22948–23196 | 249 | 83 | +1 | 22925 | ACACCG(17)AUG | |
| M | 23189–23842 | 654 | 218 | +2 | 23166 | ACACCA(17)AUG | |
| NS6 | 23842–24129 | 288 | 96 | +1 | 23790 | ACACCA(46)AUG | |
| N | 24150–25178 | 1,029 | 343 | +3 | 24137 | ACACCA(7)AUG | |
| NS7a | 25189–25623 | 435 | 145 | +1 | 25179 | ACACCA(4)AUG | |
| NS7b | 25539–25751 | 213 | 71 | +3 | 25523 | ACUCCA(10)AUG | |
| MRCoV HKU18 | 1ab | 596–19356 | 18,761 | 6,254 | +2, +1 | 64 | ACACCA(526)AUG |
| S | 19338–22991 | 3,654 | 1,218 | +3 | 19192 | ACACCA(140)AUG | |
| E | 22985–23233 | 249 | 83 | +2 | 22945 | ACACCG(34)AUG | |
| M | 23226–23882 | 657 | 219 | +3 | 23203 | ACACCA(17)AUG | |
| NS6 | 23882–24172 | 291 | 97 | +2 | 23857 | ACGCCA(19)AUG | |
| N | 24355–25395 | 1,041 | 347 | +1 | 24340 | ACACCA(9)AUG | |
| NS7a | 25407–25580 | 174 | 58 | +3 | 25396 | ACACCA(5)AUG | |
| NS7b | 25561–25932 | 372 | 124 | +1 | |||
| NS7c | 25941–26195 | 255 | 85 | +3 | 25910 | ACACCA(25)AUG | |
| NHCoV HKU19 | 1ab | 482–19323 | 18,842 | 6,281 | +2, +1 | 67 | ACACCG(409)AUG |
| S | 19305–23069 | 3,765 | 1,255 | +3 | 19156 | ACACCG(143)AUG | |
| E | 23069–23317 | 249 | 83 | +2 | 23013 | ACACCA(50)AUG | |
| M | 23310–23960 | 651 | 217 | +3 | 23211 | ACACCG(93)AUG | |
| NS6 | 23960–24238 | 279 | 93 | +2 | 23951 | ACACCU(3)AUG | |
| N | 24248–25276 | 1,029 | 343 | +2 | 24231 | ACACCU(8)AUG | |
| NS7a | 25277–25573 | 297 | 99 | +2 | 25248 | ACACCG(23)AUG | |
| NS7b | 25583–25876 | 294 | 98 | +2 | 25560 | ACACCA(17)AUG | |
| WiCoV HKU20 | 1ab | 219–18838 | 18,620 | 6,207 | +3, +2 | 60 | ACACCA(153)AUG |
| S | 18817–22455 | 3,639 | 1,213 | +1 | 18731 | ACACCU(80)AUG | |
| E | 22455–22715 | 261 | 87 | +3 | 22380 | ACACCA(69)AUG | |
| M | 22708–23358 | 651 | 217 | +1 | 22597 | ACACCG(105)AUG | |
| NS6 | 23358–23630 | 273 | 91 | +3 | |||
| N | 23646–24698 | 1,053 | 351 | +3 | 23631 | ACACCA(9)AUG | |
| NS7a | 24695–24928 | 234 | 78 | +2 | 24609 | AAACCA(80)AUG | |
| NS7b | 25218–25466 | 249 | 83 | +3 | 25177 | ACACCG(35)AUG | |
| NS7c | 25450–25716 | 267 | 89 | +1 | 25444 | ACACCGAUG | |
| NS7d | 25752–25952 | 201 | 67 | +3 | 25735 | AAACCU(11)AUG | |
| CMCoV HKU21 | 1ab | 478–19103 | 18,626 | 6,209 | +1, +3 | 63 | ACACCA(409)AUG |
| S | 19085–22729 | 3,645 | 1,215 | +2 | 18939 | ACACCA(140)AUG | |
| E | 22723–22971 | 249 | 83 | +1 | 22697 | ACACCA(20)AUG | |
| M | 22973–23779 | 807 | 269 | +2 | 22938 | ACACCA(29)AUG | |
| NS6 | 23779–24024 | 246 | 82 | +1 | 23727 | ACACCA(46)AUG | |
| N | 24052–25107 | 1,056 | 352 | +1 | 24039 | ACACCG(7)AUG | |
| NS7a | 25107–25379 | 273 | 91 | +3 | 25036 | ACACCU(65)AUG | |
| NS7b | 25391–25576 | 186 | 62 | +2 | 25379 | ACACCU(6)AUG | |
| NS7c | 25500–25916 | 417 | 139 | +2 | |||
PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21. aa, amino acid; nt, nucleotide.
Boldface indicates putative TRS sequences. The nucleotide variations are in italic.
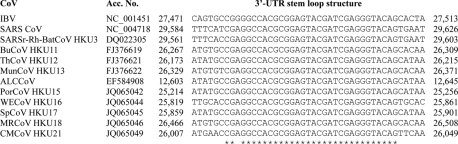
The seven novel CoVs display similar genome organizations and differ only in the number of ORFs downstream of N (Fig. 2). Their transcription regulatory sequences (TRSs) conform to the consensus motif 5′-ACACCA-3′ (Table 3), which appears to be unique to members of the genus Deltacoronavirus. Interestingly, similar to BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13, the perfect TRSs of S in the genomes of the seven novel CoVs were separated from the corresponding AUG by 80 to 145 bases (Table 3). This is in contrast to the relatively small number of bases between the TRSs for S and the corresponding AUG (range: from 0 bases in HCoV-NL63, Rhinolophus bat coronavirus HKU2 [Rh-BatCoV-HKU2], HCoV-HKU1, bovine coronavirus [BCoV], HCoV-OC43, mouse hepatitis virus [MHV], porcine hemagglutinating encephalomyelitis virus, SARS-CoV, and SARS-related Rhinolophus bat coronavirus HKU3 [SARSr-Rh-batCoV HKU3] to 52 bases in infectious bronchitis virus [IBV]) in members of Alphacoronavirus, Betacoronavirus, and Gammacoronavirus. Similar to BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13, the genomes of the seven novel CoVs have putative PLpro, which are homologous to PL2pro of Alphacoronavirus and Betacoronavirus subgroup A and PLpro of Betacoronavirus subgroups B, C, and D and Gammacoronavirus (Fig. 2). Similar to BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13, one ORF (NS6) is found between M and N of the genomes of the seven novel CoVs. On the other hand, one ORF (NS7) is present overlapping with N in PorCoV HKU15, two ORFs (NS7a and 7b) are present overlapping or downstream of N in WECoV HKU16, SpCoV HKU17, and NHCoV HKU19, three ORFs (NS7a, 7b, and 7c) are present downstream of N in MRCoV HKU18 and CMCoV HKU21, and four ORFs (NS7a, 7b, 7c, and 7d) are present overlapping or downstream of N in WiCoV HKU20. For NS7 of PorCoV, the presence of an imperfect TRS (GCACCA) and its relatively high Ka/Ks ratio (number of nonsynonymous substitutions per nonsynonymous site/number of synonymous substitutions per synonymous site) of 1.046 (data not shown) implied that this ORF may not be expressed. BLAST search revealed no amino acid similarities between these putative nonstructural proteins and other known proteins, and no functional domain was identified by PFAM and InterProScan, except that NS7a of NHCoV HKU19 was found to be homologous to the NS7a of BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13. NS7b of WiCoV HKU20 and CMCoV HKU21, and NS7d of WiCoV HKU20, were also found to be homologous to the NS3b of IBV and hypothetical protein of goose coronavirus, respectively. Transmembrane helices, predicted by TMHMM and TMpred, in putative accessory proteins downstream to the N genes in the genomes of SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21 are listed in Table S4 in the supplemental material. Each of the genomes of PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, and CMCoV HKU21 contains a stem-loop II motif (s2m) (residues 25,220 to 25,251, 25,825 to 25,856, 25,865 to 25,896, 26,472 to 26,503, and 26,013 to 26,044, respectively), a conserved RNA element downstream of N and upstream of the poly(A) tail, similar to those in IBV, TCoV, SARSr-Rh-BatCoV, and SARS-CoV, as well as other CoVs discovered in Asian leopard cat, graylag geese, feral pigeons, and mallards, for which complete genomes are not available (Fig. 3) (14, 21, 38).
Fig 3.
Multiple alignments of conserved s2m of infectious bronchitis virus (IBV), SARS-related human coronavirus (SARS CoV), SARS-related Rhinolophus bat coronavirus HKU3 (SARSr-Rh-BatCoV HKU3), BuCoV HKU11, ThCoV HKU12, MunCoV HKU13, Asian leopard cat coronavirus (ALCCoV), PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, and CMCoV HKU21. Identical nucleotides are marked by asterisks. Acc. No., accession no.
Comparison of the amino acid identities of the seven conserved replicase domains for species demarcation (ADRP, nsp5 [3CLpro], nsp12 [RdRp], nsp13 [Hel], nsp14 [ExoN], nsp15 [NendoU], and nsp16 [O-MT]) (8) among the 10 deltacoronaviruses is shown in Table S5 in the supplemental material. In all the seven domains, the amino acid sequences of PorCoV HKU15 and SpCoV HKU17 showed more than 90% identity, indicating that these two coronaviruses should be subspecies of the same species.
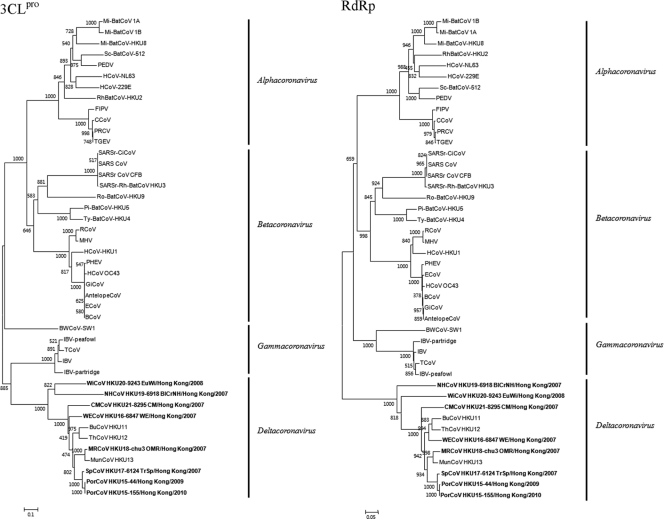
Phylogenetic analyses.
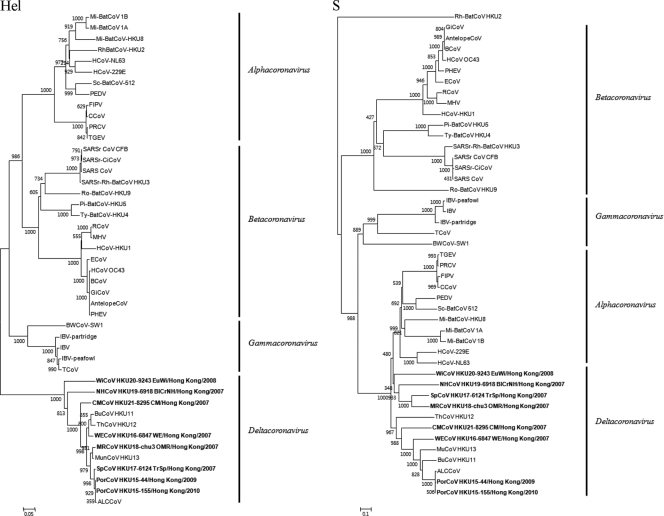
The phylogenetic trees constructed using the nucleotide sequences of the 3CLpro, RdRp, Hel, S, and N of the seven novel CoVs and other CoVs are shown in Fig. 4 and the corresponding pairwise amino acid identities are shown in Table 2. For all five genes, the seven novel CoVs possessed higher amino acid identities to each other and BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13 than to any other known CoVs with complete genomes available (Table 2). In all five trees, the seven novel CoVs were clustered with BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13 (Fig. 4). For Hel, S, and N, PorCoVs were also clustered with a CoV found in Asian leopard cat (10), for which the sequences of these genes were available (Fig. 4). There were <2% base differences between the Hel, S, and N genes of PorCoV and those of the Asian leopard cat coronavirus. Based on both phylogenetic tree analyses and amino acid differences, the seven novel CoVs as well as BuCoV HKU11, ThCoV HKU12, and MunCoV HKU13 should belong to the same genus, Deltacoronavirus.
Fig 4.
Phylogenetic analyses of 3CLpro, RdRp, helicase (Hel), S, and N proteins of PorCoV HKU15, WECoV HKU16, SpCoV HKU17, MRCoV HKU18, NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21. The trees were constructed by using the neighbor joining method using Kimura correction and bootstrap values calculated from 1,000 trees. Two hundred ninety-five, 892, 590, 802, and 249 amino acid positions in 3CLpro, RdRp, Hel, S, and N, respectively, were included in the analyses. The trees were midpoint rooted. For 3CLpro and S, the scale bar indicates the estimated number of substitutions per 10 amino acids. For RdRp and Hel, the scale bar indicates the estimated number of substitutions per 20 amino acids. For N, the scale bar indicates the estimated number of substitutions per 5 amino acids. Viruses characterized in this study are in bold. Virus name abbreviations are the same as those in the Fig. 1 legend.
Estimation of divergence dates.
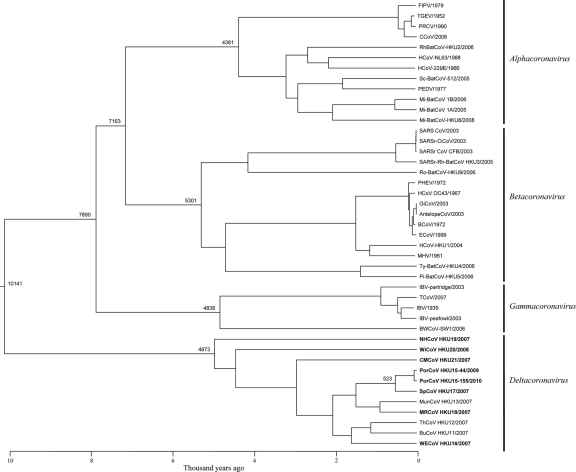
Using the Bayesian Skyline under a relaxed-clock model with an uncorrelated log-normal distribution, the mean evolutionary rate of CoVs was estimated at 1.3 × 10−4 nucleotide substitutions per site per year for the RdRp gene. Molecular clock analysis using the RdRp gene showed that the tMRCA of all CoVs was estimated at ∼8100 BC (HPDs, 20607 to 974 BC), that of Alphacoronavirus at ∼2400 BC (HPDs, 7659 to 722 BC), that of Betacoronavirus at ∼3300 BC (HPDs, 9713 to 447 BC), that of Gammacoronavirus at ∼2800 BC (HPDs, 8840 to 700 BC), and that of Deltacoronavirus at ∼3000 BC (HPDs, 9073 to 555 BC) (Fig. 5).
Fig 5.
Estimation of the time to the most recent common ancestor for Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. The time-scaled phylogeny was summarized from all MCMC phylogenies of the RdRp gene data set analyzed under the relaxed-clock model with an uncorrelated log-normal distribution in BEAST version 1.6.1. Viruses characterized in this study are in bold. The numbers indicate number of years ago. This is shown in the scale bar. Virus name abbreviations are the same as those in the legends of Fig. 1.
DISCUSSION
The diversity of CoVs in birds is comparable to that observed in bats. In the last 7 years, we and others have demonstrated a previously unrecognized diversity of CoVs in bats (4, 6, 23, 25, 26, 28, 43). More than 10 CoVs were discovered in bats, with at least nine present in our locality, and complete genome sequences are available for eight, which includes SARSr-Rh-BatCoV HKU3, Rh-BatCoV-HKU2, Miniopterus bat coronavirus 1, Miniopterus bat coronavirus HKU8, Scotophilus bat coronavirus 512, Tylonycteris bat coronavirus HKU4, Pipistrellus bat coronavirus HKU5, and Rousettus bat coronavirus HKU9 (4, 6, 25, 26, 43). Due to the similarities between bats and birds, such as their abilities to fly and high species diversity, we hypothesized that there should be previously unrecognized CoVs in birds. In our previous study and the present one, we demonstrated that there are at least nine CoVs, in addition to IBV and its close relatives, in birds (49). Potentially novel CoVs in Gammacoronavirus were also observed in another study, although complete genome sequences are not available and therefore detailed genomic and phylogenetic analysis are not possible (35). The nine CoVs discovered in the present and previous studies were found in birds of nine different families, showing host specificity. This phenomenon of host specificity is similar to that observed in bats, in which different genera are hosts of different CoVs (26, 45, 51, 52). We speculate that this diversity and host specificity of bat and bird CoVs is due to the large variety of species in bats and birds, giving rise to a large variety of cell types and receptors for the different CoVs to attach and replicate.
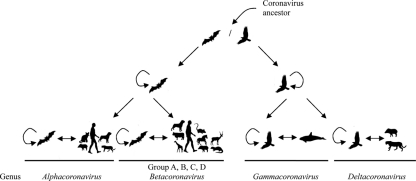
The presence of a huge diversity of bat CoVs in Alphacoronavirus and Betacoronavirus but not Gammacoronavirus and Deltacoronavirus and a huge diversity of bird CoVs in Gammacoronavirus and Deltacoronavirus but not Alphacoronavirus and Betacoronavirus supports our model of CoV evolution, in which bats are the gene source of Alphacoronavirus and Betacoronavirus and birds the gene source of Gammacoronavirus and Deltacoronavirus (Fig. 6) (52). It is not known whether the first CoVs occurred in bats and jumped to birds or vice versa. In the bat CoV lineage, the bat CoV jumped to another species of bat, giving rise to Alphacoronavirus and Betacoronavirus. These bat CoVs in turn jumped to other bat species and other mammals, including humans, with each interspecies jumping evolving dichotomously. As for the bird CoV lineage, the bird CoV jumped to another species of bird, giving rise to Gammacoronavirus and Deltacoronavirus. These bird CoVs in turn jumped to other bird species and occasionally to some mammalian species, such as whale and pig, with each interspecies jumping evolving dichotomously. Although PorCoV HKU15 was closely related to a CoV previously found in Asian leopard cats and Chinese ferret badgers, further experiments are warranted to confirm whether these viruses really replicate in the corresponding animals. Of note is that the estimation of divergence time was based on a relaxed-clock assumption with no recombination among the genomes. Since CoVs have a tendency to recombine, the estimated divergence time gives only a rough approximation of the actual divergence time. When more complete genomes of CoVs in the four different genera at different time points are available, such divergence time estimation can be performed using multiple gene loci to achieve more accurate estimation.
Fig 6.
A model of CoV evolution. CoVs in bats are the gene source of Alphacoronavirus and Betacoronavirus, and CoVs in birds are the gene source of Gammacoronavirus and Deltacoronavirus.
Both avian and mammalian CoVs are members of Deltacoronavirus, with similar genome characteristics and structures. In all the 10 members of Deltacoronavirus with complete genome sequences available, all have a very small genome size, from 25.421 (PorCoV HKU15) to 26.674 (MRCoV HKU18) kb, the smallest among all CoVs. Only one papain-like protease domain is observed in the nsp3 gene of their genomes. As for their gene contents, ORF NS6 was present between the M and N genes, and one to four ORFs were also observed downstream to the N gene. As for the TRSs, they all have the same putative TRS of ACACCA and separation of the TRS from the AUG of the S gene by a long stretch of nucleotides. Despite these similar genome characteristics among members of Deltacoronavirus, NHCoV HKU19 and WiCoV HKU20 possessed genomic features distinct from the other members of Deltacoronavirus, including the amino acids upstream of the putative cleavage sites at the junction of nsp2/nsp3, nsp3/nsp4, and nsp4/nsp5. It is also notable that NHCoV HKU19, WiCoV HKU20, and CMCoV HKU21 occupied the first three branches in the phylogenetic trees constructed using 3CLpro, Hel, RdRp, and N, indicating that they could be more ancestral than the other members. Furthermore, these three CoVs were found in large birds, including black-crowned night heron, Eurasian wigeon, and common moorhen, in contrast to BuCoV HKU11, ThCoV HKU12, MunCoV HKU13, WECoV HKU16, SpCoV HKU17, and MRCoV HKU18, which were found in small birds, including bulbuls, blackbird, gray-backed thrush, munias, Japanese white-eye, Eurasian tree sparrow, and oriental magpie robin. We speculate that the change in genome characteristics (e.g., acquisition of s2m) could have occurred during interspecies jumping of the CoV within the large birds before the jump to the small birds. Interestingly, the fact that PorCoV HKU15 and SpCoV HKU17 are the same species implies that interspecies jumping from birds to pigs may have occurred relatively recently. It is possible that a deletion of 3′ Ns7a and Ns7b had occurred during interspecies jumping from birds to pigs, which is similar to the observation of interspecies jumping of SARS-CoV from civets to humans, with the deletion of 29 bp in ORF 8 (25). As for the Asian leopard cat coronavirus, with only the Hel, S, E, M, and N gene sequences available, the sequences of these gene fragments differ from the corresponding ones in PorCoV by less than 2.1% nucleotides or 1.7% amino acids, including that for the S gene, which is responsible for receptor binding. BEAST analysis showed that the CoV jumped from birds to mammals around 523 years ago (Fig. 5). The mixing of birds, pigs, and other mammals in domestic environments and wildlife markets as well as their close contacts with humans may provide the correct environment for interspecies jumping and could subsequently pose risks of further genetic changes for adapting to human host as in the case of SARS (5). More extensive epidemiological studies in different varieties of mammalian species in other parts of the world for members of Deltacoronavirus would further improve our understanding on the diversity of this genus as well as its evolutionary history.
Supplementary Material
ACKNOWLEDGMENTS
We thank York Y. N. Chow, Health, Welfare and Food, HKSAR, The Peoples' Republic of China; Alan Chi-Kong Wong, Siu Fai Leung, Chik Chuen Lay, Thomas Sit, Elaine Lee, and Geraldine Luk of the Agriculture, Fisheries, and Conservation Department; and Clement Leung, Constance Chan, and Wing Ka Au of the Food, Environmental and Hygiene Department of the HKSAR.
We are grateful to the generous support of Hui Hoy and Hui Ming in the genomic sequencing platform and Eunice Lam for her generous donation to emerging infectious disease research. This work is partly supported by Research Grant Council grant HKU 780709 M; University Development Fund and Outstanding Young Researcher Award, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; and the Shaw Foundation.
Footnotes
Published ahead of print 25 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Apweiler R, et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bateman A, et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brian DA, Baric RS. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao J, Wu CC, Lin TL. 2008. Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res. 136:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng VC, Lau SK, Woo PC, Yuen KY. 2007. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 20:660–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu DK, Peiris JS, Chen H, Guan Y, Poon LL. 2008. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 89:1282–1287 [DOI] [PubMed] [Google Scholar]
- 7. Circella E, et al. 2007. Coronavirus associated with an enteric syndrome on a quail farm. Avian Pathol. 36:251–258 [DOI] [PubMed] [Google Scholar]
- 8. de Groot RJ, et al. 2011. Coronaviridae, p 806–828 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses, International Union of Microbiological Societies, Virology Division. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 9. Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg. Infect. Dis. 13:1295–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong BQ, et al. 2007. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 81:6920–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fouchier RA, et al. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gloza-Rausch F, et al. 2008. Detection and prevalence patterns of group I coronaviruses in bats, northern Germany. Emerg. Infect. Dis. 14:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gomaa MH, Barta JR, Ojkic D, Yoo D. 2008. Complete genomic sequence of turkey coronavirus. Virus Res. 135:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gough RE, Drury SE, Culver F, Britton P, Cavanagh D. 2006. Isolation of a coronavirus from a green-cheeked Amazon parrot (Amazon viridigenalis Cassin). Avian Pathol. 35:122–126 [DOI] [PubMed] [Google Scholar]
- 16. Guan Y, et al. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278 [DOI] [PubMed] [Google Scholar]
- 17. Hasoksuz M, et al. 2007. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 81:4981–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 72:4508–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hofmann K, Stoffel W. 1993. TMBASE - a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 20. Huang Y, Lau SK, Woo PC, Yuen KY. 2008. CoVDB: a comprehensive database for comparative analysis of coronavirus genes and genomes. Nucleic Acids Res. 36:D504–D511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jonassen CM, et al. 2005. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos). J. Gen. Virol. 86:1597–1607 [DOI] [PubMed] [Google Scholar]
- 22. Lai MM, Cavanagh D. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lau SK, et al. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 84:2808–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau SK, et al. 2006. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 44:2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lau SK, et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lau SK, et al. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lau SK, et al. 2011. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 85:11325–11337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lau SK, et al. 2010. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J. Virol. 84:11385–11394 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Li KS, et al. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213 [DOI] [PubMed] [Google Scholar]
- 30. Li W, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 31. Liu S, et al. 2005. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas). J. Gen. Virol. 86:719–725 [DOI] [PubMed] [Google Scholar]
- 32. Mardani K, Noormohammadi AH, Hooper P, Ignjatovic J, Browning GF. 2008. Infectious bronchitis viruses with a novel genomic organization. J. Virol. 82:2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marra MA, et al. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404 [DOI] [PubMed] [Google Scholar]
- 34. Mihindukulasuriya KA, Wu G, St. Leger J, Nordhausen RW, Wang D. 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82:5084–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muradrasoli S, et al. 2010. Prevalence and phylogeny of coronaviruses in wild birds from the Bering Strait area (Beringia). PLoS One 5:e13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peiris JS, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poon LL, et al. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robertson MP, Igel H, Baertsch R, Haussler D, Ares M, Jr, Scott WG. 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rota PA, et al. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399 [DOI] [PubMed] [Google Scholar]
- 40. Snijder EJ, et al. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 42. Suchard MA, Weiss RE, Sinsheimer JS. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 43. Tang XC, et al. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van der Hoek L, et al. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woo PC, et al. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woo PC, et al. 2004. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 363:841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woo PC, et al. 2006. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 80:7136–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woo PC, et al. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Woo PC, et al. 2009. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 83:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woo PC, et al. 2005. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 192:1898–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Woo PC, et al. 2006. Molecular diversity of coronaviruses in bats. Virology 351:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woo PC, Lau SK, Huang Y, Yuen KY. 2009. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 234:1117–1127 [DOI] [PubMed] [Google Scholar]
- 53. Zhang J, et al. 2007. Genomic characterization of equine coronavirus. Virology 369:92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ziebuhr J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7:412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.