Abstract
Membrane penetration by reovirus requires successive formation of two cell entry intermediates, infectious subvirion particles (ISVPs) and ISVP*s. In vitro incubation of reovirus virions with high concentration of chymotrypsin (CHT) results in partial digestion of the viral outer capsid to form ISVPs. When virions are instead digested with low concentrations of chymotrypsin, the outer capsid is completely proteolyzed to form cores. We investigated the basis for the inverse relationship between CHT activity and protease susceptibility of the reovirus outer capsid. We report that core formation following low-concentration CHT digestion proceeds via formation of particles that contain a protease-sensitive form of the μ1C protein, a characteristic of ISVP*s. In addition, we found that both biochemical features and viral genetic requirements for ISVP* formation and core formation following low-concentration CHT digestion are identical, suggesting that core formation proceeds via a particle resembling ISVP*s. Furthermore, we determined that intermediates generated following low-concentration CHT digestion are distinct from ISVPs and convert to ISVP*-like particles much more readily than ISVPs. These results suggest that the activity of host proteases used to generate ISVPs can influence the efficiency with which the next step in reovirus cell entry, namely, ISVP-to-ISVP* conversion, occurs.
INTRODUCTION
To successfully initiate infection, viruses must overcome the host membrane barrier and deliver their genomic material into host cells. For enveloped viruses, this process is well characterized and requires viral glycoprotein-mediated fusion of host and viral membranes (27, 50). In contrast, mechanisms by which nonenveloped viruses deliver their genomes across membranes are poorly understood. A variety of phylogenetically distant viruses have devised a remarkably conserved strategy to bypass host membranes. Studies on mammalian orthoreovirus (reovirus), a model nonenveloped virus, have contributed to an understanding of the general principles required for cell entry by nonenveloped viruses.
Reovirus particles contain 10 segments of double-stranded RNA (dsRNA) encapsidated within two concentric protein shells, the outer capsid and the inner core (45). Successful initiation of infection by reovirus requires delivery of the viral core into the cytoplasm by bypassing the host membrane (20). The reovirus particle undergoes a series of biochemical and structural changes to traverse the host membrane via the formation of at least two distinct intermediate particles (Table 1). The formation of the first intermediate, known as the infectious subvirion particle (ISVP), is dependent on the activity of endosomal or extracellular proteases (4, 6, 13, 18, 23, 25, 26, 41, 46, 48). These proteases completely digest the σ3 outer capsid protein and engender the cleavage of μ1 to form μ1δ and ϕ (4, 6, 23, 25) (Fig. 1). These μ1 fragments remain associated with the particle. Though ISVPs are capable of membrane interaction, penetration of host membranes requires the formation of the second reovirus entry intermediate, which is referred to as the ISVP* (15). Formation of ISVP*s is characterized by the autocleavage of μ1δ to form μ1N and δ and the release of the N-terminal μ1N fragment and the C-terminal ϕ fragment from the particles (1, 28, 39). The region of μ1 that constitutes μ1N is buried in the structure of μ1 found in ISVPs (34, 54). Thus, release of μ1N requires a massive conformational change in μ1 that requires disruption of interactions between μ1 monomers that constitute the μ1 trimer, between adjacent trimers of μ1, and between μ1 and the underlying core proteins (34, 53, 54). Once μ1N is released, it interacts with membranes to form size-selective pores, recruit virus particles to sites of pore formation, and effect membrane penetration (1, 28, 52) to deliver viral cores into the cytoplasm.
Table 1.
Alterations in reovirus outer capsid proteins during cell entry
| Outer capsid protein | Virion | ISVP | ISVP* | Core |
|---|---|---|---|---|
| σ1 | Present | Present in extended conformation | Released from virions | Absent |
| σ3 | Present | Absent due to degradation | Absent | Absent |
| μ1 | Present as μ1 | Present as particle-associated cleaved μ1 fragments μ1δ and ϕ | μ1δ autocleaved to generate μ1N and δ; δ present in particle- associated, protease-sensitive conformer; μ1N and ϕ are released from virus | Absent |
| λ2 | Present | Present | Present as altered conformer | Present as altered conformer |
Fig 1.
Schematic of the μ1 protein and its cleavage fragments. The 708-amino-acid μ1 protein is autocleaved between amino acids 42 and 43 and proteolytically cleaved by CHT between amino acids 581 and 582. The various combinations of cleavage fragments generated by these two cleavage events are shown.
Changes in the reovirus capsid described above that are required for penetration of membranes can be reproduced in vitro. Treatment of virions with high concentration of the intestinal protease chymotrypsin (CHT) results in the formation of ISVPs (8, 37, 38). Moreover, in vitro-generated ISVPs can be triggered to form ISVP*s either by high-temperature treatment or by incubation with high concentrations of monovalent cations, such as Cs+ or K+ (2, 15). While these types of studies have helped identify both the biochemical features of ISVP formation and ISVP-to-ISVP* conversion and the viral genetic determinants that control these events (2, 15, 16, 19, 21, 29, 44, 49, 53), the contribution of host factors in regulating this process is unclear. Though ISVPs can be generated in a variety of cell types and tissues by a diverse set of proteases, it is not known if the conditions under which ISVPs are generated would influence the efficiency with which ISVP* formation and membrane penetration occur.
In this study, we explored the effect of varying the concentration of CHT during disassembly of reovirus. There is an inverse relationship between the concentration of CHT and the extent of digestion of the reovirus outer capsid. Whereas treatment with high concentrations of CHT results in formation of ISVPs, exposure of virions to low concentrations of CHT results in the generation of cores (22, 31). Based on our analyses of this phenomenon, we report that the proteolytic reaction that results in formation of cores following digestion with a low concentration of CHT exhibits biochemical features reminiscent of ISVP-to-ISVP* conversion. Consistent with this idea, we found that identical viral genetic determinants, those that influence the conformational flexibility of μ1 (44), regulate monovalent cation-induced ISVP* formation and low-CHT-concentration-mediated core formation. Our results indicate that particles generated by low-concentration CHT treatment (ISVPLs) convert more readily to particles resembling ISVP*s than ISVPs produced by high-concentration CHT digestion, without the requirement for known triggers for ISVP* formation. Based on these results, we propose that the activity of host proteases that digest reovirus outer capsid proteins influences the efficiency of subsequent steps required for virus-induced membrane penetration.
MATERIALS AND METHODS
Cells.
Murine L929 (L) cells were maintained in Joklik's minimal essential medium (MEM) (Lonza) supplemented to contain 5% fetal bovine serum (FBS) (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 25 ng/ml amphotericin B (Sigma-Aldrich). BHK-T7 cells used to generate recombinant viruses were maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented to contain 5% FBS, 2 mM l-glutamine, 2% MEM amino acid solution (Invitrogen), and 1 mg/ml Geneticin (Invivogen) in alternate passages.
Viruses.
Generation of recombinant strains (rs) T1L (LLL), rsT1L/T3D M2 (DDD), rsT1L/DDL M2, rsT1L/LDL M2, rsT1L/LDD M2, rsT1L/LLD M2, rsT1L/DLD M2, and rsT1L/DLL M2, which contain a wild-type or chimeric M2 gene in an otherwise T1L background, has been previously described (44). The GenBank accession numbers of T1L and T3D M2 genes are AF490617 and EF494439, respectively. To generate viruses with single-amino-acid substitutions, plasmid pT7-T3D M2 mutagenized by QuikChange site-directed mutagenesis (Stratagene) was used in place of wild-type T3D M2 in a 4- or 10-plasmid reverse genetic system (7, 32, 33). To confirm sequences of mutant viruses, viral RNA was extracted from infected cells and subjected to reverse transcription-PCR (RT-PCR) using three sets of M2-specific primers. PCR products were resolved on Tris-acetate-EDTA agarose gels, purified, and subjected to sequence analysis. Purified reovirus virions were generated using second- or third-passage L-cell lysate stocks of reovirus as described previously (24). Viral particles were extracted from infected-cell lysates using Vertrel-XF (Dupont) (36), layered onto 1.2- to 1.4-g/cm3 CsCl gradients, and centrifuged at 187,813 × g for 4 h. Bands corresponding to virions (1.36 g/cm3) (47) were collected and dialyzed in virion storage buffer (150 mM NaCl, 15 mM MgCl2, 10 mM Tris-HCl [pH 7.4]). The concentration of reovirus virions in purified preparations was determined from an equivalence of 1 optical density (OD) unit at 260 nm equaling 2.1 × 1012 virions/ml (47).
Digestion of reovirus particles.
Although similar results were observed using virion storage buffer, all digestions were performed in a buffer containing 15 mM sodium citrate and 75 mM NaCl (pH 7.5). Virions (2 × 1011) were digested with either 7 μg/ml (low CHT) or 200 μg/ml (high CHT) of N-p-tosyl-l-lysine chloromethyl ketone-treated CHT in a total volume of 0.1 ml at 37°C for the indicated time interval (37). The specific activity of CHT used for the experiments was 54 units/mg. For digestion of a mixture of two virus strains, 1 × 1011 virions of each virus were digested under similar conditions. Aliquots of the reaction mixture were taken or the entire reaction was terminated by addition of 2 mM phenylmethylsulfonyl fluoride and incubation of reaction mixtures on ice. Digestion products were resolved by SDS-PAGE and detected by Coomassie brilliant blue staining.
Analysis of generation of ISVP*-like particles.
ISVPs or products of low-concentration CHT digestion at a concentration of 2 × 1012 particles/ml were incubated at 32°C for 20 min. For cation-triggered ISVP* formation, ISVPs at a concentration of 2 × 1012 particles/ml were incubated at 30°C for 20 min in buffer containing 300 mM NaCl or CsCl (44). The reaction mixtures were transferred to ice for 20 min and incubated with 100 μg/ml trypsin at 4°C for 30 min. Trypsin digestion was terminated by addition of SDS-PAGE loading buffer and removal of the samples to dry ice. Generation of ISVP*s was confirmed by SDS-PAGE and Coomassie brilliant blue staining.
RESULTS
Digestion of reovirus particles with a low concentration of CHT favors core formation.
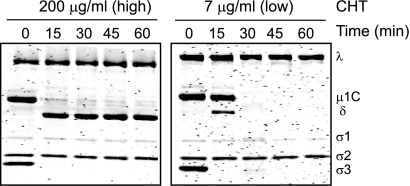
The effect of CHT on digestion of reovirus particles under different conditions has been extensively examined (8–12, 14, 31). Incubation of virions with high concentrations of CHT (100 to 200 μg/ml) results in generation of ISVPs, which retain infectivity and are transcriptionally inactive (10, 22, 31, 37). Polyacrylamide gel electrophoretic analysis of products of digestion generated by incubation with high concentrations of CHT indicates that ISVPs are generated within 15 min of digestion through degradation of the σ3 protein and nearly complete cleavage of the μ1 protein to form particle-associated δ and ϕ fragments (Fig. 2). We note that under the electrophoretic conditions used, μ1 in virions and μ1δ in ISVPs are autocleaved and therefore shown as μ1C and δ, respectively (39). We also note that ϕ is not resolved. In contrast, incubation of virions with low concentrations of CHT (7 to 14 μg/ml) results in formation of cores, which are poorly infectious but are transcriptionally active (22, 31). Core formation in the presence of low concentrations of CHT displays a distinct pattern (22, 31). The σ3 protein is removed within the first 15 min concomitantly with digestion of ∼25% of the μ1C protein to form δ and ϕ. Both uncleaved μ1C and the δ fragment then are lost through proteolysis by 30 min to generate viral cores. While core formation was observed following low-CHT-concentration digestion of three type 3 reovirus strains—Abney, Carter and Dearing (31)—the prototype type 1 reovirus strain T1L failed to form cores when digested with low concentrations of CHT (22). Moreover, this strain-specific difference was mapped to the μ1-encoding M2 gene segment. Consistent with previous observations (22, 31), we observed that rsT1L/T3D M2, a recombinant reovirus strain that contains a type 3 Dearing (T3D)-derived M2 gene in an otherwise type 1 Lang (T1L) background, forms cores when digested with low concentrations of CHT (Fig. 2). These findings confirm that core formation following digestion of reovirus particles with low concentrations of CHT is attributable to the properties of the μ1 protein.
Fig 2.
Inverse relationship between CHT concentration and reovirus digestion. Reovirus virions were incubated with high or low concentrations of CHT at 37°C. Proteolysis in aliquots of the reaction mixture was terminated at the indicated time intervals by removal of the sample to ice and addition of phenylmethylsulfonyl fluoride. The samples were resolved on 10% SDS-PAGE gels and stained with Coomassie brilliant blue. The positions of reovirus capsid proteins are shown.
A feature apparent from the time course of digestion presented in Fig. 2 is that the μ1C and δ fragments present in the particles generated by low-concentration CHT digestion become protease sensitive between 15 and 30 min. The protease sensitivity of μ1C and δ is reminiscent of the property of μ1 fragments following ISVP* formation (15). However, since the known triggers that promote formation of ISVP*s such as Cs+ or K+ ions, high temperatures, or membranes are not present in the reaction, a finding that particles resembling ISVP*s are formed during digestions of virions with low concentrations of CHT would be unexpected and intriguing. To test whether ISVP*s were indeed formed during low-concentration CHT digestion, we determined whether characteristics of the core formation reaction resemble those of the ISVP-to-ISVP* conversion reaction.
Biochemical requirements for core formation resemble those for ISVP-to-ISVP* conversion.
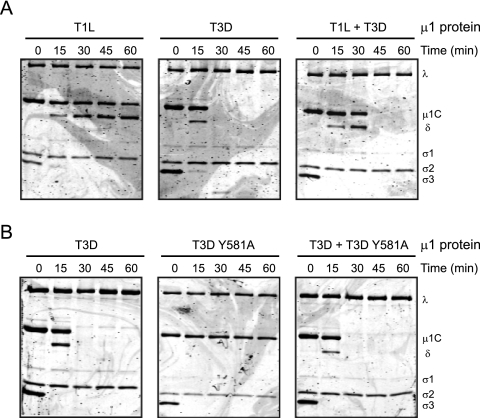
At a high particle concentration, analogous to that used in the digestion experiments above, ISVP-to-ISVP* conversion is regulated by a positive-feedback mechanism (2). Under these conditions, products of ISVP* formation act in trans to facilitate the conversion of other ISVPs to ISVP*s. To assess whether a facilitation phenomenon operates during core formation, we exploited the difference in the core-forming capacities of viruses expressing T1L and T3D μ1 following treatment with low concentrations of CHT (22). Consistent with previous observations (22), we found that low-concentration CHT treatment of a T3D μ1-containing virus led to formation of cores whereas similar treatment of a T1L μ1-containing virus did not (Fig. 3A). However, when an equimolar mixture of viruses expressing T3D μ1 and T1L μ1 was incubated with low concentrations of CHT, both viruses were converted to cores. These data suggest that analogous to the ISVP* conversion, a product of the core formation reaction may facilitate the conversion of other virions to cores.
Fig 3.
Conversion of reovirus virions to cores occurs via a facilitation mechanism dependent on δ-ϕ cleavage. (A) Equal concentrations of rsT1L virions expressing either T1L μ1 or T3D μ1 or a 1:1 mixture of the two types of virions were incubated with 7 μg/ml of CHT at 37°C. (B) Equal concentrations of rsT1L virions expressing either T3D μ1 or T3D μ1 Y581A or a 1:1 mixture of the two types of virions were incubated with 7 μg/ml of CHT at 37°C. Proteolysis in aliquots of the reaction mixture was terminated at the indicated time intervals by removal of the sample to ice and addition of phenylmethylsulfonyl fluoride. The samples were resolved on 10% SDS-PAGE gels and stained with Coomassie brilliant blue. The positions of reovirus capsid proteins are shown.
During digestion of virions to cores by low concentrations of CHT, μ1 is cleaved to δ and ϕ. Since ϕ functions with μ1N to facilitate conversion of ISVPs to ISVP*s (2), we investigated whether cleavage of μ1 at the δ-ϕ junction is required for core formation following low-concentration CHT digestion. Cleavage of μ1 by high concentrations of CHT occurs between Tyr 581 and Gly 582 (38). To determine if cleavage at Tyr 581 is required for core formation following digestion with low concentrations of CHT, we generated a virus containing a Tyr-to-Ala change at residue 581 of T3D M2 and analyzed its capacity for core formation. Consistent with results presented above, we observed that μ1 from virus expressing wild-type T3D M2 was cleaved to δ and subsequently attained a protease-sensitive conformation, leading to the formation of cores (Fig. 3B). In contrast, no δ was detected following incubation of the virus expressing T3D Y581A mutant μ1 and this virus failed to convert to cores. These data suggest that generation of cores following digestion of virions with low concentrations of CHT requires cleavage of μ1 to δ and ϕ.
One explanation for this result is that cleavage of μ1 to δ and ϕ allows for the release of ϕ, which functions in trans as a facilitating factor during formation of cores. An alternate possibility is that Y581A did not form cores, since uncleaved μ1 fails to attain a protease-sensitive conformation. To distinguish between these possibilities, we assessed whether virions of Y581A were capable of forming cores when digested in the presence of virions expressing wild-type μ1. We found that when an equimolar mixture of viruses expressing T3D μ1 and Y581A μ1 was similarly digested, both viruses converted to cores (Fig. 3B). These data indicate that in the presence of facilitating conditions, uncleaved μ1 of the Y581A mutant was capable of forming a protease-sensitive conformer and consequently converting to cores. These findings suggest that the absence of core formation by the Y581A mutant may be related to the absence of a facilitation effect produced as a consequence of cleavage of μ1 to δ and ϕ. These data therefore either reveal a purely coincidental similarity in the mechanisms underlying ISVP* and core formation or indicate that core formation may occur via formation of a particle similar to an ISVP*. To more definitively assess whether ISVP*-like particles are formed during low-concentration CHT digestion, we next assessed if the viral determinants that control ISVP-to-ISVP* conversion also influence core formation under these conditions.
Identical genetic determinants regulate core formation and ISVP* formation.
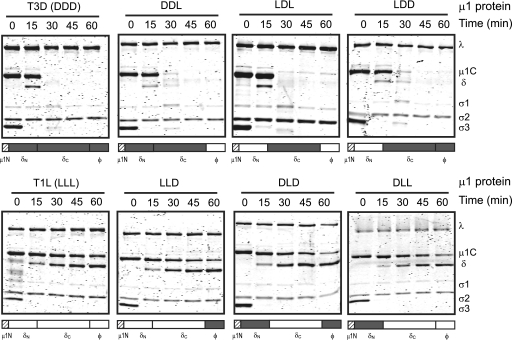
Prototype reovirus strains T1L and T3D differ in their efficiencies in undergoing ISVP-to-ISVP* conversion, and this difference in ISVP* formation is determined by the sequence of the μ1-encoding M2 gene segment (15, 44). Interestingly, the M2 gene also determines the difference in the capacity of T1L and T3D to form cores (22). We have previously shown that the presence of the C-terminal portion of the δ region of μ1 (δC) is sufficient to confer on T1L μ1 the capacity to efficiently undergo ISVP-to-ISVP* conversion (44). To test if the same region of μ1 is responsible for core formation, we employed reoviruses expressing T1L-T3D chimeric μ1 proteins (44). Viruses expressing T3D μ1 and T1L μ1 served as controls for these experiments. Consistent with previous data, low-concentration CHT digestion of virions containing T1L μ1 did not result in core formation whereas similar treatment of virions containing T3D μ1 resulted in core formation (22). Analysis of μ1 chimeric viruses, which are named by the strain origin of the N-terminal half of δ (δN), C-terminal half of δ (δC), and ϕ, indicated that viruses expressing DDL, LDL, and LDD μ1 were capable of core formation similar to that by the virus containing T3D μ1 (Fig. 4). In contrast, viruses expressing LLD, DLD, and DLL were incapable of core formation under these conditions, analogous to the virus expressing T1L μ1. Thus, these data indicate that both the efficiency for ISVP-to-ISVP* conversion and the capacity of reovirus strains to form cores following low-concentration CHT digestion are controlled by the δC domain of μ1.
Fig 4.
Strain-specific differences in core formation are governed by the δC region of μ1. Virions of rsT1L virions containing the indicated wild-type or chimeric μ1 proteins were incubated with 7 μg/ml of CHT at 37°C. Proteolysis in aliquots of the reaction mixture was terminated at the indicated time intervals by removal of the sample to ice and addition of phenylmethylsulfonyl fluoride. The samples were resolved on 10% SDS-PAGE gels and stained with Coomassie brilliant blue. The positions of reovirus capsid proteins are shown. Schematic representations of the chimeric μ1 proteins also are presented, with T3D-derived portions in gray and T1L-derived portions in white. The μ1N domain indicated by hatched bars is identical between T3D and T1L μ1 proteins.
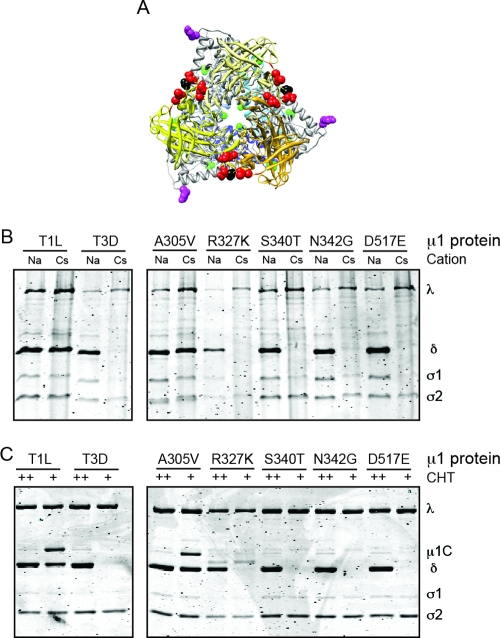
The δC domain, which forms a jelly-roll β-barrel structure in the native structure of μ1, differs at five amino acid residues (positions 305, 327, 340, 342, and 517) between T1L and T3D (34, 44) (Fig. 5A). Of these, residues 305 and 327 lie in positions that may affect interaction between two μ1 monomers within a trimer. In contrast, residues 340, 342, and 517 are solvent exposed and could influence interactions between adjacent μ1 trimers. To define if the same μ1 residues control ISVP* formation and core formation, we generated single-amino-acid substitutions in T3D M2 where we individually replaced each of the five polymorphic residues of T3D δC with a residue from T1L δC. Because it is not known how these substitutions affect ISVP* formation, we first tested the capacity of these viruses to form ISVP*s following CsCl treatment. For these experiments, ISVPs generated from each virus using high-concentration CHT digestion were incubated with CsCl prior to trypsin digestion. Sensitivity of δ to trypsin digestion signifies conversion of ISVPs to ISVP*s (15). ISVPs treated with NaCl were used as controls (15, 44). Consistent with previous results, the δ fragment of rsT1L/T3D M2 is rendered protease sensitive following incubation with CsCl whereas that of rsT1L is resistant to trypsin digestion (Fig. 5B). We found that replacement of Ala 305 in T3D μ1 with a Val from T1L was sufficient to cause T3D δ to become protease resistant (Fig. 5B). In contrast, swapping T1L for T3D residues at positions Arg 327, Ser 340, Asn 342, and Asp 517 within μ1 was not sufficient to render T3D δ trypsin resistant (Fig. 5B). These data indicate that Ala 305 is a critical regulator of ISVP-to-ISVP* conversion. To define if Ala 305 also affects the capacity for in vitro core formation, we incubated virions of each point mutant reovirus with low concentrations of CHT and assessed the products generated at 30 min following digestion using polyacrylamide gel electrophoresis (Fig. 5C). Particles of each mutant incubated with high concentrations of CHT served as controls. We found that an Ala-to-Val change at amino acid 305 in T3D μ1 results in a digestion pattern similar to that of the virus expressing T1L μ1. In contrast, viruses expressing T1L residues at Arg 327, Ser 340, Asn 342, and Asp 517 in T3D μ1 display properties similar to those of viruses expressing T3D μ1 and undergo core formation. Thus, these results suggest that along with controlling ISVP-to-ISVP* conversion, Ala 305 also serves an important function in formation of cores, possibly by affecting interactions between μ1 monomers within the trimer. Congruence in the biochemical mechanisms (Fig. 3) and similarity in the genetic requirements (Fig. 4 and 5) strongly suggest that in vitro core formation proceeds through generation of particles similar to ISVP*s.
Fig 5.
An identical viral genetic determinant regulates ISVP* formation and core formation. (A) Top view of the μ1C trimer rendered using UCSF Chimera from the crystal structure of μ1 (Protein Data Bank accession number 1JMU) is shown with δC in shades of yellow and δN in shades of blue. T1L-T3D polymorphisms within δN and δC are shown in green and red, respectively. Ala 305 and Tyr 581 (at the δ-ϕ cleavage site) are shown in black and magenta, respectively. (B) ISVPs of the indicated viruses were treated with NaCl or CsCl at 32°C for 20 min, chilled on ice for 20 min, and treated with trypsin at 4°C for 30 min. (C) Virions of the indicated viruses were incubated with 7 μg/ml (+) or 200 μg/ml (++) CHT at 37°C for 30 min. Proteolysis was terminated by removal of the sample to ice and addition of phenylmethylsulfonyl fluoride. The samples were resolved on 10% SDS-PAGE gels and stained with Coomassie brilliant blue. The positions of reovirus capsid proteins are shown.
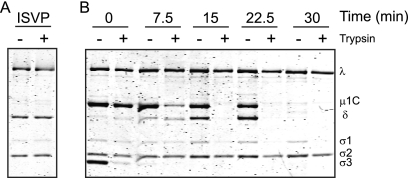
Intermediates of low-concentration CHT digestion readily form ISVP*-like particles.
It is not clear how particles with protease-sensitive μ1 may be generated following digestion with low but not high concentrations of CHT without addition of monovalent cations (Fig. 2). One hypothesis is that the precursor particles which lead to the formation of particles with ISVP*-like properties following low-concentration CHT digestion are distinct from ISVPs and undergo μ1 conformational changes under conditions different than those required for ISVP-to-ISVP* conversion. To test this idea, we terminated the proteolytic reaction at different times following addition of low concentrations of CHT and determined whether the intermediate particles formed under these conditions (ISVPLs) convert to ISVP*s more readily than to ISVPs. For these experiments, we assessed the trypsin sensitivity of the μ1 and δ fragments of ISVPLs following incubation at 32°C for 20 min. ISVPs prepared under standard conditions, by high-concentration CHT digestion, were used as controls. As expected, in the absence of additional triggers, the δ fragment of ISVPs remained resistant to trypsin digestion (Fig. 6A). In contrast, the μ1 protein and δ fragment of ISVPLs formed at 7.5, 15, and 22.5 min following low-concentration CHT digestions were partially (at 7.5 min) or completely (at 15 and 22.5 min) sensitive to trypsin treatment (Fig. 6B), indicating that μ1 domains in these particles have undergone a significant rearrangement to produce a protease-sensitive form that resembles that found in ISVP*s. The μ1 fragments of ISVPLs remained resistant to protease digestion if additional incubation at 32°C was not performed (data not shown). These data demonstrate that though the μ1 fragments in ISVPLs are not in an ISVP*-like conformation, they convert to such a conformation much more readily than ISVPs. Based on these results, we conclude that during digestion of virions with low concentrations of CHT at 37°C, ISVPLs are generated and these particles convert to ISVP*-like particles spontaneously. Because the μ1 fragments in ISVP*-like particles are protease sensitive under these conditions, continued incubation in the presence of CHT results in core formation. Thus, these data provide an explanation for the dramatic difference in the fate of reovirus particles following treatment with high and low concentrations of CHT.
Fig 6.
Particles generated by high- and low-concentration CHT digestion display differences in the efficiencies of formation of ISVP*-like particles. (A) ISVPs of rsT1L/T3D M2 incubated at 32°C for 20 min and chilled on ice for 20 min. (B) Virions digested with 7 μg/ml CHT for the indicated time intervals were incubated at 32°C for 20 min and chilled on ice for 20 min. The reactions either were left untreated (−) or were treated with trypsin (+) at 4°C for 30 min. The samples were resolved on 10% SDS-PAGE gels and stained with Coomassie brilliant blue. The positions of reovirus capsid proteins are shown.
DISCUSSION
CHT functions to uncoat reovirus in the murine intestinal tract (4, 6). In vitro studies to recapitulate digestion of reovirus outer capsid by CHT reveal a peculiar, inverse relationship between the extent of digestion and the concentration of CHT (22, 31). In this study, we sought to understand why complete cleavage of the reovirus outer capsid to form cores occurs in the presence of low but not high concentrations of CHT. Biochemical and genetic analyses of this phenomenon suggest that in vitro core formation proceeds through the formation of an ISVP*-like intermediate. Characterization of particles generated by low- and high-concentration CHT treatment indicates that intermediates of low-concentration CHT digestion assume a protease-sensitive conformation analogous to that present in ISVP* spontaneously, without the requirement of known triggers. Thus, our results explain the inverse relationship between CHT concentration and the extent of digestion of the reovirus outer capsid.
Early studies examining the effects of CHT on reovirus particles in vitro have identified a two-step uncoating process for reovirus (13). The first step is dependent on the presence of a protease and results in partial digestion of the outer capsid, leading to the formation of ISVPs. The second step does not require further protease activity and results in activation of the core-associated transcriptase. These particles have been referred to as ISVP*s in recent studies (15). While the second step can be activated by incubation of ISVPs with membranes (15, 44), a majority of studies, including our own, have used high concentrations of monovalent cations such as Cs+ to trigger these changes (1, 15, 16, 19, 21, 28, 44, 52, 53). In the present study, we found that particles generated by low-concentration CHT digestion (ISVPLs) convert much more readily to ISVP*s than do ISVPs. Generation of particles resembling ISVP*s occurs spontaneously between 22.5 and 30 min following incubation with low concentrations of CHT at 37°C. ISVP*-like particles can also be generated from ISVPLs by incubation at 32°C without the requirement of the known triggers. Because ISVPs formed by digestion of virions with high concentrations of CHT were not able to convert to ISVP*s under either of these conditions, these results suggest that the CHT concentration during ISVP formation can influence the efficiency with which μ1 undergoes conformational changes that are required for cell entry. Analogous to our findings in vitro, it is possible that ISVPs and ISVPLs will display different requirements for completing events such as ISVP* formation that allow them to penetrate membranes of host cells.
The basis for why ISVPLs convert more readily to particles resembling ISVP*s than ISVPs remains undefined. At particle concentrations used in our study, conversion of ISVPs to ISVP*s is dependent on a positive-feedback mechanism (2). During this reaction, the intrinsic stability of the particle determines the efficiency with which a fraction of the ISVPs spontaneously converts to ISVP*s and releases μ1 peptides, μ1N and ϕ. Upon accumulation, the released peptides function in trans to promote conversion of other ISVPs to ISVP*s. Promotion to ISVP*s in trans requires the release of μ1N and is most efficient when ϕ is also released. The susceptibility of target ISVPs to conversion by released peptides also contributes to the efficiency with which the particles are able to form ISVP*s. Though it is not known how the released peptides promote ISVP-to-ISVP* conversion, susceptibility to conversion also is dependent on the conformational flexibility of the particle. In the current study, we found that μ1 determinants that influence strain-specific differences in the conformational flexibility of T1L and T3D (44) govern formation of ISVP*-like particles in the presence of low concentrations of CHT (Fig. 4 and 5). Our findings also suggest that generation of ISVP*-like particles under these conditions proceeds via a facilitation phenomenon (Fig. 3). Though we did not test the requirement of the released μ1 fragments in controlling formation of ISVP*-like particles following low-concentration CHT digestion, based on the requirement for δ-ϕ cleavage in allowing formation of ISVP*-like particles and the capacity of virions with wild-type μ1 in promoting conversion of δ-ϕ cleavage-resistant virions to ISVP*-like particles, we favor the idea that released μ1 fragments also function as promoting factors during generation of ISVP*-like particles following low-concentration CHT digestion. Thus, similar factors determine the efficiency with which ISVPLs and ISVPs convert to ISVP*-like particles. Since there is no loss of promoting factors μ1N and ϕ during formation of ISVPs by high concentrations of CHT (38, 40), we do not think that the lower efficiency of ISVPs than of ISVPLs in converting to ISVP*s is related to differences in the levels of μ1N and ϕ present in ISVPs and ISVPLs. Instead, we think that the difference in the propensities for ISVP* formation by these two particles is related to the efficiency with which the μ1 fragments are released or the efficiency with which they promote conversion of target particles. Consistent with this idea, ISVPs are capable of ISVP* formation under conditions that increase μ1 conformational dynamics such as in the presence of monovalent cations or higher temperatures (2, 15, 44). Thus, we think that differences in the capacities to form ISVP*-like particles are related to the difference in the conformational flexibilities of ISVPs and ISVPLs.
How differences in CHT concentrations result in formation of particles (ISVPs and ISVPLs) with different conformational flexibilities is not known. It is possible that μ1 is cleaved at different sites when generated by incubation with high and low concentrations of CHT. Because Tyr 581 is required for μ1 cleavage under both conditions (17) (Fig. 3B), there is likely no difference in the cleavage site that leads to the generation of δ and ϕ following low- and high-concentration CHT treatment. Since the electrophoretic mobilities of δ fragments generated by low and high concentrations of CHT are indistinguishable (Fig. 2), we do not think that the difference in cleavage patterns may be related to additional proteolysis of δ under these conditions. Thus, the difference in μ1 cleavage may therefore be within ϕ. The original study that first described the generation of the ϕ fragment by proteolytic digestion of μ1 suggested that ϕ may be further cleaved by CHT near its C terminus, somewhere within the C-terminal 29 residues of the 708-amino-acid μ1 protein (38). Indeed, Tyr 698 and Tyr 706 can serve as potential CHT cleavage sites within this region of μ1 (51). It is possible that proteolysis of μ1 at one or both of these sites is sensitive to CHT concentration and therefore yields a distinct μ1 digestion pattern under different conditions. The C-terminal region of ϕ forms a hub-and-spoke structure with each of the spokes contributed by a different μ1 subunit (54). This structure is thought to be important for stabilizing the lattice formed by 200 trimers of μ1. Cleavage status of μ1 within this structure could influence the stability of the lattice and therefore affect the capacity of μ1 to undergo conformational changes to form ISVP*-like particles. An equally plausible explanation for the dissimilarity in the properties of ISVPs and ISVPLs is that a viral protein other than μ1 that is involved in generating ISVP*-like particles, such as λ2, is differentially affected by the presence of high and low CHT concentrations. Pentamers of the λ2 protein form turrets at each of the 12 icosahedral vertices and make contacts with μ1 (54). The interaction between μ1 and λ2 would need to be altered to allow for formation of ISVP*-like particles (54). Changes in μ1-λ2 interaction may occur as a consequence of alteration in the conformation of one or both of the interacting proteins. Consistent with this, λ2 is known to undergo conformational changes following high-temperature incubation, a condition that can trigger ISVP-to-ISVP* conversion (35). If similar changes occur in λ2 during digestion of virions with low concentrations of CHT, we anticipate that differential cleavage of λ2 under different concentrations of protease can have an effect on the efficiency with which ISVP*-like particles are formed.
It is not known if particles similar to ISVPLs are generated only by CHT digestion of reovirus particles in vitro or whether they are also formed in vivo. CHT functions to uncoat reovirus particles in the murine intestinal tract and generate ISVPs with properties identical to those generated in vitro by high concentrations of CHT (4, 6). We think it is likely that the amount of CHT available to act on reovirus varies with the diet of each animal (42). The gut CHT activity may also vary with the genetic makeup of each animal host. Thus, conditions similar to those that we have used for low-concentration CHT digestion may also be relevant in vivo. While CHT functions to uncoat reovirus in the murine intestinal tract, a variety of intracellular or extracellular proteases uncoat reovirus in cultured cells and host tissues (23, 25, 30, 41). Because each protease cleaves μ1 at a different site, it is possible that the ISVPs produced under each condition are different. Thus, particles that resemble ISVPLs, and convert to ISVP*s with different efficiencies, may be generated under some of these conditions. In such cases, the capacity of ISVPLs to escape further cleavage by rapidly completing steps required to cross the membrane barrier may therefore determine the efficiency with which reovirus may be able to infect that tissue.
In addition to describing alternate reovirus digestion intermediates (ISVPLs) that convert to ISVP*-like particles more efficiently than ISVPs, we think that our findings are of relevance to understanding how reovirus initiates infection in the protease-rich gastrointestinal tract. Efficient dissemination of reovirus after initiating infection of the host in the gastrointestinal tract is dependent on the generation of ISVPs by the activity of CHT (3, 4, 6). Findings presented here predict that the concentration of CHT in the murine intestinal tract and the cleavage sensitivity of the μ1 protein could have an effect on the efficiency with which reovirus initiates infection via the gastrointestinal route. This idea is in agreement with a study comparing the virulences of reovirus strains T1L and T3D following peroral inoculation. It was found that T1L replicates much more efficiently in the murine intestine than does T3D and that this property segregates with the μ1-encoding M2 gene segment (43). Furthermore, it was suggested that this difference is related to the protease susceptibilities of the μ1 proteins from these two strains.
In this study, we identified determinants that control the strain-specific differences in T1L and T3D that control the susceptibility of the μ1 protein to CHT digestion in vitro and consequently convert infectious reovirus virions to noninfectious cores. We found that viruses that contain δC derived from T3D μ1 are more sensitive to proteolytic inactivation due to the enhanced propensity of μ1 for undergoing conformational changes that lead to formation of ISVP*-like particles. In addition, our studies showed that cleavage of μ1 at Tyr 581 governs further proteolysis of the μ1 protein and resultant loss of infectivity of the particle. It is possible that the residues that control the efficiency of viral replication in the murine intestine by influencing the cleavage susceptibility of μ1 (43) are identical to those that we have identified through our in vitro studies. Although other studies have suggested that the genetic association of M2 with the difference in replication efficiencies of T1L and T3D within the intestinal tract may not be accurate due to the nature of the viruses used (5), our data provide a rationale for examining how changing the conformational flexibility and cleavage susceptibility of the μ1 protein affects reovirus replication in the intestine. These types of studies may reveal new mechanisms by which the reovirus μ1 outer capsid protein regulates viral pathogenesis.
ACKNOWLEDGMENTS
We thank members of our laboratory, Karl Boehme, Bernardo Mainou, and Tuli Mukhopadhyay for helpful suggestions and review of the manuscript.
This research was supported by the American Heart Association Midwest Affiliate Award 09SDG2140019 (P.D.) and by startup funds from Indiana University.
Footnotes
Published ahead of print 25 January 2012
REFERENCES
- 1. Agosto MA, Ivanovic T, Nibert ML. 2006. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. U. S. A. 103:16496–16501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agosto MA, Myers KS, Ivanovic T, Nibert ML. 2008. A positive-feedback mechanism promotes reovirus particle conversion to the intermediate associated with membrane penetration. Proc. Natl. Acad. Sci. U. S. A. 105:10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amerongen HM, Wilson GAR, Fields BN, Neutra MR. 1994. Proteolytic processing of reovirus is required for adherence to intestinal M cells. J. Virol. 68:8428–8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bass DM, et al. 1990. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol. 64:1830–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bodkin DK, Fields BN. 1989. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J. Virol. 63:1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bodkin DK, Nibert ML, Fields BN. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 63:4676–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boehme KW, Ikizler M, Kobayashi T, Dermody TS. 2011. Reverse genetics for mammalian reovirus. Methods 55:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borsa J, Copps TP, Sargent MD, Long DG, Chapman JD. 1973. New intermediate subviral particles in the in vitro uncoating of reovirus virions by chymotrypsin. J. Virol. 11:552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borsa J, Long DG, Copps TP, Sargent MD, Chapman JD. 1974. Reovirus transcriptase activation in vitro: further studies on the facilitation phenomenon. Intervirology 3:15–35 [DOI] [PubMed] [Google Scholar]
- 10. Borsa J, Long DG, Sargent MD, Copps TP, Chapman JD. 1974. Reovirus transcriptase activation in vitro: involvement of an endogenous uncoating activity in the second stage of the process. Intervirology 4:171–188 [DOI] [PubMed] [Google Scholar]
- 11. Borsa J, Sargent MD, Copps TP, Long DG, Chapman JD. 1973. Specific monovalent cation effects on modification of reovirus infectivity by chymotrypsin digestion in vitro. J. Virol. 11:1017–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borsa J, Sargent MD, Ewing DD, Einspenner M. 1982. Perturbation of the switch-on of transcriptase activity in intermediate subviral particles from reovirus. J. Cell. Physiol. 112:10–18 [DOI] [PubMed] [Google Scholar]
- 13. Borsa J, Sargent MD, Lievaart PA, Copps TP. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191–200 [DOI] [PubMed] [Google Scholar]
- 14. Borsa J, Sargent MD, Long DG, Chapman JD. 1973. Extraordinary effects of specific monovalent cations on activation of reovirus transcriptase by chymotrypsin in vitro. J. Virol. 11:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chandran K, Farsetta DL, Nibert ML. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chandran K, Parker JS, Ehrlich M, Kirchhausen T, Nibert ML. 2003. The delta region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361–13375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chandran K, et al. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang CT, Zweerink HJ. 1971. Fate of parental reovirus in infected cell. Virology 46:544–555 [DOI] [PubMed] [Google Scholar]
- 19. Danthi P, Coffey CM, Parker JS, Abel TW, Dermody TS. 2008. Independent regulation of reovirus membrane penetration and apoptosis by the mu1 phi domain. PLoS Pathog. 4:e1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Danthi P, et al. 2010. From touchdown to transcription: the reovirus cell entry pathway. Curr. Top. Microbiol. Immunol. 343:91–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Danthi P, et al. 2008. Reovirus apoptosis and virulence are regulated by host cell membrane penetration efficiency. J. Virol. 82:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drayna D, Fields BN. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ebert DH, Deussing J, Peters C, Dermody TS. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277:24609–24617 [DOI] [PubMed] [Google Scholar]
- 24. Furlong DB, Nibert ML, Fields BN. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Golden JW, Bahe JA, Lucas WT, Nibert ML, Schiff LA. 2004. Cathepsin S supports acid-independent infection by some reoviruses. J. Biol. Chem. 279:8547–8557 [DOI] [PubMed] [Google Scholar]
- 26. Golden JW, Schiff LA. 2005. Neutrophil elastase, an acid-independent serine protease, facilitates reovirus uncoating and infection in U937 promonocyte cells. Virol. J. 2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivanovic T, et al. 2008. Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J. 27:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jané-Valbuena J, Breun LA, Schiff LA, Nibert ML. 2002. Sites and determinants of early cleavages in the proteolytic processing pathway of reovirus surface protein σ3. J. Virol. 76:5184–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson EM, et al. 2009. Genetic and pharmacologic alteration of cathepsin expression influences reovirus pathogenesis. J. Virol. 83:9630–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joklik WK. 1972. Studies on the effect of chymotrypsin on reovirions. Virology 49:700–715 [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi T, et al. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kobayashi T, Ooms LS, Ikizler M, Chappell JD, Dermody TS. 2010. An improved reverse genetics system for mammalian orthoreoviruses. Virology 398:194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108:283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luongo CL, et al. 1997. Localization of a C-terminal region of λ2 protein in reovirus cores. J. Virol. 71:8035–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mendez II, Hermann LL, Hazelton PR, Coombs KM. 2000. A comparative analysis of freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods 90:59–67 [DOI] [PubMed] [Google Scholar]
- 37. Nibert ML, Chappell JD, Dermody TS. 1995. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved σ1 protein. J. Virol. 69:5057–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nibert ML, Fields BN. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nibert ML, Odegard AL, Agosto MA, Chandran K, Schiff LA. 2005. Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345:461–474 [DOI] [PubMed] [Google Scholar]
- 40. Nibert ML, Schiff LA, Fields BN. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 65:1960–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nygaard RM, Golden JW, Schiff LA. 2012. Impact of host proteases on reovirus infection in the respiratory tract. J. Virol. 86:1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy DM, Schneeman BO. 1981. Effect of soy protein, casein and trypsin inhibitor on cholesterol, bile acids and pancreatic enzymes in mice. J. Nutr. 111:878–885 [DOI] [PubMed] [Google Scholar]
- 43. Rubin DH, Fields BN. 1980. Molecular basis of reovirus virulence: role of the M2 gene. J. Exp. Med. 152:853–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sarkar P, Danthi P. 2010. Determinants of strain-specific differences in efficiency of reovirus entry. J. Virol. 84:12723–12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schiff LA, Nibert ML, Tyler KL. 2007. Orthoreoviruses and their replication, p 1853–1915 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 46. Silverstein SC, Astell C, Levin DH, Schonberg M, Acs G. 1972. The mechanism of reovirus uncoating and gene activation in vivo. Virology 47:797–806 [DOI] [PubMed] [Google Scholar]
- 47. Smith RE, Zweerink HJ, Joklik WK. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39:791–810 [DOI] [PubMed] [Google Scholar]
- 48. Sturzenbecker LJ, Nibert ML, Furlong DB, Fields BN. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wetzel JD, et al. 1997. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J. Virol. 71:1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilkins MR, et al. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112:531–552 [DOI] [PubMed] [Google Scholar]
- 52. Zhang L, et al. 2009. Requirements for the formation of membrane pores by the reovirus myristoylated μ1N peptide. J. Virol. 83:7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang L, Chandran K, Nibert ML, Harrison SC. 2006. Reovirus μ1 structural rearrangements that mediate membrane penetration. J. Virol. 80:12367–12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang X, et al. 2005. Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Å resolution. Structure 13:1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]