Abstract
Type I interferon (alpha/beta interferon [IFN-α/β]) stimulates the expression of interferon-stimulated gene 15 (ISG15), which encodes a ubiquitin-like protein, ISG15. Free ISG15 and ISG15 conjugates function in diverse cellular pathways, particularly regulation of antiviral innate immune responses. In this study, we demonstrate that ISG15 overexpression inhibits porcine reproductive and respiratory syndrome virus (PRRSV) replication in cell culture and that the antiviral activity of interferon is reduced by inhibition of ISG15 conjugation. PRRSV nonstructural protein 2 (nsp2) was previously identified as a potential antagonist of ISG15 production and conjugation. The protein contains a papain-like protease domain (PLP2) that plays a crucial role in the proteolytic cleavage of the PRRSV replicase polyproteins. PLP2 was also proposed to belong to the ovarian tumor domain-containing superfamily of deubiquitinating enzymes (DUBs), which is capable of inhibiting ISG15 production and counteracting ISG15 conjugation to cellular proteins. To determine whether this immune antagonist function could be selectively inactivated, we engineered a panel of mutants with deletions and/or mutations at the N-terminal border of the nsp2 PLP2-DUB domain. A 23-amino-acid deletion (amino acids 402 to 424 of the ORF1a-encoded protein) largely abolished the inhibitory effect of nsp2 on ISG15 production and conjugation, but no viable recombinant virus was recovered. A 19-amino-acid deletion (amino acids 402 to 420), in combination with a downstream point mutation (S465A), partially relieved the ISG15 antagonist function and yielded a viable recombinant virus. Taken together, our data demonstrate that ISG15 and ISGylation play an important role in the response to PRRSV infection and that nsp2 is a key factor in counteracting the antiviral function of ISG15.
INTRODUCTION
Type I interferons, such as alpha interferon (IFN-α) and IFN-β, play a critical role in antiviral innate immunity and in modulating the adaptive immune response to viral infection (39). Initially, the detection of an invading virus by the sensors of the immune system activates host protein signaling cascades, which results in the activation of transcription factors, including interferon-regulatory factor 3 (IRF3), NF-κB, and ATF-2/c JUN. The coordinated activation of these transcription factors leads to the formation of transcriptionally competent enhanceosomes in the cell nucleus to induce the expression of type I IFNs (56). After being secreted, type I IFNs bind to their receptors on adjacent cell surfaces to activate the so-called JAK-STAT signaling pathway, which induces the transcription of interferon-stimulated genes (ISGs). Among the ISGs induced by interferon, ISG15 is one of the highly expressed proteins that functions as an effector molecule in the host cell response to viral infection (4, 10, 11). ISG15 is an ubiquitin-like protein that either exists as a free molecule or is conjugated to other cellular proteins. ISG15 conjugation (ISGylation) to substrate proteins follows a process similar to that of ubiquitin conjugation by utilizing a ubiquitin-activating enzyme (UbE1L, E1) (38), a ubiquitin-conjugating enzyme (UbCH8, E2) (61), and a ubiquitin ligase (HERC5, E3) (9, 50, 54). Production of ISG15 and/or ISGylation of cellular proteins plays important roles in establishing an antiviral state in the infected cell (44, 62). ISG15 expression and ISGylation have been demonstrated to negatively regulate the replication and assembly of, for example, Sindbis virus, human immunodeficiency virus (HIV), influenza virus, herpesviruses, and Ebola virus (27, 28, 36, 37). It has been reported that overexpression of ISG15 increased the expression of host proteins that play an important role in innate immune responses, including IRF3, IFN-β, JAK2, STAT1, interleukin 8 (IL-8), PKR, and OAS (22). The antiviral molecules RIG-I, JAK-1, STAT1, PKR, and MXA were determined to be targets for ISG15 conjugation, which suggests that ISGylation of these molecules regulates their antiviral activities during infection (62). To counteract the antiviral effects of ISG15 and/or ISGylation, some viruses have evolved strategies to interfere with ISG15 function. For example, Yuan and Krug (58) reported that the influenza B virus NS1 protein targets the E1 enzyme, which subsequently prevents ISG15 conjugation. Guerra et al. (19) demonstrated that vaccinia virus E3 protein can bind to ISG15 and disrupt its antiviral activity. All these studies demonstrated the important role of ISG15 and/or ISGylation in protecting cells from viral infection.
Porcine reproductive and respiratory syndrome virus (PRRSV) is an enveloped positive-stranded RNA virus that belongs to the family Arteriviridae, order Nidovirales (45). The PRRSV genome is about 15 kb in length and contains at least 10 open reading frames (ORFs). The 3′-terminal part of the genome encodes four membrane-associated glycoproteins (GP2a, GP3, GP4, and GP5), three unglycosylated membrane proteins (E, the ORF5a protein, and M), and a nucleocapsid protein (N) (2, 15, 24, 32, 33, 34, 45, 53). The replicase gene, which consists of ORF1a and ORF1b, covers the 5′-proximal three quarters of the viral genome. Together, ORF1a and ORF1b encode two long replicase polyproteins, pp1a and the pp1ab frameshift product, which are proteolytically processed into 14 nonstructural protein (nsp) products (14, 63). Of these, nsp2 is the largest cleavage product, which is released by the autoproteolytic activity of the upstream nsp1ß toward the nsp1ß/2 site and the cleavage of the nsp2/3 site by a papain-like protease, PLP2, residing in the N-terminal domain of nsp2 (21, 47). Previous studies demonstrated that PRRSV nsp2 plays an important role in the modulation of type I IFN activity (49, 52). Sequence analysis suggested that the PRRSV nsp2 PLP2 domain may belong to the ovarian tumor domain (OTU)-containing superfamily of proteases (DUBs), of which some members were reported to possess deubiquitinating (DUB) and deISGylating activities (1, 8, 17, 23, 30). The DUB function of the PRRSV PLP2 domain was recently confirmed in our laboratories (49, 52), and its biological significance was demonstrated by its ability to inhibit type I IFN activation. We therefore refer to this arterivirus protease as PLP2-DUB in this paper.
In this study, we investigated the interaction of nsp2 with ISG15 and ISGylation and explored the potential role of ISG15 as an anti-PRRSV immune molecule. In order to eliminate or downregulate the immune antagonist function of nsp2, we used reverse genetics to construct a panel of virus mutants carrying specific deletions and/or mutations at the N-terminal border of the PLP2-DUB domain. The properties of these mutants support an important role for the antiviral activity of ISG15 in PRRSV infection and may represent a first step toward engineering a modified live virus lacking the immune antagonist function of nsp2.
MATERIALS AND METHODS
Cells and viruses.
BHK-21, HeLa, RK-13, and MARC-145 cells were cultured in modified Eagle's medium (Invitrogen, CA) containing 10% fetal bovine serum and were maintained at 37°C with 5% CO2. Porcine alveolar macrophages (PAMs) were obtained by lung lavage of 6-week-old, PRRSV-naïve piglets using the method described previously (59). The type I PRRSV strain SD01-08 (GenBank accession number DQ489311) (13) and its nsp2 mutants were used to infect MARC-145 cells or PAMs. The Sendai virus (SeV) Cantell strain was used to stimulate immune signaling.
Construction of ISG15 expression vectors.
MARC-145 cells were stimulated with 1,000 U/ml of IFN-β (PBL InterferonSource, Piscataway, NJ). At 24 h poststimulation, cells were harvested, and total RNA was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The first strand of ISG15 cDNA was synthesized from total RNA using an oligo(dT) primer and a Super Script III reverse transcriptase kit (Invitrogen). The ISG15 gene was amplified using PCR with the primer pair pFlag-ISG15-F (5′-TAGTCAATGCGGCCGCTACCATGATGAGCTGGGACCTGAAGGTG) and pFlag-ISG15-R (5′-ACCGGATCCTTAGCTCTGCCCGCCAGGCTCTGT). The PCR product was cloned into the p3xFlag vector (Sigma-Aldrich, St. Louis, MO) to generate the p3xFlag-ISG15 plasmid. To construct the ISG15 conjugation site mutation, the C-terminal binding motif LRLRGG of ISG15 was mutated to LRLRAA by site-directed mutagenesis as described previously (28). The primer pair pFlag-ISG15AA-F (5′-TAGTCAATGCGGCCGCTACCATGATGAGCTGGGACCTGAAGGTG) and pFlag-ISG15AA-R (5′-ACCGGATCCTTAGGCTGCCCGCAGGCGCAGATTCATGAAC) was used to generate the mutation. The plasmid containing the LRLRGG-to-LRLRAA mutation is designated p3xFlag-ISG15AA.
ISG15 overexpression and immunofluorescence assay.
MARC-145 cells were seeded in 12-well plates at 2 × 105 cells per well 1 day prior to transfection. Cells were transfected with 0.2, 0.4, or 0.6 μg of the p3xFlag-ISG15 or control empty vector plasmid. For each dose of plasmid DNA, cells in two parallel wells were transfected. At 48 h posttransfection, cells in one of the parallel wells were lysed in Laemmli sample buffer for Western blot analysis of the expression of ISG15. Cells in another parallel well were infected with PRRSV strain SD01-08 at a multiplicity of infection (MOI) of 1. At 24 h postinfection, the expression of PRRSV proteins was detected by immunofluorescence assay (IFA) as described in our previous publications (13, 43). Cells were stained with the PRRSV N protein-specific monoclonal antibody (MAb) SDOW17, and FITC-conjugated goat anti-mouse IgG was used as a secondary antibody.
Fluorescent-focus assay.
The method for the fluorescent-focus assay (FFA) was described previously (7, 13, 16, 49). Briefly, MARC-145 cells (2 × 104/well) were plated in 96-well plates. After 2 to 3 days of incubation, confluent cell monolayers were infected with a 2-fold or 10-fold serial dilution of the virus. At 18 h postinfection, cells were fixed using acetone-methanol (1:1 ratio) at −20°C for 30 min. Fixed cells were stained with PRRSV N protein-specific monoclonal antibody (MAb) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (MP Biomedicals, Solon, OH) was used as a secondary antibody. Fluorescent foci of infected cells were observed and counted using a phase-contrast fluorescence microscope. Virus titers were expressed in number of fluorescent-focus units per ml (FFU/ml).
In vitro ISGylation assays.
To determine the effect of ISGylation on PRRSV replication, BHK-21 cells were seeded in six-well plates at 4 × 105 cells 1 day prior to transfection. Cells were cotransfected with pCAGGS-HA-UbE1L (E1), p3xFlag-UbcH8 (E2), pcDNA-TAP-HA-HERC5 (E3), and p3xFlag-ISG15 or p3xFlag-ISG15AA. At 6 h posttransfection, cells were transfected with PRRSV full-length cDNA clone pCMV-SD01-08, which was modified from pSD01-08 plasmid (13) by using a cytomegalovirus (CMV) promoter to launch the viral RNA. Transfection was performed using Fugene HD reagent (Roche) following the manufacturer's instructions. At 24 h posttransfection, the cell culture supernatant was harvested to determine the virus titer using FFA. The cell monolayer was lysed in Laemmli sample buffer for Western blot analysis. To determine the effect of nsp2 on ISGylation, plasmids expressing the conjugation enzymes E1/E2/E3 and ISG15 were cotransfected with a plasmid expressing the wild-type PLP2-DUB domain [amino acids (aa) 386 to 578 of the ORF1a-encoded protein], nsp2 mutants, or a pCAGGS vector control in HeLa cells. Cells were stimulated with 1,000 U/ml IFN-β at 6 h posttransfection, and 24 h later, cells were harvested for Western blot analysis.
Western blot.
Cell lysates were lysed in Laemmli sample buffer and samples were heated at 95°C for 5 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a nitrocellulose membrane. After blotting, the membrane was blocked with PBST (1× phosphate-buffered saline [PBS] in 0.05% Tween 20) and 5% nonfat dry milk. The membrane was then incubated with a primary antibody for 1 h at room temperature and overnight at 4°C. After a wash with PBST, IRDye 680-conjugated goat anti-rabbit antibody and/or IRDye 800CW-conjugated goat anti-mouse antibody (Li-Cor Biosciences, Lincoln, NE) was added, and the blot was incubated for 2 h at room temperature. Imaging of the blot was performed under the appropriate excitation wavelength using a digital imaging system (Odyssey infrared imaging system; Li-Cor Biosciences). For the detection of ISG15 expression, the nitrocellulose membrane was probed with MAb F-9 or rabbit polyclonal antibody H-150 (both from Santa Cruz Biotechnology, Santa Cruz, CA). The MAb M2 (Sigma, St. Louis, MO) was used to detect the expression of Flag-tagged proteins. The expression of PRRSV nsp1β was detected by a specific MAb as described previously (6), while the expression of the PLP2-DUB domain (aa 386 to 578 of pp1a) or its mutants was detected using an nsp2-specific MAb or rabbit polyclonal antiserum generated in our laboratory (13, 49). The MAb H3 (Lambda Biotech, St. Louis, MO) was used to detect β-tubulin expression.
Quantification of ISG15 and nsp1β expression levels was performed by Western blot analysis using Adobe Photoshop, following the method described at http://rsbweb.nih.gov/ij/. For each protein lane on the Western blot, the target protein level was normalized to that of β-tubulin.
Construction of nsp2 mutants.
Each individual deletion or mutation was introduced into expression vector pCAGGS-nsp2(386–578) using a modified overlapping extension PCR technique as described previously (49). (For mutant constructs, see Fig. 5). The corresponding deletion or mutation was also introduced into the nsp2 region of a PRRSV infectious cDNA clone, pCMV-SD01-08. To analyze the effect of the deletion or mutation on the proteolytic activity of the nsp2 protease, the CD19, CD19+1, and CD23 mutations were also transferred to expression vector pLnsp2-3 to generate constructs pLnsp2-3-CD23, pLnsp2-3-CD19, and pLnsp2-3-CD19+1S. Plasmid pLnsp2-3 is a derivative of equine arteritis virus (EAV) ORF1a expression vector pL1a, in which the foreign gene is under the control of a T7 RNA polymerase promoter and an encephalomyocarditis virus internal ribosomal entry site and is followed by a downstream T7 terminator sequence (46). The EAV ORF1a sequence of pL1a was replaced by the sequence encoding PRRSV nsp2-3 (aa 386 to 1676 of PRRSV SD01-08 pp1a).
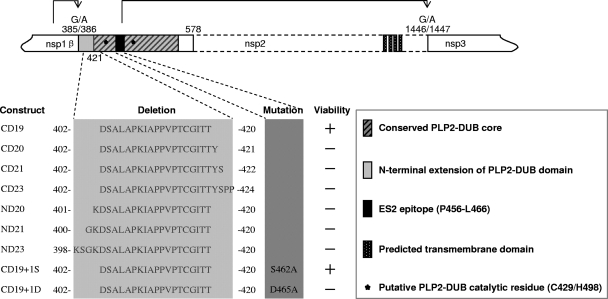
Fig 5.
Schematic diagram of PRRSV nsp2 N-terminal domain mutations and deletions. Each mutant was constructed in both the pCAGGS expression vector and in PRRSV full-length cDNA clone pCMV-SD01-08. The CD23, CD19, and CD19+1S mutations were also transferred to the pLnsp2-3 plasmid backbone. The arrowheads represent the sites being cleaved by the proteases between nsp1β/2 and nsp2/3. +, viable; −, nonviable. Amino acid numbers refer to the pp1a polyprotein sequence of PRRSV strain SD01-08 (GenBank accession number DQ489311) (13).
Luciferase reporter assay.
A dual luciferase reporter gene assay (Promega, Madison, WI) was performed according to the manufacturer's instructions. Reporter plasmid p125-Luc expresses firefly luciferase under the control of the IFN-β promoter (57). Plasmid pRL-SV40 (Promega) expresses Renilla luciferase under the control of a simian virus 40 (SV40) promoter. The luciferase reporter assay was performed as we described previously (6, 49). Briefly, HeLa cells were seeded into 24-well plates 1 day prior to transfection. Cells were cotransfected with plasmids pRL-SV40, p125-Luc, and pCAGGS-nsp2(386–578) or its mutant. The empty vector pCAGGS and plasmid pCAGGS-NS1 expressing NS1 from influenza virus A/swine/Texas/4199-2-98 were included as controls. Sendai virus [300 hemagglutinin (HA) U/ml] was used to stimulate cells at 24 h posttransfection. At 16 h poststimulation, cells were lysed and analyzed by luciferase reporter assay and Western blotting. Firefly and Renilla luciferase activities were measured with a luminometer (Synergy 2; BioTek, Winooski, VT). Values for each sample were normalized using the Renilla luciferase values. Relative luciferase activity is defined as a ratio of firefly luciferase reporter activity to Renilla luciferase activity.
Rescue of recombinant viruses.
Plasmids carrying a full-length PRRSV cDNA were used to transfect BHK-21 cells, using Fugene HD reagent (Roche, CA) following the manufacturer's instructions. To rescue the virus, cell culture supernatant obtained at 24 to 48 h posttransfection was passaged on MARC-145 cells. Two days after infection, the rescue of infectious virus was confirmed by IFA. To monitor the stability of the deletion/mutation introduced into the nsp2 region, RNA was extracted from cells infected with passage 10 of the recombinant virus using a QiaAmp viral RNA kit (Qiagen, Gaithersburg, MD). The region containing the deletion/mutation was amplified by RT-PCR and sequenced at the Iowa State University DNA sequencing facility (Ames, IA).
Growth kinetics and plaque assay.
Growth kinetics was examined by infecting MARC-145 cells with nsp2 mutants and parental virus at an MOI of 0.1. Supernatants from infected cells were collected at 12, 24, 36, 48, 60, and 72 h postinfection (hpi), and virus titers were determined by the FFA. Plaque morphologies of mutants and parental virus were compared by a plaque assay. Confluent cell monolayers were infected with a 10-fold serially diluted parental or mutant virus. At 2 hpi, medium was removed and an agar overlay was applied. After 4 days of incubation at 37°C, cells were stained by using 0.1% crystal violet.
Radioimmunoprecipitation and SDS-PAGE.
The pLnsp2-3-CD23, pLnsp2-3-CD19, and pLnsp2-3-CD19+1S plasmids were transiently expressed in RK-13 cells using the recombinant vaccinia virus/T7 polymerase expression system as described previously (46). Proteins synthesized in transfected cells were labeled from 5 to 8 h after vaccinia virus infection, using methionine-free medium and 100 μCi of [35S]methionine/ml. Cells were lysed, and radioimmunoprecipitation (RIP) analysis was performed as described previously (46). RIP was performed using a rabbit antiserum recognizing both nsp2 and nsp3 (26), and precipitated proteins were separated by SDS-PAGE. Phosphorimager screens were exposed to the gels and subsequently scanned using a Typhoon variable-mode imager (GE Healthcare). Image analysis and quantification of band intensities were performed with ImageQuant TL software (GE Healthcare).
RESULTS
Replication of PRRSV is inhibited by overexpression of ISG15 or ISG15 conjugation enzymes.
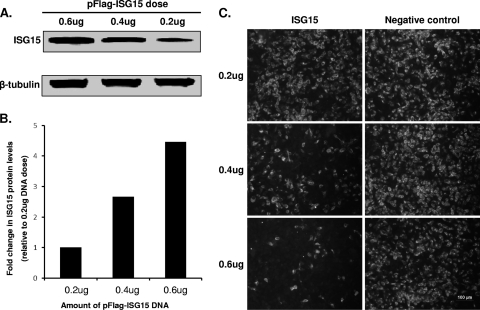
To determine the effect of ISG15 on PRRSV replication, the ISG15 gene was cloned into the p3xFlag vector and expressed in MARC-145 cells as an ISG15-Flag fusion protein. Western blot analysis with the anti-Flag antibody detected an immunoreactive band corresponding to a size of about 18 kDa (Fig. 1A), which confirmed the expression of recombinant ISG15-Flag protein in transfected cells. As shown in Fig. 1B, the ISG15 expression level correlated well with the initial transfection dose of pFlag-ISG15 plasmid DNA. The ISG15-expressing MARC-145 cells were subsequently infected with PRRSV, which revealed that overexpression of ISG15 inhibited virus replication in a dose-dependent manner (Fig. 1C). Upon transfection of 0.6 μg of pFlag-ISG15 plasmid DNA, a reduction in the number of infected cells of about 90% was observed in comparison to cells transfected with empty vector plasmid DNA, demonstrating that ISG15 overexpression inhibited PRRSV replication in MARC-145 cells.
Fig 1.
ISG15 overexpression inhibits PRRSV replication. MARC-145 cells were transfected with different doses of control vector or p3xFLAG-ISG15 plasmid DNA for 48 h. Cells were subsequently infected with PRRSV strain SD01-08 at an MOI of 1 for 24 h. (A) Western blot analysis of ISG15 expression in transfected cells. ISG15 was detected by anti-Flag antibody, and β-tubulin (loading control) was detected using a β-tubulin-specific MAb. (B) Quantitative analysis of ISG15 expression levels. Fold increase in ISG protein levels was determined by comparison with the protein expression level from cells transfected with 0.2 μg of plasmid DNA. (C) Cells were stained with PRRSV N protein specific MAb SDOW17, and FITC-conjugated goat anti-mouse IgG was used as a secondary antibody. Pictures were taken with an Olympus IX71 fluorescence microscope.
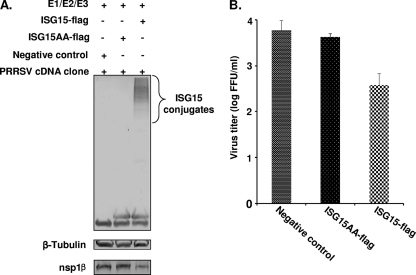
We further studied the effect of ISG15 conjugation (ISGylation) on viral replication in more detail. Initially, MARC-145 cells were cotransfected with plasmids expressing ISG15 and three conjugation enzymes (E1, E2, and E3), and the expression of ISGylated cellular proteins was assessed by Western blot analysis at 24, 36, or 48 h posttransfection. No signal was detected in 24- and 36-h samples, and only trace amounts of ISGylated cellular proteins were detected at 48 h posttransfection (data not shown). This low level of ISGylated proteins may be due to technical reasons (low efficiency of simultaneous transfection of four plasmid DNAs) or intrinsic properties of the MARC-145 cell line (see Discussion for more details). As an alternative, BHK-21 cells were used, since these cells are more easily transfected than MARC-145 cells and, most importantly, support PRRSV replication. They are commonly used in PRRSV reverse genetics for initial recovery of recombinant viruses. Therefore, in this experiment, BHK-21 cells were transfected with plasmids expressing ISG15 and three conjugation enzymes. As a control, a conjugation-negative mutant ISG15-Flag protein was expressed, in which the C-terminal LRLRGG conjugation site was changed to LRLRAA. As shown in Fig. 2A, in cells expressing the wild-type ISG15 and the three conjugation enzymes, a significant amount of ISGylated proteins was detected by Western blot analysis, while no ISG15-conjugated proteins were detected in cells transfected with a plasmid expressing the ISG15 mutant (pFlag-ISG15AA) or an empty vector plasmid (p3xFlag). After expressing ISG15 and the corresponding conjugation enzymes, cells were transfected with a CMV promoter-driven full-length cDNA clone, pCMV-SD01-08, from which the viral genome can be launched. At 24 h posttransfection, cells and culture supernatant were harvested for protein expression analysis and progeny virus titration. In Western blot analysis (Fig. 2A, bottom), a reduced level of replicase subunit nsp1β was observed in cell cultures in which ISGylation had been activated. Moreover, progeny virus titers in the cell culture supernatant were reduced about 10-fold in cells expressing wild-type ISG15 and conjugation enzymes in comparison to cells transfected with the mutant ISG15-Flag expression vector or an empty vector control (Fig. 2B).
Fig 2.
Effect of ISG15 conjugation on PRRSV replication. BHK-21 cells were transfected with plasmids that express conjugation enzymes E1/E2/E3 and Flag-tagged ISG15 or ISG15 mutant (ISG15AA). Empty vector plasmid p3xFlag was used as a negative control. At 6 h posttransfection, cells were transfected with the infectious PRRSV cDNA clone pCMV-SD01-08. Cells and supernatant were harvested at 24 h posttransfection. (A) Cell lysates were analyzed by immunoblotting using antibodies that recognize the Flag-tag of ISG15, β-tubulin, or PRRSV nsp1β. (B) Virus titers were determined by fluorescent-focus assay using cell culture supernatant. The results are mean values from four replicates, and virus titers were expressed as numbers of fluorescent-focus units per milliliter (FFU/ml).
PRRSV inhibits ISG15 production and ISGylation.
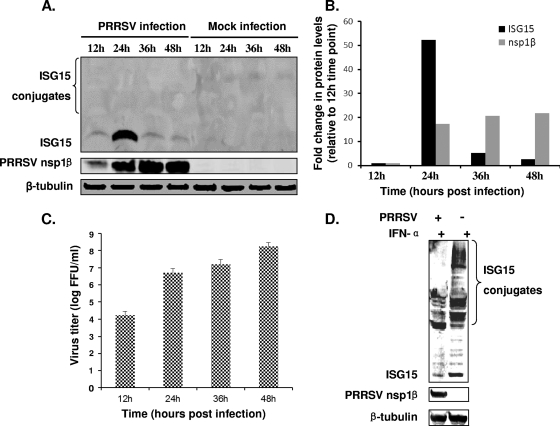
To investigate the molecular mechanism behind the inhibitory effect of ISG15 on PRRSV replication, we examined whether ISG15 expression is upregulated in PRRSV-infected cells. MARC-145 cells were infected with PRRSV strain SD01-08 at an MOI of 0.1, and cells were harvested at 12, 24, 36, and 48 hpi. The expression of endogenous ISG15 was monitored using an ISG15-specific antibody in Western blot analysis (Fig. 3A). Interestingly, a significant increase in the level of ISG15 expression was observed at 24 hpi, but 24 h later the ISG15 level had returned to near-baseline levels. In comparison to the ISG15 level at 12 hpi, there was a 52.2-fold increase by 24 hpi, but only 5.2- and 2.6-fold higher expression levels were measured at 36 hpi and 48 hpi, respectively. In contrast, the amount of PRRSV nsp1β continuously increased in virus-infected cells (Fig. 3A and B). The cell culture supernatants were harvested at different time points, and viral titers were determined by fluorescent-focus assay. The result was consistent with that of the Western blot analysis, with virus titers continuously increasing during the course of infection and the strongest increase occurring between 12 and 24 hpi (Fig. 3C). Taken together, these results suggest that PRRSV employs a mechanism to inhibit ISG15 expression during the later stages of infection (36 to 48 hpi).
Fig 3.
PRRSV suppresses ISG15 expression and conjugation. (A) MARC-145 cells were infected with MOI 0.1 of PRRSV SD01-08 or were mock infected. Cells were harvested at 12, 24, 36, or 48 hpi and analyzed by Western blotting. Membranes were probed with a specific MAb to nsp1β or ISG15. (B) Quantification of ISG15 and nsp1β protein levels at different time points postinfection. Fold change in protein levels was determined by comparing with the protein expression level at 12 hpi. (C) Cell culture supernatant was harvested at 12, 24, 36, or 48 hpi. Virus titers were determined by the fluorescent-focus assay on MARC-145 cells. (D) PAMs were infected with PRRSV SD01-08 at an MOI of 0.1 or mock infected. At 24 h postinfection, cells were stimulated with 1,000 U/ml swine IFN-α. Cells were harvested at 24 h poststimulation and analyzed by Western blotting. Membranes were probed with a specific MAb to nsp1β or tubulin (loading control) or a rabbit polyclonal antiserum to swine ISG15.
During the course of this experiment, we did not detect any ISG15-conjugated proteins in PRRSV-infected MARC-145 cells (Fig. 3A). Furthermore, we were also not able to detect ISGylated cellular proteins upon stimulation of uninfected MARC-145 cells with type I IFNs (data not shown). Since the porcine alveolar macrophage is the natural host cell for PRRSV, these cells were used to further investigate the effect of PRRSV infection on ISG15 production and ISGylation. As shown in Fig. 3D, the level of both free ISG15 and ISGylated proteins at 36 hpi was strongly reduced compared to the amounts in mock-infected cells.
PRRSV nsp2 inhibits ISG15 production and ISGylation.
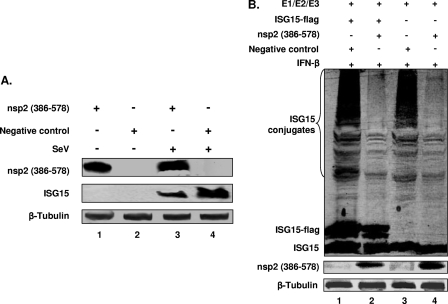
Among the PRRSV proteins, nsp2 was previously identified as a major immune antagonist (3, 17, 49). Specifically, our previous studies demonstrated that the PLP2-DUB-containing N-terminal domain of nsp2, aa 386 to 578 of the ORF1a-encoded protein, is able to inhibit IFN-β production (49, 52). In the present study, we further tested the hypothesis that the nsp2 PLP2-DUB domain affects the expression of downstream interferon-stimulated genes. In these experiments, we used the construct pCAGGS-nsp2(386–578), which expresses ORF1a-encoded aa 386 to 578, the N-terminal 193 residues of nsp2. HeLa cells were used in transient-expression experiments, since this cell line is more easily transfected and is commonly used for the analysis of innate immune responses (9, 12, 54, 61, 62). Cells were transfected with plasmid pCAGGS-nsp2(386–578) and then were stimulated by SeV infection. A 2.9-fold reduction of the ISG15 level was observed when nsp2(386–578)-expressing cells were compared to cells transfected with empty vector plasmid DNA (Fig. 4A; compare lane 3 with lane 4).
Fig 4.
The PRRSV PLP2-DUB domain inhibits ISG15 expression and conjugation. (A) HeLa cells were transfected with an expression plasmid for nsp2(386–578) or an empty plasmid vector and stimulated with 300 HA/ml of SeV at 24 h posttransfection. Cells were harvested at 24 h poststimulation, and lysates were immunoblotted for detection of ISG15 or nsp2(386–578). An anti-ISG15 MAb was used to detect ISG15 expression, and an anti-nsp2 MAb was used to detect the expression of nsp2(386–578). (B) HeLa cells were cotransfected with plasmid DNAs expressing conjugation enzymes E1/E2/E3, ISG15, and nsp2(386-578). The empty pCAGGS vector plasmid was included as a control. At 6 h posttransfection, cells were stimulated with 1,000 U/ml of IFN-β. Cells were harvested at 24 h poststimulation and analyzed by immunoblotting. The membrane was probed with anti-PRRSV nsp2 MAb to detect the expression of nsp2(386-578). Free and conjugated forms of ISG15 were detected by anti-ISG15 polyclonal antiserum. The anti-β-tubulin antibody was used to detect the expression of β-tubulin (loading control).
The effect of PRRSV PLP2-DUB expression on host protein ISGylation was further investigated. ISG15 conjugates were generated by transfecting HeLa cells with plasmids that express ISG15 and the E1, E2, and E3 enzymes. Cells were cotransfected with the PRRSV nsp2(386–578) expression vector and subsequently stimulated with IFN-β. Coexpression of PRRSV nsp2(386–578) resulted in a clear decrease in the level of ISGylated proteins (Fig. 4B), both when ISG15-Flag expression was used (compare lanes 1 and 2) and when the expression of endogenous ISG15 and ISGylated proteins was analyzed (compare lanes 3 and 4), which further confirmed that the PLP2-DUB domain has the ability to counteract ISGylation. IFN-β stimulation activates the IFN signaling pathway, which induces ISG expression. If the PLP2-DUB domain were to affect any step(s) in this pathway, we would expect to see a reduced level of (free) ISG15 in the cells. However, such a difference was not observed (Fig. 4B; compare lane 1 with lane 2 and compare lane 3 with lane 4). In contrast, when nsp2(386–578)-expressing cells were stimulated using SeV infection, the IFN production pathway was activated, and we observed a reduction of ISG15 expression (Fig. 4A; compare lane 3 with lane 4). Taken together, these results suggest that the effect of nsp2 on ISG15 expression is due to its ability to inhibit one or multiple steps in IFN production, whereas the IFN-induced signaling pathways are not affected.
Attenuation of the inhibitory effect of nsp2 on ISG15 production and ISGylation.
Having established that the PLP2-DUB domain of nsp2 has the ability to inhibit ISG15 expression and ISGylation of cellular proteins, we attempted to inactivate this function in the context of the viral replicative cycle. As outlined in Fig. 5, using reverse genetics, we constructed a panel of PRRSV mutants with a deletion and/or mutation at the N-terminal border of the predicted PLP2-DUB domain. In our previous study (49), point mutations were introduced into a B-cell epitope within the PLP2-DUB domain. Three of these mutations (D458A, S462A, and D465A) generated viable recombinant viruses, and the S462A and D465A mutants displayed a somewhat reduced ability to suppress IFN production. In the present study, we further explored whether sequences near the N-terminal border of PLP2-DUB domain can be manipulated without affecting virus replication, while at the same time reducing the activity of the protease domain as an IFN antagonist. To this end, a 19-aa region of nsp2, 402-DSALAPKIAPPVPTCGITT-420, was targeted for deletion (Fig. 5). This region is located immediately upstream of the conserved core of the PLP2-DUB domain (47), may contribute to the binding of substrates like ubiquitin and ISG15 (1), and could thus be important for the DUB and deISGylation activities of PRRSV PLP2-DUB. Furthermore, the corresponding 19-aa region in nsp2 of type II PRRSV was previously found to be nonessential for viral replication (20).
Initially, we extended this 19-aa deletion in the N- or C-terminal direction, or both, to investigate which sequence could be deleted to impair or completely block the IFN antagonist function of nsp2. Furthermore, the 19-aa deletion was combined with the previously identified S462A and D465A mutations (see above) that previously yielded recombinant viruses with a somewhat reduced ability to suppress IFN production. The resulting panel of nsp2(386–578) mutants was first expressed using the mammalian expression vector pCAGGS and then screened using a previously described IFN-β promoter-luciferase reporter assay (6, 49). As shown in Fig. 6, the influenza virus NS1, a known IFN antagonist (41, 42, 48, 58) that was used as a positive control, strongly suppressed the expression of luciferase. Compared to the wild-type PLP2-DUB domain, each of the 9 nsp2 mutants showed reduced inhibition of luciferase reporter gene expression. The CD23 deletion largely impaired the ability of the PLP2-DUB domain to inhibit IFN-β activation. The CD19 construct allowed an approximately 3-fold-higher level of reporter signal, while the CD19+1S construct allowed about a 4.5-fold-higher reporter signal level in comparison to that of the wild-type control. Since all of these nine mutations were impaired in their ability to inhibit IFN-β production, they were transferred to a PRRSV full-length cDNA clone to determine whether a viable recombinant virus containing these deletions and/or mutations could be obtained. Of all mutants tested, only constructs CD19 (aa 402 to 420 deleted) and CD19+1S (aa 402 to 420 deleted, in combination with the S462A mutation) generated viable recombinant viruses. The 421-YSPP motif immediately downstream of the initial 19-aa deletion is part of the PLP2-DUB core sequence that is highly conserved in arteriviruses (47). Deletions extending into this region were lethal, further supporting the important role of this motif in protease function and viral replication. The lethal effect of mutations extending upstream of the initial 19-aa deletion could be due to interference with cleavage of the nsp1β/2 junction.
Fig 6.
Effect of mutations and/or deletions on the ability of PLP2-DUB domain to inhibit IFN-β induction. HeLa cells were transfected with a plasmid that expresses a wild-type or mutated nsp2(386–578), pCAGGS empty vector plasmid, or a plasmid expressing influenza virus NS1 (positive control), along with the reporter plasmid p125-Luc and Renilla luciferase expression plasmid pRL-SV40. Cells were stimulated with Sendai virus at 24 h posttransfection. The luciferase activity was measured at 16 h poststimulation. Relative luciferase activity is defined as the ratio of firefly luciferase reporter activity to Renilla luciferase activity. Each data point represents a mean value from three experiments. Error bars show standard deviations of the normalized data.
We further characterized the growth properties of the vSD-CD19 and vSD-CD19+1S recombinant viruses. To investigate the stability of the 19-aa deletion and S462A mutation in the virus, the recombinant viruses were serially passaged 10 times in MARC-145 cells. Subsequently, the nsp2-coding region was amplified and sequenced, which revealed that both the 19-aa deletion and the S462A mutation could be stably maintained in the virus. The growth properties of the recombinant viruses were compared with those of their wild-type parent. Plaque assays using MARC-145 cells showed that vSD-CD19 plaques are smaller than those of wild-type virus and that vSD-CD19+1S is barely able to induce plaque formation (Fig. 7A). Close microscopic investigation of the cell monolayer showed that cytopathic effect (CPE) was observed in cells infected with vSD-CD19 and wild-type virus at 72 hpi, but CPE was not visible in cells infected with vSD-CD19+1S. Since plaque development depends on a variety of factors, including replication kinetics, virus yield and release, and cytolytic activity (40), the impaired ability of nsp2 mutants, especially vSD-CD19+1S, to form visible plaques suggested that growth rate differences between the nsp2 mutants and wild-type virus exist. Growth kinetics analysis consistently showed that both vSD-CD19 and vSD-CD19+1S were impaired in growth in MARC-145 cells (Fig. 7B). The peak viral titer for vSD-CD19 was 1.8 × 106 FFU/ml, compared to a titer of 1.0 × 107 FFU/ml for the wild-type virus. The vSD-CD19+1S maintained a very low titer throughout the time course experiment (peak viral titer, 1.0 × 103 FFU/ml).
Fig 7.
Growth characterization of recombinant PRRSVs carrying a 19-aa deletion with and without an additional S462A substitution in the nsp2 N-terminal domain. (A) Plaque morphology of recombinant viruses and wild-type virus. Confluent cell monolayers were infected with 10-fold serial dilutions of wild-type or recombinant virus. At 2 h postinfection, medium was removed and an agar overlay was applied. After 4 days incubation at 37°C, cells were stained using 0.1% crystal violet. (B) Growth kinetics of recombinant viruses in comparison with wild-type virus. MARC-145 cells were infected with each virus at an MOI of 0.5, and the amount of virus produced at 12, 24, 36, 48, 60, and 72 h postinfection was determined by FFA in MARC-145 cells. The results shown are mean values from three replicates.
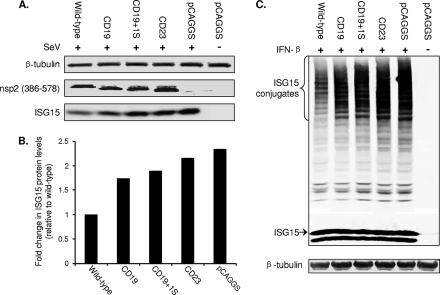
To determine whether they have a reduced ability to inhibit ISG15 production and ISGylation, mutants CD19 and CD19+1S were characterized in more detail. The wild-type [pCAGGS-nsp2(386–578)] and both mutants (pCAGGS-CD19 and pCAGGS-CD19+1S) were expressed in HeLa cells. The pCAGGS-CD23 construct was also included as a control, since this construct was most impaired in downregulating luciferase reporter gene expression (Fig. 6). We first investigated the effect of nsp2 deletions/mutations on ISG15 production. As shown in Fig. 8A, all three mutants showed a certain degree of impairment in their ability to inhibit ISG15 expression. In comparison to cells transfected with the empty vector control, ISG15 was expressed at a 2.3-fold-lower level in cells transfected with wild-type pCAGGS-nsp2(386–578) but only at 1.7-fold- and 1.8-fold-lower levels in cells transfected with pCAGGS-CD19+1S and pCAGGS-CD19, respectively. The 23-aa deletion almost completely abolished the ISG15 inhibitory function of the PLP2-DUB domain, with only a 0.2-fold difference in ISG15 expression levels compared to that of empty vector plasmid (Fig. 8B). In line with these observations, the ISGylation assay result showed that the CD23 deletion mutants had almost lost the ability to interfere with ISGylation, while this property was clearly impaired in the CD19 and CD19+1S mutants (Fig. 8C).
Fig 8.
Effect of PRRSV nsp2 N-terminal domain mutations on ISG15 expression and conjugation. (A) HeLa cells were transfected with an expression plasmid for wild-type and mutant nsp2(386–578), as indicated at the top of the panel. Cells were stimulated with 300HA/ml of SeV at 24 h posttransfection and harvested at 24 h poststimulation. Expression of ISG15 and nsp2(386–578) was analyzed by Western blot. Anti-ISG15 MAb was used to detect the expression of ISG15, and anti-nsp2 polyclonal antibody was used to detect the expression of nsp2(386–578). (B) Quantitative analysis of ISG15 protein expression. Fold increase in ISG15 levels was determined by comparing with the protein expression level from cells transfected with wild-type pCAGGS-nsp2(386–578). (C) Plasmids expressing conjugation enzymes E1/E2/E3 and ISG15 were cotransfected with a plasmid expressing the wild type nsp2(386–578), nsp2 mutant, or a pCAGGS vector control in HeLa cells. Cells were stimulated with 1,000 U/ml IFN-β at 6 h posttransfection. At 24 h poststimulation, cells were harvested for Western blot analysis. Free or conjugated form of ISG15 was detected by rabbit anti-Flag polyclonal antibody.
Effect of deletions and/or mutations in the PLP2-DUB domain on replicase polyprotein processing.
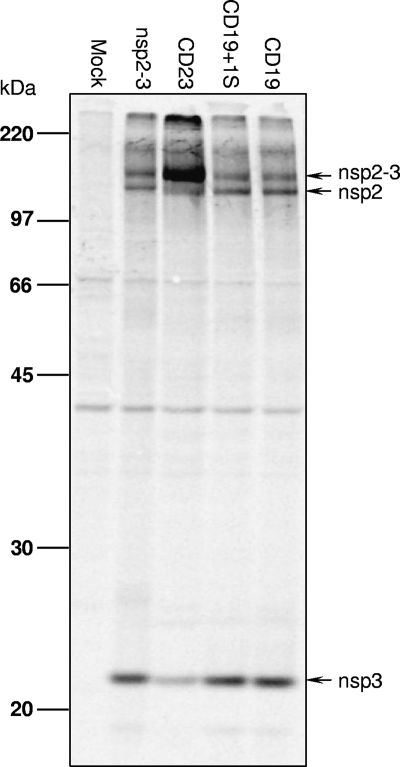
As indicated previously (47), the PLP2-DUB domain is a cysteine protease that is responsible for proteolytic cleavage of the nsp2/3 site, a critical step in arterivirus replicase polyprotein processing. To investigate whether the attenuated growth properties of the nsp2 mutant viruses could be due to an effect on the function of PLP2 in replicase maturation, the nsp2/3-coding region containing CD19 or CD19+1S was expressed in a vaccinia virus/T7 polymerase system as described previously (18). For comparison, constructs expressing wild-type (pLnsp2-3) and the 23-aa deletion mutant (pLnsp2-3-CD23) were also included in the analysis. The different nsp2-3 polyproteins were expressed by transfection of plasmid DNA into cells that had previously been infected with a recombinant vaccinia virus expressing the T7 RNA polymerase. Following radiolabeling of proteins synthesized in transfected cells, the expression and cleavage of nsp2-3 were analyzed by immunoprecipitation, SDS-PAGE, and autoradiography. As shown in Fig. 9, in comparison to the wild-type nsp2-3, mutant CD23 displayed a strongly reduced level of cleavage of the nsp2/3 site, as most of the nsp2-3 protein was detected in the uncleaved precursor form. This result demonstrated that the CD23 deletion impaired the proteolytic function of PLP2, likely explaining why no viable recombinant virus was recovered in the reverse genetic system. In contrast, for both nsp2-3-CD19 and nsp2-3-CD19+1S mutants, efficient cleavage of the nsp2/3 site was observed, since individual nsp2 and nsp3 proteins were detected in amounts quite similar to those produced by the wild-type control. The result indicates that the proteolytic function of PLP2 with regard to the nsp2/3 site is not directly affected in the mutants CD19 and CD19+1S. This suggests that the reduced growth rate (viral titer) of the vSD-CD19 and vSD-CD19+1 viruses is not caused by a basic defect in replicase polyprotein proteolysis but instead may be due to an impaired ability to counteract cellular innate immune responses.
Fig 9.
Effect of nsp2 N-terminal domain mutations on the proteolytic activity toward the nsp2/3 site. RK-13 cells were transfected with plasmids expressing wild-type or mutant nsp2-3 polyproteins, as indicated at the top of the panel. After metabolic labeling, expression products were immunoprecipitated with a rabbit antiserum recognizing both nsp2 and nsp3. Immunoprecipitated proteins were separated by SDS-PAGE and visualized by autoradiography. The positions of the nsp2-3 precursor and its cleavage products are indicated on the right.
DISCUSSION
Previous studies demonstrated that IFN-α/β treatment of host cells induces the synthesis of ISG15 and ISGylation of a large number of cellular proteins and likely plays a major role in establishing an antiviral state. Overexpression of ISG15 inhibited viral packaging and replication of HIV, Ebola virus, and Sindbis virus in type I IFN receptor-deficient mice (27, 28, 36, 37). Mice with a defect in the production of ISG15 are hyperresponsive to infection by influenza A and B virus, herpesviruses, and Sindbis virus (28). In the present study, we demonstrated that overexpression of ISG15 and ISGylation of cellular proteins also interferes with PRRSV replication. Inhibition of ISGylation activity by mutating the ISG15 conjugation site (LRLRGG to LRLRAA) increased virus titers at least 10-fold. Taken together, these results indicate that ISG15 and/or ISGylation plays a critical role in antagonizing PRRSV replication.
Viruses have evolved many strategies to counteract host immune responses. Previous studies showed that PRRSV infection suppresses or delays immune gene and protein expression. Miller et al. (35) reported that stimulation of MARC-145 cells by exogenous double-stranded RNA resulted in a significant increase in type I IFN mRNA expression. However, in the presence of PRRSV infection, type I IFN production was significantly inhibited. In this study, we further examined the effect of PRRSV on the expression of ISG15. A time course study showed that ISG15 was induced in a relatively short window postinfection (observed at 24 hpi) but returned to a minimum expression level after 24 hpi. This suggests competition between the ability of the cell to detect and respond to viral infection and the ability of the virus to counteract the cellular immune response. Between 24 and 36 hpi, PRRSV seemed to overcome the ISG15 response and replicate to higher titers. Further examination of the ISG15-conjugated cellular proteins in porcine alveolar macrophages (Fig. 3D) indicated that PRRSV has the ability to interfere with ISG15 conjugation, thus counteracting the antiviral effect of ISG15 expression and ISGylation.
Most notably, ISGylation was not observed in PRRSV-infected MARC-145 cells. We were also not able to detect the ISGylated cellular proteins upon stimulation of uninfected MARC-145 cells by type I IFNs. Previous studies showed that type I IFN stimulation did not induce ISGylation in Vero and 293T cells, although these cells showed increased levels of free ISG15 (29, 31, 60). Since MARC-145 and Vero cells are related African green monkey kidney cell lines, we suspect that they share similar mechanisms for ISG15 expression and conjugation. Furthermore, when we transfected the MARC-145 cells with plasmids expressing ISG15 and conjugation enzymes, we observed a very slow response of ISGylation (48 h posttransfection), and the expression level was very low in comparison to the other cell lines tested (BHK-21 and HeLa). As Zhang and Zhang (60) indicated, the discrepancy between ISG15 induction and conjugation could result from a lack of functional ISG15 modification enzymes or the existence of factors that inhibit these enzymes. On the other hand, we cannot exclude the possibility that the ISGylation phenomena that we observed with MARC-145 cells were due to the experimental conditions and/or sensitivity of our detection methods. Whether MARC-145 cells are actually defective in ISGylation function needs to be analyzed in more detail.
Results from this study further demonstrated that PRRSV PLP2-DUB domain plays a role in the suppression of the host innate immune responses. In our previous studies (49, 52), the PRRSV PLP2-DUB domain was demonstrated to have DUB activity. Specifically, it inhibits the activation of NF-κB, a key transcription factor for IFN-β production (49). The NF-κB activation is dependent on the polyubiquitination and degradation of IκB binding proteins. The PLP2-DUB domain reverses the polyubiquitination process of IkB-α, which results in the inhibition of NF-κB activation. In this study, we further demonstrated that the PRRSV PLP2-DUB domain possesses deISGylation activity and has the ability to interfere with ISG15 conjugation to cellular proteins. These results indicate that the PRRSV nsp2 protease targets at least two pathways of the host immune response. First, it interferes with the ubiquitination of signaling molecules in the IFN-β production pathway, such as IkB-α, resulting in the suppression of IFN-β expression and reduced activation of the downstream JACK-STAT pathway. As a consequence, the induction of ISGs, including ISG15, is impaired. Second, the PLP2-DUB domain possesses deISGylation activity and interferes with the function of a group of cellular proteins that play important roles in antiviral activities (44, 62).
The challenge now is to apply the above findings in disease control and prevention. In human medicine, such as the treatment of hepatitis C virus (HCV) infection, combined therapy, consisting of enhanced ISG15 conjugation and IFN-β treatment, exhibited improved efficacy in the clearance of a HCV subgenomic replicon from replicon cell lines. It was therefore proposed that ISG15 could be used as a therapeutic tool for combined therapy with IFN treatment against HCV infection (25). However, directly feeding or injecting animals with ISG15 would not be cost-effective in veterinary medicine. We have attempted to use recombinant PRRSV as a vector to express swine ISG15 (swISG15), but the swISG15 gene was rapidly deleted from the virus. Upon passage 3 in MARC-145 cells, almost 80% of the recombinant virus had not retained the swISG15 gene (data not shown). From an evolutionary point of view, it is not surprising that sequences that do not favor viral replication are rapidly lost from the genome. A better strategy for in vivo delivery of ISG15 needs to be developed.
Another approach could be to develop a modified live virus vaccine candidate by engineering targeted mutations and/or deletions that affect immune antagonist functions. In this study, we explored the possibility of removing (or reducing) the nsp2-mediated immune antagonist function through genetic modification of the nsp2 N-terminal domain region using reverse genetics. This approach has been used for the development of modified live virus vaccines in other viruses, such as influenza and bovine respiratory syncytial viruses (5, 41, 42, 48, 51, 55). In the present study, the mutation and/or deletion in the nsp2 N-terminal domain region generated two viable recombinant viruses (vSD-CD19 and vSD-CD19+1S). vSD-CD19 grew to a peak titer slightly lower than that of the parental virus, while vSD-CD19+1S was seriously impaired in cell culture. vSD-CD19+1S was passaged 10 times in cell culture, but its titer remained low (∼1 × 103 FFU/ml in MARC-145 cells), and no visible CPE developed in infected cells. Since the PLP2-DUB domain is also responsible for proteolytic cleavage between nsp2 and nsp3 (46, 47), we further determined whether the reduced growth of the mutant virus was due to an effect on the function of PLP2 in replicase polyproteins processing. The 23-aa deletion indeed impaired the PLP2 cleavage function, which explains why no viable recombinant virus was recovered for this mutant in the reverse genetic system. In contrast, the 19-aa deletion and S462A mutation did not show a direct effect on cleavage of the nsp2/3 site. This suggests that impaired replication of these recombinant viruses may not be directly caused by an effect on PLP2 cleavage but may rather be due to an impaired ability to suppress host cell innate immune responses. Still, we cannot exclude the possibility that the mutations tested may affect other replicative functions of the PLP2 domain, which could have an indirect effect on viral replication.
We have attempted to determine whether infection with nsp2 mutant viruses could result in improved cellular innate immune responses compared to an infection with the wild-type virus. In our previous study (49), the recombinant virus with a single S462A mutation showed a 1.5- to 2-fold reduction in the ability of nsp2 to suppress NF-κB activation, but this reduction was not as significant as we hoped. In the present study, we employed a 19-aa region at the N-terminal border of the PLP2-DUB core domain that was known to be dispensable for viral replication (20) and investigated whether combining this deletion with the S462A mutation might produce a synergistic effect in reducing the innate immune antagonist function of PLP2-DUB. Since it is uncertain whether MARC-145 cells have an intact ISGylation system (Fig. 3), we also tested these viruses in porcine alveolar macrophages. However, in order to stimulate a measurable immune response, a high virus dose is required for the initial infection, and unfortunately vSD-CD19+1S grew to an extremely low titer in macrophages (∼102 FFU/ml). For the moment, this prevents us from obtaining solid data and making an acceptable comparison with the wild-type control in terms of ISG15 expression and ISGylation levels, although several methods to increase the virus yield are still under development in our laboratory.
In the present study, we further compared the influence of wild-type and mutant PLP2-DUB on the cellular IFN-β and ISG15/ISGylation response. In particular, the CD19+1S mutant showed a reduced ability to suppress cellular innate immune responses, as was evident from increased luciferase expression levels in IFN-β promoter/luciferase reporter assays, and also from increased levels of ISG15 and ISG15-conjugated proteins in in vitro ISG15 expression and ISGylation assays. These results suggest that virus-specific immunity could be enhanced by modifying certain regions within or upstream of the PLP2-DUB domain. Follow-up studies are required to determine whether such recombinant viruses could enhance the PRRSV-specific immune response in animals and may therefore be appropriate for use in vaccine development. This study represents a step forward in elucidating the role of nsp2 in PRRS pathogenesis. A better understanding of PRRSV-host interactions and innate immune mechanisms is essential for the future development of novel antiviral therapies.
ACKNOWLEDGMENTS
We thank Ali Tas (Leiden University Medical Center) for technical assistance and Dong-Er Zhang (University of California San Diego, San Diego, CA) for helpful discussion and suggestions. Special thanks to Adolfo Garcia-Sastre (Mount Sinai School of Medicine, New York, NY) for providing the pCAGGS plasmid, Dong-Er Zhang for providing the plasmids pCAGGS-HA-UbE1L and p3xFLAG-UbcH8, Takashi Fujita (Kyoto University, Kyoto, Japan) for providing the p125-Luc plasmid, and J. M. Huibregtse (The University of Texas at Austin) for providing the pcDNA-TAP-HA-HERC5 plasmid.
This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (to Y. Fang, grant number 2007-01745).
Footnotes
Published ahead of print 18 January 2012
REFERENCES
- 1. Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. 2011. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U. S. A. 108:2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bautista EM, Meulenberg JJ, Choi CS, Molitor TW. 1996. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 141:1357–1365 [DOI] [PubMed] [Google Scholar]
- 3. Beura LK, et al. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1β modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 84:1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blomstrom DC, Fahey D, Kutny R, Korant BD, Knight E., Jr 1986. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J. Biol. Chem. 261:8811–8816 [PubMed] [Google Scholar]
- 5. Bossert B, Marozin S, Conzelmann KK. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Z, et al. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Z, et al. 2010. Immunodominant epitopes in NSP2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J. Gen. Virol. 91:1047–1057 [DOI] [PubMed] [Google Scholar]
- 8. Clementz MA, et al. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 281:4334–4338 [DOI] [PubMed] [Google Scholar]
- 10. D'Cunha J, Knight E, Jr., Haas AL, Truitt RL, Borden EC. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. U. S. A. 93:211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. D'Cunha J, et al. 1996. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J. Immunol. 157:4100–4108 [PubMed] [Google Scholar]
- 12. Durfee LA, Lyon N, Seo K, Huibregtse JM. 2010. The ISG15 Conjugation system broadly targets newly synthesized protein: implications for the antiviral function of ISG15. Mol. Cell 38:722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang Y, et al. 2006. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: expression of green fluorescent protein in the Nsp2 region. J. Virol. 80:11447–11455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 154:61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Firth AE, et al. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J. Gen. Virol. 92:1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flint SJ, Enquist LW, Racaniello VR, Skalka AM. 2009. The infectious cycle, p. 25–49 In Principles of virology, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 17. Frias-Staheli N, et al. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 4:e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han J, Liu G, Wang Y, Faaberg KS. 2007. Identification of nonessential regions of the NSP2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 81:9878–9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han J, Rutherford MS, Faaberg KS. 2009. The porcine reproductive and respiratory syndrome virus NSP2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 83:9449–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsiao NW, et al. 2010. ISG15 over-expression inhibits replication of the Japanese encephalitis virus in human medulloblastoma cells. Antiviral Res. 85:504–511 [DOI] [PubMed] [Google Scholar]
- 23. James TW, et al. 2011. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc. Natl. Acad. Sci. U. S. A. 108:2222–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 92:1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim MJ, Yoo JY. 2010. Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J. Immunol. 185:4311–4318 [DOI] [PubMed] [Google Scholar]
- 26. Kroese MV, et al. 2008. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 89:494–499 [DOI] [PubMed] [Google Scholar]
- 27. Lenschow DJ, et al. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lenschow DJ, et al. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 104:1371–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu M, Li XL, Hassel BA. 2003. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J. Biol. Chem. 278:1594–1602 [DOI] [PubMed] [Google Scholar]
- 30. Makarova KS, Aravind L, Koonin EV. 2000. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25:50–52 [DOI] [PubMed] [Google Scholar]
- 31. Malakhova O, Malakhov M, Hetherington C, Zhang DE. 2002. Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J. Biol. Chem. 277:14703–14711 [DOI] [PubMed] [Google Scholar]
- 32. Meulenberg JJ, Bende RJ, Pol JM, Wensvoort G, Moormann RJ. 1995. Nucleocapsid protein N of Lelystad virus: expression by recombinant baculovirus, immunological properties, and suitability for detection of serum antibodies. Clin. Diagn. Lab. Immunol. 2:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meulenberg JJ, Petersen-den Besten A. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 225:44–51 [DOI] [PubMed] [Google Scholar]
- 34. Meulenberg JJ, et al. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller LC, Laegreid WW, Bono JL, Chitko-McKown CG, Fox JM. 2004. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 149:2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okumura A, Lu G, Pitha-Rowe I, Pitha PM. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U. S. A. 103:1440–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okumura A, Pitha PM, Harty RN. 2008. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U. S. A. 105:3974–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pitha-Rowe I, Hassel BA, Dmitrovsky E. 2004. Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J. Biol. Chem. 279:18178–18187 [DOI] [PubMed] [Google Scholar]
- 39. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 40. Rapp F. 1964. Plaque differentiation and replication of virulent and attenuated strains of measles virus. J. Bacteriol. 88:1448–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Richt JA, Garcia-Sastre A. 2009. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top. Microbiol. Immunol. 333:177–195 [DOI] [PubMed] [Google Scholar]
- 42. Richt JA, et al. 2006. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J. Virol. 80:11009–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ropp SL, et al. 2004. Characterization of emerging European-like porcine reproductive respiratory syndrome virus isolates in the United States. J. Virol. 78:3684–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Snijder EJ, Meulenberg JJ. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79(Pt. 5):961–979 [DOI] [PubMed] [Google Scholar]
- 46. Snijder EJ, Wassenaar AL, Spaan WJ. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Snijder EJ, Wassenaar AL, Spaan WJ, Gorbalenya AE. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 270:16671–16676 [DOI] [PubMed] [Google Scholar]
- 48. Steel J, et al. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun Z, Chen Z, Lawson SR, Fang Y. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 84:7832–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takeuchi T, Inoue S, Yokosawa H. 2006. Identification and Herc5-mediated ISGylation of novel target proteins. Biochem. Biophys. Res. Commun. 348:473–477 [DOI] [PubMed] [Google Scholar]
- 51. Valarcher JF, et al. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Kasteren PB, et al. 2012. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 86:773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Nieuwstadt AP, et al. 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70:4767–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wong JJ, Pung YF, Sze NS, Chin KC. 2006. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. U. S. A. 103:10735–10740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wressnigg N, et al. 2009. Influenza B mutant viruses with truncated NS1 proteins grow efficiently in Vero cells and are immunogenic in mice. J. Gen. Virol. 90:366–374 [DOI] [PubMed] [Google Scholar]
- 56. Yie J, Senger K, Thanos D. 1999. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc. Natl. Acad. Sci. U. S. A. 96:13108–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoneyama M, et al. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3). J. Biochem. 120:160–169 [DOI] [PubMed] [Google Scholar]
- 58. Yuan W, Krug RM. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zeman D, et al. 1993. Laboratory investigation of PRRS virus infection in three swine herds. J. Vet. Diagn. Invest. 5:522–528 [DOI] [PubMed] [Google Scholar]
- 60. Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 31:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao C, et al. 2004. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. U. S. A. 101:7578–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]