Abstract
Hepatitis C virus glycoprotein E2 contains 18 conserved cysteines predicted to form nine disulfide pairs. In this study, a comprehensive cysteine-alanine mutagenesis scan of all 18 cysteine residues was performed in E1E2-pseudotyped retroviruses (HCVpp) and recombinant E2 receptor-binding domain (E2 residues 384 to 661 [E2661]). All 18 cysteine residues were absolutely required for HCVpp entry competence. The phenotypes of individual cysteines and pairwise mutation of disulfides were largely the same for retrovirion-incorporated E2 and E2661, suggesting their disulfide arrangements are similar. However, the contributions of each cysteine residue and the nine disulfides to E2 structure and function varied. Individual Cys-to-Ala mutations revealed discordant effects, where removal of one Cys within a pair had minimal effect on H53 recognition and CD81 binding (C486 and C569) while mutation of its partner abolished these functions (C494 and C564). Removal of disulfides at C581-C585 and C452-C459 significantly reduced the amount of E1 coprecipitated with E2, while all other disulfides were absolutely required for E1E2 heterodimerization. Remarkably, E2661 tolerates the presence of four free cysteines, as simultaneous mutation of C452A, C486A, C569A, C581A, C585A, C597A, and C652A (M+C597A) retained wild-type CD81 binding. Thus, only one disulfide from each of the three predicted domains, C429-C552 (DI), C503-C508 (DII), and C607-C644 (DIII), is essential for the assembly of the E2661 CD81-binding site. Furthermore, the yield of total monomeric E2 increased to 70% in M+C597A. These studies reveal the contribution of each cysteine residue and the nine disulfide pairs to E2 structure and function.
INTRODUCTION
Hepatitis C virus (HCV) is a major public health problem, with an estimated 100 to 180 million people chronically infected worldwide. The currently available therapy is an extended treatment regimen of between 24 and 48 weeks of ribavirin and pegylated interferon, with efficacy between 40 and 80% depending on the genotype, and is associated with severe side effects. HCV is a positive-sense RNA virus classified in the genus Hepacivirus within the family Flaviviridae and is grouped into six major genotypes (1 to 6) and various subtypes (a, b, c, etc.). The high degree of sequence diversity has proven a major challenge to the development of a universal vaccine to prevent HCV infection.
HCV encodes two envelope glycoproteins, E1 and E2, present as a heterodimer at the virion surface, that mediate viral attachment and fusion to facilitate virus entry. HCV cellular entry factors include the tetraspanin CD81 (25), scavenger receptor class B type 1 (SR-B1) (28), and the tight-junction membrane proteins claudins 1, 6, and 9 (12, 22, 35) and occludin (26). Several discontinuous CD81-binding motifs have been identified within E2 and are proposed to assemble during folding, including polyprotein residues Trp420, Trp437, Leu438, Leu441, Phe442, Tyr527, Trp529, Gly530, and Asp535, as well as amino acids within the region 613 to 618 (8, 24, 27). Interactions between the HCV glycoproteins and either claudin or occludin have not yet been described, although both are essential cofactors for viral entry (2, 14, 15).
Glycoproteins E1 and E2 are type I transmembrane proteins that are heavily modified during biosynthesis at 4 or 5 and 11 N-linked glycosylation sites, respectively. Expression of E1 and E2 in cis is required for the formation of the functional heterodimer that appears to undergo a slow, cooperative folding process facilitated by endoplasmic reticulum (ER) chaperones (1, 3, 5). Several heterodimerization determinants have been identified within the transmembrane domains of E1 and E2, the membrane-proximal region of E2, and the W487HY motif within the E2 ectodomain (6, 7, 10, 34). Within glycoprotein E2, an independent folding domain (polyprotein residues 384 to 661) can be efficiently expressed and secreted from cells with the retention of CD81 and SR-B1 receptor binding (23, 25, 28). Located within this receptor-binding domain (RBD) (E2661) are three discrete variable regions: the N-terminal hypervariable region 1 (HVR1) (residues 384 to 410 [H77c polyprotein numbering is used throughout]) and HVR2 (residues 474 to 482 [16, 31]) and the intergenotypic variable region (igVR) (residues 571 to 580 [21]). Both HVR2 and igVR are flanked by pairs of conserved cysteine residues, and all 3 variable regions are believed to be solvent exposed and excluded from the core domain (21). The E2 RBD is connected to the transmembrane domain (TMD) via a membrane-proximal region containing a conserved heptad repeat that appears to have structural and functional features analogous to the “stem” region of the flavivirus class II fusion protein glycoprotein E and suggested that E2 may also represent a class II fusion protein (10).
Glycoproteins E1 and E2 possess 8 and 18 cysteine residues within their respective ectodomains that are conserved across the six major genotypes (Fig. 1A). Krey et al. have recently assigned the nine disulfide bonds formed by these residues within the E2 ectodomain using trypsin proteolysis, redox chemistry, and mass spectrometry analysis (19) (Fig. 1B). The strict conservation of cysteines is indicative of the critical role disulfide bonds play in scaffolding the three-dimensional structure of proteins. Using secondary-structure prediction modeling, epitope mapping, and mutagenesis studies of the CD81-binding sites, a theoretical model of E2 was proposed based on the prefusion structure of glycoprotein E (19). In this class II model of HCV E2 (Fig. 1B), the known CD81-binding regions mapped to the interface of domains I (DI) and III. Disulfides 1 and 5 stabilize the domain I β-sheet sandwich, while disulfides 6, 7, and 8 are located within domain III. The igVR forms a “hinge” between these two domains and disulfides 5 and 6. Disulfide 7 was not formally identified in any of the tryptic digests but is assumed to form a disulfide pair. HVR1 is an N-terminal extension external to domain I. Domain II is predicted to form a relatively unstructured domain containing three short-range disulfide pairs: disulfides 2 and 3, flanking HVR2, and disulfide 4, stabilizing the candidate fusion “loop” represented by a sequence of glycine-rich hydrophobic residues between residues 502 and 520. Disulfide pair 9 is predicted to lie at the edge of domain III with C677 located within the membrane-proximal or proposed “stem” region of E2.
Fig 1.
Locations of conserved cysteines within HCV envelope glycoprotein E2. (A) Schematic diagram of the E1E2 polyprotein and the truncated E2 protein (E2661). The E1 sequence (polyprotein residues 191 to 383) is represented by light gray and E2 (polyprotein residues 384 to 746) by dark gray. The green boxes highlight the locations of conserved CD81-binding motifs, and the red boxes show the locations of the three discrete variable sequences within E2: HVR1, HVR2, and igVR, from the N to the C termini. The cylinder represents the conserved membrane-proximal heptad repeat or predicted stem region. E2661 is truncated at polyprotein residue 661 and has a C-terminal six-histidine tag. (B) Schematic representation of the disulfide-bonding pattern within the E2 glycoprotein modeled as a class II fusion protein (19). The locations of domains I, II, and III are labeled, and the location of the predicted stem region is indicated by the cylinder. The HVR1, HVR2, and igVR sequences are also labeled, and the predicted fusion peptide sequence (residues 502 to 520) within domain II is highlighted by a black dashed line. The conserved cysteine (C) residues are marked, and their corresponding disulfides, numbered 1 to 9, are indicated by a line bridging the amino acids.
In this study, we performed comprehensive alanine-scanning mutagenesis of each of the 18 cysteine residues within E2 and their corresponding disulfide bonds to further understand their roles in glycoprotein folding and function. Our findings demonstrate that all disulfides and their corresponding cysteine residues within E2 are essential for viral entry, but two disulfides, 6 (C581-C585) and 8 (C652-C677), are not essential for the assembly of the CD81-binding site. We also investigated the role of free cysteine residues in E2661 and their relationship to the extensive disulfide-mediated heterogeneity observed within preparations of soluble E2. This illustrated a complex pattern of intermolecular bonding that may have implications for the design of E2 expression systems for crystallization trials.
MATERIALS AND METHODS
Cell lines and antibodies.
HEK 293T and Huh7 cells were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum and 2 mM l-glutamine (DMF10). Monoclonal antibodies (MAbs) H53, H52, and A4 were kind gifts from Jean Dubuisson. Human hepatoma 7.5 (Huh7.5) cells were a kind gift of Charles Rice (The Rockefeller University, NY) and were maintained in DMF10 supplemented with 0.1 mM nonessential amino acids (DMF10NEA). Immunoglobulin G14 (IgG14) was purified from plasma obtained from an HIV-infected individual using protein G Sepharose (GE Healthcare) according to the manufacturer's instructions. MAb H53 has been reported to recognize a native epitope within E2 and coprecipitates noncovalently associated E1. This epitope is also independent of known CD81-binding sites and provides a broader screen for native conformational characteristics within E2. MAb 24 is a monoclonal antibody that recognizes a linear epitope within E2 (residues 411 to 423). Polyclonal antibodies to the H77c sequence of E2661 were raised in guinea pigs (Institute of Medical and Veterinary Sciences, Adelaide, Australia). Rabbit polyclonal antibodies directed against the six-histidine (His) epitope tag (anti-His; Rockland Biochemicals), fluorescence-conjugated anti-rabbit (IR-800; Rockland Biochemicals), fluorescence-conjugated anti-mouse (Alexa 680; Invitrogen), and horseradish peroxidase (HRP)-conjugated (Dako) antibodies are all commercially available.
Vectors.
The pcDNA4HisMax (Invitrogen)-based vector containing E1E2 sequences derived from the H77c genotype 1a, pE1E2, has been previously described (9). The HIV-1 luciferase reporter vector pNL4-3.LUC.R−E− was obtained from N. Landau through the NIH AIDS Research and Reference Reagent Program. The vector pJC1FLAG2(p7-NS-GLUC2A) was a kind gift from Charles Rice (The Rockefeller University, NY) and comprises the structural region (core-p7) of HCV-J6 (genotype 2a) and the nonstructural region (NS2A-NS5B) of JFH1 (genotype 2a) (20). In vitro mutagenesis was performed by standard overlap extension PCR to introduce cysteine (TGC/T)-to-alanine (GCT/C) substitutions. The inserted DNA sequences were confirmed using Big-Dye terminator chemistry and ABI automated sequencing. The pcDNA3 (Invitrogen)-based vector containing sequences encoding a truncated E2 protein (polyprotein residues 384 and 661) downstream of a tissue plasminogen activator leader sequence has also been described (pE2661) (21). Mutant E2661 sequences were amplified from the corresponding pE1E2 vectors using PCR primers to introduce a C-terminal six-His epitope tag, as well as NheI and XbaI restriction sites for insertion into pE2661. Purified DNA was transfected into HEK 293T cells using Fugene 6 (Roche) according to the manufacturer's instructions.
RIP and Western blotting.
Radiolabeling of E1E2–HIV-1-pseudotyped particles was performed in 293T cells seeded at 3.5 × 105 cells per well in six-well tissue culture dishes and cotransfected with 1 μg of pNL43.LUC.R−E− plus 1 μg of either pE1E2 or empty pcDNA4HisMax vector as previously described (9). Twenty-four hours posttransfection, the tissue culture medium was replaced with 75 μCi Trans35S-label (ICN, Costa Mesa, CA) in cysteine- and methionine-deficient DMF10 for 18 h. The tissue culture fluid was clarified and then subjected to centrifugation at 14,000 × g for 2 h at 4°C. The virus pellet was lysed in radioimmunoprecipitation (RIP) lysis buffer (0.6 M KCl, 0.05 M Tris, pH 7.4, 1 mM EDTA, 1% Triton X-100, 0.02% sodium azide) and immunoprecipitated with MAb H53 and IgG14 prior to nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and radioisotope imaging. E2 detected by MAb H53 was quantified using ImageQuant software (GE Healthcare). The intracellular expression of E1E2 was also determined by lysis of the remaining cell monolayer with Western lysis buffer (phosphate-buffered saline [PBS], pH 7.4, containing 1% Triton X-100, 1 mM EDTA, and 0.02% sodium azide), SDS-PAGE analysis, and transfer to a nitrocellulose membrane and was detected using MAb A4 (anti-E1) and MAb 24 (anti-E2) and a fluorescently conjugated goat anti-mouse Alexa 680 antibody (Invitrogen). The immunoblots were imaged using a fluorescent scanner (Odyssey; Li-Cor).
Radiolabeling of secreted E2661 was performed using 293T cells seeded at 3.5 × 105 cells per well in six-well tissue culture dishes (Nunc) and transfected with 2 μg of pE2661 or the empty pcDNA3 vector as previously described (21). Twenty-four hours posttransfection, the cells were treated with cysteine- and methionine-deficient DMF10 (MP Biomedicals) for 30 min prior to addition of 75 μCi Tran35S-label (ICN, Costa Mesa, CA) for 1 h and then transferred into serum-reduced medium (Opti-MEM; Invitrogen) for 6 h. The tissue culture fluid was clarified by centrifugation at 14,000 × g for 10 min and immunoprecipitated with MAb H53 or anti-His antibodies prior to SDS-PAGE analysis and radioisotope imaging. Expression of unlabeled E2661 for CD81-binding assays was performed by transfection of 293T cells as described above. Twenty-four hours posttransfection, the tissue culture fluid was transferred into Opti-MEM and harvested every 24 h for 72 h. The clarified tissue culture fluid was concentrated approximately 10-fold prior to reducing SDS-PAGE and transfer of proteins to a nitrocellulose membrane. Expression of E2661 was normalized by immunoblotting with anti-His and fluorescence-conjugated IR-800 antibodies. Immunoblots were analyzed using an Odyssey Li-Cor fluorescent scanner and quantification software.
E1E2-pseudotyped HIV-1 particle (HCVpp) entry assay.
Pseudotyped particle entry assays were performed as previously described (9). HEK 293T cells were cotransfected with 1 μg each of pNL43.LUC.R−E− plus either pE1E2 or empty pcDNA4HisMax vector. At 72 h posttransfection, culture supernatants were filtered (0.45 μm) and applied in triplicate to Huh7 monolayers seeded at 3 × 104 cells per well in 48-well tissue culture dishes (Nunc). At 72 h postinfection, Huh7 cells were lysed and measured for luciferase activity using the Promega luciferase substrate system and a Fluostar (BMG Labtechnologies) fitted with luminescence optics.
CD81 LEL binding.
The expression and purification of a chimera composed of maltose-binding protein (MBP) linked to the CD81 large extracellular loop (LEL) between residues 113 and 201 (MBP-LEL113-201) has been previously described and extensively used to characterize HCV E2-CD81 interactions and CD81-claudin interactions (8, 11, 14, 18). The L441M mutation within the E2 CD81-binding site was also included as a control for nonspecific binding (8). A chimera of glutathione S-transferase linked to the LEL of CD81 (residues 113 to 201) (GST-LEL) was expressed in BL21(DE3) cells. Protein was released from the cytoplasmic fraction by sonication and clarified (21,000 × g; 30 min). The binding of lysed HCVpp to GST-LEL was determined using a pulldown assay. GST-LEL bound to glutathione-Sepharose beads (GE Healthcare) was incubated with HCVpp lysed in RIP lysis buffer overnight at 4°C, followed by three washes in RIP lysis buffer and a final wash in PBS. Beads were resuspended in reducing sample buffer and subjected to SDS-PAGE and transfer to nitrocellulose for Western blotting. The amount of E2 pulled down by GST-LEL was detected with MAbs H52 and 24. The total amount of E2 present in HCVpp lysates was determined in reducing SDS-PAGE and Western blotting using MAbs H52 and 24. The specific amount of E2 capable of binding CD81 relative to wild-type glycoproteins was then calculated as follows: [(mutant E2 pulled down/total mutant E2)/(wild-type E2 pulled down/total wild-type E2)] × 100. The binding of E2661 to CD81 was determined using a solid-phase enzyme immunoassay (EIA) with 5 μg/ml of dimeric MBP-LEL113-201 immobilized to 96-well plates in PBS overnight. E2661 proteins were normalized for monomeric E2 content by Western blotting with anti-His as described above prior to addition to MBP-LEL113-201-coated plates. Bound E2 was detected using anti-His antibodies and anti-rabbit IgG-HRP conjugate (Dako) and developed with TMB (Sigma). As a protein-loading control, EIA plates were also coated with lectin derived from Galanthus nivalis agglutinin (GNA lectin; Sigma) at 0.5 μg/ml in PBS. All binding was measured as a function of absorbance at 450 nm (with 620-nm subtraction) using a Fluostar fitted with absorbance optics and calculated as a percentage of wild-type binding.
Replication and cell-free infectivity of cell culture-derived HCV.
RNA was transcribed from wild-type or mutant pJC1FLAG2(p7-NS-GLUC2A) vector as described previously (13) and transfected into Huh 7.5 cells seeded the day prior at 300,000 cells/well using DMRIE-C reagent (Invitrogen) according to the manufacturer's instructions. The ability of the RNA to replicate was monitored by measuring the luciferase activity in the tissue culture fluid at 24, 48, and 72 h for both wild-type virus and viruses encoding Cys-to-Ala mutations using the Renilla luciferase kit (Promega) according to the manufacturer's instructions. The pJC1FLAG2(p7-NS-GLUC2A) NSB5 GND mutant was included as a negative control. At 72 h posttransfection, cell-free virus was collected, filtered (0.45 μm), normalized according to luciferase activity, and used to infect naïve Huh 7.5 cells to examine the cell-free virus infectivity. Luciferase activity was measured at 72 h in both the cell lysate and tissue culture fluid using the Renilla luciferase kit (Promega) according to the manufacturer's instructions.
Lectin affinity purification of E2661 and blue-native PAGE analysis.
E2661 proteins were expressed and metabolically labeled as described above. The clarified tissue culture fluid was bound to GNA lectin-conjugated agarose beads (Vector Laboratories) overnight at 4°C. The tissue culture fluid was removed, and the remaining beads were washed in PBS. Any bound proteins were eluted in 2 bead volumes of 1 M mannose for 1 h at 4°C. A fraction of the eluent was subject to reducing SDS-PAGE analysis and radioisotope imaging to quantify relative expression using ImageQuant software (GE Healthcare). The normalized proteins were then analyzed by 4 to 16% blue-native PAGE at 4°C according to the method of Wittig et al. (33). Five micrograms each of purified thyroglobulin (660 kDa), ferritin (880/440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (45 kDa) (GE Healthcare) were used as size standard markers. The Coomassie-stained markers were marked with radioactive material prior to radioisotope imaging. Total E2661, and the proportion of each different species, was quantified using ImageQuant software (GE Healthcare).
RESULTS
Characterization of individual cysteine substitution mutations in the context of the full-length E2 glycoprotein coexpressed with E1.
The assignment of the 18 conserved cysteines to 9 disulfides in relation to the three-domain structure of E2 (DI to DIII), as proposed by Krey et al. (19), is shown in Fig. 1B. We first assessed the effects of substituting alanines for individual cysteines on E2 folding and function in the context of HCVpp-incorporated E1E2 derived from the H77c genotype 1a prototype isolate. The Cys-to-Ala substitutions abolished the ability of HCVpp to infect Huh 7 hepatoma cells, indicating that the 9 disulfides of E2 are critical for cellular entry competence (Fig. 2A). Western blot analysis of transfected cell lysates with MAb 24 (directed to E2) and MAb A4 (directed to E1) indicated that E1 and E2 were expressed at wild-type (WT) levels (Fig. 2B, Western CL). However, the use of the conformation-dependent E2-specific MAb H53 in immunoprecipitations of biosynthetically labeled HCVpp revealed that the mutations had caused defects in the glycoprotein complex (Fig. 2B, IP HCVpp). In all cases, defects in E2 expression were also observed using a polyclonal antibody raised in guinea pigs and directed toward linear and conformational epitopes in E2661 and so are not restricted to the H53 epitope (Fig. 2C). The abilities of the E2 glycoproteins incorporated into HCVpp to bind CD81 were examined using a GST-LEL pulldown assay. The E2 mutant L441M, previously shown to disrupt CD81 binding without affecting E1E2 heterodimerization or incorporation into HCVpp, served as a negative control to monitor the specificity of the E2-CD81 pulldown assay (8).
Fig 2.
Individual cysteine-to-alanine mutations in glycoprotein E2 have diverse effects on E2 folding and function in HCVpp. (A) Entry into Huh7 cells as mediated by E1E2-pseudotyped HIV-1 (HCVpp) containing E2 glycoproteins with individual cysteine-to-alanine substitution mutations. Entry of HCVpp into Huh7 cells is measured as a function of relative luciferase units (RLU). The error bars indicate standard errors (SE). (B) Maturation and incorporation into HCVpp by E2 glycoproteins containing individual cysteine-to-alanine substitution mutations. The level of intracellular expression and processing of E1 and E2 was determined in Western blots of transfected cell lysates after reducing SDS-PAGE (Western CL). Shown is nonreducing SDS-PAGE analysis of HCVpp radioimmunoprecipitated using the anti-E2 conformation-dependent MAb H53 as an indicator of E2 folding, maturation, and noncovalent heterodimerization with E1 (IP HCVpp). The amount of HCVpp produced was assessed by immunoprecipitating HCVpp with an anti-p24 antibody (p24). (C) Maturation and incorporation into HCVpp by E2 glycoproteins containing individual cysteine-to-alanine substitution mutations and CD81 LEL binding function. Shown is reducing SDS-PAGE of HCVpp radioimmunoprecipitated with polyclonal anti-E2 antibody. The amount of HCVpp produced was assessed by immunoprecipitating HCVpp with an anti-p24 antibody (p24). The ability of HCVpp containing individual Cys-to-Ala mutations to bind GST-LEL was quantitated and normalized relative to the input amount of E2 and expressed as a percentage of wild-type CD81 LEL binding function (mean ± SE; n = 2 or 3). The L441M mutation within the E2 CD81-binding site represents a control for nonspecific interactions.
Domain I.
Little or no HCVpp-associated E2 was immunoprecipitated for the disulfide 1 mutants C429A and C552A, respectively, consistent with folding and/or virion incorporation defects (Fig. 2B, IP HCVpp). A CD81 pulldown assay utilizing GST-LEL revealed the absence of CD81 LEL binding activity for both mutants (Fig. 2C). These data are consistent with the proposed roles of C429 and C552 in forming a long-range disulfide within the central beta-sandwich of domain I (Fig. 1B) (19). The mutation of C564 and C569, which form disulfide 5 at the base of domain I, resulted in discordant effects on E2 folding and function: the H53-reactive E2 and CD81 LEL binding function was retained for C569A but was absent for C564A (Fig. 2B and C). Despite the incorporation of C569A E2 into HCVpp, the mutant E2 failed to heterodimerize with E1 (Fig. 2B and C).
Domain II.
Domain II comprises disulfides 2 (C452-C459), 3 (C486-C494), and 4 (C503-C508) formed between neighboring cysteine residues in E2. The C452A mutant (disulfide 2) incorporated WT levels of E2 into HCVpp, whereas its counterpart, C459A, had a notable reduction in H53-reactive E2; both mutants failed to heterodimerize with E1 (Fig. 2B and C). CD81 LEL binding function was maintained at 100% for C452A but was reduced to 26% for C459A (Fig. 2C). Glycoprotein E2 containing a C486A mutation (disulfide 3) was incorporated into HCVpp at WT levels and retained CD81 LEL binding function. In contrast, mutation of its disulfide-bonding partner C494 to alanine resulted in greatly reduced levels of E2 incorporation into HCVpp and loss of CD81 LEL binding function. Both C486A and C494A disrupted the formation of E1/E2 heterodimers. The mutation of disulfide 4 cysteines also led to discordant effects with H53-reactive E2 in association with E1 observed for C503A but not for C508A. Both mutants lacked CD81 LEL binding function.
Domain III.
Domain III comprises disulfides 6 (C581-C585), 7 (C597-C620), and 8 (C607-C644) and is connected to the stem region via disulfide 9 (C652-C677). The participation of C597 and C620 in forming disulfide 7 has not been confirmed (19). H53-reactive E2 was obtained from HCVpp at wild-type levels for C581A and C585A (disulfide 6), but these mutants failed to heterodimerize with E1; CD81 LEL binding activity was maintained at near wild-type levels for both mutations (Fig. 2B and C). For the putative disulfide 7, C597A led to a significant reduction in E2, whereas C620A presented as an E1E2 complex (Fig. 2B and C). Both mutations reduced CD81 LEL binding function (Fig. 2C). The disulfide 8 mutation at C607A or C644A was not tolerated, as H53-reactive E2 was not detected in HCVpp and there was a large reduction in CD81 LEL binding function. The C607A and C644A mutations appear to disrupt the global conformation, as there was a large decrease in the total amount of E2 incorporated into HCVpp, as detected using polyclonal antibodies, and a lack of E1/E2 heterodimerization. Finally, while both C652A and C677A (disulfide 9) were incorporated into HCVpp at WT levels, they exhibited a reduction in heterodimerization with E1. Both mutants, however, retained CD81 LEL binding function at or near wild-type levels, consistent with their location within the E2 stem, distal to domain III and the CD81-binding region.
A subset of Cys-to-Ala mutants that retained H53 reactivity and CD81 LEL binding function were examined in the context of full-length infectious cell culture-derived HCV for the ability of viral RNA to replicate in Huh 7.5 cells following transfection and for the infectivity of virions. The results revealed that Cys-to-Ala mutations did not affect RNA replication (Fig. 3A) but abolished the infectivity of virions (Fig. 3B). These data confirm that HCV E1/E2 glycoprotein function strictly requires the presence of each of the 18 conserved cysteine residues.
Fig 3.
RNA replication and infectivity of cell culture-derived HCV bearing Cys-to-Ala mutations. (A) A subset of Cys-to-Ala mutations were inserted into the pJC1FLAG2(p7-NS-GLUC2A) vector. Replication of RNA was assessed by measuring the activity of secreted Gaussia luciferase in the supernatant fluid at 24 h (white bars), 48 h (grey bars), and 72 h (black bars) after transfection. The error bars indicate SD. (B) Tissue culture fluid collected at 72 h was filtered and applied to naïve Huh 7.5 cells using a standardized inoculum normalized for luciferase activity. To determine the infectivity of the virus, luciferase activity in the supernatant fluid was measured at 72 h. “GND” represents a replication-defective mutant of pJC1FLAG2(p7-NS-GLUC2A) containing the GND mutation in NS5B.
Simultaneous mutagenesis of cysteine pairs involved in disulfide formation.
A Cys-to-Ala scan of the HIV envelope glycoprotein gp120-gp41 complex revealed that mutations of individual cysteines did not favor a functional protein fold, whereas simultaneous Ala substitutions rescued both folding and function in 2 of 10 disulfides (29). We therefore attempted to alleviate this tendency for protein misfolding due to the presence of unpaired cysteines by simultaneous Ala replacement of each disulfide pair. We first confirmed intracellular expression and polyprotein processing of E1 and E2 for the double Cys-to-Ala mutants by Western blotting (Fig. 4A, Western CL); however, HCVpp entry activity was still absent in all cases (data not shown). The incorporation of H53-reactive E2 into HCVpp was observed for C452A-C459A (DII), C581A-C585A, and C652A-C677A (DIII), but heterodimerization with E1 was virtually absent (Fig. 4A, IP HCVpp). In these three cases, the component single mutants were also incorporated into HCVpp (Fig. 2B). Of the three mutants, only the C581A-C585A and C652A-C677A mutants retained CD81 LEL binding activity at ∼50% and ∼80% of wild-type levels, respectively (Fig. 4B). An increase in the size of E2 incorporated into HCVpp was noted for the C652A-C677A mutant, suggestive of changes in glycosylation. An H53-reactive E2 protein associated with HCVpp was not observed for the other double mutants. Reductions in the amount of E2 precipitated by H53 were similarly reflected in a reduction in the total amount of E2 immunoprecipitated using polyclonal anti-E2 immune serum, suggesting that the disulfide mutations result in defective glycoprotein incorporation into HCVpp (Fig. 4B). These data indicate that two disulfides within domain III, C581-C585 and C652-C677, are not required for efficient E2 glycoprotein incorporation into HCVpp and CD81 LEL binding function. However, HCVcc containing either the C652A-C677A or C581A-C585A mutant was not infectious despite normal levels of RNA replication (Fig. 3A and B), confirming their essential role in E1/E2 function. These data also indicate that the functional defects associated with particular individual Cys-to-Ala mutations are not alleviated by removal of the unpaired Cys following substitution of the disulfide-bonding partner.
Fig 4.
Role of disulfide pairs in folding and function of HCVpp-incorporated glycoprotein E2. (A) Maturation and incorporation into HCVpp of E2 glycoproteins containing pairwise cysteine-to-alanine substitution mutations. The levels of intracellular expression and processing of E1 and E2 were determined in Western blots of transfected cell lysates following reducing SDS-PAGE (Western CL). Shown is nonreducing SDS-PAGE analysis of HCVpp radioimmunoprecipitated with the anti-E2 conformation-dependent MAb H53 as an indicator of E2 folding, maturation, and noncovalent heterodimerization with E1 (IP HCVpp). The amount of HCVpp produced was assessed by immunoprecipitating HCVpp with an anti-p24 antibody (p24). (B) Maturation and incorporation into HCVpp by E2 glycoproteins containing individual cysteine-to-alanine substitution mutations and CD81 LEL binding function. Shown is reducing SDS-PAGE of HCVpp radioimmunoprecipitated with polyclonal anti-E2 antibody. The amount of HCVpp produced was assessed by immunoprecipitating HCVpp with an anti-p24 antibody (p24) and the level of intracellular expression. The ability of HCVpp containing individual Cys-to-Ala mutations to bind GST-LEL was quantitated and normalized relative to the input amount of E2 and expressed as a percentage of wild-type CD81 LEL binding function (mean ± standard deviation [SD]; n = 2). The L441M mutation within the E2 CD81-binding site represents a control for nonspecific interactions.
Characterization of individual cysteine and pairwise disulfide substitution mutations in the context of the E2 RBD (E2661).
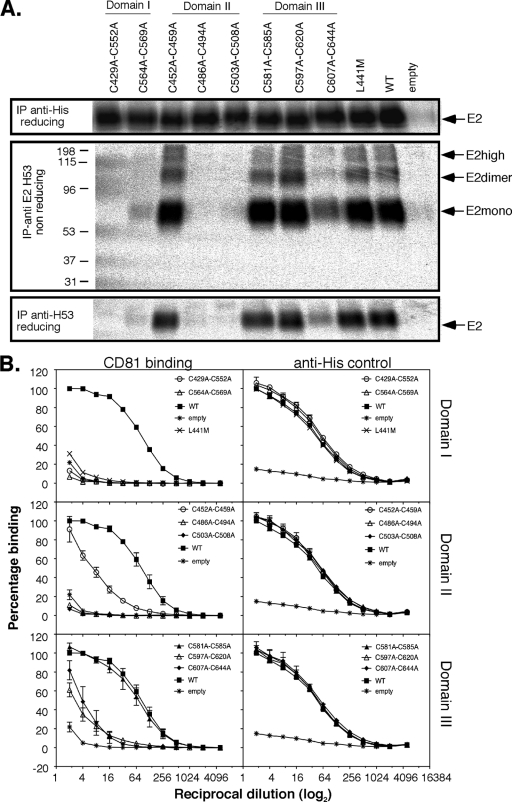
The effects of Cys-to-Ala mutations were next assessed in the context of the receptor-binding domain of E2 (residues 384 to 661 [E2661]) that folds independently of E1, encompasses the three-domain architecture described by Krey et al. (19), and retains CD81 and SR-B1 binding function. All mutants were secreted from transfected 293T cells at WT levels, as indicated by immunoprecipitation of metabolically labeled proteins with “anti-His” antibodies via the C-terminal six-His tag (Fig. 5A, top). The MAb H53 reactivity profiles of all but one of the E2661 mutants largely reflected that observed for the corresponding E1E2 mutants (Fig. 5A, middle and bottom, and Fig. 2B and C). Thus, Cys residues that are essential for H53 reactivity include C494 and C508 (DII), C552 and C564 (DI), and C607 and C644 (DIII), whereas C452, C459, C486, and C503 (DII); C569 (DI); and C581, C585, C597, C620, and C652 (DIII) are dispensable for this function (Fig. 5A, middle and bottom). An examination of the H53-reactive mutants by SDS-PAGE under nonreducing conditions (Fig. 5A, middle) revealed a ladder of bands corresponding to monomer (∼60- to 80-kDa), dimer (∼100- to 110-kDa), and higher-order species, which were also observed for the wild type. Addition of N-ethylmaleimide prior to and during lysis did not alter the proportions of monomer, dimer, and high-molecular-weight forms of E2661 observed (data not shown).
Fig 5.
Individual cysteine-to-alanine mutations have diverse effects on E2661 folding and function. (A) Expression, secretion, and folding of E2661 proteins containing individual cysteine-to-alanine substitution mutations. Shown is SDS-PAGE analysis of radiolabeled, secreted E2661 containing individual cysteine-to-alanine mutations immunoprecipitated with anti-His antibodies (top) or anti-E2 conformation-dependent MAb H53 under nonreducing (middle) and reducing (bottom) conditions. The migration of monomeric (E2mono), dimeric (E2dimer), and higher-molecular-mass (E2high) forms of E2661 under nonreducing conditions is indicated. (B) Binding to CD81 LEL by E2661 proteins containing individual cysteine-to-alanine substitution mutations. Binding of secreted E2661 containing single Cys-to-Ala mutations to MBP-LEL113-201 was detected by rabbit anti-His antibodies. The L441M mutation within the E2 CD81-binding site represents a control for nonspecific interactions. Loading controls for the same E2661 proteins are also shown (right) as captured by lectin and detected by anti-His antibody. The data are means ± SE of 2 to 4 independent experiments.
The presence of the C-terminal six-His tag in E2661 enabled CD81 LEL binding activity to be assessed independently of H53 reactivity (Fig. 5A, top, and B). Three patterns of CD81 LEL reactivity relative to H53 reactivity were discerned when the Cys-to-Ala mutants were considered according to disulfide pairing. (i) Loss of H53 reactivity following Ala substitution for at least one Cys of a disulfide pair predicted a loss of CD81 LEL binding function for both mutants: C429A and C552A (DI) and C503A and C508A (DII). (ii) The level of H53 reactivity following Ala substitution for either Cys contributing to a disulfide was predictive of the level of CD81 LEL binding function for both mutants: C452A and C459A (DII) and C581A and C585A, C597A and C620A, and C607A and C644A (DIII). (iii) One cysteine within a disulfide pair was dispensable for H53 and CD81 reactivity, while the other was essential for these functions: C569 (disulfide 5, DI) and C486 (disulfide 3, DII) were not required for acquisition of the H53 fold or CD81-binding function. Mutation of the free Cys at 652 did not affect H53 or CD81 LEL binding, suggesting that the residue is dispensable for these functions in the E2661 fold.
We again considered that the presence of an unpaired cysteine within single Cys-to-Ala mutants could lead to misfolded protein due to the formation of nonnative disulfides. We therefore asked whether simultaneous Ala replacement of Cys residues participating in disulfide formation could rescue the phenotype of defective single Cys-to-Ala mutants in the context of E2661. The MAb H53 reactivity of C452A-C459A, C581A-C585A, C597A-C620A, and C607A-C644A mutants was retained, although the CD81 LEL binding activity of the double mutants tended to be lower than that of the component single mutants (Fig. 6A and B). Notably, the single component mutations of C452A and C459A had WT CD81 LEL binding function, but the double mutant (disulfide 2) saw a 16-fold reduction in activity (Fig. 5B and 6B). Disulfide C581-C585 appears to be dispensable for H53 reactivity and CD81 LEL binding in the context of E2661 and HCVpp-incorporated E2. In contrast, the combination of single Cys-to-Ala mutations associated with loss of H53 reactivity or CD81 LEL binding function by one or both component cysteine mutations did not restore function, for example, C429A-C552A and C564A-C569A (DI) and C486A-C494A and C503A-C508A (DII) (Fig. 6A and B).
Fig 6.
Role of disulfides in E2661 folding and function. (A) Expression, secretion, and folding of E2661 proteins containing pairwise cysteine-to-alanine substitution mutations. Shown is SDS-PAGE analysis of radiolabeled, secreted E2661 containing cysteine-to-alanine mutations of the proposed disulfide pairs immunoprecipitated with anti-His antibodies (top) or anti-E2 conformation-dependent MAb H53 under nonreducing (middle) and reducing (bottom) conditions. The migration of monomeric (E2mono), dimeric (E2dimer), and higher-molecular-mass (E2high) forms of E2661 under nonreducing conditions is indicated. (B) Binding to CD81 LEL by E2661 proteins containing individual cysteine-to-alanine substitution mutations. Binding of secreted E2661 containing pairwise Cys-to-Ala mutations of the proposed disulfides to MBP-LEL113-201 was detected by rabbit anti-His antibodies. The L441M mutation within the E2 CD81-binding site represents a control for nonspecific interactions. Loading controls for the same E2661 proteins are also shown (right) as captured by lectin and detected by anti-His. The data are means ± SE of 3 independent experiments.
These data indicate that disulfides 1 (DI) and 4 (DII) are essential for maintenance of both a WT E2661 fold and CD81 LEL binding activity, while disulfide 6 (DIII) is dispensable for these functions. Disulfides 2 (DII) and 7 and 8 (DIII) are not strictly required but contribute to H53 and CD81 LEL binding activity. Formation of disulfides 3 (DII) and 5 (DI) was also not essential for folding and function, as unpaired cysteine residues at positions C494 and C564 were observed to be favorable over complete removal of their corresponding disulfides.
Characterization of free cysteines tolerated within the E2 RBD fold (E2661).
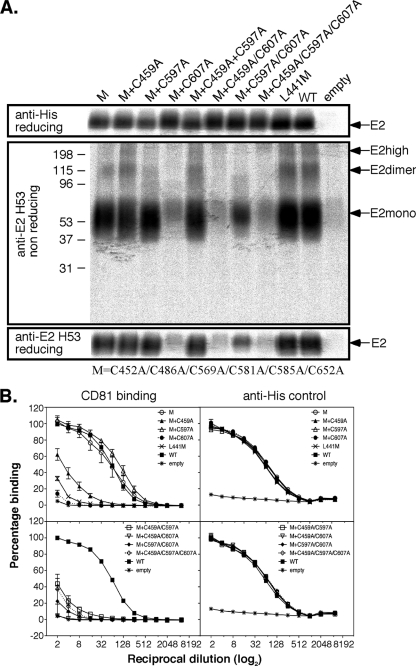
The pairwise Ala substitutions for individual cysteines involved in forming a disulfide revealed that the absence of a Cys residue at position 452, 459, 486, 569, 581, 585, or 652 is not necessarily unfavorable for E2661 folding or function. In order to examine how the E2661 fold tolerates the presence of unpaired Cys residues, we subjected E2661 mutants containing multiple Cys-to-Ala substitution mutations to phenotypic analysis. Initially, Cys-to-Ala mutations exhibiting a WT phenotype (C452A, C486A, C569A, C581A, C585A, and C652A) were combined in E2661 (designated “M”). The M mutant exhibited WT levels of H53 and CD81 LEL reactivity (Fig. 7A and B), indicating that free cysteines at multiple positions are tolerated by the E2661 fold. Further mutations, C459A, C597A, and C607A, which affected CD81 LEL binding function to various degrees (see Fig. 5B), were added individually to the M construct. The H53 and CD81 LEL binding characteristics of the M+C597A and M+C607A mutants were largely consistent with those of the individual C597A and C607A mutants: WT versus diminished activity, respectively. Notably, M+C459A exhibited a dramatic reduction in CD81 LEL binding (80%), which did not correspond to the wild-type levels of CD81 LEL binding activity of C459A in isolation. The simultaneous introduction of C459A, C597A, and C607A in various combinations to M adversely affected H53 recognition and/or CD81 LEL binding function (Fig. 7A and B). The M+C597A mutant also saw a notable reduction in the amount of “E2 high” and “E2 dimer” precipitated by H53 compared to the WT, as discussed below (Fig. 7A).
Fig 7.
Introduction of multiple cysteine-to-alanine mutations into E2661. (A) Secretion and folding of E2661 containing multiple cysteine-to-alanine substitution mutations. Shown is SDS-PAGE analysis of radiolabeled, secreted E2661 containing multiple cysteine-to-alanine mutations immunoprecipitated with anti-HIS (top) or anti-E2 conformation-dependent MAb H53 under nonreducing (middle) and reducing (bottom) conditions. The migration of monomeric (E2mono), dimeric (E2dimer), and higher-molecular-mass (E2high) forms of E2661 under nonreducing conditions is indicated. The M construct represents simultaneous mutations at positions C452, C486, C569, C581, C585, and C652. (B) Binding to CD81 LEL by E2661 proteins containing multiple cysteine-to-alanine substitution mutations. Shown is binding of secreted E2661 containing multiple Cys-to-Ala mutations to MBP-LEL113-201 as detected by rabbit anti-His antibodies. The L441M mutation within the E2 CD81-binding site represents a control for nonspecific interactions. Loading controls for the same E2661-His proteins are also shown (right) as captured by lectin and detected by anti-His antibodies. The data are means ± SE of 3 to 4 independent experiments.
These data indicate that the CD81 LEL competent fold of E2661 possesses a strikingly high level of tolerance for the presence of unpaired Cys residues at positions C459, C494, C564, and C620, as well as the removal of disulfide 6 (C581-C585) and the unpaired cysteine C652. Pairwise mutants representing all combinations of C452A, C486A, C569A, and C597A were also observed to have WT levels of H53 and CD81 LEL reactivity and therefore did not suggest a specific pattern of alternative disulfide bonding mediated between C459, C494, C564, and C620 (data not shown).
Investigating intermolecular disulfide formation between E2661 molecules.
Whidby et al. reported an increase in the monomer-to-dimer ratio of E2661 (genotype 2a) following replacement of the free Cys at position 652 with Ser (32). In contrast to these observations, we did not observe a significant decrease in E2661 dimer or higher-order oligomers for C652A following H53 immunoprecipitation and nonreducing SDS-PAGE (Fig. 5A). To rule out possible thiol-disulfide rearrangement due to sample boiling or selective immunoprecipitation by the conformation-dependent MAb H53, we purified radiolabeled E2661 proteins using GNA lectin affinity chromatography for blue-native PAGE analysis. As shown in Fig. 8A, WT and C652A E2661 proteins presented as monomeric, dimeric, trimeric, and higher-order species with almost identical electrophoretic profiles. The quantification of E2 species indicated a 10% increase in monomer for C652A relative to the WT. This increase was not due to a reduction in the dimeric or trimeric forms, but rather, conversion of higher-molecular-mass species to monomers. The electrophoretic profiles of other individual Cys-to-Ala mutants were similar to that of the wild type (data not shown). The M+C597A mutant, which retains H53 and CD81 LEL binding activities, exhibited a monomer yield increase to approximately 70% of total protein, more than double that observed for the WT and the C652A mutant with a marked reduction in all higher-molecular-mass species (Fig. 8B). These data, obtained under native conditions, are consistent with the reduction in higher-molecular-mass species detected by immunoprecipitation with MAb H53 and SDS-PAGE analysis (Fig. 7A).
Fig 8.

Analysis of the propensity of E2661 containing the minimum number of cysteine residues to form monomeric, dimeric, and higher-molecular-mass forms of E2. (A) Comparison of disulfide-linked multimers formed by E2661 containing C652A or M+C597A mutations. Shown is blue-native PAGE analysis of lectin affinity-purified radiolabeled E2661 representing either C652A, M+C597A, or WT protein as detected by radioisotope imaging (top). Reducing SDS-PAGE analysis of purified proteins as detected by radioisotope imaging was also included as a loading control (bottom; Total). The expected migration of monomeric, dimeric, trimeric, and higher-molecular-mass forms of E2661 is indicated. (B) Quantitative analysis of different E2661 oligomers indicated as detected by native PAGE. Bands corresponding in molecular mass to monomer, dimer, trimer, and higher-molecular-mass forms of E2661 were quantified using ImageQuant software, and the percentage of each species was calculated. The data shown are the averages of two independent experiments.
These data indicate that aberrant disulfides formed in the presence of C452, C486, C569, C581, C585, C597, and C652 (i.e., absent from M+C597A) contribute to the formation of high-order E2661 species.
DISCUSSION
Hepatitis C virus glycoprotein E2 performs the critical function of mediating attachment of virions to cellular receptors that include CD81 and is a target of the humoral immune response. Following attachment of HCV to cell surface receptors, viral fusion proceeds and is dependent on the association of E2 with glycoprotein E1. The biosynthetic folding pathway of E1 and E2 has not been fully elucidated. Critical to folding of a functional form of the E1E2 heterodimer are the 8 and 18 conserved cysteine residues present in E1 and E2, respectively. Glycoprotein E2 is believed to have three immunogenic/structural domains, based on the results of monoclonal antibody mapping and disulfide assignment studies that allowed E2 to be modeled on a class II fusion protein (17, 19). In this study, we performed a comprehensive mutagenesis analysis of individual cysteine residues in E2 and proposed disulfide pairs, as proposed by Krey et al. (19), in the context of both E1E2 and the isolated receptor-binding domain E2661. The results are summarized in Tables 1 and 2.
Table 1.
Phenotypes of single Cys-to-Ala mutations in E2 incorporated into HCVpp and E2661
aAs detected in Western blots of cell lysates in Fig. 2B. For all columns, unless otherwise indicated, black, 75% of wild-type levels to wild-type levels; gray, 25 to 74% of wild-type levels; white, <25% of wild-type levels.
bH53 reactivity from Fig. 2B.
cHeterodimerization from Fig. 2B and C.
dCD81 binding from Fig. 2C.
eVirus entry from Fig. 2A.
fAs detected by anti-His immunoprecipitation from Fig. 5A, top.
gH53 reactivity from Fig. 5A, middle and bottom.
hCD81 binding from Fig. 5B. Black, wild type; gray, up to 8-fold reduction, and white, >8-fold reduction in the dilution of E2 protein required to achieve 50% binding to CD81 LEL.
Table 2.
Phenotype of pairwise Cys-to-Ala mutations of disulfide bonds of E2 within HCVpp and E2661
aAs determined from Western blots of cell lysates in Fig. 4A. For all columns, unless otherwise indicated, black, 75% of wild-type levels to wild-type levels; gray, 25 to 74% of wild-type levels; white, <25% of wild-type levels.
bH53 reactivity from Fig. 4A.
cHeterodimerization from Fig. 4A and B.
dCD81 binding from Fig. 4B.
eVirus entry (data not shown).
fAs detected by anti-His immunoprecipitation from Fig. 6A top.
gH53 reactivity from Fig. 6A middle and bottom.
hCD81 binding from Fig. 6B. Black, wild type; gray, up to 8-fold reduction, and white, >8-fold reduction in the dilution of E2 protein required to achieve 50% binding to CD81 LEL.
In the context of HCVpp, each of the nine disulfides of HCV E2 and their corresponding cysteine residues were shown to be absolutely required for the formation of an entry-competent structure (Tables 1 and 2). Characterization of the effects of these mutations on E2-folding and CD81-binding activities using the conformation-dependent monoclonal antibody H53, polyclonal anti-E2 antibody, and recombinant CD81 generated similar phenotypic profiles for both HCVpp-incorporated E2 and E2661. This confirms that the disulfide arrangement of full-length E2 expressed in association with E1 and the isolated receptor-binding domain are largely similar. The notable exceptions to this were mutation of disulfide 7 or 8 within domain III that resulted in a loss of H53 reactivity and/or CD81 binding in the context of E1E2 but was partially tolerated within E2661. These data suggest that the expression of E2 containing the stem regions and TMD with glycoprotein E1 increases the susceptibility of domain III to structural changes as a result of these mutations.
The pattern of effects on H53 reactivity and CD81 LEL binding observed for the disulfide mutants also largely validated the model of E2 proposed by Krey et al. (19). For example, domain I is predicted to be comprised of 8 antiparallel β-strands stabilized through two disulfides (1 and 5). The majority of the CD81 interaction surface is located in domain I but is also proposed to partially overlap with domain III. Mutagenesis revealed disulfide 1 to be required for HCVpp incorporation of E2 and for E2661 to fold into a structure that binds conformation-dependent antibody H53 and CD81, consistent with an essential role for this relatively long-range disulfide in maintaining the structural integrity of E2. Conversely, disulfide 9 was not essential for the formation of either the H53 epitope or CD81 LEL binding, consistent with its location distal to the CD81-binding site and overlapping the proposed stem region (10). Mutation of disulfide 9 also corresponded to a loss of E1 contacts, in agreement with previous reports of heterodimerization determinants within this region (10).
In the context of HCVpp, the disulfide 6 mutant maintained H53 reactivity, heterodimerization with E1 (albeit at reduced levels), and CD81 binding but was not entry competent. It is likely that its defect occurs post-CD81 binding and may be related to conformational changes induced either by interactions with other cellular receptors, such as SR-B1, claudin 1, and/or occludin, or by low-pH-induced conformational changes associated with viral fusion. Disulfide 6 is located adjacent to the igVR, which has been proposed to act as a hinge between domains I and III (19). In flavivirus glycoprotein E, exposure to low pH changes the center of mass of domain III, displacing it 33 Å, so that the viral-membrane-anchored stem and adjoining TMD appose the target membrane-anchored fusion loop, driving membrane merging (4). Our data suggest that disulfide 6 may be essential for the igVR of E2 to perform a similar function.
Domain II extends from β-sheets D0 and E0 of domain I and is characterized by the presence of three disulfides formed through pairings of neighboring cysteine residues. The absence of long-range disulfides led to the suggestion that this domain may be a relatively flexible region within E2. Mutagenesis of disulfide pairs within domain II revealed that formation of disulfide 4 was essential for HCVpp incorporation of E2, H53 epitope assembly, and CD81-binding function. Although distal to the CD81-binding sites, disulfide 4 overlaps the predicted fusion loop. This suggests that the formation of this structure is essential for formation of the adjacent domain I/III substructures and their associated functions. In contrast, mutagenesis of disulfide 2 (C452-C459) in E2661 maintained partial reactivity to H53 and CD81 LEL, suggesting that it is less important to the structural integrity of E2. Interestingly, individual mutation of either C452 or C459 to create a free thiol at either site resulted in wild-type levels of H53 reactivity and CD81 LEL binding and was therefore more favorable than complete removal of disulfide 2. It is possible that within the prefusion HCVpp-incorporated form of E1E2, C452 and C459 exist in a reduced state or form a labile disulfide in the domain II substructure, where formation of disulfide 2 represents an end product of thiol disulfide exchange.
We also reported a high degree of tolerance for the loss of predicted disulfide bonds within E2661 folding and/or function, as disruption of disulfides 3, 4, and 5 via mutations at residues C486, C503, and C569 maintained WT H53 reactivity and/or CD81 LEL binding. However, discordant effects were observed upon mutation of their disulfide-bonding partners—C494, C508, and C564, respectively—which saw the loss of these functions. Pairwise mutation of these residues corresponding to their predicted disulfides did not alleviate this loss-of-function phenotype, indicating that the presence of a free thiol was unlikely to be responsible for misfolding via the formation of aberrant disulfides, as previously described in a similar cysteine mutagenesis study of the HIV gp120-gp41 complex (29). This would indicate that the Cys-to-Ala mutation at these sites was detrimental to local conformation within E2, resulting in the loss of H53 and CD81 binding. Removal of the hydrophilic side chain by mutation to an alanine may have contributed to this phenotype. Introduction of serine rather than alanine at these positions may reduce alanine-induced conformational defects and clarify the roles of these cysteines in E1E2 function.
The minimal number of cysteine residues required for E2661 to maintain WT biosynthesis, H53 reactivity, and CD81 binding was delineated using a phenotypic-mutagenesis approach. Simultaneous introduction of C452A, C486A, C569A, C597A, C581A, C585A, and C652A (M+C597A) resulted in the expression of wild-type levels of E2661 that maintained H53 and CD81-binding activity. This suggested that C459, C494, C564, and C620 exist as free thiols, as there was no evidence that these residues engaged in a specific, alternative pattern of disulfide bonding. Consistent with the presence of free thiols, the M+C597A mutant also displayed a reduced propensity to form higher-molecular-weight species, resulting in a significant increase in the amount of monomeric E2661 secreted from transfected cells that was not observed for any of the individual mutants. Together, these data indicate that free thiols at positions 459, 494, 564, and 620 are tolerated in a CD81-binding-competent E2661 structure and that their predicted disulfide-bonding partners (C452, C486, C569, and C597) are available to engage in a complex pattern of aberrant intermolecular disulfide bonds during E2661 biosynthesis. As the cysteine mutagenesis results obtained with E2661 largely reflect the results obtained in HCVpp-incorporated E2, it is possible that these cysteine residues also include determinants for disulfide-mediated intermolecular contacts between E2 molecules, as well as between E1 and E2, as has been recently described on the surfaces of the mature HCV virions (30). Furthermore, it has recently been proposed that both E1 and E2 contain free thiol groups and that alkylation of virions prevents virus entry (13). However, after attachment to cells, the virus becomes resistant to alkylation, suggesting that postattachment disulfide isomerization occurs (13). It is possible that C452, C486, C569, and C597 participate in labile inter- and intramolecular disulfide-bonding arrangements that are critical for viral entry competence.
In summary, the results of this study provide new insights into E1E2 structure and suggest that HCVpp-incorporated E2 and soluble E2661 share similar disulfide-bonding arrangements. We have reported that the formation of a CD81-competent structure of E2661 strictly requires the presence of three disulfides across each of the three predicted domains: C429-C552A (DI), C503-C508 (DII), and C607-C644 (DIII). For the remaining cysteines, the disulfide can either be removed (C581-C585 or C652-C677) or unpaired (C459, C494, C564, or C620) without affecting this basic conformational requirement. Our data also suggest that the presence of unpaired thiols within E2 may reflect a mechanism for intra- or intermolecular disulfide exchange. In addition, the removal of C452, C486, C569, C581, C585, C597, and C652 is, somewhat unexpectedly, favorable to the formation of larger amounts of functional monomeric E2661 and may signal a new approach to synthesizing larger amounts of soluble E2 as a lead candidate for crystallization studies and the resolution of a three-dimensional structure.
ACKNOWLEDGMENTS
This work was supported by NHMRC project grants 543113, 603735, and 433913. H.E.D. was the recipient of an NHMRC R. D. Wright Biomedical Research Fellowship, and K.M. was the recipient of a Dora Lush Postgraduate Scholarship.
We sincerely thank Jean Dubuisson for the provision of reagents and CSL Limited for assistance with the production of MAb 24.
Footnotes
Published ahead of print 25 January 2012
REFERENCES
- 1. Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633–64212615904 [Google Scholar]
- 2. Benedicto I, et al. 2009. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J. Virol. 83:8012–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brazzoli M, et al. 2005. Folding and dimerization of hepatitis C virus E1 and E2 glycoproteins in stably transfected CHO cells. Virology 332:438–453 [DOI] [PubMed] [Google Scholar]
- 4. Bressanelli S, et al. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choukhi A, Ung S, Wychowski C, Dubuisson J. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciczora Y, et al. 2005. Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J. Gen. Virol. 86:2793–2798 [DOI] [PubMed] [Google Scholar]
- 7. Ciczora Y, Callens N, Penin F, Pecheur EI, Dubuisson J. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 81:2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drummer HE, Boo I, Maerz AL, Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J. Virol. 80:7844–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drummer HE, Maerz A, Poumbourios P. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385–390 [DOI] [PubMed] [Google Scholar]
- 10. Drummer HE, Poumbourios P. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279:30066–30072 [DOI] [PubMed] [Google Scholar]
- 11. Drummer HE, Wilson KA, Poumbourios P. 2002. Identification of the hepatitis C virus e2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans MJ, et al. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 13. Fraser J, Boo I, Poumbourios P, Drummer HE. 2011. Hepatitis C virus (HCV) envelope glycoproteins e1 and e2 contain reduced cysteine residues essential for virus entry. J. Biol. Chem. 286:31984–31992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris HJ, et al. 2010. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 285:21092–21102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris HJ, et al. 2008. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J. Virol. 82:5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kato N, et al. 1992. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 189:119–127 [DOI] [PubMed] [Google Scholar]
- 17. Keck ZY, et al. 2004. Hepatitis C virus e2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J. Virol. 78:9224–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitadokoro K, et al. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krey T, et al. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6:e1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marukian S, et al. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCaffrey K, Boo I, Poumbourios P, Drummer HE. 2007. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J. Virol. 81:9584–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meertens L, et al. 2008. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J. Virol. 82:3555–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michalak JP, et al. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299–2306 [DOI] [PubMed] [Google Scholar]
- 24. Owsianka AM, et al. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pileri P, et al. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 26. Ploss A, et al. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roccasecca R, et al. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scarselli E, et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Anken E, et al. 2008. Only five of 10 strictly conserved disulfide bonds are essential for folding and eight for function of the HIV-1 envelope glycoprotein. Mol. Biol. Cell 19:4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vieyres G, et al. 2010. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J. Virol. 84:10159–10168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weiner AJ, et al. 1991. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180:842–848 [DOI] [PubMed] [Google Scholar]
- 32. Whidby J, et al. 2009. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J. Virol. 83:11078–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wittig I, Braun HP, Schagger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
- 34. Yi M, Nakamoto Y, Kaneko S, Yamashita T, Murakami S. 1997. Delineation of regions important for heteromeric association of hepatitis C virus E1 and E2. Virology 231:119–129 [DOI] [PubMed] [Google Scholar]
- 35. Zheng A, et al. 2007. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 81:12465–12471 [DOI] [PMC free article] [PubMed] [Google Scholar]