Abstract
West Nile virus (WNV) recently became endemic in the United States and is a significant cause of human morbidity and mortality. Natural WNV strain infections do not induce stress granules (SGs), while W956IC (a lineage 2/1 chimeric WNV infectious clone) virus infections produce high levels of early viral RNA and efficiently induce SGs through protein kinase R (PKR) activation. Additional WNV chimeric viruses made by replacing one or more W956IC genes with the lineage 1 Eg101 equivalent in the W956IC backbone were analyzed. The Eg-NS4b+5, Eg-NS1+3+4a, and Eg-NS1+4b+5 chimeras produced low levels of viral RNA at early times of infection and inefficiently induced SGs, suggesting the possibility that interactions between viral nonstructural proteins and/or between viral nonstructural proteins and cell proteins are involved in suppressing early viral RNA synthesis and membrane remodeling during natural WNV strain infections. Detection of exposed viral double-stranded RNA (dsRNA) in W956IC-infected cells suggested that the enhanced early viral RNA synthesis surpassed the available virus-induced membrane protection and allowed viral dsRNA to activate PKR.
INTRODUCTION
West Nile virus (WNV) is maintained in nature in a mosquito-bird transmission cycle and has recently become endemic in the United States. Humans and horses are occasionally infected but are dead-end hosts (10, 35). Infections in humans are usually asymptomatic, but some individuals develop fever and/or flu-like symptoms, and a few of these progress to central nervous system disease (10, 35). WNV belongs to the family Flaviviridae, genus Flavivirus, which includes other mosquito-borne human pathogens such as dengue virus, yellow fever virus (YFV), and Japanese encephalitis virus. The WNV genome is a positive-sense, single-stranded RNA of ∼11,000 nucleotides (nt) that contains a 5′ cap but no 3′ poly(A) tail (35). It encodes a single polyprotein that is co- and posttranslationally processed by both host and viral proteases into three structural (capsid [C], membrane [M], and envelope [E]) and seven nonstructural (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) proteins (35). Most of the more than 70 strains of WNV are classified in either lineage 1 or lineage 2 (5, 6, 28, 33). Lineage 1 WNV strains are endemic in northern Africa, Europe, the Middle East, and parts of Asia and the Americas, while lineage 2 WNV strains are typically found in sub-Saharan Africa (5, 6, 28, 33).
In response to several types of cellular stress, eukaryotic translation initiation factor 2 alpha (eIF2α) Ser51 is phosphorylated by one of four kinases: protein kinase R (PKR), activated by double-stranded RNA (dsRNA) (viral infection); heme-regulated inhibitor kinase (HRI), activated by oxidative stress; PKR-like endoplasmic reticulum kinase (PERK), activated by unfolded proteins in the endoplasmic reticulum (ER); and general control nonrepressed 2 (GCN2), activated by amino acid starvation (2). Phosphorylation of eIF2α reduces the amount of available eIF2-GTP-tRNA complex and thus inhibits translation initiation, resulting in polysome disassembly through ribosome runoff. RNA-binding proteins such as TIA-1, TIAR, and G3BP can then bind to the 5′ end of the mRNA with a stalled initiation complex and aggregate with other mRNA-bound proteins, forming microscopically visible stress granules (SGs) (2, 3). SGs recruit many additional proteins, including RNA-stabilizing proteins (2, 9). Although cap-dependent translation is suppressed in stressed cells, translation from internal ribosome entry site (IRES) elements and alternative open reading frames (ORFs), such as those in the mRNAs of heat shock proteins, chaperones, and some transcription factors (i.e., ATF4), still occurs (51). If a cell survives the stress, SGs dissociate and the mRNAs resume translation (9). However, if a cell is unable to recover and enters apoptosis, stalled mRNAs in SGs are transported to and degraded by processing bodies (PBs) (46, 48). Some virus infections have been shown to suppress SG formation (15, 38, 54). Lineage 1 WNV Eg101 infections in BHK cells were previously reported not to induce SGs and to inhibit SG induction by arsenite treatment (17). A subsequent study showed that infections with the lineage 2/1 infectious chimeric W956IC virus activated PKR, while WNV Eg101 infections did not (16).
In the present study, SG induction by infections with additional natural lineage 1 or lineage 2 WNV strains and induction by infections with W956IC were compared in BHK cells. None of the natural lineage 1 or lineage 2 WNV strains tested induced SGs by 24 h after infection, while W956IC virus infections efficiently induced SGs starting at 12 h after infection. All SG-positive, WNV-infected cells had elevated levels of phosphorylated eIF2α (p-eIF2α). PKR was identified as the kinase phosphorylating eIF2α in WNV-infected cells. Data from mapping experiments done with a set of WNV chimeric viruses associated SG formation with two different combinations of nonstructural proteins, suggesting that interactions between viral nonstructural proteins and/or between viral nonstructural proteins and cell proteins regulate the level of early viral RNA synthesis. Increased levels of SG-positive, chimeric virus-infected cells correlated with higher levels of early viral RNA synthesis and the presence of viral dsRNA that was not protected by intracellular membranes.
MATERIALS AND METHODS
Cells.
All cell lines used were maintained at 37°C in 5% CO2. Baby hamster kidney 21 strain W12 (BHK) cells (49) and a C3H/He mouse embryo fibroblast (MEF) cell line (44) were maintained in minimal essential medium (MEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS), glutamine, and 10 μg/ml gentamicin. C57BL/6, PERK+/+, PKR−/−, PERK−/−, GCN2−/−, and HRI−/− MEFs were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS, glutamine, and 10 μg/ml gentamicin. The PERK−/−, GCN2−/−, and HRI−/− cells are allelic knockouts, while the PKR−/− cells express a nonfunctional PKR due to a deletion of the RNA-binding domain located in exons 2 to 4 (57). The PERK+/+, PERK−/−, GCN2−/−, and HRI−/− MEFs were provided by David Ron (New York University School of Medicine, New York, NY), and the PKR−/− MEFs were provided by Robert Silverman (Lerner Research Institute, Cleveland Clinic, Cleveland, OH).
Viruses.
WNV Eg101 was prepared by infecting BHK cell monolayers at a multiplicity of infection (MOI) of 0.1 and harvesting culture fluid at 32 h after infection. Clarified culture fluid was aliquoted and stored at −80°C. W956IC RNA (1 μg/2 × 106 cells) in vitro transcribed as previously described (7) was transfected into BHK cells using DMRIE-C according to the manufacturer's protocol (Invitrogen). Clarified culture fluid harvested at 32 h after infection was aliquoted and stored at −80°C. Aliquots of WNV NY99, Tx113, B956, Mg78, and SPU were provided by Robert Tesh (University of Texas Medical Branch, Galveston, TX), and virus stocks were grown in BHK cells as described for Eg101. The titers of these stocks were as follows (in PFU/ml): Eg101, 1 × 108; W956IC, 5 × 107; NY99, 1 × 108; Tx113, 5 × 107; B956, 1 × 107; Mg78, 3 × 106; and SPU, 7 × 107. Virus infectivity was assessed by plaque assay on BHK monolayers as previously described (45).
Construction of chimeric viruses.
The construction of the 956D117B3/Eg101 infectious clone SP6WNEg3′/Xba, here referred to as W956IC, was described previously (56). The capsid, NS1, NS2a, NS3-NS4a, NS4b, and NS5 lineage 2 gene regions or combinations of these gene regions were replaced in the W956IC backbone with the corresponding lineage 1 genes from Eg101. RNA was purified from WNV Eg101, and specific cDNA fragments were amplified by reverse transcription-PCR (RT-PCR) using specific primers and a SuperScript one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen). The primers were designed based on the Eg101 sequence (GenBank accession no. AF260968). Nucleotide changes, which did not alter the amino acid sequence, were included in some primers to introduce unique restriction sites present in the W956IC sequence. The nucleotide coordinates indicated below are from the 956D117B3 sequence (GenBank accession no. M12294). To create a W956IC chimera with a capsid gene from Eg101 (Eg-C), an 89- to 484-nt fragment containing a unique BglII site was amplified from Eg101 RNA, and a 465- to 918-nt fragment containing an MfeI site was amplified from W956IC; a BglII-MfeI fragment was obtained by overlapping PCR and cloned into W956IC. Additional chimeras were created by amplifying the following fragments from Eg101 RNA and cloning them into W956IC: Eg-NS1, a MunI (nt 2393)-NsiI (nt 3513) fragment (this clone contains sequence encoding the C-terminal 23 amino acids [aa] of E); Eg-NS2a+2b+3N (referred to as Eg-NS2a+2b), an NsiI (nt 3513)-BstB1 (nt 5135) fragment (this clone contains sequence encoding Eg101 NS2a, NS2b, and the first 178 aa of NS3); Eg-NS3C+4aN (referred to as Eg-NS3+4a), a BstB1 (nt 5135)-SphI (nt 6777) fragment (this clone contains the C-terminal 439 aa of NS3 and the N-terminal 105 aa of NS4a); Eg-NS4aC+4bN (referred to as Eg-NS4b), an SphI (nt 6777)-FseI (nt 7021) fragment (this clone contains sequence encoding the C-terminal 42 aa of NS4a and the N-terminal 40 aa of NS4b); and Eg-NS4bC+5 (referred to as Eg-NS4b+5), an FseI (nt 7021)-AvrII (nt 8901) fragment (this clone contains sequence encoding the C-terminal 214 aa of NS4b as well as NS5). For the Eg-NS1+3C+4aN (referred to as Eg-NS1+3+4a) chimera, a MunI (nt 2393)-NsiI (nt 3513) fragment was amplified from Eg101 and cloned into the Eg-NS3+4a chimera, and for the Eg-NS1+4bC+5 (referred to as Eg-NS1+4b+5) chimera, a MunI (nt 2393)-NsiI (nt 3513) fragment was amplified from Eg101 and cloned into the Eg-NS5 chimera. All of the chimeric clones were validated by sequencing. The sequences of the primers used in this study are available upon request.
Confocal microscopy.
Cells were rinsed in phosphate-buffered saline (PBS) and then fixed for 10 min at room temperature in 4% paraformaldehyde in PBS. After fixation, cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. After blocking in 5% horse serum in PBS for 1 h at room temperature, the cells were incubated with antibody. Virus-infected cells were detected with anti-dsRNA antibody (English & Scientific Consulting, Szirak, Hungary). SGs were detected with anti-G3BP antibody (Ras-GAP-SH3-binding protein) (Abcam and Sigma-Aldrich). Anti-Dcp1a antibody (a gift from J. Lykke-Anderson, University of Colorado, Boulder, CO) was used to detect PBs. Alexa Fluor 488- or Alexa Fluor 594-labeled secondary antibodies (Invitrogen) were diluted in blocking buffer containing 0.5 μg/ml Hoechst 33258 dye (Molecular Probes) to detect nuclei. Images were taken using an LSM 510 confocal microscope (Zeiss, Oberkochen, Germany) with either a 63× water immersion or a 100× oil immersion objective. Images were analyzed with Zeiss LSM Image Browser software.
SG-positive infected-cell quantification.
A wide-field immunofluorescence microscope with a 40× objective was used to acquire images of large fields of WNV-infected cells stained with anti-dsRNA antibody to detect infected cells and anti-G3BP to detect SG-positive, infected cells at various times after infection and/or treatment as indicated in the figures. The percentage of SG-positive, infected cells was calculated by counting 100 cells per field from four or more fields per experiment. Quantification was done using the percentages obtained from three separate experiments, and statistical significance was determined at a 95% confidence interval.
Quantification of viral RNA.
In vitro-transcribed viral genome RNA of a known concentration was serially diluted and used to generate a standard curve (y = −0.2472x + 10.751, where “x” represents the cycle threshold [CT]) by real-time RT-PCR. This standard curve was then used to determine the absolute amount of viral RNA in each cell extract sample. BHK cells were infected with a strain of WNV at an MOI of 1 and lysed 12 h after infection. Total cell RNA was purified using TriReagent (Molecular Research Center) according to the manufacturer's protocol and quantified by spectrophotometry at an optical density at 260 nm (OD260). The mean CT value for each RNA sample was determined by real-time RT-PCR using a probe targeting the WNV NS1 gene. The mean CT value was plotted on the standard curve to estimate the number of viral genome copies per 1 μg of total RNA in each cell extract sample.
Western blotting.
Western blot analyses were performed as previously described (44). Primary antibodies used were anti-p-PKR (T451) (Millipore), anti-PKR (SCBT), anti-p-eIF2α (Ser51) and anti-eIF2α (Cell Signaling), and anti-actin (Abcam). The secondary antibodies were anti-rabbit horseradish peroxidase (HRP) and anti-mouse HRP (Cell Signaling). All antibodies were diluted in 5% nonfat dry milk, except for anti-p-PKR and anti-p-eIF2α, which were diluted in 5% bovine serum albumin (BSA) in TBS+Tween 20 buffer (50 mM Tris, 150 mM NaCl, pH 7.6, and 0.05% Tween 20).
Plaque assay.
Virus titers were determined as previously described (45).
Cell viability assay.
Confluent monolayers in 96-well plates were infected with W956IC at an MOI of 1. Virus was adsorbed for 1 h at room temperature, and the cells were then incubated in fresh media containing different concentrations of 2-aminopurine (2-AP) or dimethyl sulfoxide (DMSO) (solvent control) for 24 h at 37°C in 5% CO2. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay to assess cell viability was done using a CellTiter 96 nonradioactive cell proliferation assay kit (Promega) according to the manufacturer's protocol.
RESULTS
Efficiency of SG formation induced by different strains of WNV.
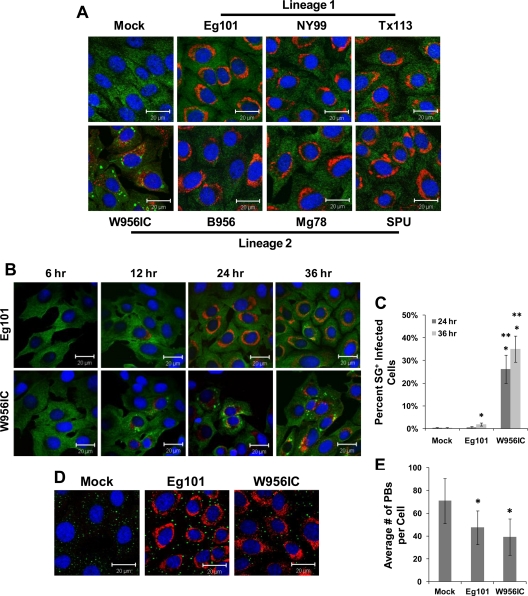
It was previously reported that lineage 2-based W956IC virus infections induced PKR phosphorylation while lineage 1 WNV Eg101 infections did not (16). WNV Eg101 infections were previously reported not to induce SGs (17). To determine whether different strains of WNV varied in the efficiency with which they induced SGs, BHK cells were infected with WNV lineage 1 strain Eg101, NY99, or Tx113, with WNV lineage 2 strain B956, Mg78, or SPU, or with lineage 2/1-based chimeric WNV infectious clone W956IC virus. Cells were fixed at 36 h after infection and analyzed by confocal microscopy. Less than 3% of the infected cells were SG positive by 36 h after infection with each of the lineage 1 and 2 WNV strains tested (Fig. 1A), confirming our previous results (17). In contrast, W956IC infections induced SGs in ∼30% of the infected BHK cells by 36 h after infection (Fig. 1A and C).
Fig 1.
Stress granule induction by different strains of WNV. (A) BHK cells were infected with a lineage 1 or lineage 2 WNV strain at an MOI of 1, fixed at 36 h after infection, and analyzed by confocal microscopy. Anti-G3BP antibody (green) was used to detect SGs, anti-dsRNA antibody (red) to detect WNV-infected cells, and Hoechst dye (blue) to detect nuclei. Merged images are shown. (B) BHK cells were infected with either Eg101 (lineage 1) or W956IC (lineage 2/1-based infectious clone) virus at an MOI of 1 and analyzed for SGs and dsRNA at the indicated times after infection as described for panel A. Merged images are shown. (C) The total number of infected cells and the number of SG-positive, infected cells were counted, and the percentage of Eg101- or W956IC-infected cells containing SGs was determined as described in Materials and Methods. *, significance compared to mock-infected cells at a 95% confidence interval (P < 0.05); **, significance compared to Eg101-infected cells at a 95% confidence interval (P < 0.05). (D) BHK cells were infected with either Eg101 or W956IC at an MOI of 1 and analyzed for PBs. Anti-Dcp1a antibody (green) was used to detect PBs, anti-dsRNA antibody (red) to detect virus-infected cells, and Hoechst dye (blue) to detect nuclei. Merged images are shown. (E) PBs were counted to determine the average number per infected cell. *, significance compared to mock-infected cells at a 95% confidence interval (P < 0.05). Bars indicate ± standard deviations (SD).
The time course of SG formation in Eg101- and W956IC-infected cells was next analyzed. BHK cells were infected with either WNV Eg101 or W956IC (MOI of 1) and then fixed and analyzed for SGs at various times after infection. Less than 0.5% of the mock-infected cells contained SGs at either 24 or 36 h after infection (Fig. 1B and C). No SG-positive, WNV Eg101-infected cells were detected at either 6 or 12 h after infection, whereas SG-positive W956IC-infected cells were observed by 12 h. By 24 h and 36 h, respectively, less than 1% and less than 3% of Eg101-infected cells contained SGs (Fig. 1B and C). In contrast, ∼25% of the W956IC-infected cells were SG positive at 24 h after infection, and ∼35% were SG positive by 36 h (Fig. 1B and C). Differential efficiency of SG induction by Eg101 and W956IC virus infections was also observed in C3H/He MEFs, C57BL/6 MEFs, and HEK 293T cells (data not shown). Although the numbers of SG-positive infected cells and the times at which they initially appeared differed among the cell lines tested, W956IC infections consistently induced SGs at earlier times after infection in a higher number of infected cells than did Eg101 infections.
During stress, stalled pre-mRNA complexes are held in SGs, and when the fate of the cell is decided, either mRNA translation is reinitiated or the mRNAs are transferred to PBs for degradation (2, 8, 46). Alterations in PB number occur in response to various stress stimuli (48), and our lab previously reported that the number of PBs in WNV Eg101- and dengue 2 virus-infected BHK cells was reduced by 24 h after infection (17). To determine the effect of a W956IC virus infection on PBs, W956IC-infected BHK cells were fixed at 36 h after infection and analyzed by confocal microscopy. Mock-infected cells contained an average of ∼70.93 PBs/cell, whereas Eg101-infected cells had ∼47.5 PBs/cell and W956IC-infected cells had ∼39.03 PBs/cell (Fig. 1E). The results showed that both Eg101 and W956IC infections reduced PB numbers in infected cells, with a slightly greater reduction seen in the W956IC-infected cells. PB numbers were also reduced in BHK cells infected with the NY99, Tx113, B956, Mg78, and SPU strains of WNV compared to numbers in mock-infected cells (data not shown).
SG formation in WNV-infected cells is PKR dependent.
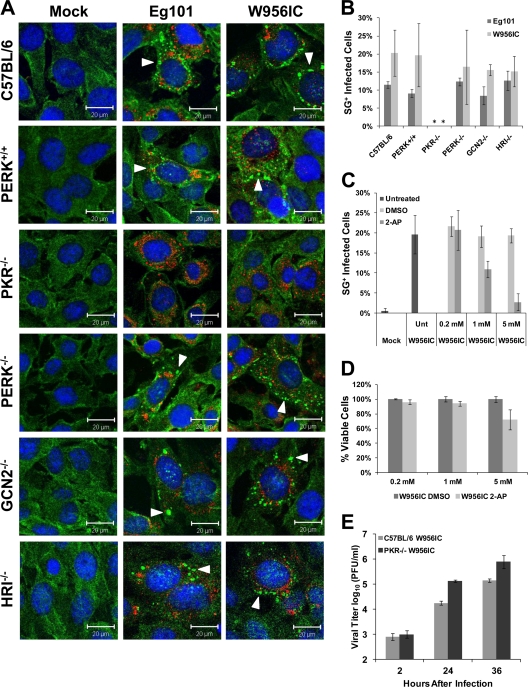
The phosphorylation of eIF2α, which leads to SG formation in mammals, is mediated by four kinases, PKR, PERK, GCN2, and HRI (2). To determine which of these four eIF2α kinases mediates SG induction in WNV-infected cells, the eIF2α kinase knockout MEF cell lines PKR−/−, PERK−/−, GCN2−/−, and HRI−/− (24, 26, 57) were mock infected or infected with WNV Eg101 or W956IC at an MOI of 1 and then fixed at 24 h after infection and analyzed by confocal microscopy. C57BL/6 (B6) and PERK+/+ (27) MEFs that express all four of the eIF2α kinases were used as controls. SGs were detected in Eg101- and W956IC-infected control, PERK−/−, GCN2−/−, and HRI−/− MEFs (Fig. 2A). Quantification of the number of SG-positive, infected cells indicated that the control, PERK−/−, GCN2−/−, and HRI−/− MEF cultures each contained 8 to 12% SG-positive, Eg101-infected cells or 15 to 20% SG-positive, W956IC-infected cells at 24 h (Fig. 2B). However, no SGs were observed in PKR−/− cells infected with either virus (Fig. 2A and B), indicating that SG induction in WNV-infected cells is PKR dependent. Unlike BHK cells, which were reported not to produce or respond to type I interferon (IFN) (4, 31, 32, 41), MEFs are type I IFN competent. It was previously reported that signaling by type I IFN induces elevated levels of both PKR and phosphorylated PKR in MEFs (16, 47). The increased levels of SGs observed in WNV-infected MEFs compared to those in IFN-incompetent BHK cells are therefore likely to be due to activation of PKR by IFN. However, W956IC infection still induced higher levels of SG-positive, infected MEFs than Eg101 infection. BHK cells were used for the remainder of the experiments in this study to avoid a contribution of IFN to PKR activation.
Fig 2.
Stress granule formation induced by WNV infection is PKR dependent. (A) Control MEFs (C57BL/6 and PERK+/+) and eIF2α kinase knockout MEFs (PKR−/−, PERK−/−, GCN2−/−, and HRI−/−) were infected with WNV Eg101 or W956IC at an MOI of 1, fixed at 24 h after infection, and analyzed by confocal microscopy. SGs were detected with anti-G3BP antibody (green), WNV-infected cells with anti-dsRNA antibody (red), and nuclei with Hoechst dye (blue). Arrowheads indicate SG-positive cells. Merged images are shown. (B) The total number of infected cells and the number of SG-positive, infected cells were counted, and the percentage of infected cells containing SGs was determined as described in Materials and Methods. *, significance compared to PERK+/+ control cells for the indicated infection at a 95% confidence interval (P < 0.05). (C) BHK cells were treated with different concentrations of 2-AP or DMSO (vehicle control) starting immediately after a 1-h adsorption of W956IC virus (MOI of 1), fixed at 24 h after infection, and analyzed by confocal microscopy for SGs and dsRNA as described for panel A. The percentage of SG-positive, infected cells was determined. (D) An MTT assay, performed as described in Materials and Methods, was used to assess the viability of BHK cells infected with W956IC and treated with different concentrations of 2-AP or DMSO. (E) Wild-type C57BL/6 and PKR−/− MEFs were infected with W956IC (MOI of 1). Virus yields were measured by plaque assay in BHK cells at the indicated times after infection. Bars indicate ± SD.
As an alternative means of showing that PKR is the kinase responsible for the formation of SGs in WNV-infected cells, BHK cells were incubated with 2-aminopurine (2-AP), a purine analog that is a potent PKR inhibitor (29, 37). BHK cells were infected with W956IC virus (MOI of 1) for 1 h and then incubated in media containing 2-AP or DMSO (solvent control) for 24 h. While approximately 20% of the W956IC-infected cells were SG positive in untreated, DMSO-treated, and 0.2 mM 2-AP-treated cultures, only 10% of W956IC-infected cells were SG positive in the 1 mM 2-AP-treated cultures, and 3% were SG positive in 5 mM 2-AP-treated cultures (Fig. 2C). An MTT assay showed a <4% decrease in cell viability after 0.2 mM 2-AP treatment, a <6% decrease after 1 mM 2-AP treatment, and an ∼25% decrease after 5 mM 2-AP treatment of W956IC-infected cells (Fig. 2D). These results suggest that the dose-dependent decrease in SG formation observed with 2-AP treatment was not due to a decrease in cell viability and further confirm that PKR is the kinase responsible for SG formation during infection.
To determine whether the increased SG formation by W956IC virus infections had a negative effect on virus production, wild-type C57BL/6 and PKR−/− MEFs were infected with W956IC virus at an MOI of 1. Clarified culture fluids collected at various times after infection were analyzed for viral infectivity by plaque assay. The yields of W956IC virus from PKR−/− MEFs were significantly higher than those from the control MEFs (Fig. 2E). These results suggest that PKR activation leading to SG formation during WNV infections has an antiviral effect, but the previously reported involvement of PKR in IFN-β production in WNV-infected cells could also contribute to the decrease in virus production observed (20).
W956IC infections induce higher levels of phosphorylated PKR and phosphorylated eIF2α in BHK cells at early times after infection.
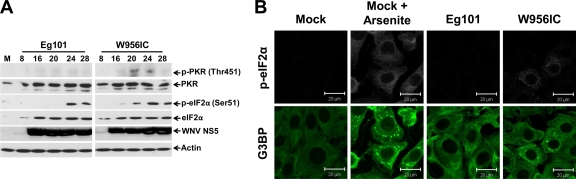
Consistent with the higher number of SG-positive cells in W956IC-infected BHK cultures, higher levels of activated PKR (phosphorylated at Thr451) and phosphorylated eIF2α in these cultures than in WNV Eg101-infected cultures are also expected. Cell lysates collected at various times after infection with WNV Eg101 or W956IC were analyzed by Western blotting for total and phosphorylated PKR and eIF2α. PKR phosphorylation was not detected in any of the WNV Eg101-infected lysates; however, phosphorylated PKR was detected in the W956IC samples from 16 through 28 h after infection (Fig. 3A). eIF2α phosphorylation in W956IC-infected cells was detected starting at 16 h after infection (Fig. 3A). Because a more sensitive Western blot detection system was used in this study than in our previous study (17), some phosphorylated eIF2α was detected at 24 and 28 h after infection with WNV Eg101. In contrast, similar levels of total eIF2α were detected by 16 h after infection with either virus. The increase in phosphorylated eIF2α at later times of infection correlated with the increase in SG-positive, WNV Eg101-infected BHK cells from <1% at 24 h to <3% at 36 h after infection. Increased eIF2α phosphorylation in Eg101-infected BHK cells in the absence of PKR phosphorylation suggested that this was due to the activation of another eIF2α kinase.
Fig 3.
W956IC infection induces PKR and eIF2α phosphorylation. (A) Western blot analysis of BHK cells infected with WNV Eg101 or W956IC at an MOI of 1. Lysates were collected in radioimmunoprecipitation assay (RIPA) buffer at the indicated times (hours) after infection and analyzed using antibodies to phosphorylated PKR (Thr541), PKR, phosphorylated eIF2α (Ser51), eIF2α, and actin. M, mock. (B) BHK cells were infected with WNV Eg101 or W956IC at an MOI of 1, fixed at 24 h after infection, and analyzed by confocal microscopy. Anti-G3BP antibody (green) was used to detect SGs, and anti-p-eIF2α antibody (white) was used to detect p-eIF2α. More than 95% of the cells were infected.
To determine whether SG-positive, infected cells contained elevated levels of phosphorylated eIF2α, BHK cells were infected with WNV Eg101 or W956IC, fixed at 24 h after infection, and analyzed by confocal microscopy. The level of p-eIF2α was increased in all cells that were SG positive (Fig. 3B). However, not all p-eIF2α-positive cells contained SGs (data not shown), suggesting that there may be a threshold level of p-eIF2α needed for SG induction or that additional cell factors/conditions may also be involved in SG formation.
Mapping the viral determinants involved in enhanced SG induction by W956IC virus infection.
The W956IC cDNA plasmid is very stable in bacteria and has been a useful tool for recovering WNV genomes with engineered mutations (7). The W956IC cDNA was constructed from a highly passaged WNV B956 virus stock, designated 956D117B3 (56). The 956D117B3 sequence differs from that of B956 by 32 single nucleotide substitutions within the coding region (55, 56). Although 956D117B3 cDNA was used to construct the majority of the W956IC infectious clone, the C terminus of the NS5 gene through the 3′ untranslated region (UTR) was amplified from WNV Eg101 genomic RNA (56). To identify viral components associated with less efficient SG induction, additional chimeric W956IC genomes were made by replacing one or more W956IC genome regions with the Eg101 equivalent as described in Materials and Methods. Eg101 sequence was used because it was hypothesized that the different phenotype of W956IC might be due to the presence of the 3′ end of the NS5 gene and that adding additional Eg101 regions to “match” the C-terminal region of NS5 would restore the natural virus phenotype. Eg-C (capsid), Eg-NS1, Eg-NS2a+2b, Eg-NS3+4a, Eg-NS4b, Eg-NS4b+5, Eg-NS1+3+4a, and Eg-NS1+4b+5 chimeric viruses were constructed (Fig. 4). All of the chimeric WNV infectious clones constructed contained the 3′ UTR from Eg101, and all but the Eg-NS4b+5 and Eg-NS1+4b+5 clones contained a 956D117B3/Eg101 hybrid NS5 gene. The capsid gene was replaced by Eg101 sequence in the Eg-C chimera. These sequences differed by 12 aa. The C-terminal 23 aa of E through the end of NS1 were replaced with Eg101 sequence in the Eg-NS1 chimera. One of the 23 E aa and 31 of the 351 NS1 aa differed between Eg101 and W956IC. The Eg-NS2a+2b chimera contained Eg101 sequence encoding all of NS2a and NS2b as well as the N-terminal 178 aa of NS3. The Eg101 and W956IC sequences differed at 26 of 230 NS2a aa, 2 of 130 NS2b aa, and 4 of 178 N-terminal NS3 aa. The Eg-NS3+4a chimera contained Eg101 sequence encoding the C-terminal 439 aa of NS3 and the N-terminal 105 aa of NS4a. Twenty-two NS3 aa and 8 NS4a aa differed between Eg101 and W956IC. The Eg-NS4b chimera contained Eg101 sequence encoding the C-terminal 42 aa of NS4a and the N-terminal 40 aa of NS4b. One NS4a aa and 12 NS4b aa differed between Eg101 and W956IC. The Eg-NS4b+5 chimera contained Eg101 sequence encoding the C-terminal 214 aa of NS4b and the N-terminal 410 aa of NS5. Nine NS4b aa and 24 N-terminal NS5 aa differed between Eg101 and W956IC. The C-terminal region of NS5 in W956IC was already Eg101 sequence and contained no amino acid changes.
Fig 4.
Schematic representation of the WNV W956IC-Eg101 chimera genomes. Eg101 sequences are indicated by dark-gray and W956IC sequences by light-gray shading. The majority of the W956IC genome sequence is from 956D117B3, but the C terminus of NS5 and the 3′ UTR are from Eg101 (56). Additional chimeric viruses were created by replacing one or more W956IC gene regions with the Eg101 equivalent on the W956IC backbone. The number of amino acid differences between Eg101 and W956IC is indicated above each replaced gene segment.
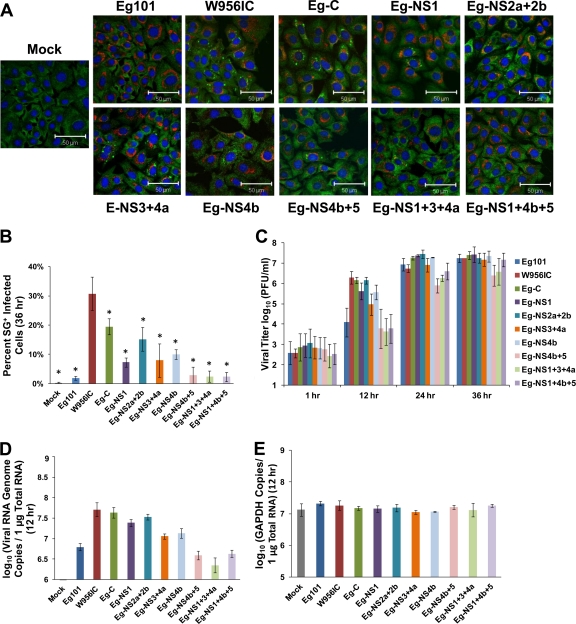
BHK cells were infected with one of the chimeric viruses, fixed at 36 h after infection, and analyzed for SGs by confocal microscopy. Consistent with data shown in Fig. 1, WNV Eg101 infections induced SGs in <3% of infected cells while W956IC infections induced SGs in ∼30% of the infected cells at this time after infection (Fig. 5A and B). The Eg-NS1 (∼7%), Eg-NS2a+2b (∼15%), Eg-NS3+4a (∼8%), Eg-NS4b (∼10%), and Eg-C (∼20%) chimeric virus infections induced intermediate numbers of SG-positive, infected cells by 36 h after infection, while the Eg-NS4b+5 (<3%), Eg-NS1+3+4a (<3%), and Eg-NS1+4b+5 (<3%) chimeric virus infections induced a low number of SG-positive, infected cells similar to the result seen with Eg101 infections (Fig. 5A and B). The data indicate that either the replacement of the C-terminal region of NS4b together with the N-terminal region of NS5 or the combination of the NS1, NS3, and NS4a gene regions with Eg101 sequence recreated the WNV Eg101 SG phenotype.
Fig 5.
Higher levels of early viral RNA synthesis correlate with increased early virus yields and more SG-positive, infected cells. (A) BHK cells were infected with a chimeric virus at an MOI of 1, fixed at 36 h after infection, and analyzed by confocal microscopy. SGs were detected with anti-G3BP antibody (green), WNV-infected cells with anti-dsRNA antibody (red), and nuclei with Hoechst dye (blue). Merged images are shown. (B) The total number of infected cells and the number of SG-positive, infected cells were counted, and the percentage of infected cells containing SGs was determined as described in Materials and Methods. *, significance compared to W956IC at a 95% confidence interval (P < 0.05). (C) BHK cells were infected with a chimeric virus at an MOI of 1, and virus yield was determined by plaque assay at the indicated times after infection. (D) BHK cells were infected with a chimeric virus at an MOI of 1, and total cell RNA was extracted and purified at 12 h after infection. The number of viral genome copies in each sample was determined by real-time RT-PCR using probes targeting the viral NS1 gene as described in Materials and Methods. (E) The amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA in each sample was measured by real-time RT-PCR as an internal control for sample variation. Bars indicate ± SD.
The growth kinetics of the various chimeric viruses was analyzed to determine whether the differential SG phenotypes were due to differences in the growth efficiencies of these viruses. BHK cells were infected with WNV Eg101, W956IC, or one of the chimeric viruses at an MOI of 1, and culture fluid was collected at various times after infection. Eg101 and W956IC infections produced similar virus yields at 24 and 36 h after infection (Fig. 5C). However, W956IC infections produced significantly more progeny virus than Eg101 infections at 12 h after infection (Fig. 5C). The yields produced by Eg-C, Eg-NS1, Eg-NS2a+2b, Eg-NS3+4a, and Eg-NS4b chimeric virus infections at 12 h were similar to that produced by a W956IC infection, while the 12-h yields from Eg-NS4b+5, Eg-NS1+3+4a, and Eg-NS1+4b+5 infections were comparable to that from an Eg101 infection (Fig. 5C). Virus yields from the Eg-C, Eg-NS1, Eg-NS2a+2b, Eg-NS3+4a, and Eg-NS4b chimeric virus infections at 24 and 36 h were similar to those of both the Eg101 and the W956IC infections (Fig. 5C). However, the Eg-NS4b+5 and Eg-NS1+3+4a chimeric virus infections produced slightly reduced virus yields at 24 and 36 h after infection. Although the yield of Eg-NS1+4b+5 virus was slightly decreased at 24 h, levels comparable to those seen with Eg101 and W956IC virus infections were observed by 36 h (Fig. 5C). These data indicate that the replication efficiencies of none of the chimeric viruses were significantly compromised.
Since PKR is activated by double-stranded regions of viral RNA (19), the levels of early intracellular viral RNA replication were assessed in Eg101-, W956IC-, and chimeric virus-infected cells. BHK cells were infected with WNV Eg101, W956IC, or a chimeric virus at an MOI of 1, and total cellular RNA was collected at 12 h after infection. The number of viral genome copies per 1 μg of total RNA was quantified as described in Materials and Methods. Consistent with the 12-h viral yields, W956IC infections synthesized the highest levels of viral RNA by 12 h after infection, while Eg101 infections synthesized the lowest (Fig. 5D). The Eg-C, Eg-NS1, Eg-NS2a+2b, Eg-NS3+4a, and Eg-NS4b chimeric virus infections produced higher levels of early intracellular viral RNA, as did a W956IC infection, while the Eg-NS4b+5, Eg-NS1+3+4a, and Eg-NS1+4b+5 infections produced lower levels of early viral RNA similar to those seen with an Eg101 infection (Fig. 5D). The results indicate that more efficient SG induction in infected cells correlates with increased levels of viral RNA synthesis at early times after infection. The data further indicate that a single viral protein is not responsible for regulating early viral RNA synthesis in cells infected with natural WNV strains or involved in inducing SGs.
Enhanced early viral RNA synthesis in W956IC-infected cells results in cytoplasmic exposure of viral dsRNA.
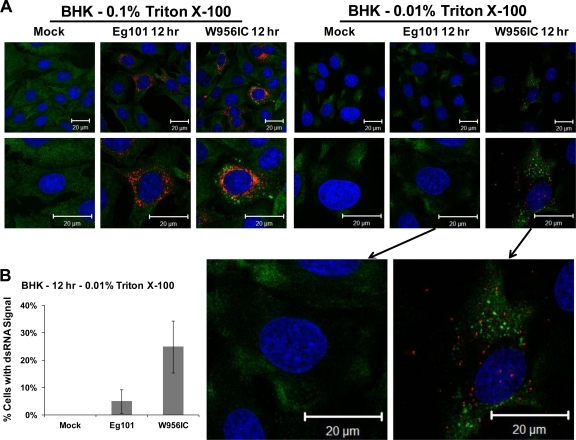
Viral dsRNA in infected cells is sensed by the pathogen recognition receptors (PRRs) PKR, retinoic acid inducible gene I (RIG-I), and melanoma differentiation factor 5 (MDA5) (19, 30). Replicating flavivirus RNAs are thought to evade detection by cell RNA sensors by localization within ER membrane vesicles (42). It was previously reported that treating cells with 0.01% Triton X-100 permeabilizes the plasma membrane but leaves the intracellular ER membranes intact, while 0.1% Triton X-100 solubilizes both plasma and ER membranes (39). Differential membrane permeabilization was used to assess viral dsRNA membrane association at early times after infection. BHK cells were infected with WNV Eg101 or W956IC at an MOI of 5 and fixed at 12 h after infection. After 4% paraformaldehyde fixation, cells were permeabilized with either 0.1% or 0.01% Triton X-100 and analyzed for the presence of viral dsRNA and SGs by confocal microscopy. Viral dsRNA was detected in the majority of Eg101- and W956IC-infected cells permeabilized with 0.1% Triton X-100 (Fig. 6A). Most cells in the W956IC-infected cultures appeared to have higher levels of viral dsRNA than the cells in Eg101-infected cultures. After permeabilization with 0.01% Triton X-100, viral dsRNA was detected in ∼25% of the W956IC-infected cells but in only ∼4% of the Eg101-infected cells (Fig. 6B). The W956IC-infected cells that contained elevated levels of exposed viral dsRNA also contained SGs (Fig. 6A), suggesting that when viral dsRNA is not protected by cytoplasmic membranes, it can activate PKR, leading to eIF2α phosphorylation and SG formation. Why a maximum of ∼35% of W956IC-infected cells have high levels of early viral RNA is currently not known. It is possible that additional cell factors may be involved in the regulation of early viral RNA synthesis.
Fig 6.
Exposed viral dsRNA was detected in 0.01% Triton X-100-permeabilized W956IC-infected, but not Eg101-infected, cells. (A) BHK cells were infected with WNV Eg101 or W956IC at an MOI of 5, fixed at 12 h after infection, permeabilized with either 0.1% or 0.01% Triton X-100, and analyzed by confocal microscopy. Anti-dsRNA antibody (red) was used to detect viral dsRNA, anti-G3BP antibody (green) to detect SGs, and Hoechst dye (blue) to detect nuclei. Merged images are shown. (B) The total number of cells and the number of dsRNA signal-positive cells from 0.01% Triton X-100-treated cells were counted, and the percentage of cells containing detectable dsRNA was determined. Bars indicate ± SD.
DISCUSSION
The cytoplasmic PRR PKR can be activated by viral dsRNA. Activated PKR phosphorylates eIF2α on Ser51, which attenuates cap-dependent translation and usually leads to the formation of SGs. Because the WNV genome is capped, its translation is expected to be inhibited by eIF2α phosphorylation. In contrast, because members of other flavivirus genera, such as pestiviruses and hepatitis C virus, initiate translation from an IRES, their translation is not expected to be affected by eIF2α phosphorylation (43). Another reason why flavivirus infections would benefit from not inducing the formation of SGs is our previous finding that the SG-nucleating proteins TIA-1 and TIAR facilitate plus-strand viral RNA synthesis (18, 34). Although infections with natural lineage 1 and lineage 2 strains of WNV did not activate PKR, infections with the chimeric W956IC virus did, and SG induction by W956IC virus infections was shown to be dependent on PKR. A maximum of ∼35% of W956IC virus-infected cells were SG positive. Previous reports of SG induction through PKR activation also showed that SGs formed in only a subset of the cells in an infected culture (25, 37, 52). Similar to our results with natural and chimeric strains of WNV, infections with a respiratory syncytial virus (RSV) chimeric virus (LeC) induced a higher number of SG-positive, infected cells than natural RSV strain infections through the activation of PKR (22, 25, 36, 37). Although many types of viruses have evolved mechanisms to inhibit PKR activation (19), a previous study from our lab showed that WNV Eg101 infections do not actively inhibit PKR activation but instead do not activate PKR (16). The slight increase in SG-positive, Eg101-infected type I IFN-nonresponsive BHK cells observed at later times of infection (from ∼1% at 24 h to ∼3% by 36 h) could be due to PERK activation caused by the induction of an unfolded protein response (UPR). Natural WNV infections were previously reported to induce the UPR at later times of infection (1). The UPR can activate PERK, another of the eIF2α kinases, and activated PERK was previously detected in WNV-infected cells at later times of infection (1).
The chimeric junction in the W956IC genome produces a 956D117B3/Eg101 hybrid NS5. This hybrid NS5 did not cause a reduction in the replication efficiency of the W956IC virus, since the yields from Eg101 and W956IC infections in BHK cells were comparable at 24 to 36 h and the chimeric W956IC virus synthesized RNA more efficiently at early times after infection than did Eg101. The increased levels of early viral RNA synthesis observed in W956IC-infected cells correlated with an increase in PKR activation and SG formation. The replication of flavivirus RNA in infected cells was previously reported to be biphasic (11, 12). Genomic and complementary minus-strand RNAs are replicated symmetrically at low levels until about 10 to 12 h after infection. Thereafter, RNA replication increases exponentially and is asymmetric, with genome RNA synthesis predominating. The exponential increase in viral RNA replication occurs after sufficient virus-induced proliferation and remodeling of ER membranes has occurred (35). Flavivirus infections as well as some other types of positive-sense RNA virus infections form vesicles in ER membranes that open to the cytoplasm through a small pore and serve as the sites of exponential viral RNA synthesis. Viral nonstructural proteins as well as cell proteins have been reported to be involved in cytoplasmic membrane remodeling (13, 21, 53). Data from several studies suggest that these vesicles protect replicating viral RNA from detection by cytoplasmic sensors (13, 21, 42, 50, 53).
How flavivirus RNA replication is differentially regulated during the early and late phases of viral RNA synthesis is not known. It was previously shown that cyclization of the genome through base pairing between 3′ and 5′ RNA sequences was required for RNA replication (7, 23, 58). Even though in the chimeric W956IC genome, the 3′ cyclization, downstream of the AUG codon (DAR), and upstream of the AUG codon (UAR) sequences were from Eg101 and the 5′ cyclization, DAR, and UAR sequences were from W956IC, all of the long-distance 5′-3′ base pairing interactions were completely conserved (data not shown). It has also been reported that the WNV 5′ UTR interacts with the NS5 protein, which contains an N-terminal methyltransferase domain and a C-terminal RNA-dependent RNA polymerase domain. Alteration of this RNA-protein interaction dramatically affected the replication efficiency of WNV (14). The WNV Eg101 and W956IC 5′ UTRs differ by only 1 nt, located in a position that was not previously shown to be involved in the interaction with NS5, suggesting that this substitution would not affect viral RNA replication efficiency. It is possible that the viral nonstructural protein complexes assume different conformations during the early and late phases of viral RNA replication. The replication complex conformations could be mediated by interactions with different cell partners and/or different cellular membrane microenvironments. The three WNV chimeras that displayed low levels of early viral RNA synthesis and of SG induction similar to those seen with Eg101 were Eg-NS4b+5, Eg-NS1+3+4a, and Eg-NS1+4b+5. Alignment of the sequences of the nonstructural regions of the Eg101 and W956IC genomes revealed 31 aa differences in NS1, 22 in NS3, 8 in NS4a, 9 in NS4b, and 24 in the N-terminal region of NS5. The majority of the 140 aa differences between the Eg101 and W956IC sequences shown in Fig. 4 are conservative changes. However, the amino acid differences at five sites, 1 in NS1 (G291E), 2 in NS4a (G17V and A54V), 1 in NS4b (A100V), and 1 in the N-terminal region of NS5 (V206I), were unique to the W956IC and 956D117B3 proteins, compared to the other WNV strains tested in the current study. Analysis of other West Nile virus strains in the Flavitrack database (40) showed that the NS1 G291E, NS4a G17V, and NS4b A100V amino acid differences were unique to W956IC and 956D117B3 viruses but that the NS4a A54V amino acid difference was an A54I substitution in the Rabensburg isolate (WNe97CZmNmX/1-3433) and that the NS5 V206I amino acid substitution was also observed in the WNe03AZAmXxX_1623/1-3433 virus strain. The unique change in NS4b was located at the C terminus of transmembrane domain 2, while the unique NS5 change was in the methyltransferase region. None of the 5 unique changes were located in a known enzymatic motif. However, regions between known motifs may contain contact sites for interactions between replication complex proteins or between these proteins and cell proteins. Each of the unique amino acid changes is being tested individually and in combination for an effect on early viral replication efficiency and SG induction. The finding that chimeras that encode either the Eg101 C-terminal region of NS4b and a complete Eg101 NS5 or the Eg101 NS1, NS3, and NS4a proteins produced low levels of early viral RNA synthesis suggests that changing particular amino acids in either the C-terminal region of NS4b and the N-terminal region of NS5 or in the combined NS1, NS3, and NS4a region can rescue the “natural” WNV strain phenotype of Eg101 in the context of the W956IC genome with an N-terminal NS5 region. These data support the hypothesis that the conformation of the replication complex plays a role in downregulating RNA synthesis early during natural WNV infections. A previous study that mapped flavivirus nonstructural proteins involved in inhibiting IFN signaling also did not identify a single viral nonstructural protein responsible for this function (1). The current study does not rule out the possibility that genome RNA folding alterations could also play a role in regulating early viral RNA synthesis.
It is currently not known how viral dsRNA is protected from cytoplasmic RNA sensors during early low-level symmetric viral RNA synthesis. The results of the present study indicate that essentially all of the viral dsRNA present at 12 h in the majority of Eg101-infected cells was associated with cytoplasmic membranes, while some of the dsRNA in ∼25% of W956IC virus-infected cells was not. This finding suggests that natural strains of WNV are able to carefully match the level of RNA replication with the degree of cytoplasmic membrane remodeling. When this coordination is disregulated, such as in W956IC-infected cells, the higher levels of early RNA replication exceed the capacity of the available virus-induced membrane protection, exposing some of the viral dsRNA and resulting in PKR activation, eIF2α phosphorylation, and SG formation. Additional studies with IFN-competent cells infected with W956IC virus showed that the increased early viral RNA synthesis also results in the induction of higher early levels of type I IFN. A manuscript describing these results is in preparation.
ACKNOWLEDGMENTS
This work was supported by developmental grant U54 AI 057157 (SE-DP-001) to M.A.B. from the Southeastern Regional Centers for Excellence for Biodefense and Emerging Infections and by Public Health Service research grant AI048088 to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. S.C.C. was supported by a Molecular Basis of Disease Fellowship from Georgia State University.
Footnotes
Published ahead of print 18 January 2012
REFERENCES
- 1. Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 85:2723–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 [DOI] [PubMed] [Google Scholar]
- 3. Anderson P, Kedersha N. 2002. Stressful initiations. J. Cell Sci. 115:3227–3234 [DOI] [PubMed] [Google Scholar]
- 4. Andzhaparidze OG, Bogomolova NN, Boriskin YS, Bektemirova MS, Drynov ID. 1981. Comparative study of rabies virus persistence in human and hamster cell lines. J. Virol. 37:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bakonyi T, Hubalek Z, Rudolf I, Nowotny N. 2005. Novel flavivirus or new lineage of West Nile virus, central Europe. Emerg. Infect. Dis. 11:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakonyi T, et al. 2006. Lineage 1 and 2 strains of encephalitic West Nile virus, central Europe. Emerg. Infect. Dis. 12:618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basu M, Brinton MA. 2011. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5′-3′ cyclization sequence basepairs are viable. Virology 412:220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brinton MA. 2002. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371–402 [DOI] [PubMed] [Google Scholar]
- 11. Chu PW, Westaway EG. 1985. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 140:68–79 [DOI] [PubMed] [Google Scholar]
- 12. Cleaves GR, Ryan TE, Schlesinger RW. 1981. Identification and characterization of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology 111:73–83 [DOI] [PubMed] [Google Scholar]
- 13. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 14. Dong H, Zhang B, Shi PY. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381:123–135 [DOI] [PubMed] [Google Scholar]
- 15. Dougherty JD, White JP, Lloyd RE. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 85:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elbahesh H, Scherbik SV, Brinton MA. 2011. West Nile virus infection does not induce PKR activation in rodent cells. Virology 421:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emara MM, Liu H, Davis WG, Brinton MA. 2008. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J. Virol. 82:10657–10670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811 [DOI] [PubMed] [Google Scholar]
- 20. Gilfoy FD, Mason PW. 2007. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 81:11148–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groskreutz DJ, Babor EC, Monick MM, Varga SM, Hunninghake GW. 2010. Respiratory syncytial virus limits alpha subunit of eukaryotic translation initiation factor 2 (eIF2alpha) phosphorylation to maintain translation and viral replication. J. Biol. Chem. 285:24023–24031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hahn CS, et al. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33–41 [DOI] [PubMed] [Google Scholar]
- 24. Han AP, et al. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanley LL, et al. 2010. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology 406:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harding HP, et al. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099–1108 [DOI] [PubMed] [Google Scholar]
- 27. Harding HP, et al. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell 7:1153–1163 [DOI] [PubMed] [Google Scholar]
- 28. Hayes EB, et al. 2005. Epidemiology and transmission dynamics of West Nile virus disease. Emerg. Infect. Dis. 11:1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Y, Conway TW. 1993. 2-Aminopurine inhibits the double-stranded RNA-dependent protein kinase both in vitro and in vivo. J. Interferon Res. 13:323–328 [DOI] [PubMed] [Google Scholar]
- 30. Kato H, et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kramer MJ, et al. 1983. Cell and virus sensitivity studies with recombinant human alpha interferons. J. Interferon Res. 3:425–435 [DOI] [PubMed] [Google Scholar]
- 32. Lam V, Duca KA, Yin J. 2005. Arrested spread of vesicular stomatitis virus infections in vitro depends on interferon-mediated antiviral activity. Biotechnol. Bioeng. 90:793–804 [DOI] [PubMed] [Google Scholar]
- 33. Lanciotti RS, et al. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298:96–105 [DOI] [PubMed] [Google Scholar]
- 34. Li W, et al. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 36. Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE., Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindquist ME, Mainou BA, Dermody TS, Crowe JE., Jr 2011. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology 413:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mertens E, et al. 2010. Viral determinants in the NS3 helicase and 2K peptide that promote West Nile virus resistance to antiviral action of 2′,5′-oligoadenylate synthetase 1b. Virology 399:176–185 [DOI] [PubMed] [Google Scholar]
- 40. Misra M, Schein CH. 2007. Flavitrack: an annotated database of flavivirus sequences. Bioinformatics 23:2645–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagai Y, Ito Y, Hamaguchi M, Yoshida T, Matsumoto T. 1981. Relation of interferon production to the limited replication of Newcastle disease virus in L cells. J. Gen. Virol. 55:109–116 [DOI] [PubMed] [Google Scholar]
- 42. Overby AK, Popov VL, Niedrig M, Weber F. 2010. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 84:8470–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robert F, et al. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP. Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell 17:4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scherbik SV, Stockman BM, Brinton MA. 2007. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J. Virol. 81:12005–12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Su Q, et al. 2006. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 103:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vaheri A, Sedwick WD, Plotkin SA, Maes R. 1965. Cytopathic effect of rubella virus in RHK21 cells and growth to high titers in suspension culture. Virology 27:239–241 [DOI] [PubMed] [Google Scholar]
- 50. van Hemert MJ, et al. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 101:11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Venticinque L, Meruelo D. 2010. Sindbis viral vector induced apoptosis requires translational inhibition and signaling through Mcl-1 and Bak. Mol. Cancer 9:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Welsch S, et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 55. Yamshchikov G, et al. 2004. An attenuated West Nile prototype virus is highly immunogenic and protects against the deadly NY99 strain: a candidate for live WN vaccine development. Virology 330:304–312 [DOI] [PubMed] [Google Scholar]
- 56. Yamshchikov VF, Wengler G, Perelygin AA, Brinton MA, Compans RW. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294–304 [DOI] [PubMed] [Google Scholar]
- 57. Yang YL, et al. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 373:1–13 [DOI] [PubMed] [Google Scholar]