Abstract
Proteolytic activation of the fusion protein of the highly pathogenic Nipah virus (NiV F) is a prerequisite for the production of infectious particles and for virus spread via cell-to-cell fusion. Unlike other paramyxoviral fusion proteins, functional NiV F activation requires endocytosis and pH-dependent cleavage at a monobasic cleavage site by endosomal proteases. Using prototype Vero cells, cathepsin L was previously identified to be a cleavage enzyme. Compared to Vero cells, MDCK cells showed substantially higher F cleavage rates in both NiV-infected and NiV F-transfected cells. Surprisingly, this could not be explained either by an increased F endocytosis rate or by elevated cathepsin L activities. On the contrary, MDCK cells did not display any detectable cathepsin L activity. Though we could confirm cathepsin L to be responsible for F activation in Vero cells, inhibitor studies revealed that in MDCK cells, cathepsin B was required for F-protein cleavage and productive replication of pathogenic NiV. Supporting the idea of an efficient F cleavage in early and recycling endosomes of MDCK cells, endocytosed F proteins and cathepsin B colocalized markedly with the endosomal marker proteins early endosomal antigen 1 (EEA-1), Rab4, and Rab11, while NiV F trafficking through late endosomal compartments was not needed for F activation. In summary, this study shows for the first time that endosomal cathepsin B can play a functional role in the activation of highly pathogenic NiV.
INTRODUCTION
Nipah virus (NiV) is a zoonotic paramyxovirus that causes severe encephalitic and respiratory diseases in humans and animals. Due to the lack of established antiviral therapeutics and vaccines, NiV is classified as a biosafety level 4 (BSL-4) agent. During the first outbreak beginning in 1998 in Malaysia, NiV was transmitted from Pteropus fruit bats to pigs and then to humans (12, 13). Since then, NiV has reemerged in Bangladesh and in India, causing encephalitis with mortality rates up to 80% (10). NiV transmission occurs via the respiratory route, and virus replication in vivo is mostly observed in epithelial and endothelial cells. The systemic infection of endothelia is accompanied by vasculitis and is a hallmark of NiV infection in all species (60, 79). In humans, widespread infection of small blood vessels in the central nervous system (CNS) resulting in severe damage of the microvasculature is thought to be the basis for the development of encephalitis (27, 40).
Successful NiV entry into host cells is accomplished by the concerted action of the two viral envelope glycoproteins. After binding of the attachment protein G to ephrin B2/B3 receptors on the cell surface (6, 50, 51), the fusion protein F in cooperation with the G protein promotes fusion of the viral envelope and cell membranes, leading to virus entry. After productive NiV replication, newly synthesized F and G proteins are expressed on the surface of the infected cell and can trigger cell-to-cell fusion with receptor-bearing neighboring cells, resulting in the formation of multinucleated syncytia (68). To gain fusion competence, the NiV F protein, which is synthesized in host cells as inactive precursor F0, must be cleaved by cellular proteases to generate a fusion-active, disulfide-linked F1-F2 heterodimer (48). Unlike cleavage of most other paramyxovirus F proteins, NiV F processing depends on the transport of precursor NiV F0 to the cell surface and subsequent endocytosis mediated by a tyrosine-based internalization signal in its cytoplasmic tail (525YSRL) (18, 77). Within the endolysosomal compartment, F0 is then ubiquitously activated by pH-dependent proteases at its monobasic cleavage site (arginine 109) (16, 48). After cleavage and release of the hydrophobic peptide at the N terminus of F1, the fusion-active F1-F2 form is recycled to the cell surface, where it can induce syncytium formation or is incorporated into budding virus particles. The ubiquitously expressed cysteine protease cathepsin L has conclusively been proven to be the NiV F-activating protease in Vero cells (53). In agreement with the precise NiV F activation at the monobasic cleavage site, cathepsin L had been shown to specifically process host cell proteins at dibasic and monobasic cleavage sites in endosomal compartments (74, 80). So far, other cathepsins have not been thought to be involved in NiV F activation. This study however, provides strong evidence that F cleavage in Madin-Darby canine kidney (MDCK) cells depends on cathepsin B, another pH-dependent and ubiquitously expressed cathepsin that can cleave substrates at monobasic cleavage sites (2). Using cathepsin L- and B-specific inhibitors, we confirmed the previously reported dependence of F activation on cathepsin L in Vero cells. Yet in MDCK cells, cathepsin L activity was not detectable, whereas cathepsin B was highly expressed. In contrast to Vero cells, F cleavage and NiV replication in MDCK cells were almost completely blocked by the cathepsin B inhibitor NS134P. We furthermore showed that block of transport from early to late endosomes by nocodazole did not affect fusion activity, suggesting that F cleavage does not require trafficking through late endosomal compartments but rather occurs within early-recycling endosomal compartments of MDCK cells. Supporting this view, endocytosed F proteins and cathepsin B substantially colocalized with early endosomal antigen 1 (EEA-1)-, Rab4-, and Rab11-positive endosomes. Together, these data demonstrate for the first time that besides cathepsin L, cathepsin B can function as a NiV-activating protease within the endosomal compartment.
MATERIALS AND METHODS
Cell culture.
MDCK cells were maintained in Eagle's minimal essential medium (MEM; Gibco) supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Vero cells (African green monkey kidney), HeLa cells, 293 cells, Huh7 cells, porcine aortic endothelial cells (PAECs) (20), and mouse embryo fibroblasts (MEFs) derived from wild-type (wt), cathepsin L-knockout (L−/−), cathepsin B-knockout (B−/−), or double-knockout (L−/−/B−/−) mice (65) were grown in Dulbecco's modified MEM (DMEM; Gibco) containing 10% FCS, penicillin, and streptomycin. Primary porcine brain microvascular endothelial cells (PBMECs) were cultured in medium 199 (Gibco) supplemented with 20% FCS and antibiotics.
Virus infections.
All experiments with live NiV were performed under BSL-4 conditions at the Institute of Virology, Philipps University of Marburg. The NiV strain used in this study was isolated from human and propagated as described earlier (48). For NiV infection studies, confluent monolayers of Vero, MDCK cells, or MEFs were infected with NiV at a multiplicity of infection (MOI) of 0.2, 0.5, or 2. After incubation for 1 h at 37°C, input virus was removed by extensive washings and cells were cultured in the absence or presence of inhibitors with DMEM without FCS for 24 h at 37°C.
Virus titers in the supernatant were determined by the 50% tissue culture infective dose (TCID50) method on Vero cells (57).
Plasmids and transfections.
The cDNA fragment spanning the F gene of the NiV genome (GenBank accession number AF212302) was cloned into the pczCFG5 vector as described earlier (45). To allow detection of the F protein with commercially available antibodies, a tagged version of the F protein was established by inserting the amino acids YPYDVPDYA of the influenza virus hemagglutinin (HA), known as the HA tag, at the C terminus (45). Plasmids encoding cyan fluorescent protein (CFP)- or green fluorescent protein (GFP)-tagged Rab genes (Rab4-CFP, Rab11-CFP, Rab7-GFP) (14) were a kind gift of R. Jacob (Marburg, Germany).
Vero and MDCK cells were transfected by using the cationic lipid transfection reagent Lipofectamine 2000 (Invitrogen).
Inhibitors.
Cathepsin L activity was inhibited by incubation with 20 μM CatLIII (Calbiochem), and cathepsin B activity was inhibited by addition of 10 or 20 μM NS134P (62). Both cathepsins were blocked by 20 μM the membrane-permeant pan-cysteine cathepsin inhibitor E64d (Sigma). Nocodazole (5 μM; Sigma) was used to analyze the effect of disrupted microtubuli on NiV replication. All chemicals were dissolved in dimethyl sulfoxide (DMSO).
Western blot analysis.
NiV-infected cell lysates were collected at 24 h postinfection (p.i.) by scraping the cells into phosphate-buffered saline (PBS) containing 1% sodium dodecyl sulfate (SDS). Proteins were separated on a 12% reducing SDS-gel and transferred onto nitrocellulose. Membranes were then incubated with an antibody directed against the NiV F cytoplasmic tail (17), followed by probing with biotin-labeled anti-rabbit immunoglobulins (Amersham) and horseradish peroxidase-conjugated streptavidin (Amersham). Proteins were visualized with an enhanced chemiluminescent system (SuperSignalWest Pico; Pierce). Band intensities were quantified using Tina (version 2.0) software. To analyze F processing in transfected cells, medium was changed at 8 h after transfection, and then cells were further incubated in the absence or presence of inhibitors. At 24 h posttransfection (p.t.), cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 10 mM EDTA, 10 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, 50 units/ml aprotinin, and 20 mM Tris-HCl, pH 8.5), followed by centrifugation for 45 min at 100,000 × g, and F proteins were immunoprecipitated using a polyclonal anti-HA antibody (H6908; Sigma). After addition of a suspension of protein A–Sepharose CL-4B (Sigma), immune complexes were washed with RIPA buffer and suspended in reducing sample buffer for separation on a 12% SDS-gel. Then, proteins were transferred onto nitrocellulose and probed with a monoclonal anti-HA antibody (HA.11; Covance) and IRDye 800 infrared dye antimouse antibody (LI-COR). Protein bands were detected and quantified using a LI-COR Odyssey imaging system. The amount of F1 protein as a percentage of total F protein (F1 plus F0) was calculated to yield percent cleavage.
Metabolic labeling.
For pulse-chase analysis, MDCK or Vero cells transiently expressing NiV F proteins were incubated at 24 h p.t. for 40 min with medium lacking cysteine and methionine, followed by incubation with medium containing [35S]cysteine and -methionine (Promix; Perkin Elmer) at a final concentration of 100 μCi/ml for 10 min (pulse). Then, labeling medium was replaced by serum-free nonradioactive medium and cells were incubated at 37°C for 2 or 3 h with or without the inhibitor CatLIII, NS134P, or nocodazole. After labeling, cells were extensively washed with PBS, followed by cell lysis in RIPA buffer. F proteins were then immunoprecipitated as described above and separated on a 12% polyacrylamide gel under reducing conditions. Dried gels were subjected to autoradiography and analyzed with a BAS1000 Bio-Image analyzer (Fuji).
Quantitative endocytosis assay.
Endocytosis rates of NiV F protein in different cell lines were quantified as described earlier (47). Briefly, Vero and MDCK cells transiently expressing NiV F were surface labeled with cleavable disulfide-linked NHS-SS-biotin (Pierce) at 4°C. Then, cells were shifted to 37°C for 0, 5, or 15 min to allow endocytosis to occur. After rapid cooling to 4°C, biotin still bound to the cell surface was cleaved by incubation with 50 mM 2-mercaptoethanesulfonic acid (MESNA; Sigma). To determine the total amount of biotinylated protein, one sample was neither incubated at 37°C nor reduced with MESNA (control). After cell lysis, F proteins were immunoprecipitated as described above, separated on an SDS-gel under nonreducing conditions, and transferred to nitrocellulose. Biotinylated proteins were detected with horseradish peroxidase-conjugated streptavidin (Amersham), visualized with the enhanced chemiluminescence system, and quantified densitometrically. Mean internalization rates per min were calculated as the difference of the ratio of intracellular biotinylated protein after 5, 10, and 15 min of internalization and total F protein (control) divided by 5, 10, and 15, respectively.
Cathepsin activity assays.
Confluent cell monolayers were lysed with 300 μl CytoBuster reagent (Novagen) for 5 min at room temperature. Cell lysates were centrifuged at 4°C for 7 min at 14,000 × g and then stored at 4°C. Enzyme activity of 10 μg of total protein was measured using InnoZyme cathepsin L and B activity kits (Calbiochem) following the instructions of the supplier. Fluorescence was measured with a Perkin Elmer LS55 luminescence spectrometer at excitation and emission wavelengths set at 370 nm and 450 nm, respectively. For each sample, enzymatic activity was measured in duplicate and indicated in relative fluorescence units (RFU).
Fusion assays.
To determine the effects of cathepsin inhibitors on NiV glycoprotein-mediated cell-to-cell fusion, syncytium formation was quantified as described earlier (46). Vero and MDCK cells were cotransfected with NiV F- and G-encoding plasmids. Inhibitors at a concentration of 20 μM were added at 2.5 h p.t., and cells were fixed and stained with 1:10-diluted Giemsa staining solution 18 h later. To quantify fusion, five randomly chosen microscopic fields for each sample were photographed. The number of nuclei in single, nonfused cells and the number of nuclei in syncytia were counted and averaged. The relation of the total number of nuclei and the number of nuclei in syncytia per microscopic field indicated the percent cell fusion.
Colocalization studies.
For colocalization studies of NiV F with early endosomes or Rab proteins, antibody uptake assays were performed as described previously (18). Briefly, at 24 h p.t., NiV F-expressing MDCK cells were incubated with a polyclonal guinea pig anti-NiV serum (1:1,000; kindly provided by H. Feldmann) for 1 h at 4°C. After stringent washing, cells were shifted to 37°C for 10 or 30 min to allow endocytosis to occur. Then, surface-bound primary antibodies were blocked with a peroxidase-conjugated secondary antibody (1:50; Dako). Following fixation with 4% paraformaldehyde (PFA) for 20 min and permeabilization with 0.2% Triton X-100–PBS, internalized primary antibodies were probed with Alexa Fluor (AF) 568-conjugated secondary antibodies (1:200; Molecular Probes). For costaining of early endosomes, an antibody against EEA-1 (1:100; BD Biosciences) was added after fixation and permeabilization for 1 h at 4°C. Bound primary antibodies were detected with AF 488-conjugated secondary antibodies (1:250; Molecular Probes). To analyze colocalization of endocytosed NiV F with marker proteins of the recycling and late endosomal compartments, pczCFG5-NiV-F was cotransfected with plasmids encoding either Rab4-CFP, Rab11-CFP, or Rab7-GFP. At 18 h p.t., antibody uptake assays were performed as described above.
For colocalization studies of endogenous cathepsin B with marker proteins of endosomal compartments, MDCK cells were transfected with Rab-encoding plasmids. Cells were fixed and permeabilized at 18 h p.t. After incubation with an anti-cathepsin B antibody from goat (1:100; Neuromics), primary antibodies were probed by consecutive incubation with AF 568-conjugated secondary antibodies (1:200). Costaining of EEA-1 was performed as described above. Representative images were recorded with a confocal laser scanning microscope (Leica SP5).
To visualize active cathepsin B, Rab11-CFP-transfected MDCK cells were seeded on 8-well glass-bottomed μ slides (Ibidi). At 16 h p.t., cells were washed with PBS and incubated with the cell-permeant cathepsin B-specific Magic Red fluorogenic substrate MR-(RR)2 (Immunochemistry Technologies, LLC). Optimal substrate concentration and incubation times were titrated in comparison to those for 293 cells, which have low cathepsin B activity. Finally, the fluorogenic substrate was used at a concentration 10-fold lower than that recommended by the manufacturer, and the incubation time was reduced to 15 min at 37°C. After rinsing with PBS, red fluorescence generated as a result of intracellular cathepsin B-mediated cleavage and CFP fluorescence were directly examined in live cells by confocal microscopy (Leica SP5).
Immunostaining of NiV-infected cells.
For the visualization of NiV-induced syncytia in MEFs at 48 h p.i., infected cells were inactivated and fixed with 4% PFA for 48 h and then permeabilized with methanol-acetone and stained with a polyclonal guinea pig anti-NiV serum (1:1,000). Primary antibodies were detected with rhodamine-conjugated secondary antibodies (1:200), and nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI).
For the analysis of NiV-induced cell-to-cell fusion in the presence of nocodazole, cells were infected in the absence or presence of 5 μM nocodazole (added at 8 h p.i.). At 24 h p.i., infected cells were fixed, permeabilized, and incubated with primary anti-NiV antibodies as described above. Primary antibodies were detected with AF 488-conjugated secondary antibodies (1:250) and nuclei were counterstained with DAPI. Staining of microtubules was performed using mouse anti-α-tubulin (1:1,000; Sigma) and AF 568-conjugated anti-mouse IgG. Images were recorded using a Zeiss Axiovert 200 M microscope.
RESULTS
NiV F cleavage efficiency varies between different cell types.
As for all paramyxoviruses, NiV production from infected host cells and virus spread via cell-to-cell fusion depend among other factors on the efficiency of F cleavage by host cell proteases. However, the NiV proteolytic activation pathway differs from that for other systemically replicating paramyxoviruses, for example, measles virus (MV). F proteins of MV are activated by the ubiquitous Golgi apparatus protease furin during transport to the cell surface, resulting in almost complete MV F cleavage (>95%) (16, 38). In contrast, the inactive precursor NiV F0 is transported to the cell surface and must then undergo clathrin-mediated endocytosis to get in contact with its activating protease in the endolysosomal compartment. After pH-dependent cleavage, fusion-active F1-F2 must then recycle back to the plasma membrane before cell-to-cell fusion can occur. As shown previously by others and us, F cleavage efficiencies differ significantly depending on the cell line (1, 7, 24, 48, 53). We found F processing rates to vary between 20 and 70% in cells from different species and tissues or in cell lines stably transfected with NiV glycoproteins. Interestingly, MDCK cells, well-established epithelial cells used to study virus replication in polarized epithelia (39, 78), showed consistently higher F cleavage rates than other cell types, including Vero cells, which are generally used to propagate NiV and in which cathepsin L was identified to be the NiV F-activating protease (53). A representative experiment showing F cleavage in virus-infected Vero and MDCK cells is presented in Fig. 1A. Here, cells were infected with NiV at an MOI of 0.2 to 0.5. At 24 h p.i., infected cells were lysed and inactivated, and Western blot analysis under reducing conditions was performed using a specific antiserum directed against the NiV F cytoplasmic tail. For quantification of the cleavage efficiency, the percentage of the F1 subunit was determined. Vero cells displayed an F cleavage of about 55%, whereas MDCK cells showed a processing rate of 75%. To investigate if the differences are also seen in the absence of virus infection, F cleavage in lysates of cells transfected with a NiV F-encoding plasmid was analyzed at 24 h after transfection. The Western blot analysis displayed in Fig. 1B revealed similar differences in cleavage efficiencies. Again, MDCK cells showed larger amounts of F1 cleavage products than Vero cells (Fig. 1C).
Fig 1.
NiV F cleavage efficiency in Vero and MDCK cells. (A) Vero and MDCK cells were infected with NiV at an MOI of 0.2 or 0.5. At 24 h p.i., cells were lysed. Samples were subjected to SDS-PAGE under reducing conditions, transferred to nitrocellulose, and probed with NiV F-specific antibodies and peroxidase-conjugated secondary antibodies. Proteins were visualized by enhanced chemiluminescence and quantified densitometrically. (B and C) Cells were transfected with a NiV F-encoding plasmid (pczCFG5-NiV-F). At 24 h p.t., F proteins were immunoprecipitated from cell lysates and subjected to Western blot analysis using an Odyssey infrared imaging system. (C) The amounts of F0 and F1 proteins were quantified from four independent experiments, and the amount of F1 protein as a percentage of total F protein (F1 plus F0) was calculated to yield the percent cleavage (% F1).
NiV F endocytosis rates do not significantly differ in Vero and MDCK cells.
Due to the fact that F endocytosis is an indispensable prerequisite for activation by endosomal cathepsins, different internalization rates could be the reason for the various efficiencies in F processing. We therefore analyzed NiV F uptake by a quantitative biotin-based endocytosis assay (77). For that purpose, F-expressing cells were surface labeled with NHS-SS-biotin and then incubated for 0, 5, or 15 min at 37°C to allow endocytosis to occur. Surface-remained biotin was then removed by incubation with 50 mM MESNA. To quantify the total amount of surface-biotinylated F, one sample was neither incubated at 37°C nor reduced with MESNA (control). After immunoprecipitation, F proteins were separated by SDS-PAGE under nonreducing conditions and transferred to nitrocellulose, and biotinylated proteins were detected with peroxidase-conjugated streptavidin. As shown in Fig. 2, the F internalization rates in Vero and MDCK cells were comparable (2.1 to 2.3% per min). Hence, differences in endocytosis kinetics are unlikely the reason for the substantial variations in F processing. Supporting the idea that minor variations in endocytosis rates did not affect overall processing of the F protein, no correlations between F cleavage and internalization rates were seen in other cell lines (e.g., 293, Huh-7, or HeLa cells or PAECs; data not shown).
Fig 2.
Endocytosis rate of NiV F in Vero and MDCK cells. At 24 h p.t., NiV F-expressing cells were surface labeled with cleavable NHS-SS-biotin at 4°C and then shifted to 37°C for the times indicated to allow endocytosis to occur. Subsequently, cell surface proteins were reduced with MESNA at 4°C. To determine the total amount of surface-biotinylated proteins, samples were compared to cells that were neither incubated at 37°C nor treated with MESNA (control). Following cell lysis, F proteins were immunoprecipitated and samples were separated on a 12% SDS-gel under nonreducing conditions and transferred to nitrocellulose. Biotinylated proteins were then detected with peroxidase-conjugated streptavidin and chemiluminescence. The control lane represents 50% of the total amount of biotinylated F proteins. After quantification of three independent experiments, the internalization rate was calculated and is indicated as percent per min.
Cathepsin L and B activities differ principally between Vero and MDCK cells.
Differences in the endocytosis rates did not explain the more pronounced F processing rates in MDCK cells compared to Vero cells. We thus hypothesized that variations in cleavage efficiencies might be the result of differences in the activity of cathepsin L, the endolysosomal protease shown to mediate NiV F cleavage (53). To evaluate this idea, we performed cathepsin L activity assays with lysates from Vero and MDCK cells using commercial InnoZyme cathepsin assays. Confirming previous data, cathepsin L activity in Vero cells was very high (52), but to our surprise, we did not detect any cathepsin L activity in MDCK cells (Fig. 3A). The efficient F processing, despite the lack of any detectable cathepsin L activity, strongly hints to a functional role of another endosomal protease for F activation. Besides cathepsin L, the only other cysteine protease known to be widely expressed in the endolysosomal compartment and shown to process substrates at basic cleavage sites is cathepsin B (2). Since cathepsins B and L have been shown to have redundant or synergistic functions in several viral infections (11, 19, 36, 64), we wanted to evaluate the role of cathepsin B in NiV infection. In agreement with reports that cathepsin B is the most abundant cysteine protease in most cell types (32, 73), we detected high cathepsin B activities in MDCK cells (Fig. 3B). Though cathepsin expression patterns and activities are differently regulated across various tissues and cells (26, 31, 35, 67), there is no cell type described so far completely lacking either cathepsin L or cathepsin B. We thus assume that MDCK cells are not cathepsin L deficient but display activities too low to detect. The idea of a potential role for cathepsin B in NiV F cleavage was further supported by the finding that several cell types aside from MDCK cells which readily support NiV F cleavage displayed very low cathepsin L activities (Fig. 3C). In line with the described high cathepsin B and low cathepsin L expression in endothelial cells (30), both endothelial cell types tested, primary PBMECs and PAECs, highly resemble MDCK cells in their overall cathepsin L and B activities.
Fig 3.
Cathepsin L and B activities. Cathepsin L (A) and cathepsin B (B) enzyme activities in Vero and MDCK cell lysates are plotted as relative fluorescence units (RFU). (C) Cathepsin L and B activities in different human (293, HeLa, Huh7) and porcine (PBMECs, PAECs) cell types are shown.
F cleavage, cell-to-cell fusion, and production of infectious virus in Vero and MDCK cells are differently affected by specific cathepsin L and B inhibitors.
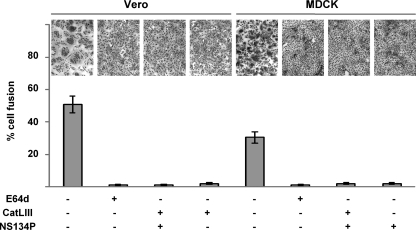
To directly assess the role of cathepsin B in NiV F activation, we performed inhibitor studies using cathepsin L (CatLIII) and cathepsin B (NS134P) inhibitors (62). To analyze the effect of the inhibitors on cleavage of transiently expressed NiV F protein, MDCK and Vero cells were metabolically labeled at 24 h after transfection with [35S]cysteine and -methionine for 10 min. Then, labeling medium was removed and cells were further incubated at 37°C for 2 h in the presence of 20 μM CatLIII, 10 μM NS134P, or both inhibitors. F proteins were then immunoprecipitated with an F-specific antiserum and separated by SDS-PAGE under reducing conditions. In agreement with the higher steady-state cleavage rates observed in the Western blot analysis (Fig. 1), cleavage efficiency at 2 h after labeling was higher in control MDCK cells than Vero cells (41% and 32%, respectively). Supporting our idea of different protease usage in the two cell types, F cleavage in Vero cells was drastically reduced to 4% and 3% in the presence of the cathepsin L inhibitor (Fig. 4, Vero cells with CatLIII) and with the addition of cathepsin L and B inhibitors (Vero cells with CatLIII and NS134P), respectively. Inhibition of cathepsin B with NS134P barely affected F processing. In contrast to Vero cells, CatLIII had no influence on F processing in MDCK cells, while cathepsin B inhibition alone (Fig. 4, MDCK cells with NS134P) or the combination of both inhibitors efficiently reduced F cleavage to 3% and 0.5%, respectively. In agreement, syncytium formation upon F and G coexpression, which directly depends on functional F cleavage, was effectively inhibited by CatLII in Vero cells and by NS134P in MDCK cells. Syncytium formation was blocked as efficiently by the single inhibitors selective for cathepsin L or cathepsin B as by the combination of the two inhibitors or the pan-cysteine cathepsin inhibitor E64d (Fig. 5). These findings clearly support the idea that cathepsin B is the F-activating protease in MDCK cells, whereas cathepsin L represents the key protease in Vero cells.
Fig 4.
Effect of specific cathepsin L and B inhibitors on NiV F cleavage. Vero and MDCK cells were transfected with pczCFG5-NiV-F. At 24 h p.t., cells were metabolically labeled with 35S-labeled Promix for 10 min and then incubated for 2 h in the absence (−) or presence (+) of 20 μM CatLIII, 10 μM NS134P, or both inhibitors. F proteins were immunoprecipitated from cell lysates and analyzed by autoradiography after separation on a 12% SDS-gel under reducing conditions. The amount of F1 protein (% F1) as a percentage of total F protein (F1 plus F0) was calculated to yield percent cleavage.
Fig 5.
Effect of cathepsin inhibitors on NiV glycoprotein-mediated fusion activity. Vero and MDCK cells were cotransfected with NiV G- and F-encoding plasmids. Cathepsin inhibitors at a final concentration of 20 μM were added at 2.5 h p.t. Eighteen hours later, syncytium formation was visualized by Giemsa staining. Quantification (percent cell fusion) was performed as described in Materials and Methods. Magnification, ×100.
We further analyzed the effects of the inhibitors on NiV infection. Since cleaved and fusion-active F proteins are essentially required for the production of infectious virus, we infected Vero and MDCK cells with NiV in the presence of either one or both cathepsin inhibitors and titrated the amount of infectious virus released into the cell supernatant at 24 h p.i. Confirming previous data (17), virus titers in Vero cell supernatants were reduced by more than 3 log steps in the presence of CatLIII (Table 1). As expected, cathepsin B inhibition had no effect. In contrast to Vero cells, infectious virus production in NiV-infected MDCK cells was not affected by CatLIII but was drastically reduced by the cathepsin B inhibitor NS134P (Table 1). These findings again demonstrate that protease usage differs in the two cell types. Cathepsin L represents the main activating protease in Vero cells, whereas NiV F cleavage in MDCK cells predominantly depends on cathepsin B.
Table 1.
Effect of cathepsin inhibitors on production of infectious NiVa
| Inhibitor | Virus titer (TCID50/ml) |
|
|---|---|---|
| Vero cells | MDCK cells | |
| Control | 4 × 105 | 1.2 × 104 |
| CatLIII (20 μM) | 2.5 × 102 | 1.5 × 104 |
| NS134P (10 μM) | 4.5 × 105 | 7 × 102 |
| CatLIII + NS134P | <2 × 102 | <2 × 102 |
Vero and MDCK cells were infected with NiV at MOIs of 0.2 and 0.5, respectively. After 1 h virus adsorption at 37°C, cells were washed to remove input virus and then further incubated with the indicated inhibitors. At 24 h p.i., virus titers in the supernatant were determined by the TCID50 method.
NiV replication is completely blocked only in cathepsin L and B double-deficient MEFs.
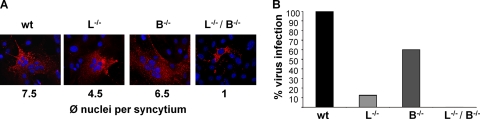
To further evaluate the impact of cathepsin L and B on NiV infection, MEFs derived from wt, L−/−, B−/−, or L−/−/B−/− mice were infected with NiV at an MOI of 2. At 48 h p.i., NiV infection was visualized by immunostaining (Fig. 6A). Syncytium formation was observed in wt, B−/−, and L−/− MEFs, with the average amount of nuclei per syncytium declining from wt to L−/− MEFs. Nevertheless, absence of either cathepsin L or cathepsin B alone did not completely abolish functional F cleavage, as measured by cell-to-cell fusion. Syncytium formation was completely prevented only in the absence of both cathepsins L and B (L−/−/B−/−). To quantify virus production, virus titers in the MEF supernatants were determined (Fig. 6B). In agreement with the reduced syncytium formation, production of infectious viruses was slightly impaired by knockout of cathepsin B (B−/−), whereas knockout of cathepsin L (L−/−) led to a more enhanced reduction. A complete block of production of infectious viruses was observed only in L−/−/B−/− MEFs. Even if the knockout of cathepsin L has a more prominent effect on NiV replication, these data support the idea that cathepsin B can functionally activate NiV in the absence of cathepsin L.
Fig 6.
NiV infection in cathepsin-knockout MEFs. MEFs derived from wild-type (wt), cathepsin L-knockout (L−/−), cathepsin B-knockout (B−/−), or double-knockout (L−/−/B−/−) mice were infected with NiV. (A) At 48 h p.i., NiV infection was visualized by immunostaining with a polyclonal anti-NiV serum and rhodamine-conjugated secondary antibodies, and nuclei were counterstained with DAPI. ϕ, average. (B) To quantify virus release, supernatants were collected at 48 h p.i., and virus titers were determined by the TCID50 method. One representative example from three independent experiments is shown. Virus titers in wild-type MEFs were set at 100%.
Microtubule-dependent transport to late endosomes is not needed for F processing and NiV-mediated cell-to-cell fusion.
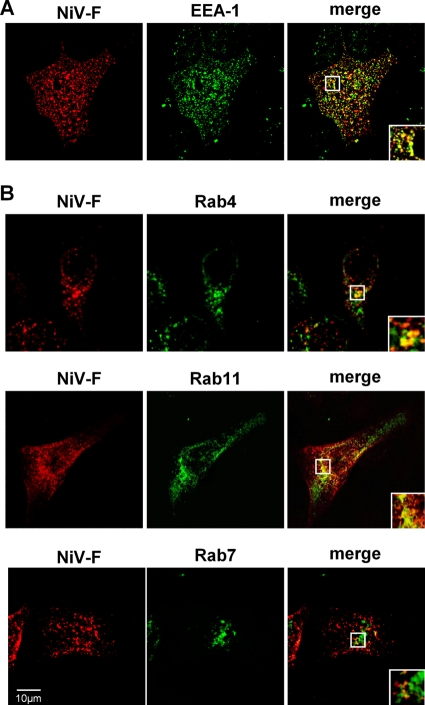
Efficient processing of NiV F by cathepsin B in MDCK cells requires a colocalization of cathepsin B and endocytosed F in the same endosomal compartments of MDCK cells. To evaluate this idea, we first determined which endosomes are reached by NiV F proteins after endocytosis. For this purpose, we performed an antibody uptake assay with F-expressing MDCK cells and analyzed the colocalization with EEA-1. At 24 h p.t., surface-expressed NiV F was labeled with a NiV-specific antiserum at 4°C, and then cells were shifted to 37°C for 10 min to allow endocytosis to occur. Subsequently, surface-bound primary antibodies were blocked with a peroxidase-conjugated antibody. After fixation and permeabilization, internalized F proteins were probed with AF 568-conjugated secondary antibodies. Early endosomes were stained with an EEA-1-specific and an AF 488-conjugated secondary antibody. As expected, F proteins substantially colocalized with early endosomes after 10 min of internalization (Fig. 7A). To further monitor endosomal F trafficking, NiV F was coexpressed with Rab4-CFP (marker for rapidly recycling endosomes [44]), Rab11-CFP (marker for recycling endosomes [44]), and Rab7-GFP (marker for late endosomes [22]). After surface labeling of the F proteins, endocytosis was allowed to proceed for 30 min. Then, surface-bound antibodies were blocked and internalized F proteins were detected as described above. Figure 7B clearly shows a prominent colocalization of endocytosed NiV F with Rab4 and Rab11, indicating that F proteins are mainly present in early and recycling endosomes after 30 min of endocytosis. F barely colocalized with Rab7, showing that only a few F proteins had entered late endosomal compartments (Fig. 7B).
Fig 7.
Intracellular localization of internalized NiV F proteins in MDCK cells. (A) At 24 h p.t., an antibody uptake assay with NiV F-expressing MDCK cells was performed. Cells were incubated with a polyclonal anti-NiV serum for 1 h at 4°C and then incubated for 10 min at 37°C. Following endocytosis, surface-bound primary antibodies were blocked by incubation with a peroxidase-conjugated secondary antibody. After fixation and permeabilization, internalized F proteins were detected with AF 568-conjugated secondary antibodies. Early endosomes were stained with an anti-EEA-1 antibody and AF 488-conjugated secondary antibodies. (B) MDCK cells were cotransfected with pczCFG5-NiV-F and Rab4-CFP-, Rab11-CFP-, or Rab7-GFP-encoding plasmids. At 18 h p.t., surface-expressed F proteins were labeled with the anti-NiV serum. Then, endocytosis was allowed to proceed for 30 min at 37°C. Internalized F proteins were detected as described above. Images were recorded with a confocal laser scanning microscope (SP5; Leica). Insets represent expansions of the boxed regions of the merged images.
To demonstrate that F is not only concentrated but also cleaved in early-recycling endosomes, microtubule (MT)-dependent protein transport from early to late endosomes was disrupted by the MT-depolymerizing agent nocodazole (4, 34). MT disruption was monitored by staining untreated MDCK cells (control) and cells treated with 5 μM nocodazole for 2 h with an anti-α-tubulin antibody. Figure 8A demonstrates that addition of nocodazole was effective in destroying the MT skeleton. To determine if the block of transport to late endosomes influences F processing, MDCK cells expressing NiV F were metabolically labeled for 10 min and then incubated for a further 3 h in normal medium (control) or in the presence of 5 μM nocodazole. After cell lysis, F proteins were immunoprecipitated, separated under reducing conditions, and then subjected to autoradiography. As shown in Fig. 8B, F cleavage was not significantly affected by nocodazole, indicating that F processing did not depend on the transport to late endosomes. To finally see if the cleaved F protein is biologically active and can induce cell-to-cell fusion in the absence of functional transport from early to late endosomes, MDCK cells were infected with NiV. At 8 h p.i., medium was replaced by medium without (control) or with nocodazole. Sixteen hours later, cells were fixed and NiV infection was visualized by immunostaining with a polyclonal anti-NiV serum. The result shown in Fig. 8C reveals that syncytium formation was not blocked by nocodazole treatment, demonstrating that disruption of MT blocking vesicle transport from early to late endosomes had no impact on the generation and surface expression of fusion-active F proteins in NiV infection.
Fig 8.
Influence of nocodazole on NiV F processing and NiV infection. (A) MDCK cells were treated for 2 h with 5 μM nocodazole (+ nocodazole) or left untreated (control). Then, cells were fixed, permeabilized, and incubated with mouse anti-α-tubulin antibodies and AF 568-labeled anti-mouse IgG. Nuclei were visualized by DAPI staining. (B) NiV F-expressing MDCK cells were metabolically labeled with 35S-labeled Promix for 10 min and then incubated for 3 h in the absence (control) or presence of 5 μM nocodazole (+ nocodazole). F proteins were immunoprecipitated from cell lysates and analyzed by autoradiography after separation on a 12% SDS-gel under reducing conditions. (C) MDCK cells were infected with NiV at an MOI of 0.5. At 8 h p.i., medium was replaced by fresh medium (control) or by medium containing 5 μM nocodazole (+ nocodazole). At 24 h p.i., cells were fixed and permeabilized. Virus-induced syncytium formation was detected by incubation with a polyclonal anti-NiV serum and AF 488-conjugated secondary antibodies.
Cathepsin B predominantly localizes in the F cleavage compartments of MDCK cells.
Since we have shown that F cleavage in MDCK cells occurs in early or recycling endosomes and are proposing that F cleavage is mediated by cathepsin B, we finally wanted to investigate the intracellular distribution of cathepsin B in MDCK cells. We thus performed colocalization studies of endogenous cathepsin B with endosomal marker proteins. As shown in Fig. 9A (EEA-1), cathepsin B was detected in early endosomes. Colocalization with Rab4- and Rab11-positive recycling endosomes was even more pronounced (Fig. 9B). To confirm that endosomal vesicles contain active cathepsin B, Rab11-expressing cells were incubated with a cathepsin B substrate linked to Magic Red. MR-(RR)2 represents an indicator dye that fluoresces only when the peptide substrate is cleaved by cathepsin B and thus stains intracellular compartments with catalytically active cathepsin B. In live MDCK cells, intensive punctate Magic Red staining which partly overlapped with Rab11-positive recycling endosomes was detected (Fig. 9C). These findings substantiate the idea that efficient F processing in MDCK cells is due to cathepsin B protease expression in the (early) recycling endosomal compartments where F cleavage takes place.
Fig 9.
Subcellular localization of endogenous cathepsin B in MDCK cells. (A) MDCK cells were fixed and permeabilized. Early endosomes were visualized with an EEA-1-specific primary antibody and AF 488-conjugated secondary antibodies. Endogenous cathepsin B was detected with anti-cathepsin B antibodies and AF 568-conjugated secondary antibodies. (B) MDCK cells were transfected with Rab4-CFP- or Rab11-CFP-encoding plasmids. At 18 h p.t., cells were fixed and endogenous cathepsin B was stained as described above. Images were recorded with a confocal laser scanning microscope (SP5; Leica). Insets represent expansions of the boxed regions of the merged images. (C) To visualize catalytically active cathepsin B, Rab11-CFP-expressing cells were incubated with the fluorogenic cathepsin B substrate Magic Red MR-(RR)2. Live cells were directly examined by confocal microscopy.
DISCUSSION
Proteolytic activation of the NiV F protein depends on clathrin-mediated endocytosis and subsequent cleavage at the monobasic cleavage site (R109) by a pH-dependent endosomal protease (18, 48). Cathepsin L has been conclusively shown to act as an F cleavage enzyme using Vero cells as a model cell line (53). Here, we show that in contrast to Vero cells, F activation in MDCK cells does not depend on cathepsin L but rather requires active cathepsin B. Since MDCK cells did not display any detectable cathepsin L activity, the different protease usage in these cells strongly argues for a redundant or alternative functional role of cathepsin B in NiV activation. The finding that functional F cleavage and NiV infection in MDCK cells were not prevented by nocodazole and the prominent colocalization of internalized F and cathepsin B with EEA-1, Rab4, and Rab11 strengthen the idea that NiV activation by cathepsin B occurs within early and recycling endosomes of MDCK cells and does not require trafficking through late endosomal compartments.
Despite a pronounced activity of cathepsin B in Vero cells, we did not see a significant effect of the cathepsin B inhibitor NS134P on NiV F cleavage (Fig. 4) and virus release (Table 1). This is in principal agreement with the finding of Pager et al. (53), reporting that cathepsin L is the only protease responsible for F activation in Vero cells. In contrast to MDCK cells, in which cathepsin B was found to functionally process NiV F0, the readily expressed cathepsin B appeared not to be involved in NiV activation in Vero cells. As proposed by Pager et al. (53), this might be due to an incorrect cleavage by cathepsin B in Vero cells. An alternative explanation could be a different compartmentalization of cathepsin B in Vero cells that does not allow sufficient contact with the NiV F protein or pH conditions in endosomes that prevent a proteolytic processing by cathepsin B at R109.
Cathepsins L and B are ubiquitous cysteine proteases of the papain superfamily not only involved in bulk protein turnover in the lysosomal compartment but also critically engaged in proteolytic processing of specific substrates in endosomes like hormones, growth factors, and protein antigens (see reference 59 and references therein). Even though these processing steps require precise cleavage, cathepsins exhibit a broad substrate and cleavage site specificity, albeit some prefer certain amino acids to others in the target sequence. For example, cathepsin L prefers hydrophobic amino acids in P2 and basic residues in P1 (56). In contrast, cathepsin B preferentially cleaves after two basic amino acid residues in P1 and P2, but efficient cleavage still takes place if P2 consists of a hydrophobic amino acid (23). Analysis of the P4-P2′ NiV F cleavage site by the software tool PEPS (Prediction of Endopeptidase Substrates [37]), which is based on known cathepsin B or cathepsin L cleavage sites in full-length protein substrates, revealed prediction scores of the NiV F cleavage site of 0.165 and 0.2 for cathepsin L and cathepsin B, respectively. Both scores indicate a more than 1,000-fold likelihood of NiV F cleavage by cathepsin B or L than by a random amino acid sequence. The major difference in the structures of cathepsins L and B is that the latter contains a so-called occluding loop that covers part of the catalytic center, thereby limiting access of substrate proteins for endoproteolytic cleavage. Recently, the cleavage specificities of cathepsins B and L on naturally occurring peptides have been reassessed by the proteomic approach PICS (proteomic identification of protease cleavage sites [5, 63]). Remarkably, the PICS cleavage site logo at pH 6 for cathepsin B contains 6 of the 8 amino acids present in the P4-P4′ NiV F cleavage site. The P3′ glycine in the NiV F site is especially typical for the cathepsin B endoprotease activity, because this small amino acid is not sterically hindered by the occluding loop. This enables access of the NiV F protein to the catalytic center of the protease and determines the specific cleavage site. In contrast, the PICS profile for cathepsin L highlights only 4 of the 8 residues present in the P4-P4′ NiV F cleavage site. These structure-substrate considerations indicate that cathepsin B, in addition to cathepsin L, is well suited for selective processing of NiV F. Although the amino acid sequence of the substrate itself strongly dictates its cleavage, specificity of cleavage is orchestrated by the biochemical environment, the set of cleaving enzymes, the time that a protein substrate resides within the same compartment, and the change of endosomal pH from almost neutral to acidic (29). Due to different pH optima, proteases are active only at certain stages in the endolysosomal compartment (32, 42). While specific cleavage by cathepsin L is optimal at a slightly acidic pH of 5.5 to 6 and the protease is unstable under neutral conditions (43, 70, 72), cathepsin B has a wider pH optimum (pH 4.5 to 7.0) (61, 71, 75). Activity of cathepsin B endopeptidase is maximal between pH 6 and 7 (3, 55), thereby strongly supporting our model that cathepsin B-mediated NiV F cleavage occurs in early and recycling endosomes (pH 5.9 to 6.5) rather than in late endosomal compartments (pH 5.0 to 6.0).
Even though the substrate specificity and pH dependence of cathepsins L and B are not fully identical, they share many substrates. The idea of a partial redundancy of both enzymes was supported by studies with knockout mice. Here, the loss of one protease can be compensated for by the remaining protease (9, 65). Knockout mice deficient in only cathepsin B or cathepsin L presented only a few phenotypic alterations (58), whereas double-knockout mice lacking both cathepsin L and cathepsin B died early due to CNS atrophy (21). We also observed a redundant function of cathepsins L and B when we infected MEFs from knockout mice. Even though syncytium formation and release of infectious NiV in cathepsin L-knockout MEFs were drastically reduced and the reduction was more pronounced than that in MEFs lacking cathepsin B, productive virus replication was completely blocked only in the absence of both cathepsin L and cathepsin B. This again supports our view that both cathepsins have the ability to produce fusion-active NiV F proteins and further strengthens our model that cathepsin B can play an important role in NiV activation in cells lacking sufficient cathepsin L activity.
Redundant functions of cathepsins L and B, especially if one cathepsin is lacking, were also reported for the proteolytic processing of other viral proteins. For example, Ebola virus (EBOV) entry into Vero cells and MEFs requires glycoprotein digestion by cathepsins B and L (11, 64). However, cathepsin L-deficient human monocyte-derived dendritic cells also supported EBOV entry, thus indicating that cathepsin L is dispensable for infection in these cells (41). In Moloney murine leukemia virus (MLV) infection, virus entry is facilitated by cathepsin B, which cleaves the surface unit SU in the early endosome (36). While inhibition of cathepsin L had no effect on infection in cells expressing both cathepsin B and cathepsin L, a block of cathepsin L significantly inhibited MLV infection in cathepsin B-deficient NIH 3T3 cells (81). From this it was concluded that both cathepsin B and cathepsin L can support MLV infection, with cathepsin B being the favored protease. These data are consistent with our idea of an alternative usage of cathepsins L and B for NiV activation, depending on cell type and availability of the respective protease. Recent studies on reovirus infections also highlighted distinct roles of cathepsins in vivo. While reoviruses are known to utilize cathepsins B, S, and L for disassembly of the outer capsid to initiate productive replication, only cathepsin L was required for efficient virus growth in mouse brains in vivo (19, 25, 28). The contribution of the two cathepsins for cell or organ tropism of NiV infection in vivo might be an interesting question for the future.
Currently, no approved antiviral treatment for NiV infection is available, even though a variety of approaches have been investigated (for a review, see reference 76). A more detailed understanding of NiV activation by host cell proteases could be a further starting point for developing antivirals acting against cellular targets. Due to the impact of several cathepsins in pathophysiological processes, including cancer, osteoarthritis, and autoimmune disorders, some promising results for the effectiveness of cathepsin inhibitors in tumor invasion, in osteoporosis prevention, and as immunomodulators have been obtained (15, 66, 69). Short-term inhibition of cathepsins in the context of a potentially lethal Nipah virus infection might be a clinically feasible option. Since our data indicate that NiV can principally use cathepsins L and B and as it is not yet clear which protease is predominantly used in in vivo infections, antiviral strategies should include inhibitors blocking both cathepsins.
Besides the restricted expression of cellular receptors, limited expression of activating proteases is an important factor for virus spread and cell tropism, thereby determining pathogenesis in vivo (8, 33, 38, 49, 54). Our results showing that NiV can use two broadly expressed cathepsins, however, suggest that protease expression does not restrict NiV spread in vivo. The ability to utilize either cathepsin L or cathepsin B likely allows the virus to principally replicate in any receptor-positive cell type.
ACKNOWLEDGMENTS
We are grateful to Heinz Feldmann (Hamilton, MT) and Ralf Jacob (Marburg, Germany) for providing anti-NiV serum and Rab constructs, respectively.
L.S. was supported by a fellowship of the Jürgen-Manchot-Stiftung. This work was supported by grants of the German Research Foundation (DFG) to A.M. (GK 1216/2 and SFB 593/TP B11).
Footnotes
Published ahead of print 25 January 2012
REFERENCES
- 1. Aguilar HC, et al. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J. Virol. 81:4520–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almeida PC, et al. 2000. Hydrolysis by cathepsin B of fluorescent peptides derived from human prorenin. Hypertension 35:1278–1283 [DOI] [PubMed] [Google Scholar]
- 3. Aronson NN, Jr, Barrett AJ. 1978. The specificity of cathepsin B. Hydrolysis of glucagon at the C-terminus by a peptidyldipeptidase mechanism. Biochem. J. 171:759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bayer N, et al. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biniossek ML, Nägler DK, Becker-Pauly C, Schilling O. 2011. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J. Proteome Res. 10:5363–5373 [DOI] [PubMed] [Google Scholar]
- 6. Bonaparte MI, et al. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U. S. A. 102:10652–10657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bossart KN, et al. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 76:11186–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bottcher E, et al. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brix K, Dunkhorst A, Mayer K, Jordans S. 2008. Cysteine cathepsins: cellular roadmap to different functions. Biochimie 90:194–207 [DOI] [PubMed] [Google Scholar]
- 10. Chadha MS, et al. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chua KB, et al. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257–1259 [DOI] [PubMed] [Google Scholar]
- 13. Chua KB, et al. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435 [DOI] [PubMed] [Google Scholar]
- 14. Cramm-Behrens CI, Dienst M, Jacob R. 2008. Apical cargo traverses endosomal compartments on the passage to the cell surface. Traffic 9:2206–2220 [DOI] [PubMed] [Google Scholar]
- 15. Deaton DN, Tavares FX. 2005. Design of cathepsin K inhibitors for osteoporosis. Curr. Top. Med. Chem. 5:1639–1675 [DOI] [PubMed] [Google Scholar]
- 16. Diederich S, Dietzel E, Maisner A. 2009. Nipah virus fusion protein: influence of cleavage site mutations on the cleavability by cathepsin L, trypsin and furin. Virus Res. 145:300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diederich S, Thiel L, Maisner A. 2008. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology 375:391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diederich S, Moll M, Klenk HD, Maisner A. 2005. The Nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 280:29899–29903 [DOI] [PubMed] [Google Scholar]
- 19. Ebert DH, Deussing J, Peters C, Dermody TS. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609–24617 [DOI] [PubMed] [Google Scholar]
- 20. Erbar S, Diederich S, Maisner A. 2008. Selective receptor expression restricts Nipah virus infection of endothelial cells. Virol. J. 5:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Felbor U, et al. 2002. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc. Natl. Acad. Sci. U. S. A. 99:7883–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng Y, Press B, Wandinger-Ness A. 1995. Rab 7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 131:1435–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fineschi B, Miller J. 1997. Endosomal proteases and antigen processing. Trends Biochem. Sci. 22:377–382 [DOI] [PubMed] [Google Scholar]
- 24. Garner OB, et al. 2010. Endothelial galectin-1 binds to specific glycans on Nipah virus fusion protein and inhibits maturation, mobility, and function to block syncytia formation. PLoS Pathog. 6:e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Golden JW, Bahe JA, Lucas WT, Nibert ML, Schiff LA. 2004. Cathepsin S supports acid-independent infection by some reoviruses. J. Biol. Chem. 279:8547–8557 [DOI] [PubMed] [Google Scholar]
- 26. Gong Q, Chan SJ, Bajkowski AS, Steiner DF, Frankfater A. 1993. Characterization of the cathepsin B gene and multiple mRNAs in human tissues: evidence for alternative splicing of cathepsin B pre-mRNA. DNA Cell Biol. 12:299–309 [DOI] [PubMed] [Google Scholar]
- 27. Hooper P, Zaki S, Daniels P, Middleton D. 2001. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 3:315–322 [DOI] [PubMed] [Google Scholar]
- 28. Johnson EM, et al. 2009. Genetic and pharmacologic alteration of cathepsin expression influences reovirus pathogenesis. J. Virol. 83:9630–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jordans S, et al. 2009. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem. 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joyce JA, et al. 2004. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell 5:443–453 [DOI] [PubMed] [Google Scholar]
- 31. Katunuma N, Kakegawa H, Matsunaga Y, Nikawa T, Kominami E. 1993. Different functional share of individual lysosomal cathepsins in normal and pathological conditions. Agents Actions Suppl. 42:195–210 [DOI] [PubMed] [Google Scholar]
- 32. Kirschke H, Barrett AJ, Rawlings ND. 1995. Proteinases 1: lysosomal cysteine proteinases. Protein Profile 2:1581–1643 [PubMed] [Google Scholar]
- 33. Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39–43 [DOI] [PubMed] [Google Scholar]
- 34. Kok JW, Hoekstra K, Eskelinen S, Hoekstra D. 1992. Recycling pathways of glucosylceramide in BHK cells: distinct involvement of early and late endosomes. J. Cell Sci. 103:1139–1152 [DOI] [PubMed] [Google Scholar]
- 35. Kominami E, Tsukahara T, Bando Y, Katunuma N. 1985. Distribution of cathepsins B and H in rat tissues and peripheral blood cells. J. Biochem. 98:87–93 [DOI] [PubMed] [Google Scholar]
- 36. Kumar P, Nachagari D, Fields C, Franks J, Albritton LM. 2007. Host cell cathepsins potentiate Moloney murine leukemia virus infection. J. Virol. 81:10506–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lohmüller T, et al. 2003. Toward computer-based cleavage site prediction of cysteine endopeptidases. Biol. Chem. 384:899–909 [DOI] [PubMed] [Google Scholar]
- 38. Maisner A, et al. 2000. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J. Gen. Virol. 81:441–449 [DOI] [PubMed] [Google Scholar]
- 39. Maisner A, Klenk H, Herrler G. 1998. Polarized budding of measles virus is not determined by viral surface glycoproteins. J. Virol. 72:5276–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maisner A, Neufeld J, Weingartl H. 2009. Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb Haemost. 102:1014–1023 [DOI] [PubMed] [Google Scholar]
- 41. Martinez O, et al. 2010. Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cell. Microbiol. 12:148–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mason RW. 1995. Lysosomal metabolism of proteins. Subcell. Biochem. 27:159–190 [DOI] [PubMed] [Google Scholar]
- 43. Mason RW, Massey SD. 1992. Surface activation of pro-cathepsin L. Biochem. Biophys. Res. Commun. 189:1659–1666 [DOI] [PubMed] [Google Scholar]
- 44. Maxfield FR, McGraw TE. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132 [DOI] [PubMed] [Google Scholar]
- 45. Moll M, Kaufmann A, Maisner A. 2004. Influence of N-glycans on processing and biological activity of the Nipah virus fusion protein. J. Virol. 78:7274–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moll M, Klenk HD, Maisner A. 2002. Importance of the cytoplasmic tails of the measles virus glycoproteins for fusogenic activity and the generation of recombinant measles viruses. J. Virol. 76:7174–7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moll M, Klenk HD, Herrler G, Maisner A. 2001. A single amino acid change in the cytoplasmic domains of measles virus glycoproteins H and F alters targeting, endocytosis, and cell fusion in polarized Madin-Darby canine kidney cells. J. Biol. Chem. 276:17887–17894 [DOI] [PubMed] [Google Scholar]
- 48. Moll M, Diederich S, Klenk HD, Czub M, Maisner A. 2004. Ubiquitous activation of the Nipah virus fusion protein does not require a basic amino acid at the cleavage site. J. Virol. 78:9705–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nagai Y. 1995. Virus activation by host proteinases. A pivotal role in the spread of infection, tissue tropism and pathogenicity. Microbiol. Immunol. 39:1–9 [DOI] [PubMed] [Google Scholar]
- 50. Negrete OA, Chu D, Aguilar HC, Lee B. 2007. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J. Virol. 81:10804–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Negrete OA, et al. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405 [DOI] [PubMed] [Google Scholar]
- 52. Pager CT, Dutch RE. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12714–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pager CT, Craft WW, Jr, Patch J, Dutch RE. 2006. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346:251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peeters BP, de Leeuw OS, Koch G, Gielkens AL. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J. Virol. 73:5001–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Polgar L, Csoma C. 1987. Dissociation of ionizing groups in the binding cleft inversely controls the endo- and exopeptidase activities of cathepsin B. J. Biol. Chem. 262:14448–14453 [PubMed] [Google Scholar]
- 56. Puzer L, et al. 2004. Comparative substrate specificity analysis of recombinant human cathepsin V and cathepsin L. Arch. Biochem. Biophys. 430:274–283 [DOI] [PubMed] [Google Scholar]
- 57. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. J. Hyg. (Lond.) 27:493–497 [Google Scholar]
- 58. Reinheckel T, Deussing J, Roth W, Peters C. 2001. Towards specific functions of lysosomal cysteine peptidases: phenotypes of mice deficient for cathepsin B or cathepsin L. Biol. Chem. 382:735–741 [DOI] [PubMed] [Google Scholar]
- 59. Reiser J, Adair B, Reinheckel T. 2010. Specialized roles for cysteine cathepsins in health and disease. J. Clin. Invest. 120:3421–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rockx B, et al. 2011. Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J. Virol. 85:7658–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roederer M, Bowser R, Murphy RF. 1987. Kinetics and temperature dependence of exposure of endocytosed material to proteolytic enzymes and low pH: evidence for a maturation model for the formation of lysosomes. J. Cell. Physiol. 131:200–209 [DOI] [PubMed] [Google Scholar]
- 62. Schaschke N, et al. 2002. Epoxysuccinyl peptide-derived cathepsin B inhibitors: modulating membrane permeability by conjugation with the C-terminal heptapeptide segment of penetratin. Biol. Chem. 383:849–852 [DOI] [PubMed] [Google Scholar]
- 63. Schilling O, Overall CM. 2008. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26:685–694 [DOI] [PubMed] [Google Scholar]
- 64. Schornberg K, et al. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sevenich L, Pennacchio LA, Peters C, Reinheckel T. 2006. Human cathepsin L rescues the neurodegeneration and lethality in cathepsin B/L double-deficient mice. Biol. Chem. 387:885–891 [DOI] [PubMed] [Google Scholar]
- 66. Sever N, Filipic M, Brzin J, Lah TT. 2002. Effect of cysteine proteinase inhibitors on murine B16 melanoma cell invasion in vitro. Biol. Chem. 383:839–842 [DOI] [PubMed] [Google Scholar]
- 67. Soderstrom M, et al. 1999. Cathepsin expression during skeletal development. Biochim. Biophys. Acta 1446:35–46 [DOI] [PubMed] [Google Scholar]
- 68. Tamin A, et al. 2002. Functional properties of the fusion and attachment glycoproteins of Nipah virus. Virology 296:190–200 [DOI] [PubMed] [Google Scholar]
- 69. Thurmond RL, Sun S, Karlsson L, Edwards JP. 2005. Cathepsin S inhibitors as novel immunomodulators. Curr. Opin. Investig. Drugs 6:473–482 [PubMed] [Google Scholar]
- 70. Turk B, et al. 1999. Acidic pH as a physiological regulator of human cathepsin L activity. Eur. J. Biochem. 259:926–932 [DOI] [PubMed] [Google Scholar]
- 71. Turk B, et al. 1994. Human cathepsin B is a metastable enzyme stabilized by specific ionic interactions associated with the active site. Biochemistry 33:14800–14806 [DOI] [PubMed] [Google Scholar]
- 72. Turk B, Dolenc I, Turk V, Bieth JG. 1993. Kinetics of the pH-induced inactivation of human cathepsin L. Biochemistry 32:375–380 [DOI] [PubMed] [Google Scholar]
- 73. Turk B, Turk V, Turk D. 1997. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol. Chem. 378:141–150 [PubMed] [Google Scholar]
- 74. Turk D, Guncar G. 2003. Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr. D Biol. Crystallogr. 59:203–213 [DOI] [PubMed] [Google Scholar]
- 75. Tycko B, Maxfield FR. 1982. Rapid acidification of endocytic vesicles containing alpha 2-macroglobulin. Cell 28:643–651 [DOI] [PubMed] [Google Scholar]
- 76. Vigant F, Lee B. 2011. Hendra and Nipah infection: pathology, models and potential therapies. Infect. Disord. Drug Targets 11:315–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. 2005. Endocytosis of the Nipah virus glycoproteins. J. Virol. 79:3865–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Weise C, et al. 2010. Tyrosine residues in the cytoplasmic domains affect sorting and fusion activity of the Nipah virus glycoproteins in polarized epithelial cells. J. Virol. 84:7634–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wong KT, et al. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 161:2153–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yasothornsrikul S, et al. 2003. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. U. S. A. 100:9590–9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yoshii H, et al. 2009. Cathepsin L is required for ecotropic murine leukemia virus infection in NIH 3T3 cells. Virology 394:227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]