Abstract
RIG-I and mda-5 are activated by viral RNA and stimulate type I interferon production. Laboratory of genetics and physiology 2 (LGP2) shares homology with RIG-I and mda-5 but lacks the CARD domains required for signaling. The V proteins of paramyxoviruses limit interferon induction by binding mda-5 and preventing its activation; however, they do not bind RIG-I and have not been considered inhibitors of RIG-I signaling. Here we uncover a novel mechanism of RIG-I inhibition in which the V protein of parainfluenzavirus type 5 (PIV5; formerly known as simian virus type 5 [SV5]) interacts with LGP2 and cooperatively inhibits induction by RIG-I ligands. A complex between RIG-I and LGP2 is observed in the presence of PIV5-V, and we propose that this complex is refractory to activation by RIG-I ligands. The V proteins from other paramyxoviruses also bind LGP2 and demonstrate LGP2-dependent inhibition of RIG-I signaling. This is significant, because it demonstrates a general mechanism for the targeting of the RIG-I pathway by paramyxoviruses.
INTRODUCTION
Virus infection stimulates innate immune responses in the host, among which the production of type I interferon (IFN) plays a critical role in restricting virus replication, via the upregulation of a large number of IFN-stimulated genes, and through the modulation of subsequent adaptive immune responses (reviewed in reference 37). IFN induction is triggered by the recognition of pathogen-associated molecular patterns (PAMPs), principally those associated with viral nucleic acids, which are seen as foreign by cellular pattern recognition receptors (PRRs; reviewed in reference 21). The RNA helicases RIG-I and mda-5 are the two best-characterized cytoplasmic PRRs; RIG-I preferentially recognizes RNA molecules with an uncapped 5′-triphosphate in a short region of blunt double-stranded RNA (dsRNA), while mda-5 recognizes longer molecules of dsRNA which need not be 5′-triphosphorylated (13, 16, 32, 33, 42, 44; reviewed in reference 43). Viral RNA binds to the C-terminal regulatory domain (RD) of RIG-I or mda-5, and it is this region that confers specificity of PAMP recognition, although the central RNA helicase domain may also be involved in binding longer RNA molecules (22, 24, 28, 49, 52). RNA binding causes a conformational change which promotes oligomerization and allows interaction of the N-terminal CARD domains of RIG-I or mda-5 with the mitochondrial adapter protein IPS-1 (also known as VISA, MAVS, and CARDIF). This initiates a signaling cascade leading to activation and nuclear translocation of the transcription factors IRF-3 and NF-κB, which are needed to turn on transcription of the IFN-β gene.
While the interactions between synthetic PAMPs and RIG-I or mda-5 have been well characterized, the types of PAMPs recognized by RIG-I and mda-5 during viral infections are less clear. Mice lacking mda-5 showed no IFN induction in response to the picornavirus encephalomyocarditis virus (EMCV) (10, 17), consistent with the generation of a dsRNA replicative intermediate for this virus; RIG-I0/0 mice showed severely impaired IFN induction in response to a range of both positive- and negative-stranded RNA viruses (17), leading to the view that RIG-I was the only PRR essential for IFN induction by the majority of RNA viruses. However, it has become clear that, in many cases, both mda-5 and RIG-I are important for mounting full IFN responses. For example, both helicases contribute to IFN induction in infections by the paramyxoviruses measles virus (MeV) (14) and Sendai virus (SeV) (11, 17). The nature of PAMPs generated during infection remains largely unknown. Paramyxoviruses appear to generate a range of concentrations of the mda-5 protoligand dsRNA during the course of an infection (5, 53), while RIG-I ligands are potentially generated by the uncapped 5′-triphosphorylated genomic RNA or the leader RNA. Interestingly, recent work has demonstrated that RNA from the LII region of the L gene of parainfluenzavirus type 5 (PIV5) can induce IFN-β through an mda-5- and RNaseL-dependent pathway (26). Paramyxoviruses are also well known as efficient inducers of IFN as a result of the generation of defective interfering (DI) particles in working stocks (6, 15, 18, 35, 47); these DI particles can be either internal deletions or copyback genomes, and the latter have been shown to be potent activators of RIG-I (4, 48).
A third RNA helicase, laboratory of genetics and physiology 2 (LGP2), shares significant homology with RIG-I and mda-5 within the helicase domain but lacks the CARD domains and is therefore thought incapable of signaling directly. Initial cell culture experiments suggested that LGP2 acted as a negative regulator of IFN induction due to its ability to bind dsRNA but not trigger downstream signaling (20, 38, 54). However, the generation of LGP2 knockout mice revealed a more complex role for LGP2 in regulation of the IFN response. Consistent with a negative regulatory role for LGP2 in IFN synthesis, LGP2°/° mouse embryonic fibroblasts (MEFs) showed increased levels of IFN-β mRNA in response to poly(I · C) and vesicular stomatits virus (VSV), and the mice were more resistant to VSV infection than their wild-type (wt) counterparts (51). However, more surprisingly, LGP2°/° macrophages made less IFN in response to EMCV than wild-type cells, virus titers were higher, and the mice were more sensitive to EMCV infection. This suggested that LGP2 could play both positive and negative roles in IFN induction and that there may be distinct effects depending on the virus and whether it is recognized by RIG-I (as seen with VSV) or mda-5 (as seen with EMCV). More recently, another study has suggested that LGP2 can also be a positive regulator for some viruses that activate RIG-I (40).
Viruses have evolved numerous mechanisms to counteract the IFN response, and their ability to do this effectively is critical to their ability to establish an infection and often influences their pathogenicity. Paramyxoviruses are known to block both type I IFN signaling and type I IFN induction (reviewed in reference 37). In the case of parainfluenza virus type 5 (PIV5; formerly known as SV5), both of these functions are a property of the multifunctional V protein. In contrast to the wild-type virus, which induces negligible amounts of IFN-β, PIV5 lacking a functional V protein induces large amounts of IFN-β and grows very poorly in IFN-competent cells (12, 35). Studies on other paramyxoviruses have also confirmed the importance of the V protein for virulence and the control of IFN production (see, for example, references 30, 36, and 41). The V protein limits IFN induction by binding to the helicase domain of mda-5, preventing dsRNA binding and consequent oligomerization and activation of mda-5 (2, 7, 8); in contrast, V does not bind to RIG-I directly or inhibit IFN induction by overexpressed RIG-I (7). Given the observations revealing that paramyxoviruses are able to activate both mda-5 and RIG-I, the reported mda-5-specific targeting by the V proteins would potentially leave these viruses prone to IFN induction through the activation of RIG-I. In at least some cases, additional mechanisms have been described whereby paramyxoviruses inhibit RIG-I function; thus, the C protein of SeV is able to inhibit RIG-I (48), while the W protein of Nipah virus acts to inhibit IRF-3 (46). However, many paramyxoviruses do not encode C or W proteins; to date, no general mechanism for RIG-I inhibition has been described for paramyxoviruses. Here we show that the V proteins of all paramyxoviruses tested interact directly with LGP2, confirming and extending previous observations (31). The physiological significance of this interaction was previously unknown. We show that this interaction inhibits the activation of RIG-I by specific PAMPs. These results are significant, because they demonstrate a general mechanism for the targeting of the RIG-I pathway by paramyxoviruses.
MATERIALS AND METHODS
Plasmids.
The IFN-ß promoter reporter plasmid pIFΔ(−116)lucter, the constitutive ß-galactosidase reporter plasmid pJatLacZ, the expression vector pEFplink2, pEF.mda-5 and pEF.RIG-I, pEF.Flag.mda-5 and pEF.Flag.RIG-I, pEF.mda-5ΔN, pEF.RIG-IΔN, and expression vectors for paramyxovirus V and P proteins and deletion mutants of V have been previously described (8). To create a plasmid expressing the C terminus of PIV5-V with a V5 tag, pEF.V5.PIV5-VΔN, a BamHI-XbaI fragment encoding amino acids 125 to 222 of PIV5-V, was cut out of pEF.PIV5-V and cloned between the BamHI and XbaI sites of pEF.V5.plink2. pEF.RIGCARD (a plasmid expressing only the CARD domains of RIG-I) was constructed by inserting a fragment encoding amino acids 1 to 226 between the NcoI and EcoRI sites of pEFplink2. A cDNA clone for human LGP2 was obtained from the IMAGE Consortium (clone 4865798), and the open reading frame (ORF) was cloned into pEFplink2 derivatives permitting expression of N-terminally V5- and Flag-tagged inserts to create pEF.Flag.LGP2 and pEF.V5.LGP2. pEF.Flag.LGP2(K634E) and pEF.Flag.LGP2ΔIV were generated by PCR site-directed mutagenesis using standard methods.
For yeast two-hybrid assays, cDNAs were cloned into pGBKT7 or pGADT7 (Clontech) for expression of proteins as GAL4 DNA-binding domain (DBD) or GAL4 activation domain (AD) fusions, respectively. pGBKT7.RIG-I, pGBKT7.mda-5(676-816), and pGADT7 plasmids containing cDNAs from the paramyxovirus V proteins, have been previously described (8). pGADT7.PIV5-VΔN104 and pGADT7.PIV5-VΔC174 were created from pEF.PIV5-VΔN104 and pEF.PIV5-VΔC174 (35). pGBKT7.LGP2 was created by cloning the LGP2 cDNA into the NcoI and EcoRI sites of pGBKT7. A PciI-BspHI fragment encoding the N-terminal 145 amino acids of LGP2 was cloned into the NcoI site of pGBKT7 to create pGBKT7.LGP2(1-145). A fragment encoding amino acids 327 to 465 of LGP2 was generated by PCR incorporating an NcoI site at the 5′ end, which was then cloned into the NcoI site of pGBKT7. The C terminus of LGP2 (amino acids 465 to 678) was cloned as an NcoI (partial)-EcoRI fragment. pGBKT7.mda-5(676-816)ΔIV was created by joining two PCR products through an engineered XhoI site. The first PCR product encoded amino acids 676 to 719 and contained an XhoI site immediately after the A719 codon, and the second encoded amino acids 732 to 816, with an XhoI site immediately 5′ of the Y732 codon. The resulting product contained sequences encoding two amino acids (Leu and Glu) in place of the deleted sequences encoding amino acids 720 to 731 derived from the XhoI site. Similarly, pGBKT7.LGP2(327-465)ΔIV was created by joining a fragment encoding amino acids 327 to 368 to a fragment encoding amino acids 381 to 465 through an XhoI site, replacing amino acids 369 to 380 with Leu and Glu. LGP2 with helicase motif IV replaced with the equivalent sequence from RIG-I [LGP2(IV)R] was made by replacing an AccI-BmrI fragment with a synthesized DNA molecule (Eurofins) containing the desired sequence by which amino acids 369 to 380 of LGP2 were replaced with amino acids 630 to 640 of RIG-I. pGBKT7.RIG-I(225-925) was previously described in reference 8, and pGBKT7.RIG-I(225-925)(IV)L was created by replacing a BamHI-NciI fragment with a synthesized sequence (Eurofins) in which amino acids 630 to 640 of RIG-I were replaced with amino acids 369 to 380 of LGP2.
Cells, transfections, and siRNAs.
HEK293 (ATCC CRL-1573) Vero (ATCC CCL-81) and A549/pr(IFN-β).GFP (6) cells were maintained in Dulbecco's modification of Eagle's combined medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Transfections were carried out using linear polyethyleneimine (PEI) (Polysciences Inc., Warrington, PA) (molecular weight [MW], ∼25,000) or Lipofectamine (Invitrogen) under standard conditions. Luciferase and β-galactosidase assays were carried out 48 h after transfection, and luciferase activity was corrected to β-galactosidase activity. The data presented in each of the figures represent the means of the results of at least three independent experiments, with error bars shown. Statistical significance was determined by paired t tests. Small interfering RNAs (siRNAs) corresponding to LGP2 were composed of an equimolar combination of two LGP2-specific siRNAs from Qiagen (catalog no. S100470288 and S100470295), the mda-5 siRNA was Hs_IFIH1_2 (Qiagen catalog no. S100445851), the RIG-I siRNA was Hs_DDX58_1 (Qiagen catalog no. S100361809), and the control siRNA corresponded to an unrelated protein, LRRC37A (Qiagen catalog no. S100622874).
Virus infections and inductions.
Where indicated, cells were infected at a multiplicity of infection (MOI) of 5 with the W3A strain of PIV5 in DMEM plus 2% FBS. Induction of cells with poly(I · C) was carried out as described previously (7). Poly(dA-dT) (Sigma; catalog no. P0883) was transfected into cells by the use of PEI, and RNA from influenza virus-infected cells and RIG-Ipan RNA were transfected using Lipofectamine (Invitrogen). RNA from influenza virus-infected cells was made by infecting a confluent 9-cm-diameter plate of HEK293 cells with Influenza A/Duck/Singapore/97 (a gift from J. McCauley, NIMR, London, United Kingdom) in 3.5 ml of serum-free DMEM at an MOI of 5. Cells were incubated for 2 h, after which a further 6.5 ml of DMEM plus 2% FBS was added and the mixture was incubated for a further 6 h before harvesting. RIG-Ipan RNA was synthesized from an oligonucleotide template (5′-GACACACACACACACACACACAAAAAAAGTGTGTGTGTGTGTGTGTGTCTATAGTGAGTCGTATTA-3′; MWG-Eurofins) hybridized to a T7 promoter oligonucleotide (5′-TAATACGACTCACTATAG-3′; MWG-Eurofins) by the use of a T7 polymerase in vitro transcription kit (MegaShortScript; Ambion) and gel purified; the ability of this material to induce IFN expression is completely abolished by treatment with alkaline phosphatase.
Immunofluorescence and fluorescence microscopy.
For immunofluorescence experiments, cells grown on glass coverslips were washed in phosphate-buffered saline (PBS) and then fixed using 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Immunofluorescence was carried out using the anti-Pk antibody to detect PIV5 P/V and goat anti-mouse IgG:Texas Red (AbD Serotec). Texas Red and green fluorescent protein (GFP) were visualized using a Nikon Eclipse TS100 fluorescence microscope. Fluorescence-activated cell sorter (FACS) analysis was performed on a Beckman-Coulter Cytomics FC500 cytometer, analyzing for GFP as described previously (6); PIV5 infection was monitored using an anti-NP monoclonal antibody as the primary antibody and goat anti-mouse IgG:RPE (AbD Serotec 103009) as the secondary antibody.
Coimmunoprecipitation assays and immunoblotting.
To make cell extracts, 6-cm-diameter dishes of transfected cells were washed in PBS and then lysed in 500 μl of lysis buffer (50 mM Tris-HCl pH [7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40). A 100-μl volume of extract was used for coimmunoprecipitation assays using mouse monoclonal antibodies against the Flag (Sigma F3165) or V5 (anti-Pk) tags. Complexes were collected on protein A-Sepharose beads (GE Healthcare), washed thrice with 1 ml of lysis buffer, and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Proteins were separated by SDS-PAGE, and immunoblotting was performed using monoclonal antibodies to the Flag or V5 tags (as described above), the myc tag (Sigma clone 9E10; catalog no. SAB4300605), or α-tubulin (Sigma catalog no. T9026) or polyclonal antibodies to LGP2 (Abcam; catalog no. ab67270) or mda-5 (AT113) (Alexis Biochemicals; catalog no. ALX-210-935). Bound primary antibodies were detected using either horseradish peroxidase (HRP)-conjugated sheep anti-mouse Ig or donkey anti-rabbit whole antibody (GE Healthcare).
Yeast two-hybrid assays.
Combinations of GAL4 DBD and GAL4 AD fusion plasmids were introduced into Saccharomyces cerevisiae strain PJ69-4α (or CG1945 where indicated) by the use of standard methods. Double transformants were selected on synthetic dropout (SD) medium lacking leucine and tryptophan (SD-L-W) and subsequently streaked onto SD-L-W medium also lacking histidine (SD-L-W-H) and containing 2 to 20 mM 3-aminotriazole (3-AT). Growth was monitored for 4 to 10 days at 30°C.
RESULTS
The V protein of parainfluenza virus 5 (PIV5) interacts with LGP2.
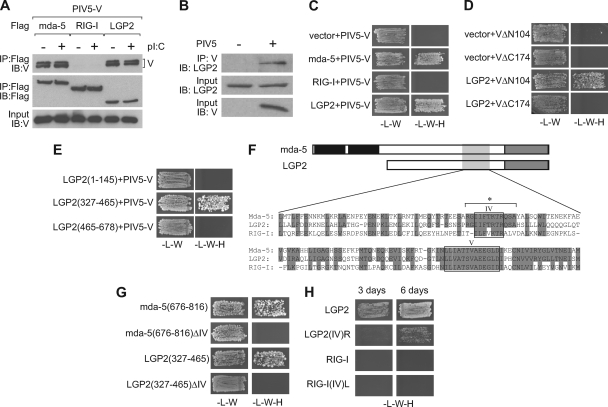
We have previously shown that the V proteins of 13 paramyxoviruses interact with mda-5, blocking its activation by dsRNA and consequently inhibiting IFN production (2, 7, 8). However, none of the V proteins tested were able to interact with RIG-I or block signaling in response to overexpressed RIG-I. Since the third member of this family, LGP2, exhibits greater homology to mda-5 than RIG-I, we tested whether the PIV5 V protein could also interact with LGP2. Indeed, we found that PIV5-V coimmunoprecipitated with both mda-5 and LGP2, but not with RIG-I, in extracts from transiently transfected cells (Fig. 1A) and also that LGP2 could be coprecipitated with the V protein in PIV5-infected cells (Fig. 1B). These interactions were not affected by treatment of the cells with dsRNA (Fig. 1A). We then confirmed this interaction using the yeast two-hybrid assay (Fig. 1C) and extended the results to show that the unique C-terminal region comprising amino acids 175 to 222 of PIV5-V, but not the N-terminal 104 amino acids, were required for binding LGP2 (Fig. 1D). This is the same region of PIV5-V that is required for binding to mda-5. Our studies of mda-5 identified amino acids 676 to 816 of mda-5 as sufficient to bind the V proteins from all 13 paramyxoviruses (7); thus, to determine whether the corresponding section of LGP2 was also sufficient to bind PIV5-V, we generated plasmids expressing N-terminal (amino acids 1 to 145), central (amino acids 327 to 465), and C-terminal (amino acids 465 to 678) fragments of LGP2 and assessed their ability to bind to PIV5-V in a yeast two-hybrid assay. The central fragment comprising amino acids 327 to 465 is equivalent to amino acids 676 to 816 of mda-5, and this was sufficient to bind PIV5-V (Fig. 1E), indicating that V targets the same region of both helicases.
Fig 1.
The V protein of PIV5 interacts with LGP2. (A) HEK293 cells were transfected with pEF.PIV5-V and pEF.Flag.mda-5, pEF.Flag.RIG-I, or pEF.Flag.LGP2. At 30 h after transfection, cells were either mock treated or induced with poly(I · C) for 8 h prior to harvesting. Cell extracts were subjected to immunoprecipitation (IP) with anti-Flag, and proteins present in the immunoprecipitate were analyzed by immunoblotting (IB) using anti-Flag or an antibody against the V protein. IB was also carried out on the cell extracts to confirm expression of the V protein. (B) HEK293 cells transfected with pEF.Flag.LGP2 were mock infected or infected with PIV5 for 12 h prior to harvesting. Extracts were subjected to IP with an antibody against V, and proteins present in the immunoprecipitate were analyzed by IB with anti-LGP2. Expression of LGP2 and V was confirmed by IB. (C, D, E, G, and H) Interactions between protein pairs were investigated using the yeast two-hybrid assay. Yeast cells were transformed with (C) empty pGBKT7 vector or pGBKT7 expressing mda-5, RIG-I, or LGP2 as a GAL4 DBD fusion and a plasmid expressing PIV5-V as a GAL4 AD fusion; (D) empty pGBKT7 vector or pGBKT7 expressing LGP2 as a GAL4 DBD fusion and a plasmid expressing PIV5-V lacking the N-terminal 104 amino acids (VΔN104) or the C-terminal 48 amino acids (VΔC174) as GAL4 AD fusions; (E) the indicated fragments of LGP2 as a GAL4 DBD fusion and a plasmid expressing PIV5-V as a GAL4 AD fusion; (G) amino acids 676 to 816 of mda-5, mda-5(676-816) with motif IV deleted (ΔIV), or amino acids 327 to 465 of LGP2 or LGP2(327-465) with motif IV deleted as GAL4 DBD fusions and a plasmid expressing PIV5-V as a GAL4 AD fusion; or (H) LGP2, LGP2 with motif IV replaced with the corresponding sequence from RIG-I [LGP2(IV)R], the helicase domain of RIG-I (amino acids 225 to 925), or the helicase domain of RIG-I with motif IV replaced with the corresponding sequence from LGP2 [RIG-I(IV)L] as GAL4 DBD fusions and a plasmid expressing PIV5-V as a GAL4 AD fusion. Transformants were initially selected on SD-L-W and then streaked onto SD-L-W and SD-L-W-H with 3-AT at 5 mM (C, E, and G), 10 mM (H), or 20 mM (D). (F) Sequence alignment of mda-5, LGP2, and RIG-I within the region of mda-5 and LGP2 that binds PIV5-V. Amino acids present in mda-5 conserved in LGP2 and RIG-I are highlighted. IV and V denote helicase motifs IV and V, and * indicates the region encompassing domain IV that is deleted or swapped in the ΔIV and (IV)R/L constructs.
Mda-5, RIG-I, and LGP2 are characterized as DExH RNA helicases by the presence of six conserved signature motifs designated I to VI. The region containing the PIV5-V protein binding site on mda-5 and LGP2 spans motifs IV and V, which are proposed to play a role in RNA binding (3) (Fig. 1F). Since motif V is completely conserved between mda-5, LGP2, and RIG-I, it is unlikely to be critical for PIV5-V binding, as PIV5-V does not bind to RIG-I. However, motif IV and the residues immediately to either side of it are conserved between mda-5 and LGP2 but are more divergent in RIG-I, so we decided to analyze the role of motif IV in PIV5-V binding. Deletion of motif IV abolished binding of PIV5-V to both mda-5(676-816) and LGP2(327-465) (Fig. 1G). Replacement of the LGP2 IV motif with that of RIG-I [LGP2(IV)R] impaired, but did not abolish, binding to PIV5-V, while the reciprocal replacement [RIG-I(IV)L] failed to confer PIV5-V binding to RIG-I (Fig. 1H). Thus, motif IV of the helicase domain is necessary but not sufficient to determine the specificity of PIV5-V recruitment.
PIV5-V inhibits IFN induction by both mda-5 and RIG-I in the presence of LGP2.
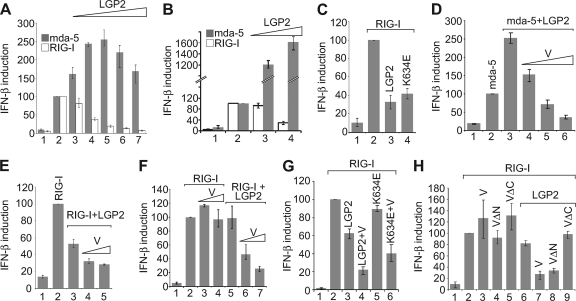
Parisien et al. also recently reported an interaction between paramyxovirus V proteins and LGP2 but did not report any consequences of this interaction for IFN induction (31). To characterize the behavior of LGP2, we first looked at the effects of LGP2 on IFN induction by overexpression of RIG-I or mda-5 (i.e., in the absence of added PAMPs). In agreement with previous studies (29, 34, 39), we found that overexpression of LGP2 stimulated IFN induction by mda-5 in Vero cells but, in contrast, inhibited IFN induction by RIG-I (Fig. 2A). We repeated these experiments in HEK293 cells and obtained similar results with respect to RIG-I inhibition, although the magnitude of activation of mda-5 was considerably higher than that observed in Vero cells (Fig. 2B). The expression of mda-5 and RIG-I was confirmed by immunoblotting, and we also verified that increasing levels of LGP2 plasmid correlated with increasing levels of LGP2 protein (see Fig. S1A and S1B in the supplemental material). One trivial explanation for the inhibition of RIG-I by LGP2 is that overexpression of LGP2 sequesters an endogenous RNA ligand that is responsible for the activation of the ectopically expressed RIG-I. To eliminate this possibility, we constructed a vector expressing a K634E mutant form of LGP2; this alteration has been previously shown to completely abolish dsRNA binding (23, 34). This point mutant is expressed to a level similar to that of the wild-type LGP2 (see Fig. S1C in the supplemental material) and was just as effective as the wild-type LGP2 in its ability to inhibit RIG-I (Fig. 2C).
Fig 2.
PIV5-V inhibits IFN induction by mda-5 and RIG-I in the presence of LGP2. (A) Vero cells were transfected with pIFΔ(−116)lucter, pJatlacZ, 80 ng of pEF.mda-5 or pEF.RIG-I, and 0 ng (lanes 1 and 2), 1 ng (lane 3), 10 ng (lane 4), 100 ng (lane 5), 200 ng (lane 6), or 388 ng (lane 7) of pEF.LGP2. Total amounts of plasmid DNA were kept equal within experiments by adding empty pEFplink2 vector. (B) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, 2 ng of pEF.mda-5 or pEF.RIG-I, and 0 ng (lanes 1 and 2), 10 ng (lane 3), or 400 ng (lane 4) of pEF.LGP2. (C) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), pEF.RIG-I (lanes 2 to 4), and a plasmid expressing either LGP2 or LGP2(K634E). (D) Vero cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), 80 ng of pEF.mda-5 (lanes 2 to 6), 40 ng of pEF.LGP2 (lanes 3 to 6), and 40, 80, or 160 ng of pEF.PIV5-V (lanes 4 to 6). (E) Vero cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), 80 ng of pEF.RIG-I (lanes 2 to 5), 10 ng of pEF.LGP2 (lanes 3 to 5), and 10 ng or 50 ng of pEF.PIV5-V (lanes 4 and 5). The additional inhibition observed in the presence of LGP2 and V was statistically significant (i.e., P < 0.05). (F) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), 2 ng of pEF.RIG-I (lanes 2 to 7), 10 ng of pEF.LGP2 (lanes 5 to 7), and 10 ng (lanes 3 and 6) or 100 ng (lanes 4 and 7) of pEF.PIV5-V. The additional inhibition observed in the presence of LGP2 and V was statistically significant (i.e., P < 0.05). (G) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), pEF.RIG-I (lanes 2 to 6), and plasmids expressing LGP2, LGP2(K634E), or PIV5-V. (H) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), 2 ng of pEF.RIG-I (lanes 2 to 9), 10 ng of pEF.LGP2 (lanes 6 to 9), and 20 ng of pEF.PIV5-V (V), pEF.PIV5-VΔN125 (VΔN), or pEF.PIV5-VΔC174 (VΔC).
We then studied the effect of expressing PIV5-V on the ability of LGP2 to regulate overexpressed mda-5 or RIG-I. Consistent with the ability of the V protein to inhibit IFN induction by binding to mda-5, we found that the capacity of LGP2 to stimulate IFN induction by mda-5 was inhibited by PIV5-V in a dose-dependent manner (Fig. 2D). Increasing levels of PIV5-V protein were confirmed by immunoblotting (see Fig. S1D in the supplemental material). More interestingly, when PIV5-V was cotransfected with LGP2 and RIG-I, the inhibition of RIG-I by LGP2 was enhanced by the presence of the V protein (Fig. 2E). To confirm this observation, we repeated the experiment in HEK293 cells using limited concentration of a LGP2 plasmid chosen to have little or no effect on RIG-I activity. Figure 2F shows that the V protein could significantly inhibit overexpressed RIG-I; importantly, consistent with our previous results (7), this inhibition was not seen in the absence of coexpressed LGP2. The V protein could also cooperate with the LGP2 K634E mutant in inhibiting RIG-I (Fig. 2G), confirming that dsRNA binding is not needed for inhibition of RIG-I.
Since the C terminus of the PIV5-V protein is sufficient to bind LGP2 (see Fig. 1D), we investigated whether this region of PIV5-V is also sufficient for inhibition of RIG-I. RIG-I was cotransfected with LGP2 and full-length PIV5-V, PIV5-VΔN (which lacks 125 amino acids of the N terminus), or PIV5-VΔC (which lacks 48 amino acids of the C terminus). PIV5-VΔN, but not PIV5-VΔC, was able to inhibit RIG-I in the presence of LGP2 as effectively as full-length PIV5-V (Fig. 2H) despite similar expression levels (see Fig. S1E in the supplemental material), thus demonstrating that the C terminus of PIV5-V is sufficient both to bind LGP2 and to inhibit RIG-I in an LGP2-dependent manner. This function is distinct from the effect of V on mda-5, since cooperative inhibition of RIG-I signaling by LGP2 and V was observed in the presence of an siRNA against mda-5 (see Fig. S2 in the supplemental material).
LGP2 and PIV5-V cooperatively inhibit IFN induction by RIG-I ligands.
We previously demonstrated that signal transduction in response to overexpressed RIG-I could not be antagonized by paramyxovirus V proteins (7). The results described above indicate that this situation can be altered by coexpression of LGP2, implying that, in our previous experiments, overexpression of RIG-I may have overcome LGP2-mediated inhibition by V simply by disrupting the RIG-I/LGP2 ratio. An important question to arise from this is whether V proteins can antagonize RIG-I activation in response to specific PAMPs under conditions in which RIG-I and LGP2 levels are not manipulated. We did not previously test this because of the lack of knowledge about the distinction between mda-5 and RIG-I ligands, but several PAMPs have recently been characterized that activate only RIG-I. RNA purified from influenza virus-infected cells (16) and poly(dA-dT) (1, 9) have both been demonstrated to induce IFN expression in a RIG-I-dependent manner with little or no dependence on mda-5. In addition, we synthesized by in vitro transcription a short panhandle RNA that we predicted would be a potent activator of RIG-I (RIG-Ipan RNA). To confirm the selectivity of these PAMPs for RIG-I, we used specific siRNAs or the expression of dominant-negative forms of RIG-I and mda-5 (see Fig. S3 in the supplemental material). Only the RIG-I siRNA and the dominant-negative form of RIG-I (RIG-IΔN), and not the mda-5 siRNA or the dominant-negative form of mda-5 (mda-5ΔN), were able to inhibit IFN induction by poly(dA-dT), influenza virus RNA, and RIG-Ipan RNA. Furthermore, IFN induction by overexpressed RIG-I, but not mda-5, was stimulated upon addition of these RIG-I-specific PAMPs.
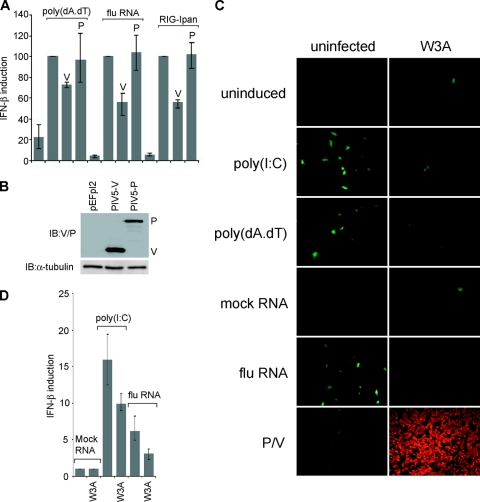
We therefore tested whether the V protein of PIV5 was able to antagonize induction by these ligands. Figure 3A and B show that expression of PIV5-V, but not PIV5-P, inhibited IFN induction by all three RIG-I-specific PAMPs. We also tested PIV5 proteins NP, SH, M, F, HN, and L, but none of these were able to limit induction by poly(dA-dT) in HEK293 cells (data not shown), indicating that V is the only PIV5 protein able to inhibit IFN induction through the RIG-I pathway. To confirm the inhibition of IFN induction by RIG-I ligands in the context of a virus infection, we sought to determine whether preinfection of cells with PIV5 would limit the amount of IFN induced by subsequent challenge with a RIG-I ligand. To monitor IFN induction, we used a stable A549 cell line containing a GFP reporter gene under the control of the IFN-β promoter [A549/pr(IFN-β).GFP] (6). Challenging mock-infected cells with poly(I · C) or the RIG-I activators (poly(dA-dT) or RNA from influenza virus-infected cells resulted in a significant induction of GFP-positive cells (Fig. 3C). Infection with the W3A strain of PIV5 gave a much smaller number of GFP-positive cells, despite effectively all cells being infected. PIV5 preinfection was able to reduce the number of cells responding to both mda-5 and RIG-I stimulation to background levels (FACS analyses of influenza virus RNA samples as a representative data set demonstrated that the presence of productive PIV5 infection lowers the number of cells responding to the influenza virus RNA RIG-I ligand by about 80%), suggesting that wt PIV5 infection is able to potently inhibit IFN-β induction in response to stimulation of either mda-5 or RIG-I. We have confirmed this in transient-transfection assays of HEK293 cells (Fig. 3D).
Fig 3.
PIV5-V inhibits IFN induction by RIG-I ligands. (A) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, and pEFplink2, pEF.PIV5-V, or pEF.PIV5-P. At 30 h after transfection, cells were induced with poly(dA-dT), RNA purified from influenza virus-infected cells, or RIG-Ipan RNA for 16 h prior to harvesting for luciferase and β-galactosidase assays. (B) Expression of PIV5-V and -P was confirmed by immunoblotting with an anti-V/P antibody. (C) A549/pr(IFN-β).GFP cells were mock infected or infected with the W3A strain of PIV5 at an MOI of 5 for 24 h. Cells were then induced with poly(I · C), poly(dA-dT), RNA from uninfected cells (mock RNA), or RNA from influenza virus-infected cells (flu RNA) for a further 24 to 48 h. Expression of GFP was visualized by fluorescence microscopy. To verify that the majority of cells had been infected, PIV5 P/V protein expression was visualized by immunofluorescence using the anti-Pk antibody. (D) HEK293 cells were transfected with pIFΔ(−116)lucter and pJatlacZ. At 24 h after transfection, cells were either mock infected or infected with PIV5 W3A at an MOI = 5 and then induced 6 h later with either poly(I · C) or influenza virus RNA for 16 h prior to harvesting for luciferase and β-galactosidase assays.
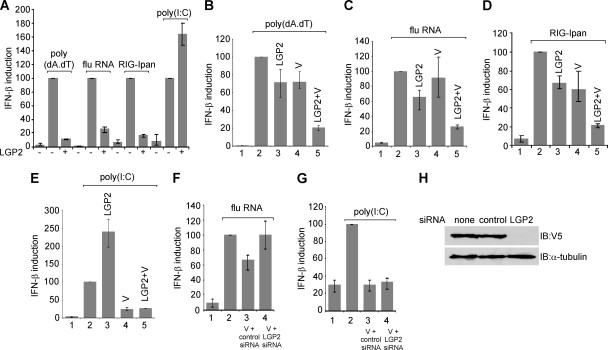
We next investigated the effects of LGP2 expression on the induction of IFN by defined ligands. Consistent with its role as an inhibitor of RIG-I and an activator of mda-5, the expression of exogenous LGP2 inhibited IFN induction by poly(dA-dT), influenza virus RNA, and RIG-Ipan RNA and stimulated induction by poly(I · C) (Fig. 4A). To examine the consequences of PIV5-V expression with respect to the LGP2-mediated effects on induction, we used a limiting concentration of LGP2 plasmid in order to observe the expected cooperativity between PIV5-V and LGP2. Under these conditions, the induction of reporter activity by poly(dA-dT), RNA from influenza virus-infected cells, or RIG-Ipan RNA was only moderately inhibited by LGP2 alone and was either unaffected or slightly inhibited by PIV5-V alone (Fig. 4B, C, and D). However, the combination of PIV5-V and LGP2 resulted in significant inhibition of IFN induction by the RIG-I-dependent inducers. Therefore, PIV5-V is able to synergize with LGP2 to limit IFN induction in response to RIG-I ligands. In the case of the mda-5 ligand poly(I · C), LGP2 stimulated IFN induction whereas the PIV5-V protein alone was sufficient to achieve a 75% reduction in IFN-β promoter activity (Fig. 4E). In the presence of both PIV5-V and LGP2, the result was the same as for PIV5-V alone; thus, activation by LGP2 was overcome by PIV5-V, but no further reduction in activity was observed.
Fig 4.
LGP2 and PIV5-V cooperatively inhibit IFN induction by RIG-I ligands. (A) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, and either pEFplink2 or pEF.LGP2. At 30 h after transfection, cells were induced with poly(dA-dT), RNA from influenza virus-infected cells, RIG-Ipan RNA, or poly(I · C) for a further 16 h prior to harvesting for luciferase and β-galactosidase assays. (B to E) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, and either pEFplink2 (lanes 1 and 2) or plasmids expressing LGP2 or PIV5-V. At 30 h after transfection, cells were induced with poly(dA-dT) (B), RNA from influenza virus-infected cells (C), RIG-Ipan RNA (D), or poly(I · C) (E) for a further 16 h prior to harvesting for luciferase and β-galactosidase assays. The additional inhibition observed in the presence of LGP2 and V was statistically significant (i.e., P < 0.05). (F and G) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lanes 1 and 2), or pEF.PIV5-V (lanes 3 and 4) with either a mixture of 2 siRNAs directed against LGP2 or an siRNA directed against an unrelated transcript (control siRNA). At 30 h after transfection, cells were induced with either (F) RNA from influenza virus-infected cells or (G) poly(I · C) for a further 16 h prior to harvesting for luciferase and β-galactosidase assays. (H) To verify the effectiveness of the LGP2 siRNAs, HEK293 cells were transfected with a plasmid expressing V5-tagged LGP2 along with no siRNA, a control siRNA, or the LGP2 siRNAs. Cell extracts were immunoblotted with an antibody against the V5 tag and α-tubulin as a loading control.
Next we used sufficient PIV5-V to significantly inhibit the induction of IFN in the absence of additional LGP2 and then investigated whether the PIV5-V-mediated inhibition could be disrupted by an siRNA targeted to endogenous LGP2. Figure 4F shows that the inhibition of IFN induction by influenza virus RNA was dependent on the presence of endogenous LGP2, since it was specifically abolished by an siRNA directed against LGP2; in contrast, PIV5-V was able to inhibit IFN induction by the mda-5 agonist, poly(I · C), when either the control or LGP2 siRNAs were used (Fig. 4G).
LGP2 binding and RIG-I inhibition are general properties of paramyxovirus V proteins.
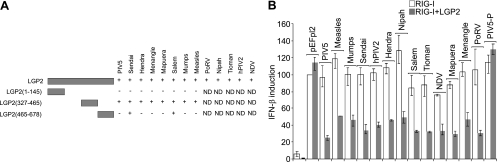
We then tested the V proteins from a number of other paramyxoviruses for their ability to interact with LGP2. We found that, in addition to PIV5-V, the V proteins from Sendai, Hendra, Menangle, Mapuera, Salem, mumps, measles, Nipah, Tioman, and Newcastle disease viruses and human PIV2 (hPIV2) and porcine rubulavirus (PoRV) were all able to interact with LGP2 (Fig. 5A).
Fig 5.
All paramyxovirus V proteins interact with LGP2 and inhibit RIG-I in an LGP2-dependent manner. (A) A further 12 paramyxovirus V proteins were tested for their ability to interact with LGP2 in a yeast two-hybrid assay. Yeast cells were transformed with a plasmid expressing full-length LGP2 or the indicated fragments of LGP2 as GAL4 DBD fusion proteins and a plasmid expressing the indicated paramyxovirus V protein as a GAL4 AD fusion protein. Positive transformants were selected on SD-L-W and then streaked onto SD-L-W and SD-L-W-H plus 5 to 20 mM 3-AT to assay for an interaction. Due to some background transactivation being observed with some V proteins in the yeast strain PJ69-4α, two-hybrid assays with hPIV2, Nipah, Tioman, and NDV V proteins were carried out with yeast strain CG1945 in which no background transactivation was observed. A plus sign denotes a positive interaction, and a minus sign denotes no interaction. ND, not determined. (B) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lanes 1 and 2), 2 ng of pEF.RIG-I (white bars), or 2 ng of pEF.RIG-I or 10 ng of pEF.LGP2 (grey bars), and pEFplink2 or a plasmid expressing the V protein from the indicated paramyxovirus or the P protein from PIV5 (PIV5-P).
Amino acids 327 to 465 of LGP2, which were sufficient to bind PIV5-V (see Fig. 1E), were also sufficient to bind at least eight of the viral V proteins, indicating that the binding site for all V proteins is conserved and is also shared between mda-5 and LGP2. Parisien et al. also reported an interaction between V proteins and LGP2 and mapped it to amino acids 351 to 479, which is in close agreement with our data (31). The V proteins from Sendai and Salem viruses could also independently bind to the C-terminal fragment encoding amino acids 465 to 678 of LGP2. Interestingly, we had previously observed that, while all the V proteins bound to amino acids 676 to 816 of mda-5, the V proteins from PIV5 and from Menangle and Salem viruses also made additional contacts with other parts of the protein (8). Although Salem virus V binds to additional sites within both mda-5 and LGP2, they appear to be different, since it binds to a site N-terminally oriented to the conserved binding site in mda-5 (amino acids 287 to 458) and not to the C terminus.
Next, we examined the ability of these other V proteins to enhance inhibition of RIG-I by LGP2. As previously observed, none of these V proteins alone have any significant effect on IFN induction by RIG-I overexpression (Fig. 5B, white bars); however, when they were cotransfected with a small amount of LGP2 plasmid, all 13 V proteins effectively inhibited RIG-I signaling (Fig. 5B, gray bars). In contrast, the PIV5 P protein, which does not interact with LGP2 (data not shown), does not inhibit RIG-I in the presence of LGP2 (Fig. 5B, final lane).
Mechanism of inhibition of LGP2 by paramyxovirus V proteins.
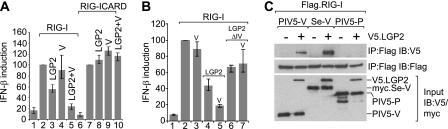
We next sought to characterize the mechanism of cooperative inhibition of RIG-I by LGP2 and PIV5-V. As discussed above, we have excluded the possibility that the effect is a consequence of sequestration of an RNA ligand. To determine whether PIV5-V and LGP2 limit activation of RIG-I through the helicase domain or the CARD domain, we compared the abilities of PIV5-V and LGP2 to block IFN induction by overexpression of full-length RIG-I or the CARD domains only. In contrast to the effects observed with full-length RIG-I, cotransfection of LGP2 did not inhibit induction by RIG-I CARD in either the presence or the absence of PIV5-V (Fig. 6A). This demonstrates that the inhibitory effect of LGP2 is operating at the level of RIG-I activation and is dependent on the helicase domain of RIG-I.
Fig 6.
Paramyxovirus V proteins inhibit RIG-I by interacting with LGP2 and promoting the formation of inactive complexes between RIG-I and LGP2. (A) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lanes 1 and 6), a plasmid expressing RIG-I (lanes 2 to 5), a plasmid expressing just the CARD domains of RIG-I (lanes 7 to 10), and plasmids expressing either LGP2 or PIV5-V as indicated. (B) HEK293 cells were transfected with pIFΔ(−116)lucter, pJatlacZ, pEFplink2 (lane 1), 2 ng of pEF.RIG-I (lanes 2 to 7), and either pEF.LGP2 or pEF.LGP2ΔIV and pEF.PIV5-V where indicated. (C) HEK293 cells were transfected with a plasmid expressing RIG-I with a Flag tag, pEFplink2 (lanes 1, 3, and 5) or a plasmid expressing LGP2 with a V5 tag (lanes 2, 4, and 6), and a plasmid expressing PIV5-V (lanes 1 and 2), the V protein from Sendai virus with a myc tag (myc.Se-V) (lanes 3 and 4), or the P protein from PIV5 (PIV5-P) (lanes 5 and 6). Cell extracts were subjected to immunoprecipitation (IP) with an antibody against the Flag tag, and proteins present in the immunoprecipitate were analyzed by immunoblotting (IB) with an antibody to the V5 or Flag tags. Cell extracts were also blotted using antibodies to the V5 and myc tags to confirm expression of all proteins.
We have shown above that PIV5-V binding to LGP2 is completely disrupted by a deletion of 12 amino acids in motif IV (see Fig. 1G). We tested whether this mutation in LGP2 was sufficient to abolish the inhibition of RIG-I in the presence or absence of PIV5-V. Interestingly, this mutant retained a limited ability to inhibit RIG-I in the absence of PIV5-V (Fig. 6B); however, in contrast to wild-type LGP2, PIV5-V was unable to enhance the inhibition of RIG-I by LGP2ΔIV. Therefore, the ability of V to interact with LGP2 is critical to its ability to inhibit RIG-I.
Finally, we looked to see whether we could detect an interaction between RIG-I and LGP2 and, if so, whether it is affected by the V protein. Figure 6C shows that LGP2 coimmunoprecipitated with RIG-I in extracts from cells expressing the V proteins from PIV5 or Sendai virus but not in extracts from cells expressing the P protein from PIV5. Thus, V proteins promote an interaction between RIG-I and LGP2, and we propose that this interaction results in inhibition of IFN induction through the RIG-I pathway.
DISCUSSION
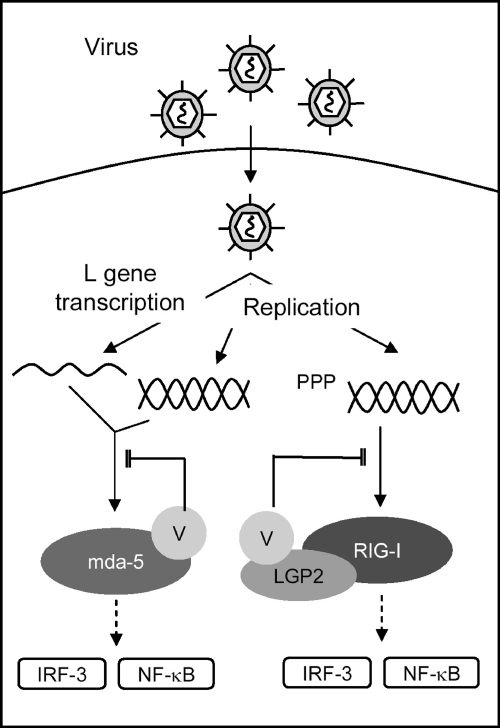
Paramyxoviruses evade the innate immune response by encoding proteins that inhibit both IFN induction and IFN signaling. We have previously shown that the V proteins of all paramyxoviruses tested block IFN induction by binding to mda-5 and inhibiting its activation by dsRNA (2, 7, 8). Although this feature is clearly effective for blocking IFN induction through the mda-5 pathway, paramyxoviruses have been shown to activate RIG-I, raising the question of whether they also encode mechanism(s) for at least limiting RIG-I activity. Here we describe a novel mechanism of inhibition of IFN induction by paramyxovirus V proteins by which they are able to inhibit RIG-I activation in a manner that depends upon their interaction with LGP2. Using PIV5-V as a model system, we show that the V protein is capable of promoting the formation of a complex between RIG-I and LGP2 which renders RIG-I unable to respond to RIG-I-specific PAMPs. We propose a model in which the multifunctional V protein is able to inhibit both mda-5-dependent and RIG-I-dependent IFN induction through the formation of distinct inactive complexes (Fig. 7). The inhibition of RIG-I is mechanistically distinct from the inhibition of mda-5 in that the PRR (RIG-I) is not a direct target.
Fig 7.
Inhibition of IFN induction by paramyxovirus V proteins. Paramyxovirus V proteins block both mda-5-dependent and RIG-I-dependent IFN induction. Replication potentially generates ligands for both mda-5 and RIG-I, and transcription of the PIV5 L gene has been shown to generate a ligand for mda-5 (26). Inhibition of mda-5 occurs through a direct interaction between V and mda-5 which prevents ligand binding and consequent activation of the signaling pathway leading to IRF-3 and NF-κB activation. V proteins inhibit RIG-I signaling by interacting with LGP2 and promoting the formation of an inhibitory complex between RIG-I and LGP2 which is refractory to activation by viral RNA.
Our previous work (7) showed that V proteins do not bind directly to RIG-I and do not inhibit IFN induction by RIG-I overexpression; however, in the earlier experiments the overexpressed RIG-I would have been in considerable excess over the endogenous LGP2 level and therefore likely insensitive to LGP2-mediated repression. It may be significant that a considerable excess of LGP2 plasmid is required to see effective inhibition of RIG-I signaling (see Fig. 2A and B). Thus, it is interesting to speculate that LGP2 may not be a very potent inhibitor of RIG-I in the absence of V and that the role of V may be to increase the stability of the LGP2/RIG-I complex, thus promoting the inhibitory action of LGP2. It also seems likely that the magnitude of inhibition would be proportional to the endogenous LGP2 levels. Both LGP2 and RIG-I are IFN-inducible genes, and it is interesting to speculate that the V-mediated inhibition of RIG-I activation may become more significant as the levels of these proteins rise. Such a scenario could be extremely beneficial to viruses.
Several mechanisms have been proposed for the observed inhibition of RIG-I by LGP2. First, since LGP2 can bind dsRNA, it may act to sequester PAMPs from recognition by RIG-I (38, 54); this is an unlikely explanation of both the V-dependent and V-independent inhibitions we observed here, because inhibition was still observed with a mutant form of LGP2 that is unable to bind to RNA. Additionally, we have been unable to reverse the inhibition by the addition of excess RIG-I ligand (data not shown). Second, it has been proposed that LGP2 interacts with IPS-1 and inhibits IFN induction by competing with IκBε (IKKε) for binding to IPS-1 (20). This is similarly unlikely as an explanation for our observations, since the block is upstream of RIG-I activation whereas both IKKε and IPS-1 are downstream of RIG-I. The third hypothesis is that LGP2 blocks RIG-I activation through an inhibitory interaction between the RD of LGP2 and RIG-I (39); we think this is a likely scenario, and we hypothesize that this is an intrinsically weak interaction that is stabilized by paramyxovirus V proteins. We also speculate that the LGP2/RIG-I complex forms in the absence of RNA and is disrupted by ligand binding to either RIG-I or LGP2 but is refractory to disruption in the presence of the V protein. We note that, to date, we have been unable to provide convincing evidence of a triple complex containing PIV5-V, LGP2, and RIG-I by immunoprecipitation. It is possible that the reactive epitopes that we use for immunoprecipitation are not efficiently recognized in triple complexes or that the binding of antibody to the triple complex disrupts the association of LGP2/RIG-I with respect to PIV5-V. Alternatively, the effect of PIV5-V on the affinity of the LGP2/RIG-I association may be indirect (i.e., through an intermediate protein) or may be a consequence of modification of one or both components. The resolution of this issue awaits further experimentation.
In addition to the novel aspect of inhibition of RIG-I activation, we observed that IFN induction by the combination of mda-5 and LGP2 could be completely blocked by overexpression of PIV5-V. This result is not surprising in that we have previously shown that paramyxovirus V proteins are potent inhibitors of mda-5 (2, 7, 8). However, the observations that V proteins bind LGP2, and that LGP2 stimulates mda-5 activity, raise interesting issues, namely, whether activation of mda-5 is dependent on LGP2 as a cofactor (at least in response to some PAMPs) and, if so, whether the primary inhibitory role of the V protein is acting through LGP2 rather than mda-5. To distinguish between these possibilities, it would be necessary to generate a combination of LGP2 and V that is unable to interact while preserving the interaction between these proteins and mda-5. The binding of a common motif in the V proteins to a common epitope in mda-5 and LGP2 (31 and our results) makes this a difficult issue to resolve. Additionally, we have observed that LGP2 mutants that show impaired binding to PIV5-V also failed to stimulate mda-5 activity (data not shown), suggesting that sequences required for V binding are also involved in mda-5 activation. This may provide the rationale behind the targeting of this particular sequence by the V protein, as it disrupts several important aspects of mda-5/LGP2 function, namely, dsRNA binding, homo- and heterodimer formation, and the synergy between mda-5 and LGP2.
In contrast to the data presented here, a recent report suggests that LGP-2 may play a role in stimulating both mda-5 and RIG-I activity (40). This is an intriguing observation, not least because the effect is not seen with all viruses; notably, induction by influenza virus (thought to be entirely dependent upon the generation of RIG-I ligands) is unaffected by the loss of LGP2. Additionally, the effects are cell type specific (LGP2 appears to stimulate IFN induction in MEFs but to suppress induction in HEK293 cells). It is suggested that LGP2 plays a role in recognizing infecting RNA virus particles by actively removing proteins from viral ribonucleoprotein (RNP) complexes or unwinding complex RNA structures to facilitate recognition of viral RNA (40). An attractive model to reconcile these disparate conclusions could be that LGP2 is bound weakly to RIG-I in the absence of PAMP and that it helps to process incoming PAMPs and present these in such a way as to stimulate RIG-I; paramyxovirus V proteins could interact with LGP2 and stabilize LGP2/RIG-I in a nonproductive complex. The ability of LGP2 to stimulate mda-5 is probably due to a distinct mechanism. LGP2 does not appear to be an inhibitor of mda-5 and associates with mda-5 only in response to dsRNA (unpublished data). Since LGP2 appears to be a much more avid dsRNA binding protein than mda-5 (54 and data not shown), we suggest that LGP2 stimulates mda-5 function by either delivering the ligand to mda-5 or promoting the formation of LGP2/mda-5 higher-order complexes that then promote downstream signaling; the binding of paramyxovirus V proteins to the RNA binding domains of both LGP2 and mda-5 would prevent this activation.
The data presented here suggest that paramyxoviruses have evolved a pleiotropic mechanism for limiting IFN induction, since the interaction of the V proteins with LGP2 can inhibit the activation of both mda-5 and RIG-I, albeit with apparently distinct mechanisms. This elegant solution extends our knowledge on the mechanisms by which RNA viruses evade innate immune responses, but it is unlikely to be the only strategy employed by paramyxoviruses to block the induction of IFN. We note that the V proteins of paramyxoviruses have also been implicated as inhibitors of TBK1 and IKK and the p65 subunit of NF-κB (25, 45). More strikingly, the respiroviruses hPIV1, hPIV3, and bPIV3 do not encode functional V proteins and appear to evade IFN production by a combination of C protein functions and limitation of the production of PAMPs (5, 19, 27, 30, 50).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust (grant AL087751/B).
We thank Craig Ross for RNA purified from influenza virus-infected cells and RIG-Ipan RNA, Isabelle Crevel for technical assistance, and Alan Johnstone for advice on FACS analysis.
Footnotes
Published ahead of print 1 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ablasser A, et al. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrejeva J, et al. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bamming D, Horvath CM. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284:9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boonyaratanakornkit J, et al. 2011. The C proteins of human parainfluenza virus type 1 limit double-stranded RNA accumulation that would otherwise trigger activation of MDA5 and protein kinase R. J. Virol. 85:1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen S, et al. 2010. Heterocellular induction of interferon by negative-sense RNA viruses. Virology 407:247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Childs K, et al. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- 8. Childs KS, Andrejeva J, Randall RE, Goodbourn S. 2009. Mechanism of mda-5 inhibition by paramyxovirus V proteins. J. Virol. 83:1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gitlin L, et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gitlin L, et al. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He B, et al. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15–32 [DOI] [PubMed] [Google Scholar]
- 13. Hornung V, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997 [DOI] [PubMed] [Google Scholar]
- 14. Ikegame S, et al. 2010. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 84:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnston MD. 1981. The characteristics required for a Sendai virus preparation to induce high levels of interferon in human lymphoblastoid cells. J. Gen. Virol. 56:175–184 [DOI] [PubMed] [Google Scholar]
- 16. Kato H, et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 18. Killip MJ, et al. 2011. Failure to activate the IFN-beta promoter by a paramyxovirus lacking an interferon antagonist. Virology 415:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Komatsu T, Takeuchi K, Gotoh B. 2007. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes Infect. 9:954–962 [DOI] [PubMed] [Google Scholar]
- 20. Komuro A, Horvath CM. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar H, Kawai T, Akira S. 2011. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30:16–34 [DOI] [PubMed] [Google Scholar]
- 22. Li X, et al. 2009. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch. Biochem. Biophys. 488:23–33 [DOI] [PubMed] [Google Scholar]
- 23. Li X, et al. 2009. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 284:13881–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu C, Ranjith-Kumar CT, Hao L, Kao CC, Li P. 2011. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 39:1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu LL, Puri M, Horvath CM, Sen GC. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269–14276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luthra P, Sun D, Silverman RH, He B. 2011. Activation of IFN-beta expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U. S. A. 108:2118–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malur AG, Hoffman MA, Banerjee AK. 2004. The human parainfluenza virus type 3 (HPIV 3) C protein inhibits viral transcription. Virus Res. 99:199–204 [DOI] [PubMed] [Google Scholar]
- 28. Moon H, Choe J. 2009. Crystallization and preliminary crystallographic studies of human RIG-I in complex with double-stranded RNA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:648–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murali A, et al. 2008. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J. Biol. Chem. 283:15825–15833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakatsu Y, et al. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82:8296–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parisien JP, et al. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pichlmair A, et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001 [DOI] [PubMed] [Google Scholar]
- 33. Pichlmair A, et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pippig DA, et al. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 37:2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poole E, He B, Lamb RA, Randall RE, Goodbourn S. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33–46 [DOI] [PubMed] [Google Scholar]
- 36. Ramachandran A, Horvath CM. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 38. Rothenfusser S, et al. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268 [DOI] [PubMed] [Google Scholar]
- 39. Saito T, et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Satoh T, et al. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schaap-Nutt A, et al. 2011. Identification of human parainfluenza virus type 2 (HPIV-2) V protein amino acid residues that reduce binding of V to MDA5 and attenuate HPIV-2 replication in nonhuman primates. J. Virol. 85:4007–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schlee M, et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmidt A, Endres S, Rothenfusser S. 2011. Pattern recognition of viral nucleic acids by RIG-I-like helicases. J. Mol. Med. 89:5–12 [DOI] [PubMed] [Google Scholar]
- 44. Schmidt A, et al. 2009. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 106:12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schuhmann KM, Pfaller CK, Conzelmann KK. 2011. The measles virus V protein binds to p65 (RelA) to suppress NF-kappaB activity. J. Virol. 85:3162–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strahle L, Garcin D, Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111 [DOI] [PubMed] [Google Scholar]
- 48. Strähle L, et al. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 81:12227–12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahasi K, et al. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J. Biol. Chem. 284:17465–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Cleve W, et al. 2006. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology 352:61–73 [DOI] [PubMed] [Google Scholar]
- 51. Venkataraman T, et al. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455 [DOI] [PubMed] [Google Scholar]
- 52. Wang Y, et al. 2010. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 17:781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoneyama M, et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.