Abstract
Previous studies characterized two types of replication-defective human immunodeficiency virus type 1 (HIV-1) integrase mutants: class I, which are specifically blocked at the integration step, and class II, which harbor additional virion production and/or reverse transcription defects. Class I mutant enzymes supported little if any metal ion-dependent 3′-processing and DNA strand transfer activities in vitro, whereas class II enzymes displayed partial or full catalytic function in studies with simplified assay designs, suggesting that defective interaction(s) with heterologous integrase binding proteins might underlie the class II mutant viral phenotype. To address this hypothesis, class I and II mutant enzymes were interrogated under expanded sets of in vitro conditions. The majority failed to catalyze the concerted integration of two viral DNA ends into target DNA, highlighting defective integrase function as the root cause of most class II in addition to all class I mutant virus infection defects. One mutant protein, K264E, in contrast, could support the wild-type level of concerted integration activity. After accounting for its inherent reverse transcription defect, HIV-1K264E moreover formed preintegration complexes that supported the efficient integration of endogenous viral DNA in vitro and normal levels and sequences of 2-long terminal repeat-containing circle junctions during acute infection. K264E integrase furthermore efficiently interacted in vitro with two heterologous binding partners, LEDGF/p75 and reverse transcriptase. Our results underscore the physiological relevance of concerted integration assays for tests of integrase mutant function and suggest that the K264E mutation disrupts an interaction with an intranuclear integrase binding partner that is important for HIV-1 integration.
INTRODUCTION
Key players in retroviral integration are the viral integrase (IN) protein, a component of virion particles, and the ends of the linear viral DNA (vDNA) made by reverse transcription. IN functions in the context of the preintegration complex (PIC), a large nucleoprotein complex (7, 88) that can support vDNA integration into heterologous target DNA in vitro (10, 42, 45). IN initially processes the vDNA ends adjacent to conserved CA 3′ dinucleotides, which liberates a pGpTOH dinucleotide from each end of human immunodeficiency virus type 1 (HIV-1) (94). IN then uses the processed 3′ vDNA termini to cut opposing strands of target DNA in a concerted fashion, which joins the vDNA ends to chromosomal 5′ phosphates (11, 41, 48). The resulting gapped recombination intermediate, with unjoined vDNA 5′ ends, is repaired by host cell machinery to generate the integrated provirus (see reference 33 for a recent overview of retroviral integration).
Numerous cellular factors can interact with HIV-1 IN (see references 114 and 118 for reviews), and one in particular, lens epithelium-derived growth factor (LEDGF)/p75, has been shown to play an important role during HIV-1 infection (reviewed in references 35 and 97). IN is composed of three evolutionarily conserved domains interconnected by flexible linkers. Conserved His and Cys residues within the N-terminal domain (NTD) bind zinc, whereas invariant Asp and Glu residues within the catalytic core domain (CCD) comprise the D,D-35-E active site, which coordinates Mg2+ ions for catalysis (reviewed in references 33 and 72). The C-terminal domain (CTD) can engage different ligands, including DNA (39, 120, 125), reverse transcriptase (RT) (55, 123, 130), and the SIP1/Gemin2 host factor important for reverse transcription (50, 90), whereas LEDGF/p75 binding determinants are comprised by the NTD and CCD (53, 81).
Results of biochemical experiments have indicated that a tetramer of HIV-1 IN catalyzes the concerted integration of two vDNA ends (46, 51, 71, 92), findings that are consistent with recent X-ray crystal structures of functional prototype foamy virus (PFV) IN-vDNA complexes or intasomes (52, 82). PFV IN efficiently integrates relatively short, oligonucleotide vDNA substrates into target DNA in a concerted fashion in vitro (115), whereas similar reaction conditions support aberrant integration of single vDNA ends by HIV-1 IN (13, 53). Various parameters, including the conditions under which IN is purified (108), length of vDNA substrate (70), and binding ligands, such as LEDGF/p75 (53, 93), can increase the yield of concerted HIV-1 integration products in vitro.
Studies that initially mutagenized the IN coding region of pol revealed the importance of the viral recombinase for HIV-1 replication (66, 106, 109). From this start, it was evident some mutations affected more than just integration. For example, a CCD linker insertion mutant failed to release virus from transfected cells (109). Subsequent work confirmed virus assembly/virion morphology defects for some HIV-1 IN mutants (2, 12, 38, 43, 60, 63, 67, 73, 77, 78, 89, 96, 100, 107, 111, 126), whereas other mutants displayed defects in core uncoating (9), reverse transcription (3, 15, 40, 67, 75–78, 80, 84, 89, 102, 104, 107, 113, 124, 126, 130), or PIC nuclear import (3, 57, 59). In contrast, other mutations, typified by changes of D,D-35-E active site residues, solely blocked the integration step (2, 38, 61, 66, 67, 79, 84, 105, 111, 124). We previously defined two phenotypic classes of replication-defective HIV-1 IN mutant viruses to distinguish those specifically blocked at integration (class I) from those that behave pleiotropically (class II) (31). Due to the specific block, a hallmark of class I IN mutant viral infection is transient accumulation of unintegrated nuclear vDNA (2, 38, 61, 67, 79, 84, 105, 124), akin to the phenotype observed for wild-type (WT) HIV-1 in the presence of IN strand transfer inhibitors (INSTIs) (54). Reverse transcription is the most common pleiotropic defect associated with class II IN mutant viruses (31, 32).
Central to understanding the mechanism of IN mutant virus behavior is analysis of enzyme catalytic function, and recombinant INs derived from class I and II mutant viruses have accordingly been analyzed for integration activities in vitro. A separate IN activity, disintegration (26), which is unlikely to be relevant during virus infection, has nonetheless been shown to be an important parameter because it, unlike 3′ processing and DNA strand transfer, can be catalyzed by the isolated CCD (14, 65). Certain amino acid substitutions of active site D,D-35-E residues ablate 3′-processing, DNA strand transfer, and disintegration activities (37, 68, 117), highlighting the requirement for divalent metal ion coordination by the CCD for IN catalysis. In contrast, enzymes derived from class II mutant viruses support 3′-processing and DNA strand transfer activities, some even at the level of WT IN (INWT) (15, 37, 40, 60, 63, 77, 78, 80, 98, 99, 102, 117). The infectivity defect of IN mutant viruses can be rescued by supplying INWT in trans as a Vpr fusion protein (47, 127), and class II IN mutant fusion proteins, moreover, effectively trans-complement class I IN mutant virus infection defects (6, 47, 77, 78, 80). Taken together, these results highlight the catalytic competence of class II IN mutant proteins and, by extension, that defects in auxiliary functions, for example interaction(s) with critical protein binding partners, might underlie the observed infection defects (6, 15, 29, 49, 59, 78, 128, 130). To address this hypothesis, 12 recombinant IN proteins derived from class II mutant viruses were analyzed alongside class I mutant control proteins IND64N and INW235E in a variety of assays, including the most stringent test for IN function in vitro, the concerted integration assay. Our findings revealed that defective IN function underlies the majority of class II mutant virus infection defects and moreover highlight K264E as a mutation that potentially disrupts an interaction with a heterologous factor important for integration.
MATERIALS AND METHODS
Plasmid DNAs.
C-terminally His6-tagged IN proteins derived from clade B strain HIV-1NL4-3 were expressed from bacterial expression vector pKBIN6Hthr (86). Mutations were introduced by PCR using Pfu Ultra DNA polymerase (Agilent Technologies, Inc., Santa Clara, CA), and sequences of IN mutant expression vectors were verified by Sanger sequencing. The plasmid pGST-RT, which encodes a fusion between glutathione S-transferase (GST) and the p66 subunit of RT (91), was a kind gift from Jeroen van Wamel and Ben Berkhout, University of Amsterdam.
Plasmids encoding full-length HIV-1NL4-3 and single-round luciferase reporter derivatives of WT and IN mutants K215A/K219A, D64N/D116N (78), D167K (102), and K264E (77) were previously described.
Protein expression and purification.
Escherichia coli strain BL21(DE3) or its PC2 derivative (21) transformed with IN expression constructs was grown for 16 h at 30°C. The next day, bacteria subcultured at 1:10 in 2 liters of LB broth containing 100 μg/ml ampicillin were grown at 30°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8, at which time IN expression was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested after 4 h of induction at 28°C. The bacterial pellet was resuspended in ice-cold buffer A {25 mM Tris-HCl (pH 7.4), 1 M NaCl, 7.5 mM 3-[(3-cholamidopropyl) dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPS)]} containing 25 mM imidazole–0.5 mM phenylmethanesulfonylfluoride (PMSF) and sonicated. After centrifugation for 15 min at 60,000 × g, the supernatant was incubated with 1 ml of buffer A–25 mM imidazole–equilibrated Ni2+-nitrilotriacetic acid (Ni-NTA)–agarose beads (Qiagen, Valencia, CA) at 4°C for 1 h. The beads were washed twice with 20 volumes of buffer A–25 mM imidazole, followed by 30 volumes of buffer A–35 mM imidazole. IN was eluted with buffer A–200 mM imidazole. IN-containing fractions diluted with 3 volumes of 25 mM Tris-HCl (pH 7.4)–7.5 mM CHAPS were injected into a 5-ml HiTrap heparin column (GE Healthcare, Piscataway, NJ), and bound proteins were eluted with a linear gradient of 0.25 M to 1 M NaCl in 25 mM Tris-HCl (pH 7.4)–7.5 mM CHAPS using an AKTA 10 purifier. Immediately after elution, 10 mM dithiothreitol (DTT) was added to each fraction, and the NaCl concentration was adjusted to 1 M. The His tag was removed from IN by using 40 U of thrombin (Sigma-Aldrich, St. Louis, MO) per mg of protein for 3 h at room temperature, which left the heterologous LVPR sequence at each C terminus. After removal of thrombin by incubation with Benzamidine beads (Novagen, Madison, WI) and ultrafiltration using 9-kDa molecular weight cutoff Pierce concentrators (Thermo Fisher Scientific, Waltham, MA), IN was dialyzed overnight against buffer D (25 mM Tris-HCl [pH 7.4], 1 M NaCl, 7.5 mM CHAPS) containing 10% glycerol (wt/vol)–10 mM DTT. Protein concentration was determined by spectrophotometry, and aliquots flash-frozen in liquid N2 were stored at −80°C. Quantitative image analysis (FluorChem FC2; Alpha Innotech, San Leandro, CA) of Coomassie-stained gels revealed each IN preparation to be minimally 90% pure.
E. coli strain BL21 transformed with pGST-RT grown overnight at 37°C in LB–100 μl/ml ampicillin was diluted 1:10 the next day into 2 liters, and IPTG was added to a final concentration of 1 mM after reaching an OD600 of 0.8. After 3 h at 37°C, harvested cells resuspended in 50 ml of buffer B (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM PMSF, 5 mM DTT) were sonicated at 50 W using 10-s pulses followed by 20 s of cooling over 10 min. The supernatant recovered after centrifugation for 15 min at 60,000 × g was incubated with 2 ml of glutathione-Sepharose 4 Fast Flow (GE Healthcare) at room temperature for 2 h, after which the beads were washed with 500 ml of phosphate-buffered saline containing 5 mM DTT. After washing with 20 ml of buffer B, protein was eluted with 20 ml of buffer E (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 5 mM DTT, 20 mM reduced glutathione, Complete protease inhibitor cocktail [Roche Applied Science, Indianapolis, IN]) at room temperature. Protein concentration was determined by spectrophotometry following concentration and dialysis against buffer C (20 mM Tris-HCl [pH 7.4], 120 mM NaCl, 1 mM EDTA, 10% glycerol, 5 mM DTT, Complete protease inhibitor cocktail). Aliquots flash-frozen in liquid N2 were stored at −80°C.
Recombinant IND64N (27), LEDGF/p75 (116), and GST fused to the C-terminal domain of LEDGF/p75 (GST-LEDGF326-530) (23) proteins expressed in bacteria were purified as previously described.
DNA substrates.
Substrates that mimicked the HIV-1 U5 vDNA end were used to measure IN 3′-processing, DNA strand transfer, and disintegration activities. To prepare the 21-mer processing substrate, the 3′ recess in annealed AE117 (5′-ACTGCTAGAGATTTTCCACAC)/AE150 (5′-GTGTGGAAAATCTCTAGCAG) was filled in with [α-32P]TTP (3,000 Ci/mmol; PerkinElmer, Waltham, MA) using Sequenase version 2.0 T7 DNA polymerase (GE Healthcare) (41). The 30-mer substrate was similarly prepared from annealed AE143 (5′-ACTGCTAGAGATTTTCCACACTGACTAAAA)/AE191 (5′-TTTTAGTCAGTGTGGAAAATCTCTAGCAG) DNA. To prepare the preprocessed duplex for DNA strand transfer, AE155 (5′-TTTTAGTCAGTGTGGAAAATCTCTAGCA) labeled with [γ-32P]ATP (3,000 Ci/mmol; PerkinElmer) using T4 polynucleotide kinase (GE Healthcare) was annealed with AE143. The branched Y-mer disintegration substrate was prepared by annealing 5′-end-labeled AE157 (5′-GAAAGCGACCGCGCC) with equimolar amounts of AE146 (5′-GGACGCCATAGCCCCGGCGCGGTCGCTTTC), AE117 (5′-ACTGCTAGAGATTTTCCACAC), and AE156 (5′-GTGTGGAAAATCTCTAGCAGGGGCTATGGCGTCC). For each substrate, unincorporated radionuclide was removed by passing the labeled duplexes through Bio-Spin 6 columns (Bio-Rad, Richmond, CA) equilibrated with 10 mM Tris-HCl (pH 8.0)–20 mM NaCl–0.1 mM EDTA.
Concerted integration of two vDNA ends into target DNA requires relatively long substrate DNA in the absence of LEDGF/p75 (53, 70). A 517-bp substrate was accordingly prepared from plasmid pU3U5 (25) following digestion with ScaI and BclI. The gel-purified DNA after 5′-end labeling was separated from unincorporated radionuclide by spin column chromatography. AE3653 (5′-CCTTTTAGTCAGTGTGGAAAATCTCTAGCA) annealed to AE3652 (5′-ACTGCTAGAGATTTTCCACACTGACTAAAAGG) yielded the preprocessed 32-mer U5 end for LEDGF/p75-dependent integration assays (27, 53).
Recombinant IN activity assays.
The 3′-processing reaction mixture (20 μl) contained 25 mM morpholinepropanesulfonic acid (pH 7.2), 50 mM NaCl, 10 mM MgCl2 or MnCl2, 5 μM ZnSO4, 10 mM DTT, 0.5 μM IN, and 5 nM labeled DNA. Reaction mixtures were incubated at 37°C for 1 h, and reactions were stopped by addition of an equal volume of sequencing gel sample buffer (95% formamide, 10 mM EDTA, 0.003% xylene cyanol, 0.003% bromophenol blue) and boiling for 2 min prior to fractionation through denaturing 20% polyacrylamide gels. Products were visualized using a Storm 820 PhosphorImager and quantified using ImageQuant version 1.2 (GE Healthcare). Radiolabeled vDNA strand transfer and disintegration reactions were conducted under the same conditions, except that products were fractionated through 15% sequencing gels and disintegration activity was assessed only in the presence of MnCl2.
Concerted integration reaction mixtures (100 μl) contained 20 mM HEPES (pH 7.0), 100 mM NaCl, 10 mM MgCl2, 25 μM ZnCl2, 5 mM DTT, 10% polyethylene glycol 6000 (PEG 6000), 10% dimethyl sulfoxide (DMSO), 25 to 100 nM IN, 3.0 nM labeled 517-bp vDNA, and 1.5 nM pGEM-3 plasmid DNA. Reaction mixtures were incubated at 37°C for 2 h, and reactions were stopped by addition of 25 mM EDTA–0.5% Na dodecyl sulfate (SDS). Products deproteinized by digestion with proteinase K and precipitated with ethanol were analyzed by electrophoresis through 1.5% agarose–TAE (40 mM Tris base, 20 mM acetate, 1 mM EDTA) gels containing 0.1% SDS. Products, visualized in dried gels, were quantified using ImageQuant version 1.2.
LEDGF/p75-dependent integration reactions (36-μl volume) were begun by mixing 0.55 μM vDNA with 0.3 μg pGEM-3 in 35.4 mM NaCl, 5.5 mM MgSO4, 11 mM DTT, 4.4 μM ZnCl2, and 22 mM HEPES-NaOH (pH 7.4). IN (2 μl) in dilution buffer (750 mM NaCl, 10 mM DTT, 25 mM Tris-HCl [pH 7.4]) was added, and after 15 min at room temperature LEDGF/p75 (2.0 μl) was added. Reactions (mixtures for which contained final IN and LEDGF/p75 concentrations of 0.8 μM and 0.6 μM, respectively) after 1 h at 37°C were stopped by addition of EDTA and SDS to final concentrations of 25 mM and 0.5%, respectively, and deproteinization with 30 μg proteinase K (Roche Applied Sciences) for 1 h at 37°C. DNAs recovered following precipitation with ethanol and separation on 1.5% agarose–TAE gels were stained with ethidium bromide (EtBr; 0.5 μg/ml). DNA strand transfer activity was quantified using Alpha Innotech FluorChem FC2 software or ImageQuant version 1.2 for assays that included radiolabeled vDNA.
LEDGF/p75-IN binding assays.
Capture of soluble IN (1 μg; 300 nM) by GST-LEDGF326-530 (0.5 μg; 100 nM) or control GST (0.26 μg; 100 nM) prebound to glutathione-Sepharose beads (10 μl) was performed essentially as previously described (23, 102) in 100 μl PD buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 5 mM DTT, 0.1% NP-40) for 2 h at 4°C; 5 μg of bovine serum albumin (BSA) was included as an additional specificity control. Beads collected by settling for 20 min in the absence of centrifugation were washed with 700 μl PD buffer 4 times, with intermittent 20-min periods for settling. Proteins eluted with SDS gel sample buffer by boiling were fractionated through Novex bis-Tris–10% polyacrylamide gels. IN levels in gels stained with Sypro orange (Invitrogen Corp., Carlsbad, CA) were quantified by using Alpha Innotech FluorChem FC2.
Ni-NTA beads (20 μl, settled volume) prewashed with PDA buffer (25 mM bis-Tris [pH 6.85], 150 mM NaCl, 2 mM MgCl2, 25 mM imidazole, 0.1% NP-40, Complete protease inhibitor cocktail) were added to reaction mixtures (100 μl) containing 5.8 μg LEDGF/p75 (0.95 μM), 0.7 μg IN-His6 (0.2 μM), and 5 μg BSA. Following incubation at 4°C for 4 h with gentle agitation, beads washed four times with PDA buffer without centrifugation were mixed with 2× SDS gel sample buffer containing 200 mM imidazole and boiled, and resulting supernatants were fractionated through Novex bis-Tris–10% polyacrylamide gels. LEDGF/p75 levels in gels stained with Sypro orange were quantified by using Alpha Innotech FluorChem FC2.
RT-IN binding assay.
IN (4.8 μl of a 2.9-μg/ml stock in 200 μl buffer D) was precleared by centrifugation as follows, to reduce nonspecific aggregation during GST-RT binding assays. After incubation for 15 min on ice and spinning at 15,000 × g for 15 min at 4°C, the concentration of IN in the supernatant was determined by FluorChem FC2 imaging of Sypro orange-stained SDS-polyacrylamide gels. GST-RT pulldown of precleared IN was performed as described above for GST-LEDGF326-530, except that 15 μg of GST-RT (1.7 μM) was used in place of the LEDGF fusion protein, IN was used at 3 μg (0.9 μM), and control reaction mixtures contained 1 μM GST protein.
IN cross-linking.
Protein cross-linking was performed as previously described (64).
Cells, viruses, and infections.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented to contain 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin, while SupT1 CD4+ T cells were grown in RPMI 1640 medium modified to contain the same supplements. WT and IN mutant HIV-1NL4-3 were produced by transfecting HEK293T cells with full-length molecular clones, whereas single-round derivatives carrying the luciferase reporter gene were constructed by cotransfection with a vesicular stomatitis virus glycoprotein G (VSV-G) expression vector (78). Levels of virus production from transfected HEK293T cells were assessed in an exogenous RT assay (38, 78). Infectivities were quantified as levels of luciferase enzyme relative light units (RLU), normalized to the total protein concentration in the cell extracts (78, 85).
Viral DNA analyses.
PICs extracted from SupT1 cells (4 × 107) at 7 h after acute infection with 15 ml of HEK293T cell supernatant were reacted with pTZ18U/PL target DNA, and purified DNAs were analyzed in duplicate by quantitative real-time PCR (Q-PCR) to determine levels of strand transfer reaction products as described previously (42, 80). In brief, background values obtained from parallel Q-PCRs that omitted pTZ18U/PL-specific primers were subtracted from the samples that contained them. IN mutant activity was ascribed alongside an endpoint diluted HIV-1WT standard curve, and specific strand transfer activity was in turn normalized to the level of vDNA substrate in each reaction mixture, determined by parallel Q-PCR measures of late reverse transcription (LRT) products (42, 80).
To monitor the formation of vDNA species during acute infection, SupT1 cells (4 × 106) spinoculated with 2 × 107 RT-counts per minute of VSV-G-pseudotyped WT or IN mutant reporter viruses for 2 h at room temperature were lysed at multiple time points postinfection, and purified DNAs were analyzed in duplicate by Q-PCR to determine levels of LRT, 2-long terminal repeat (LTR)-containing circles, and integration products as previously described (42, 85).
WT and IN mutant 2-LTR circle junctions (CJs) amplified by nested PCR were cloned and sequenced essentially as previously described (83). In brief, 1 μg of cellular DNA prepared 24 h after infection was amplified (50 μl) in triplicate by using Pfu Ultra DNA polymerase using the primer pair AE4395 (5′-GCACCATCCAAAGGTCAGTGGATATCTG)/AE4450 (5′-GCCTGGGAGCTCTCTGGCTAA). After 5 min at 95°C, reactions were cycled 35 times at 95°C (15 s), 50°C (1 min), and 72°C (30 s), followed by a final 5-min extension at 72°C. Resulting DNAs (5 μl) were amplified by nested PCR (50 μl) in triplicate using the primers AE2948 (5′-AACTAGGGAACCCACTGCTTAAG)/AE4394 (5′-GTGTGTGGTAGATCCACAGATCAAGG) under first-round reaction conditions. Pooled DNAs concentrated by precipitation with ethanol and purified by using a SNAP UV-free gel purification kit (Invitrogen Corp.) were cloned using the Zero Blunt TOPO kit (Invitrogen Corp.). Plasmid DNA sequences obtained from the M13 reverse primer were parsed for LTR sequence content and aligned by eye.
RESULTS
Experimental strategy.
HIV-1 IN proteins derived from class II mutant viruses can efficiently trans-complement the infectivity defects of class I mutant viruses and support appreciable levels of enzyme activity in vitro, suggesting that some if not most class II viral infection defects may lie in the inability of the mutant IN to properly engage host cell cofactors (6, 15, 29, 49, 59, 78, 128, 130). Our initial goal was to test this hypothesis by systematically evaluating representative class II IN mutant protein activities under a variety of conditions that included the concerted integration of two vDNA ends, a relatively underutilized stringent measure of HIV-1 IN activity (51, 70).
Missense mutations throughout the IN coding region can elicit the class II mutant viral phenotype (31, 32), and 12 representative proteins that we and others had previously analyzed under simplified integration reaction conditions were selected for study. The mutant INs harbored amino acid substitutions in the CCD (E69R, V165A, R166A, D167K, Q168A, K186Q, and R199A), the CTD (R228A, K258A, and K264E), or the CCD-CTD interdomain linker (Q214L/Q216L and K215A/K219A) (72) (Table 1). Glu69, Val165, Arg166, Asp167, and Gln168 in particular help to form the LEDGF/p75 binding pocket at the CCD-CCD dimer interface (15, 22, 102), whereas Lys186 forms an intermolecular salt bridge with NTD residue Glu11, which is important for IN tetramerization (5, 51, 86). Arg199, Lys219, Arg228, and Lys264 contribute to vDNA binding (64, 129), while Lys258 forms part of the CTD binding interface with RT (123). The interdomain linker mutations, which target part of a putative bipartite nuclear localization signal (49), yield variable levels of nuclear import defects (3, 78, 95), with more robust defects observed when the mutations are combined with additional IN changes in some (49, 59), but not all, (95) studies. The reported single-round infectivities of the corresponding mutant viruses varied from undetectable (<0.01%) to approximately 6% of WT for HIV-1D167K (Table 1). Two proteins derived from class I IN mutant viruses were used as controls: IND64N, harboring a conservative change of active site residue Asp64 (27, 37), and INW235E, with a change of a conserved CTD residue (67, 68). The IN proteins, expressed in E. coli, were purified to ≥90% homogeneity by using Ni-NTA and cation exchange affinity chromatography methods.
Table 1.
HIV-1 IN mutants included in this study
| Change(s) | Classa | Proposed function(s) | Infectivityb | Reference(s) |
|---|---|---|---|---|
| D64N | I | Active site | 0.06 (0.03)c | 77 |
| E69R | II | LEDGF binding | <0.01 | 102 |
| V165A | II | LEDGF binding | <0.01 | 102, 114 |
| R166A | II | LEDGF binding | <0.01 | 102 |
| D167K | II | LEDGF binding | 5.7 (0.1) | 102 |
| Q168A | II | LEDGF binding | 0.8 (0.1) | 102 |
| K186Q | II | Multimerization | 0.23 (0.28) | 78 |
| R199A | II | DNA binding | 0.24 (0.13) | 129 |
| Q214L/Q216L | II | Nuclear import | 0.11 (0.05) | 78 |
| K215A/K219A | II | Nuclear import, DNA binding | 0.02 (0.02) | 78, 129 |
| R228A | II | DNA binding | 0.18 (0.25) | 77 |
| W235E | I | Multimerization | 0.02 (0.01) | 77 |
| K258A | II | RT binding | 0.09 (0.01) | 77 |
| K264E | II | DNA binding | 0.11 (0.00) | 77 |
Phenotypic replication defect class.
Percent single-round luciferase reporter virus activity relative to WT HIV-1NL4-3, with the standard deviation shown in parentheses.
Value is for the double active site mutant HIV-1D64N/D116N.
3′ processing of vDNA ends.
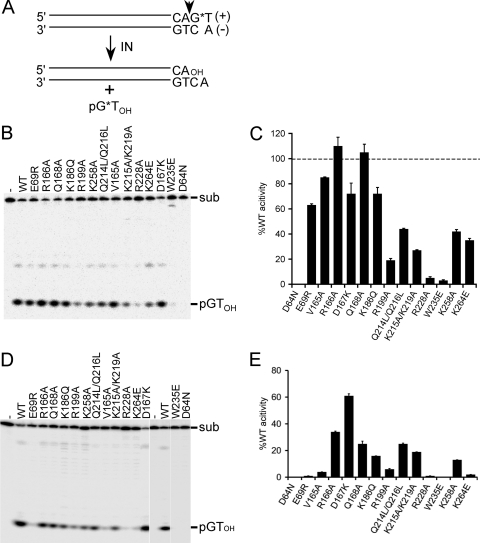
The 3′-processing substrates, which modeled the HIV-1 U5 vDNA end, were labeled by incorporating radionuclide T at the plus-strand terminus, allowing reactions to be monitored through formation of labeled pGTOH cleavage product in denaturing polyacrylamide gels (Fig. 1A and B) (41, 121). INWT activity was defined as the percent substrate converted to product, and mutant activities were quantified relative to INWT values (Fig. 1C; results are summarized in Table 2). Divalent metal ion is an essential catalytic cofactor, and because mutant activities were typically reported using Mn2+ and a 21-bp vDNA substrate (37, 40, 78, 102), these conditions were initially used to facilitate comparisons between studies. The class II IN mutant proteins supported a wide range of function under these conditions, which ranged from a low of 5% of WT activity for INR228A to a high of 110% for INR166A (Fig. 1B and C; Table 2). These results agree well with our own studies (78, 102, 129) as well as other previous work (117).
Fig 1.
3′-processing substrate design and IN activities. (A) Blunt-ended vDNA substrate, highlighting the subterminal CA 3′ dinucleotide that is conserved among all retroviruses. IN processes the indicated phosphodiester bond (vertical arrowhead), releasing labeled pGTOH dinucleotide. *, positions of 32P label. (B) Polyacrylamide gel image of Mn2+-dependent 3′-processing reactions, highlighting the 21-bp vDNA substrate (sub) and pGTOH cleavage product. IN was omitted from the reaction mixture loaded in the first lane (-). (C) Mutant 3′-processing activities of the products shown in panel B, plotted as the percent INWT function. Results are means ± standard errors of the means (SEM) for two independent experiments. (D) The same experiment as in panel B, except reaction mixtures contained MgCl2 and the vDNA substrate was 30 bp long. (E) Cumulative Mg2+-dependent 3′-processing activities expressed as the percent INWT function ± SEM for two (most mutants) to four (INW235E and IND64N) independent experiments.
Table 2.
Activities of IN mutant proteinsa
| Change(s) in protein | Relative activitya |
||||
|---|---|---|---|---|---|
| 3′ processing |
DNA strand transfer |
Disintegration | |||
| Mn2+ | Mg2+ | Mn2+ | Mg2+ | ||
| D64N | − | −b | −b | −b | − |
| E69R | 63 (1.0) | 1.0 (0.0) | 64 (1.5) | 3.0 (0.5) | 53 (5.5) |
| V165A | 85 (0.5) | 4.0 (0.0) | 150 (7.3)c | 24 (3.6) | 35 (3.0) |
| R166A | 110 (7.0) | 34 (0.5) | 136 (5.0) | 94 (17) | 56 (7.0) |
| D167K | 72 (8.5) | 61 (1.5) | 144 (4.5) | 119 (7.0) | 81 (26) |
| Q168A | 105 (6.5) | 25 (2.0) | 155 (11) | 71 (14) | 41 (12) |
| K186Q | 72 (5.0) | 16 (0.0) | 13 (6.0) | 28 (5.0) | 76 (4.0) |
| R199A | 19 (1.5) | 6.0 (0.5) | 4.0 (1.5) | 2.0 (0.0) | 329 (63) |
| Q214L/Q216L | 44 (0.5) | 25 (0.5) | 27 (4.5) | 3.0 (0.5) | 34 (9.5) |
| K215A/K219A | 27 (0.5) | 19 (0.0) | − | − | 57 (14) |
| R228A | 5.0 (1.0) | 1.0 (0.0) | 3.0 (2.5) | − | 139 (25) |
| W235E | 3.0 (0.5) | −b | 2.0 (0.5)b | −b | 41 (0.5) |
| K258A | 42 (1.5) | 13 (0.0) | 33 (2.5) | 3.0 (1.5) | 152 (8.5) |
| K264E | 35 (1.5) | 2.0 (0.0) | 37 (8.3)d | − | 49 (3.5) |
Relative to INWT, which in each case was set to 100%. Values are means and standard errors of the means (in parentheses) for 2 independent experiments unless otherwise indicated. −, undetectable (<1% of INWT activity).
Based on 4 experiments.
Based on 5 experiments.
Based on 3 experiments.
The class I IND64N mutant protein failed to support the formation of detectable reaction products, as expected (27, 37, 78). The activity of INW235E was, however, unexpectedly low (3% of INWT) (Table 2), as this enzyme was previously reported to support WT levels of Mn2+-dependent 3′-processing and DNA strand transfer activities (68). Two additional preparations of this mutant were therefore made from a sequence-reverified bacterial expression vector. Because all three preparations displayed the same activity profile, we concluded that INW235E is defective for 3′-processing and DNA strand transfer activities (see below).
The concentration of Mg2+ in the human body is several orders of magnitude greater than that of Mn2+ (74), indicating that Mg2+ is the physiologically relevant IN catalytic cofactor during HIV-1 infection. Mg2+-dependent 3′-processing activities were accordingly evaluated next. Likely due to the requirement for IN-vDNA contacts distal from the reactive vDNA end (44), somewhat longer substrates, in the range of 30 to 35 bp, support robust IN function in the presence of Mg2+ (36), and a 30-bp substrate was accordingly used here. Relative IN activities were decreased across the board under these conditions compared to prior conditions, with greater-than-10-fold differences noted for INE69R, INV165A, and INK264E (Fig. 1, compare panels D versus B and panels E versus C; Table 2). These results are consistent with previous findings that certain vDNA mutations preferentially affect Mg2+-dependent over Mn2+-dependent IN activities (44).
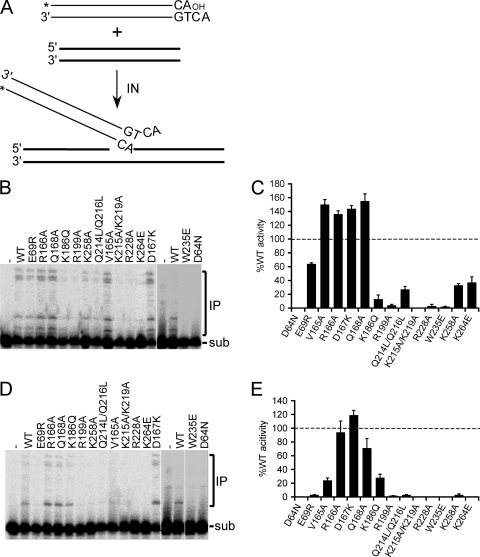
Single vDNA end strand transfer activities.
The DNA strand transfer substrate was labeled at the 5′ end of the strand that becomes joined to target DNA, such that the products of single vDNA integration were detected as a ladder of bands above the substrate in sequencing gels (Fig. 2A and B). To enable quantification of strand transfer activity independently from 3′ processing, the substrate lacked the terminal GT dinucleotide that is normally removed by hydrolysis (Fig. 1A). Mn2+-dependent DNA strand transfer activities generally mimicked relative levels of 3′-processing activities, although two of the mutants (INK186Q and INK215A/K219A) were at least 5-fold more defective at DNA joining (compare Fig. 2B and 1B; Table 2). In line with the results of 3′-processing assays, most of the class II mutant enzymes were defective for Mg2+-dependent DNA strand transfer activity: only INR166A, IND167K, and INQ168A displayed >50% relative activity, with INK186Q and INV165A harboring about 25 to 30% of INWT function (Fig. 2D and E; Table 2).
Fig 2.
DNA strand transfer assay and IN mutant activities. (A) Precleaved 30-mer vDNA substrate, indicating the position of 5′-end 32P label (*). A second oligonucleotide, which served as target acceptor DNA, is indicated by the bold lines. (B) Gel image highlighting migration positions of substrate (sub) DNA and Mn2+-dependent integration products (IP). (C) Mean activities ± standard errors of the means (SEM) for three (INK264E), four (INW235E and IND64N), five (INV165A), or two (all other mutants) independent experiments under the panel B reaction conditions, plotted as the percent INWT activity. (D) Gel image of Mg2+-dependent DNA strand transfer activities. (E) Cumulative Mg2+-dependent activities expressed as the percent INWT function ± SEM for two to four (INW235E and IND64N) independent experiments.
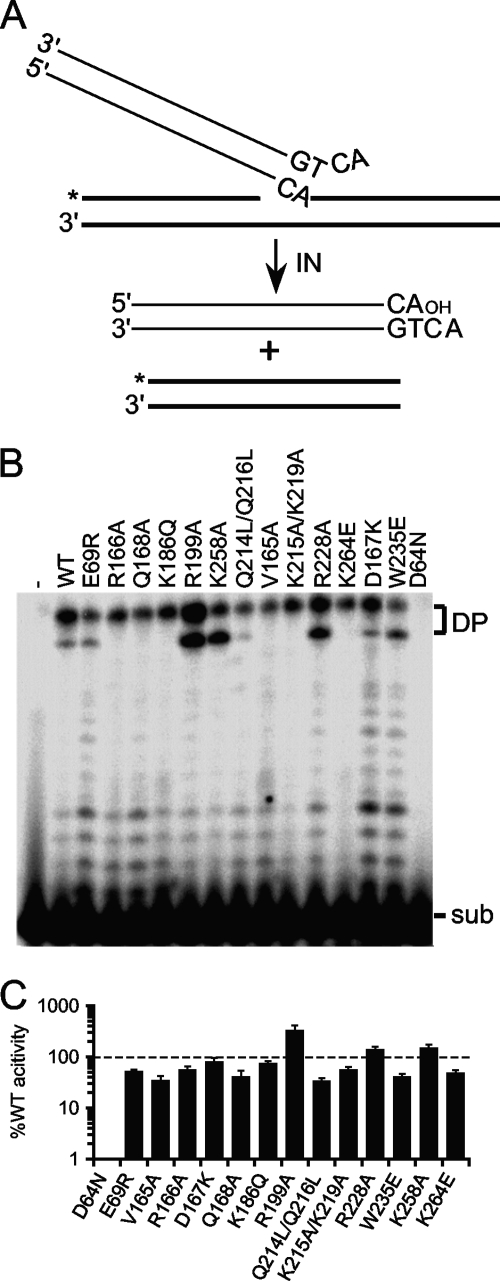
IN mutant disintegration activities.
The isolated HIV-1 CCD is sufficient to catalyze disintegration activity (14), and NTD and CTD mutant proteins defective for 3′-processing and DNA strand transfer activities can accordingly support appreciable levels of disintegration activity (37, 40, 68, 117). Because INW235E and INR228A supported only about 2 to 5% Mn2+-dependent 3′-processing and DNA strand transfer activities (Table 2), their disintegration functions were evaluated alongside the rest of the IN proteins. The disintegration substrate mimics the product of single U5 vDNA end integration, with label placed at the 5′ end of the disrupted target DNA strand (26). Nucleophilic attack by the corresponding 3′-OH group on the vDNA pA-target DNA phospho-diester bond liberates the vDNA end and produces a joined, labeled 30-bp strand (Fig. 3A). Consistent with our previous report (37), active site mutant IND64N displayed a very low level (<1%) of INWT disintegration activity. Class II IN mutant activities were noticeably more robust, ranging from a low of 34% for INQ214L/Q216L to a high of 329% for INR199A (Fig. 3B and C; Table 2). Because INW235E and INR228A supported about 41% and 139% activity, respectively, we conclude that the active sites of these enzymes are catalytically competent and that the CTD mutations render the proteins effectively unable to catalyze vDNA 3′-end processing or strand transfer activities. Functional enzyme active sites underscore the ability of class II IN mutant proteins as well as INW235E to efficiently trans-complement the infection defects of class I IN CCD mutant viruses (6, 47, 77, 78, 80).
Fig 3.
Disintegration substrate design and IN activities. (A) Substrate illustration, highlighting vDNA (thin lines) and target DNA (bold lines) components as well as the position of the 32P label (*). (B) Gel image showing the migration positions of disintegration substrate (sub) and reaction products (DP). IN was omitted from the reaction mixture loaded in lane 1; the remaining lanes contained the indicated IN proteins. (C) Mean disintegration activities ± standard errors of the means for two independent experiments, expressed as the percent INWT function.
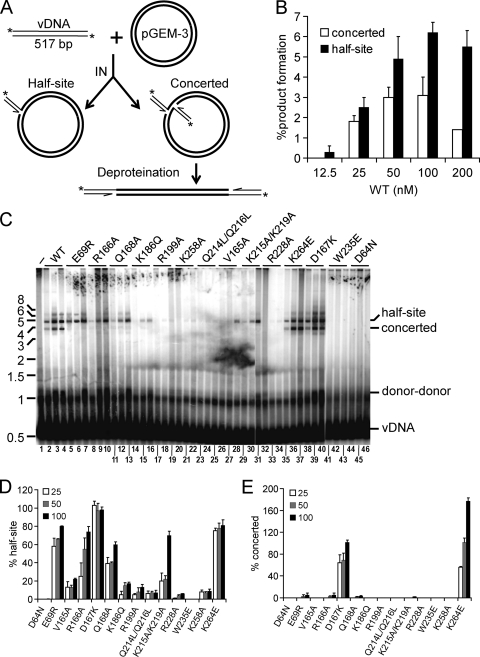
Concerted vDNA integration activities.
Unlike PFV IN (115), HIV-1 IN is unable to efficiently integrate relatively short vDNA substrates into target DNA in a concerted fashion in the absence of added binding factors (13, 53). For reasons that are not entirely clear, long vDNA substrates, in the range of several hundred base pairs, support concerted HIV-1 IN integration activity under certain reaction conditions (70). We therefore generated a 5′-end-labeled 517-bp restriction enzyme fragment that harbored the complete, blunt-ended U5 sequence at one of its ends to assess concerted integration activity. Circular pGEM-3 target DNA (2,867 bp) was included in the mixture to monitor the course of the reaction: integration of a single vDNA end into this molecule yielded a tagged nicked circle with relatively slow mobility in agarose gels, whereas concerted integration of two vDNA ends after deproteination yielded a population of approximately 3.9-kb linearized pGEM-3 fragments (Fig. 4A to C). Preliminary experiments with varied concentrations of INWT in the reaction mixture were conducted to discern optimal levels of integration activity. Consistent with prior reports (70), relatively minor fractions of the vDNA substrate, ∼3% and 6%, were converted to the concerted and half-site products, respectively, under optimal conditions (50 and 100 nM INWT) (Fig. 4B and C, lanes 1 to 4). IN mutant activities were accordingly compared to INWT at protein concentrations of 25, 50, and 100 nM. Resultant mutant half-site and concerted integration activities were independently quantified as the percent INWT function at each protein concentration (results are summarized in Table 3).
Fig 4.
Concerted integration assay design and IN mutant activities. (A) Schematic showing 5′-end-labeled blunt U5 substrate (thin lines) and circular plasmid target (bold lines) DNAs, as well as outcomes of single-end versus concerted vDNA integration. *, positions of 32P label. (B) Results of preliminary titration experiments (n = 2) with the indicated INWT concentrations, quantified as the percentages of substrate converted to half-site and concerted integration reaction products. (C) Phosphorimage of agarose gel, highlighting migration positions of the 517-bp vDNA substrate and integration products. IN was omitted from the reaction mixture loaded in lane 1; the remaining reaction sets contained the indicated IN protein at 25 nM, 50 nM, or 100 nM (left to right). Positions of mass standards (in kb) are indicated to the left. (D and E) Quantitation of IN mutant half-site (D) and concerted (E) integration activities, expressed as a percentage of INWT function (means ± standard errors of the means for two independent experiments).
Table 3.
Relative activities of HIV-1 IN proteins
| Mutant protain | Relative integration activity,a 517-bp vDNA (nM IN) |
LEDGF-dependent integration |
LEDGF-IN interactionb | RT-IN interaction | Tetramer formation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Half-site |
Concerted |
Half-site | Concerted | ||||||||
| 25 | 50 | 100 | 25 | 50 | 100 | ||||||
| D64N | − | − | − | − | − | − | − | − | 108 (6.2) | 71 (5.4)c | 110 (2.4) |
| E69R | 58 (8.8) | 66 (0.3) | 80 (0.2) | − | 3 (2.7) | 5 (3.4) | 44 (8.6) | 1.0 (0.6) | 18 (2.5) | 74 (8.4)c | 48 (4.2) |
| V165A | 13 (6.3) | 13 (2.4) | 22 (1) | − | − | − | 92 (12) | 9.0 (0.1) | 26 (4.4) | 57 (8.4)c | 65 (8.4) |
| R166A | 25 (14.8) | 55 (12.3) | 74 (5.6) | − | 3 (0.7) | 5 (3.6) | 80 (8.4) | 11 (1.2) | 7 (3.1) | 70 (5.8)c | 48 (8.0) |
| D167K | 103 (4.4) | 97 (8.1) | 98 (3) | 64 (15.1) | 69 (12.6) | 102 (3.9) | 76 (6.5) | 36 (2.7) | 68 (9.4) | 49 (8.7)d | 97 (3.2) |
| Q168A | 39 (6.9) | 40 (0.8) | 60 (2.8) | − | 2 (0.4) | 3 (1.2) | 77 (9.4) | 10 (0.8) | 70 (0.4) | 70 (1.7)c | 14 (2.4) |
| K186Q | 5 (3.6) | 15 (2.9) | 17 (1.7) | − | − | − | 67 (13) | 68 (0.3) | 64 (9.7) | 67 (3.7)c | 45 (6.1) |
| R199A | 5 (1) | 8 (4.5) | 13 (3.2) | − | − | − | 57 (7.1) | 1.0 (1.0) | 68 (16) | 46 (3.2)c | 70 (5.4) |
| Q214L/Q216L | 6 (2.9) | 7 (1.6) | 7 (2.9) | − | − | − | 86 (13) | 49 (0.8) | − | 50 (0.8)c | 96 (7.9) |
| K215A/K219A | 20 (8.8) | 22 (3.5) | 70 (4.4) | − | − | 2 (0.1) | 103 (6.6) | 86 (16) | 85 (11) | 77 (7.3)c | 124 (8.7) |
| R228A | 1 (0.1) | 4 (0.8) | 6 (0) | − | − | − | 6.0 (6.0) | − | 67 (8.7) | 29 (7.5)c | 83 (9.1) |
| K258A | 8 (2.2) | 7 (1.2) | 9 (2.1) | − | − | − | 27 (1.8) | − | 85 (8.8) | 28 (2.1)c | 52 (4.0) |
| K264E | 75 (3) | 78 (5.3) | 81 (6.1) | 56 (1.5) | 102 (7.5) | 177 (6.1) | 26 (6.4) | − | 96 (5.3) | 90 (15)d | 107 (1.8) |
| W235E | − | − | − | − | − | − | − | − | 112 (1.6) | −d | 26 (1.4) |
Relative to INWT, which was set to 100%. Values are means, with standard errors of means in parentheses for 2 independent experiments unless otherwise indicated. −, undetectable (<1% of INWT activity).
Results of GST-LEDGF326-530 pulldown (Fig. 10A and B).
Based on 3 experiments.
Based on 4 experiments.
A subset of the mutant proteins, including INE69R, INR166A, IND167K, INQ168A, INK215A/K219A, and INK264E, supported relatively robust (>50% of INWT) half-site DNA strand transfer activity (Fig. 4C, lanes 1 to 13, 29 to 31, and 35 to 40), with class I IN mutant control proteins IND64N and INW235E predictably inactive (lanes 41 to 46) (results are summarized in Fig. 4D and Table 3). Negligible concerted integration activities (2 to 5% of INWT activity at 50 nM and 100 nM [Fig. 4E and Table 3]) rather strikingly accompanied INE69R, INR166A, INQ168A, and INK215A/K219A, indicating that these mutations perturb a function(s) that is central to the concerted integration of two vDNA ends under these conditions. The remaining class II mutants displayed half-site integration activities that ranged from a low of 1% for INR228A to 22% for INV165A without revealing detectable levels of concerted integration activity (Fig. 4C and Table 3). Two of the class II mutants, IND167K and INK264E, in contrast, supported robust levels of concerted integration (Fig. 4C, lanes 35 to 40, and E; Table 3).
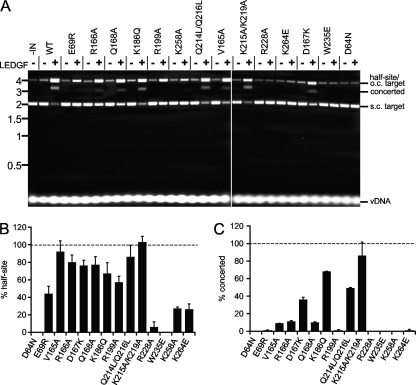
DNA binding proteins HMGA1, HMGB1, and HIV-1 nucleocapsid (NC) have been shown to significantly stimulate HIV-1 IN concerted integration activity in some (16, 56) but not all (70) studies. The lentiviral IN binding protein LEDGF/p75 (21, 24, 30, 114) has also shown variable effects on concerted integration activity, which at low nanomolar vDNA concentrations depended on protein concentrations (93) and/or the order by which they were added to the reaction mixture (101). Increasing reactant concentrations to similar submicromolar levels of IN, LEDGF/p75, and oligonucleotide vDNA substrate, in contrast, revealed potent stimulation of concerted integration activity (53). We therefore next evaluated IN mutant responses under conditions that promote efficient LEDGF/p75-dependent concerted integration activity (53). Due to relatively high levels of vDNA substrate in these mixtures and the reaction efficiency, substrates and products were initially visualized following EtBr staining of agarose gels.
Integration of a single 32-bp vDNA end into pGEM-3 yields a tagged circular product that migrates at the same position as nicked plasmid DNA circles isolated from E. coli, whereas concerted integration in this case yielded a population of approximately 3-kb linear reaction products (53) (Fig. 5A, first three lanes). In the absence of LEDGF/p75, INWT catalyzed the formation of a low level of half-site integration products without detectable concerted integration activity (Fig. 5A, lanes 1 and 2) (27, 53). LEDGF/p75 significantly stimulated IN activity, such that the brunt of the supercoiled pGEM-3 target DNA was consumed, yielding palpable increases in half-site and concerted integration reaction products (lanes 1 to 3). The effects of LEDGF/p75 on IN mutant half-site (Fig. 5B) and concerted (Fig. 5C) integration activities were accordingly quantified as a percentage of the INWT responses. IND64N and INW235E were each predictably defective for both activities, whereas LEDGF/p75 stimulated a small amount of INR228A half-site activity in some but not all experiments (Table 3). Half-site integration activities of the other class II IN mutant proteins were stimulated reasonably well by the host factor to levels that varied from about 25% (INK258A and INK264E) to 100% (INK215A/K219A) of INWT levels (Fig. 5A and B; Table 3). Akin to the previous experiment (Fig. 4), mutant concerted integration activities varied more so than their corresponding half-site activities. For example, the concerted integration activities of INE69R, INV165A, INR199A, INK258A, and INK264E were less than 10% of their half-site values (Fig. 5; Table 3). INK215A/K219A and INK186Q, in contrast, catalyzed fairly similar levels of half-site and concerted vDNA integration under these reaction conditions.
Fig 5.
LEDGF/p75-dependent integration activities. The assay design was similar to that shown in Fig. 4A, but here we used a significantly shorter (32-bp versus 517-bp) vDNA. (A) Agarose gel image revealing migration positions of vDNA substrate, supercoiled (s.c.) and open circular (o.c.) pGEM-3, as well as half-site and concerted integration reaction products. IN and/or LEDGF/p75 was omitted from reaction mixtures as indicated above the gel image. Migration positions of mass standards (in kb) are shown to the left. (B) Half-site integration activities of IN mutant proteins relative to INWT (set to 100%); means ± standard errors of the means for two independent experiments are shown. (C) IN mutant concerted integration activities relative to INWT from 2 experiments.
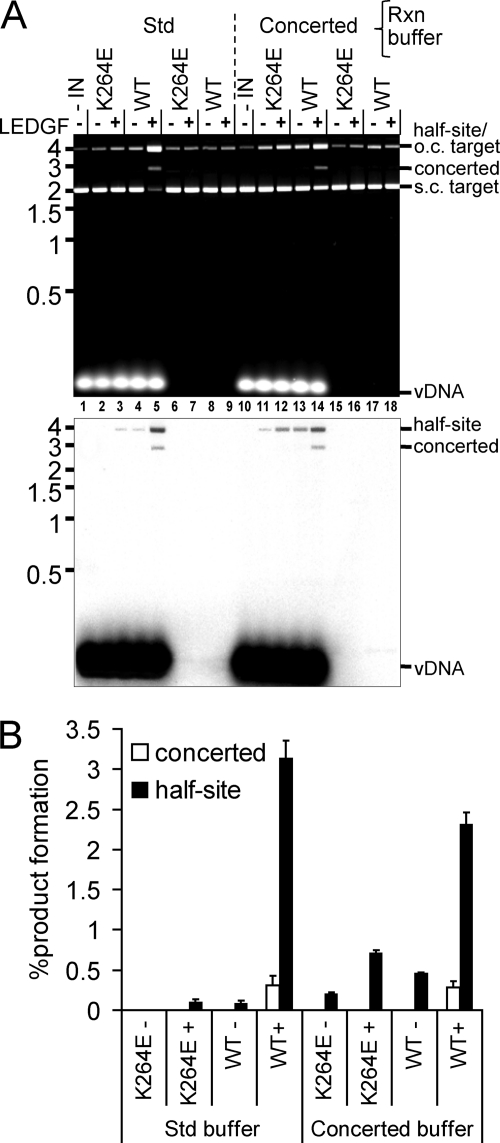
INK264E stood out as a protein that behaved rather differently under different reaction conditions: it 3′ processed and supported the robust concerted integration of 517-bp vDNA ends, yet failed to integrate precleaved 32-bp vDNA in a concerted fashion in the presence of LEDGF/p75 (Fig. 4 and 5). INK264E-mediated 3′ processing and integration of oligonucleotide vDNA substrate, moreover, was largely Mn2+ dependent (Fig. 1 and 2; Table 2), yet its robust response to 517-bp vDNA occurred in the presence of Mg2+ (Fig. 4). To corroborate these findings, protein repurified after resequencing the expression construct was analyzed, and this confirmed the results. Aside from the specific divalent metal ion or vDNA substrate, the oligonucleotide-based assays (Fig. 1 to 3) were conducted under identical buffer conditions. In contrast, for the concerted integration assay (Fig. 4) we used noticeably lower levels of IN protein and included DMSO and the molecular crowding agent PEG (70, 92, 101). To see if these parameters contribute to differences in INK264E activities, 3′ processing of the 30-mer oligonucleotide substrate was assessed under the buffer conditions used for Fig. 4. Although this significantly suppressed INWT activity, INK264E nonetheless appeared equally active (data not shown). LEDGF/p75-dependent integration activities were also assessed under these buffer conditions. To increase the sensitivity of the assay, radiolabeled vDNA was incorporated at 5% of the total substrate concentration (Fig. 6). INWT converted approximately 0.3% and 3% of the precleaved 32-bp substrate to concerted and half-site integration products, respectively, in the presence of LEDGF/p75 under both sets of reaction conditions (Fig. 6A, lanes 5 and 14; quantified in panel B). Although the conditions that included PEG and DMSO (denoted as concerted buffer in Fig. 6) marginally stimulated INK264E half-site integration (Fig. 6A, bottom panel, compare lanes 11 and 12 to lanes 2 and 3), concerted integration activity remained undetectable. We therefore inferred that although INK264E prefers the buffer conditions used for the experiment in Fig. 4, its robust concerted integration activity seems particularly dependent on an aspect(s) of the relatively long vDNA substrate (Fig. 4 and 6).
Fig 6.
LEDGF/p75-dependent INWT and INK264E integration activities under different reaction buffer conditions. (A) EtBr stained image (upper panel) and phosphorimage (lower panel) for integration reactions conducted under standard (Std) conditions (Fig. 5) or with the same protein, vDNA, and pGEM-3 concentrations under the Fig. 4 reaction conditions (“concerted”). Migration positions of substrates and reaction products are indicated to the right of the gel panels, whereas mass standards (in kb) are shown on the left. (B) Quantitation of panel A phosphorimaging results for 2 independent experiments (means ± standard errors of the means).
In vitro activities of PICs extracted from acutely infected cells.
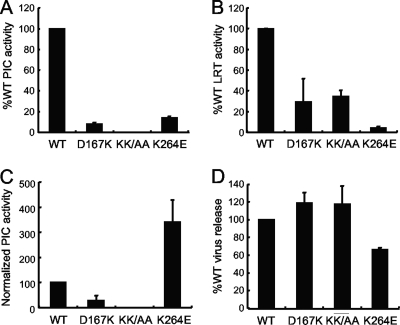
Our results clarified that some class II IN mutants, in particular INE69R, failed to support concerted integration despite being able to robustly integrate single vDNA ends under a variety of reaction conditions (Tables 2 and 3). We therefore concluded that defective IN function underscores the HIV-1E69R infection defect (102). To further address the link between purified IN activities and HIV-1 biology, we next characterized the profiles of a subset of mutants that supported robust concerted integration in the context of virus infection. HIV-1K215A/K219A and HIV-1K264E were selected due to the relatively robust responses of these INs in one of the utilized assays (Table 3). As HIV-1D167K retained about 6% of WT infectivity (Table 1) and IND167K catalyzed concerted vDNA integration under both sets of assay conditions, it was included for comparison. SupT1 T cells 7 h after acute infection with WT or IN mutant virus were lysed, and extracted PICs were incubated with an excess of plasmid target DNA in the presence of Mg2+ ions. Deproteinized extracts were queried by different Q-PCR assays to ascertain levels of reverse transcription that occurred prior to cell lysis and subsequent in vitro integration activities (42, 80). Of note, the Q-PCR PIC activity assay monitors the formation of U5 vDNA end-target DNA junctions and therefore does not specifically assess the concerted integration of vDNA U3 and U5 ends (42, 80).
PIC analyses rely on relatively high-multiplicity infections, which were accomplished using viral supernatants freshly harvested from HEK293T cells (42). Although this approach side-stepped normalization of input WT and IN mutant viral inocula based on RT activity or p24 content, the experiment was internally controlled through parallel assessment of vDNA substrate levels in each of the cell extracts. HIV-1D167K and HIV-1K264E PICs supported the formation of strand transfer reaction products, whereas HIV-1K215A/K219A did not (Fig. 7A). Each virus also revealed a reverse transcription defect, which under these infection conditions varied from about 4% of WT for HIV-1K264E to about 35% for HIV-1K215A/K219A (Fig. 7B). Specific PIC activities, calculated by normalizing integration activity to corresponding levels of viral substrate DNA, revealed that HIV-1D167K supported ∼26% of HIV-1WT function, whereas HIV-1K264E integration activity exceeded that of the WT (Fig. 7C). Assessment of the number of viral particles in HEK293T cell supernatants prior to SupT1 cell infection revealed an ∼35% release defect for HIV-1K264E relative to the WT (Fig. 7D). Correction for this difference revealed the normalized level of HIV-1K264E reverse transcription to be ∼6% of the WT.
Fig 7.
PIC formation and in vitro integration activities. (A) PIC activities in extracts prepared from cells infected with the indicated viruses. KK/AA, K215A/K219A. (B) The same experiment as in panel A, except the Q-PCR detected the corresponding levels of vDNA substrate in the integration reaction mixtures. (C) PIC activities from panel A, normalized for levels of vDNA substrate (from panel B). Standard errors were calculated as M1/M2√(SEM12/M12 + SEM22/M22), where M1 is the average PIC activity, M2 is the average vDNA level, SEM1 is the standard error of the mean of PIC activity measurements, and SEM2 is the SEM from vDNA measurements. (D) Relative levels of RT activity in HEK293T cell supernatants prior to SupT1 cell infection.
DNA synthesis profiles of IN mutant viruses.
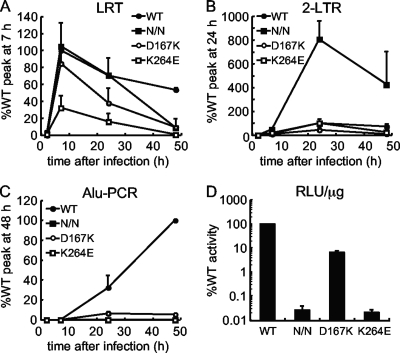
The results of the previous experiment revealed cytoplasmic HIV-1K264E PICs, which formed, admittedly, at significantly lower levels than HIV-1WT, nevertheless supported efficacious integration activity in vitro. Single-round HIV-1 constructs defective for replicative spread were utilized next to assess the fates of WT and IN mutant DNAs during acute infection. The use of such constructs previously revealed approximate 5- to 10-fold reverse transcription defects for HIV-1D167K (102) and HIV-1K264E (77), similar to the responses observed in the previous experiment. Because pseudotyping with the VSV-G envelope glycoprotein can significantly stimulate levels of class II IN mutant reverse transcription relative to the WT control (9), the heterologous envelope glycoprotein was employed to boost the comparatively low level of HIV-1K264E DNA synthesis. Cell extracts prepared at 2, 7, 24, and 48 h after infection were queried by different Q-PCR assays to determine products of LRT, 2-LTR circles, which can serve as a marker for PIC nuclear import, and integration (Alu-PCR). HIV-1D64N/D116N was used as a catalytically inactive IN mutant control.
Consistent with prior findings (9), VSV-G significantly stimulated relative levels of class II HIV-1K264E and HIV-1D167K IN mutant reverse transcription, to ∼32% and 85% of HIV-1WT, respectively, at 7 h postinfection (Fig. 8A, compare with 7B), whereas HIV-1D64N/D116N behaved as the WT. The formation of HIV-1WT 2-LTR circles peaked at 24 h after infection, with the level of class I mutant HIV-1D64N/D116N product expectedly significantly outweighing that of the WT (Fig. 8B) (75). HIV-1K264E formed circles at a level that was indistinguishable from the WT, whereas HIV-1D167K circles formed at about 45% of the WT (Fig. 8B). HIV-1D167K integration peaked at about 6.5% of the WT level, equal to the relative infectivity of this virus (Fig. 8C and D). Infectivity under these conditions was therefore directly proportional to the level of chromosomal vDNA integration. Consistently, the levels of HIV-1K264E and HIV-1D64N/D116N integration were below the detection limit of the assay (Fig. 8C and D).
Fig 8.
Kinetics of WT and IN mutant viral DNA synthesis and integration. (A) LRT products of the indicated VSV-G-pseudotyped viruses (N/N, D64N/D116N) were amplified from cell extracts obtained 2, 7, 24, or 48 h after infection. Values are expressed as the peak WT product formation, which occurred at 7 h. (B) 2-LTR circle Q-PCR product levels, expressed as the percent WT value, which was set to 100% at the peak (24 h postinfection). (C) Integrated proviruses assessed as nested Alu-PCR. The peak WT value at 48 h postinfection was set to 100%. (D) Normalized levels of single-round viral infectivities, expressed in RLU per μg of total cell protein. Compiled data are from two independent experiments (means ± standard errors of the means).
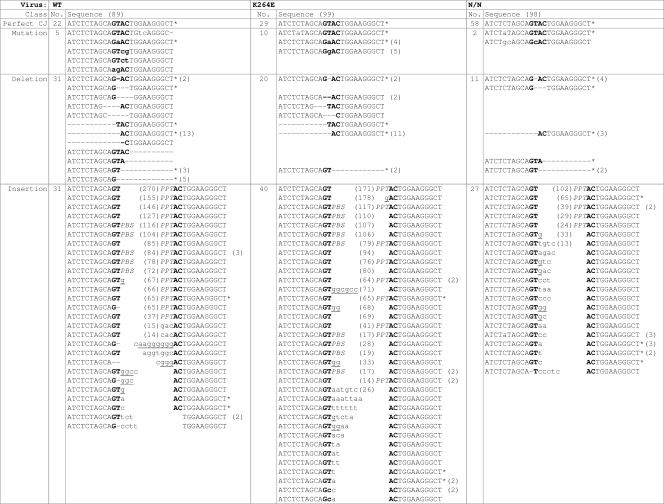
HIV-1K264E effectively formed 2-LTR circles, indicating that this infection was blocked after PIC nuclear entry (Fig. 8B). The sequence of the associated 2-LTR circle junction (CJ), which results from the ligation of U3 and U5 vDNA ends by the cellular nonhomologous DNA end-joining repair pathway (69), is accordingly a metric of vDNA end integrity (62). Certain INSTIs (110) as well as the relatively inhospitable environment of quiescent CD4+ T cells (119) significantly increased the frequency of 2-LTR CJ deletions, indicative of reduced vDNA end integrity prior to ligation. HIV-1WT, HIV-1K264E, and HIV-1D64N/D116N 2-LTR CJs from acutely infected cells were accordingly cloned and sequenced to assess relative end integrity of these vDNAs. Resulting sequences were parsed into four categories, including (i) the perfect CJ sequence, ATCTCTAGCAGTACTGGAAGGGCT (boldface corresponds to the GT dinucleotide that is normally removed by IN from the U5 plus-strand and U3 minus-strand termini, respectively, prior to end ligation); (ii) mutation, which harbored one or more base pair substitutions within this sequence; (iii) deletion, with a minimum of 1 bp absent; (iv) insertion. Approximately 25% of the WT sequences were perfect CJs, with deletion and insertion sequences accounting for about 35% each (Fig. 9; Table 4). Most insertions contained an intact copy of the polypurine tract (PPT) or primer binding site (PBS) sequence, indicative of aberrant primer removal by RNase H during reverse transcription (62, 87). As expected (110), the lack of IN activity significantly increased the frequency of perfect CJ sequences recovered from HIV-1D64N/D116N-infected cells, with a notable decrease in the number of deletions (Table 4 and Fig. 9). Because the frequency of HIV-1K264E deletion junctions was likewise lower than with the WT, we inferred that the ends of nuclear HIV-1K264E DNA were no more prone to degradation than were the WT ends (Fig. 9; Table 4).
Fig 9.
Sequences of WT and IN mutant viral 2-LTR CJs. Sequences were grouped into four classes (perfect CJ, mutation, deletion, and insertion), as defined in the text. Numbers for each class are indicated below the viral names (N/N, D64N/D116N), with total numbers of sequences obtained for each virus tabulated above the sequences (shown in parentheses). The GTAC that represents the pGT dinucleotide normally processed by IN from the U5 plus strand and U3 minus strand is indicated in bold type. Mutations are indicated in lowercase letters, and deletions are shown as dashes. Numbers to the right of sequences (in parentheses) indicate the frequency at which the mutation was determined for that virus. *, sequence detected with >1 virus. PBS and PPT in the insertion category represent perfect matches to the viral primer binding site (GGCGCCCGAACAGGGAC) and polypurine tract (AAAAGAAAAGGGGGG), respectively, whereas underlined bases mark partial identities to these sequences. Internal numbers in parentheses mark additional lengths of DNA insertion, which in many cases extended from the PBS and/or PPT (62).
Table 4.
Sequence frequencies among WT and IN mutant 2-LTR circle junctionsa
| Sequence class | CJ frequency |
||
|---|---|---|---|
| WT | K264E | N/Nb | |
| Perfect CJ | 0.25 | 0.29 | 0.59 |
| Mutation | 0.06 | 0.10 | 0.02 |
| Deletion | 0.35 | 0.20 | 0.11 |
| Insertion | 0.35 | 0.40 | 0.28 |
Sequences are illustrated in Fig. 9.
N/N, D64N/D116N.
Interactions with heterologous binding partners. (i) LEDGF/p75.
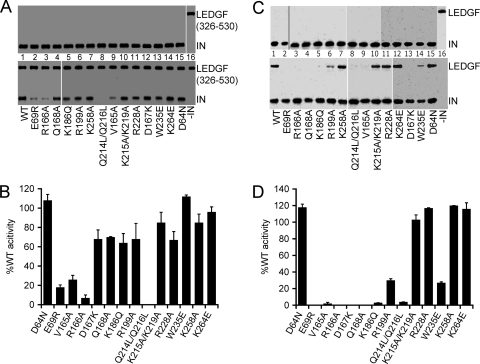
The results of the work described above highlighted that HIV-1K264E forms active cytoplasmic PICs and accesses cell nuclei during acute infection. As inferred from 2-LTR CJ sequences, the mutant vDNA ends, moreover, appeared grossly similar to the WT, indicating that they were not prone to preferential degradation by host nucleases (119). We therefore hypothesized that the HIV-1K264E infection defect might result from the inability of INK264E to interact with a nuclear protein important for vDNA integration. A specific interaction between IN and LEDGF/p75 dictates both the frequency and distribution of lentiviral integration (35, 97), and LEDGF/p75, moreover, failed to stimulate the concerted integration activity of INK264E in vitro (Fig. 5 and 6). The LEDGF/p75 binding profile of INK264E was therefore compared to the WT and other IN mutants in reciprocal pulldown assays. In one format, soluble IN was captured by bead-bound GST-LEDGF326-530 protein (23). As previously reported (102), some mutations in the LEDGF/p75 binding pocket, like E69R, V165A, and R166A, impacted the host factor interaction more significantly than other pocket mutations, such as D167K and Q168A (Fig. 10A and B). The Q214L/Q216L CCD-CTD interdomain linker mutation significantly perturbed the interaction, whereas the overlapping K215A/K219A mutation, interestingly, did not. Consistent with prior work that indicated that the NTD and CCD fully account for the affinity of IN for LEDGF/p75 (53, 81), each of the CTD mutant proteins, including INK264E, was efficiently recovered with GST-LEDGF326-530 (Fig. 10A and B). We previously reported that a reciprocal Ni-NTA pulldown assay mediated by C-terminally His6-tagged IN revealed some LEDGF/p75 binding defects not evident from the GST assay (102). Accordingly IND167K, INQ168A, and INK186Q each failed to appreciably capture LEDGF/p75 under this condition (compare Fig. 10C and D with A and B, respectively). Because INK264E, by contrast, retained its efficient interaction with the host factor (Fig. 10C and D), we conclude that the K264E mutation does not obviously perturb the IN-LEDGF/p75 interaction.
Fig 10.
LEDGF/p75 binding activities of IN mutant proteins. (A) GST-LEDGF326-530 pulldown of WT and IN mutant proteins. The top and bottom gel images reveal relative levels of input (50% of total reaction mixture) and bead-bound proteins (100% of pellet), respectively. IN was omitted from the pulldown reaction mixture loaded in lane 16. (B) Quantitation of IN mutant recovery for 2 experiments, expressed as averages ± standard errors of the means. Pulldown assays performed with GST-bound beads failed to reveal a detectable signal for any of the IN proteins (data not shown). (C and D) Results for assays akin to those shown in panels A and B, except Ni-NTA pulldown of soluble LEDGF/p75 was mediated by the indicated His6-tagged WT or IN mutant protein.
(ii) RT.
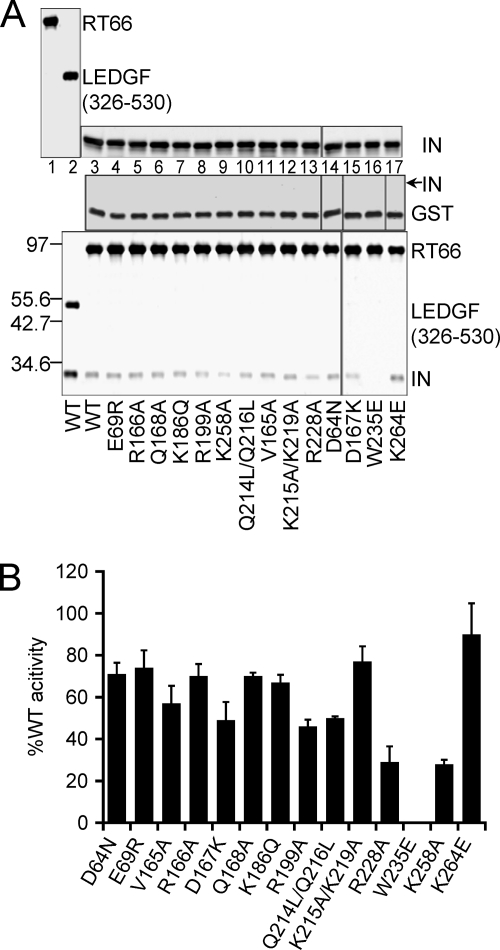
The IN CTD mediates a direct interaction with RT (55, 123, 130), and RT can, moreover, stimulate IN DNA strand transfer activity under certain assay conditions (55). The RT-IN interaction is furthermore ablated by the IN mutation K258A, suggesting that loss of RT binding affinity might dictate class II IN mutant reverse transcription defects (123). Binding assays were therefore performed to assess the affinity of RT for the different IN mutant proteins.
Preliminary experiments with IN-His6 prebound to Ni-NTA beads failed to specifically pull down heterodimeric p66/p51 RT protein, as RT was occasionally recovered on beads lacking IN. Because the isolated p66 subunit bound HIV-1 IN as efficiently as the heterodimer (55), GST-RT(p66) protein was tested next in a GST pulldown format. Preliminary experiments again revealed evidence of nonspecificity due to occasional recovery of IN on GST-loaded beads, which was remedied by first preclearing the IN proteins by centrifugation after 15 min of preincubation in pulldown assay buffer. Consistent with the approximate 61 nM (123) and 11 nM (112) affinity constants reported for the RT-IN and LEDGF/p75-IN interactions, respectively, GST-LEDGF326-530 in a parallel control sample pulled down more IN than did GST-RT (Fig. 11A, bottom panel, lanes 2 and 3). Our assay format, moreover, confirmed that the K258A mutation lowered the apparent affinity of IN for RT (Fig. 11A, lane 9; results quantified in panel B). The R228A mutation reduced binding to a similar extent as K258A, whereas, interestingly, the class I mutation W235E ablated binding (lanes 13 and 16). Because INK264E was efficiently pulled down by GST-RT (Fig. 11; Table 3), neither the HIV-1K264E reverse transcription nor integration defect was attributable to reduced affinity of the mutant IN for RT.
Fig 11.
Binding of RT to WT and IN mutant proteins. (A) Top panels show input levels of GST-RT (lane 1, 50% of reaction input), positive control GST-LEDGF326-530 (lane 2, 50% of input), and WT and mutant IN proteins (20% of input). BSA (not shown) was included in all assays at 5 μg. Middle and bottom panels, levels of IN proteins pulled down by GST alone and GST-RT, respectively (100% of pellet fractions). The expected position of IN in the middle panels is indicated with an arrow. (B) Quantification of panel A GST-RT pulldowns based on Alpha Innotech FluorChem FC2 imaging for 4 (IND167K, INK264E, and INW235E) or 3 independent experiments (remaining IN proteins).
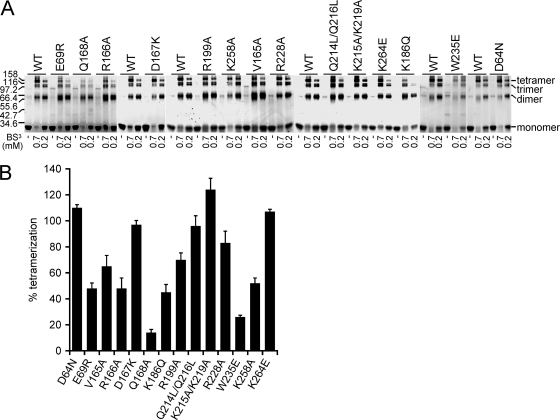
Mutational effects on IN multimerization.
HIV-1 IN tends to form dimers and tetramers in solution, which can be monitored through the use of covalent cross-linking reagents (4, 24, 34, 46, 51, 64, 71). Cross-linking was accordingly utilized to assess self-association properties of mutant proteins by quantifying the extent of tetramer formation relative to INWT. Four general patterns emerged from this analysis (Fig. 12). A few of the class II mutant proteins, INV165A, INR199A, and INR228A, displayed marginal defects that ranged from about 15% to 35% reductions in cross-linking efficiency. Four mutants, INE69R, INR166A, INK186Q, and INK258A, formed tetramers about half as efficiently as INWT, whereas INQ168A and the class I mutant INW235E were more defective, yielding about 14% and 26% relative tetramer formation, respectively (Fig. 10A and B; Table 3). In contrast, IND64N, IND167K, INQ214L/Q216L, INK215A/K219A, and INK264E formed tetramers as efficiently as the WT.
Fig 12.
IN-IN cross-linking. (A) IN proteins were electrophoresed after incubation in the absence of cross-linker (-) or in the present of 0.7 or 0.2 mM bis(sulfosuccinimidyl) suberate (BS3) as indicated. INWT was repeatedly analyzed to afford the positive-control metric in each gel that contained an IN mutant protein. (B) Percent tetramer formation versus INWT (means ± standard errors of the means) for two independent cross-linking experiments.
DISCUSSION
In vitro activity assays and HIV-1 IN mutant biology.
Class II IN mutant viruses harbor a variety of replication defects that can seemingly span the entire replication cycle, from particle assembly and release to uncoating, reverse transcription, PIC nuclear import, and integration (recently reviewed in references 8 and 32). Class I mutant enzymes that carry alterations in IN active site residues typically do not support detectable levels of 3′-processing or DNA strand transfer activities in vitro, whereas numerous INs derived from class II mutant viruses were catalytically proficient under assay conditions that did not distinguish the cutting and joining of single vDNA ends from the concerted integration of two ends, as occurs during virus infection. Class II IN mutant enzymes, moreover, efficiently trans-complemented the infection defect inherent to class I mutant viruses (6, 47, 77, 78, 80), suggesting that an underlying defect of class II mutant viruses might be the inability of the associated IN protein to properly engage critical binding partners. Here we addressed this overarching hypothesis by employing a number of in vitro assays that included tests for the concerted integration of two vDNA ends into target DNA.
Assays that utilized denaturing polyacrylamide gel electrophoresis to separate strand transfer reaction products and hence did not distinguish half-site from concerted integration activity revealed that Mn2+ supported more robust IN mutant activities than Mg2+ (Fig. 2 and Table 2). Mg2+ is the likely physiologically relevant IN cofactor, and class II mutants INR166A, IND167K, and INQ168A displayed relatively robust Mg2+-dependent DNA strand transfer activities, whereas INV165A and INK186Q were about 25 to 30% as active as INWT. As PICs derived from HIV-1V165A-infected cells did not support a detectable level of in vitro integration activity (80), we concluded that assays that query Mg2+-dependent single vDNA end strand transfer activity, although more relevant than those with designs that instead utilize Mn2+, do in some, but importantly not all, cases yield results reflective of viral IN mutant function. This conclusion was underscored by the behaviors of INE69R, INR166A, INQ168A, and INK215A/K219A in the concerted integration assay (Fig. 4): each enzyme supported robust levels of half-site integration activity yet negligible levels of concerted integration activity. Three of these mutations, E69R, Q168A, and R166A, reduced inherent IN multimerization by approximately 2- to 7-fold (Fig. 12; Table 3), suggesting that defective tetramerization in part accounts for the inability to effectively integrate two vDNA ends in concerted fashion. LEDGF/p75, which has been shown to enhance lentiviral IN tetramerization (51, 86), accordingly stimulated the concerted integration activities of INK186Q, INQ214L/Q216L, and INK215A/K219A (Fig. 4 and 5). IND25K and INK188D were similarly shown to be preferentially defective for the concerted integration of a 500-bp vDNA substrate and IN tetramerization yet efficiently stimulated by LEDGF/p75 to integrate 32-bp vDNA in a concerted fashion (51). We, however, noted that LEDGF/p75 binding was neither necessary nor sufficient to stimulate this function: the host factor efficiently bound INK264E whereas binding to INQ214L/Q216L was not detected (Fig. 10), yet LEDGF/p75 stimulated INQ214L/Q216L but not INK264E concerted integration activity (Fig. 5 and Table 3). Because LEDGF/p75-IN binding was assessed in the absence of DNA, it is likely prudent to exercise caution in applying the results of these assays to activity-based measures. Aside from Fig. 4, we moreover note that our assays were conducted at single protein concentrations optimized based on INWT responses, perhaps also limiting the interpretations of IN mutant behaviors.
Although our study encompassed only a modest subset of previously characterized class II mutant INs, the findings nevertheless clarified that the vast majority are defective for concerted integration activity when queried with a relatively long vDNA substrate (Fig. 4). Although these results do not rule out that defective binding of IN to auxiliary factors may contribute to the specificity of different class II mutant viral phenotypes, we argue that the root cause of the associated infectivity defect is in most cases a lack of IN catalytic function compared to loss of heterologous factor binding capacity. We extend this conclusion to speculate that non-active site epitopes that predictably encompass the slew of class II IN mutant changes will at least in some cases afford novel opportunities for development of allosteric IN inhibitors.
The IN-RT interaction and class II mutant viral phenotype.
The most common pleiotropic defect associated with the class II IN mutant viral phenotype occurs at reverse transcription (32). IN and RT proteins have accordingly been shown to directly interact and, under certain conditions, stimulate each other's in vitro activities (28, 55). The interaction is furthermore mediated by the IN CTD and the K258A mutation, which we previously reported to reduce DNA synthesis by more than 10-fold (77), ablated the CTD-RT interaction (123). Three CTD mutant proteins in addition to INK258A were assessed here for RT binding in an affinity pulldown assay. INR228A, derived from a virus that also displayed a greater-than-10-fold reverse transcription defect (77), interestingly showed a similar RT binding defect as INK258A, with an approximate 70% reduction from the INWT (Fig. 11B; Table 3). HIV-1K264E also possesses a reverse transcription defect, which varied from about 3- to 16-fold depending on certain aspects of experimentation (Fig. 7 and 8) (77). Yet, INK264E interacted with RT as efficiently as any mutant tested, to about 90% of the level of INWT (Table 3). The only mutation tested that ablated the RT-IN interaction, moreover, was W235E, based on a virus that does not display a reverse transcription defect (19, 67). Hehl et al. previously noted normal levels of RT binding to INW235E (55), whereas Ishikawa et al. (58), like us, reported complete loss of binding. Although our observation could be due to a global CTD structural perturbation that speculatively negatively impacted INW235E self-association (Fig. 12), our data nonetheless highlight that caution should be exercised in interpreting RT-IN mutant binding affinity and causation of class II IN mutant viral reverse transcription defects.
The K264E IN mutation and HIV-1 replication.
A subset of class II IN mutant proteins that efficiently catalyzed concerted integration activity in vitro, including IND167K, INK215A/K219A, and INK264E, was analyzed for their abilities to support integration during acute HIV-1 infection. The utilized PIC assay for IN DNA strand transfer activity measured levels of vDNA U5 end joining and thus did not monitor concerted integration activity per se. PICs derived from replication-defective U5-end mutant viruses that effectively processed the U3 vDNA end failed to integrate the solo processed end in vitro, indicating that HIV-1 tightly couples the functionality of both vDNA ends to IN DNA strand transfer activity during infection (18). Consistent with the residual infectivity, HIV-1D167K PICs supported detectable levels of in vitro integration activity. Unexpectedly, HIV-1K264E PIC activity appeared to exceed that of the WT (Fig. 7). Although HIV-1K264E was defective for reverse transcription, the approximate 3- to 16-fold reduction cannot fully account for the greater-than-1,000-fold defect in virus infectivity (77) (Fig. 8). Because the frequencies of U3 or U5 end deletions in HIV-1K264E 2-LTR CJs were lower than the corresponding WT value (Fig. 9; Table 4), we inferred that the mutant vDNA ends were no more susceptible to degradation by host nucleases than was the WT (119). Lys264 contributes to vDNA binding (129), yet INK264E supported INWT activity under certain reaction conditions (Fig. 4 and 7), seemingly discounting defective vDNA binding as the underlying cause of the mutant infection defect. We therefore speculate that INK264E is defective for interacting with an intranuclear factor that plays an important role in HIV-1 integration. Consistent with this interpretation, the side chain of Lys264 is solvent accessible in the X-ray crystal structure of the IN two-domain CCD-CTD fragment (20) as well as in the HIV-1 intasome model that was built in silico from this and other composite molecules (64). It appears that our study in one case would support the initial hypothesis that class II IN mutations perturb interactions with binding partners that are essential for HIV-1 replication.
The LEDGF/p75 chromatin-associated protein, which plays a critical role in HIV-1 integration, engages the IN NTD and CCD with little or no contribution from the CTD (81), and INK264E concordantly displayed WT binding affinity for the host factor in vitro (Fig. 10). The CTD, by contrast, supplies the primary binding epitope for RT (55, 130), and the polymerase can stimulate IN catalytic function (55). Our findings, though, revealed that INK264E retained the WT binding affinity for RT (Fig. 11), seemingly dispensing a role for the polymerase in the underlying viral reverse transcription or integration defect. Based on levels of vDNA synthesis during infection, we would have expected HIV-1K264E to yield an approximate 3-fold increase in 2-LTR circles relative to the WT if the mutant behaved similarly to catalytically deficient class I mutant viruses, like HIV-1D64N/D116N (Fig. 8A and B) and HIV-1W235E (67). It therefore seems possible that HIV-1K264E is marginally (∼3-fold) impaired for PIC nuclear import. Because Lys264 reportedly contributes to importin α3 binding affinity (59), it could be instructive to test the effect of the K264E change on this interaction.
Residue 264 is one of three CTD lysines acetylated by p300 (17), which forms a binding site for the transcriptional corepressor KAP1 and recruitment of the histone deacetylase HDAC1 (1). Although we have not specifically investigated the role of the K264E change on IN-KAP1-HDAC1 complex formation, we speculate that this is unlikely to be the root cause of the associated virus infection defect. Acetylation-defective IN supported significantly less DNA strand transfer activity in vitro than the corresponding modified enzyme (17), yet HIV-1K264E PICs were at least as active as those derived from WT HIV-1NL4-3 (Fig. 7). Another host factor that has been reported to engage HIV-1 IN through its CTD and stimulate catalytic function is EED (122), although follow-up work has indicated EED plays a primary role in controlling virus egress compared to influx and integration (103). Additional work is required to determine if the K264E mutation specifically disrupts interactions with KAP1, EED, importin α3, or possibly an as-yet-uncharacterized IN binding factor. Due to the severity of HIV-1K264E infection and integration blocks (Fig. 8), the identification of the presumed binding partner(s) could lead to new therapeutic opportunities to interfere with a critical virus-host interaction.
ACKNOWLEDGMENTS
We are indebted to Jeroen van Wamel and Ben Berkhout for pGST-RT plasmid DNA, Duane Grandgenett for purified INWT protein and pGEM-3 plasmid DNA, and Peter Cherepanov for INWT protein, PC2 cells, and critical review of the manuscript.
This work was funded by NIH grant R37AI039394 and the Harvard University Center for AIDS Research (CFAR), an NIH-funded program (P30AI060354) that is supported by the following NIH Institutes and Centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM.
The contents of this article and mention of commercial products or organizations in no way reflect the opinions or endorsement by the U.S. Government.
Footnotes
Published ahead of print 25 January 2012
REFERENCES
- 1. Allouch A, et al. 2011. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9:484–495 [DOI] [PubMed] [Google Scholar]
- 2. Ansari-Lari MA, Donehower LA, Gibbs R. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332–335 [DOI] [PubMed] [Google Scholar]
- 3. Ao Z, Fowke K, Cohen E, Yao X. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bera S, Pandey KK, Vora AC, Grandgenett DP. 2009. Molecular interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J. Mol. Biol. 389:183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berthoux L, Sebastian S, Muesing MA, Luban J. 2007. The role of lysine 186 in HIV-1 integrase multimerization. Virology 364:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouyac-Bertoia M, et al. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025–1035 [DOI] [PubMed] [Google Scholar]
- 7. Bowerman B, Brown PO, Bishop JM, Varmus HE. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469–478 [DOI] [PubMed] [Google Scholar]
- 8. Briones MS, Chow SA. 2010. A new functional role of HIV-1 integrase during uncoating of the viral core. Immunol. Res. 48:14–26 [DOI] [PubMed] [Google Scholar]
- 9. Briones MS, Dobard CW, Chow SA. 2010. Role of human immunodeficiency virus type 1 integrase in uncoating of the viral core. J. Virol. 84:5181–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown PO, Bowerman B, Varmus HE, Bishop JM. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347–356 [DOI] [PubMed] [Google Scholar]
- 11. Brown PO, Bowerman B, Varmus HE, Bishop JM. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. U. S. A. 86:2525–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bukovsky A, Göttlinger H. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bushman FD, Craigie R. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: Specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U. S. A. 88:1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. U. S. A. 90:3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Busschots K, et al. 2007. Identification of the LEDGF/p75 binding site in HIV-1 integrase. J. Mol. Biol. 365:1480–1492 [DOI] [PubMed] [Google Scholar]
- 16. Carteau S, Gorelick RJ, Bushman FD. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cereseto A, et al. 2005. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 24:3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H, Engelman A. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen H, Wei S-Q, Engelman A. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 274:17358–17364 [DOI] [PubMed] [Google Scholar]
- 20. Chen JC-H, et al. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. U. S. A. 97:8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cherepanov P. 2007. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35:113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cherepanov P, Ambrosio ALB, Rahman S, Ellenberger T, Engelman A. 2005. From the cover: structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U. S. A. 102:17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cherepanov P, Devroe E, Silver PA, Engelman A. 2004. Identification of an evolutionarily-conserved domain in LEDGF/p75 that binds HIV-1 integrase. J. Biol. Chem. 279:48883–48892 [DOI] [PubMed] [Google Scholar]
- 24. Cherepanov P, et al. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372–381 [DOI] [PubMed] [Google Scholar]
- 25. Cherepanov P, et al. 1999. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 27:2202–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chow SA, Vincent KA, Ellison V, Brown PO. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723–726 [DOI] [PubMed] [Google Scholar]
- 27. Dar MJ, et al. 2009. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobard CW, Briones MS, Chow SA. 2007. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 81:10037–10046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dvorin JD, et al. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emiliani S, et al. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280:25517–25523 [DOI] [PubMed] [Google Scholar]
- 31. Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411–426 [DOI] [PubMed] [Google Scholar]
- 32. Engelman A. 2011. Pleiotropic nature of HIV-1 integrase mutations, p 67–81 In Neamati N. (ed), HIV-1 integrase: mechanism and inhibitor design. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 33. Engelman A. 2010. Reverse transcription and integration, p 129–159 In Kurth R, Bannert N. (ed), Retroviruses: molecular biology, genomics and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 34. Engelman A, Bushman FD, Craigie R. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Engelman A, Cherepanov P. 2008. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4:e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Engelman A, Craigie R. 1995. Efficient magnesium-dependent human immunodeficiency virus type 1 integrase activity. J. Virol. 69:5908–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Engelman A, Craigie R. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Engelman A, Hickman AB, Craigie R. 1994. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 68:5911–5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Engelman A, Liu Y, Chen H, Farzan M, Dyda F. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol. 71:3507–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221 [DOI] [PubMed] [Google Scholar]
- 42. Engelman A, Oztop I, Vandegraaff N, Raghavendra NK. 2009. Quantitative analysis of HIV-1 preintegration complexes. Methods 47:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Englund G, Theodore TS, Freed EO, Engleman A, Martin MA. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Esposito D, Craigie R. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Farnet CM, Haseltine WA. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 87:4164–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Faure A, et al. 2005. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 33:977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fletcher TM, III, et al. 1997. Complementation of integrase function in HIV-1 virions. EMBO J. 16:5123–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fujiwara T, Mizuuchi K. 1988. Retroviral DNA integration: structure of an integration intermediate. Cell 54:497–504 [DOI] [PubMed] [Google Scholar]
- 49. Gallay P, Hope T, Chin D, Trono D. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 94:9825–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. 2006. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J. Virol. 80:5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hare S, et al. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 5:e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hare S, et al. 2009. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5:e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hazuda DJ, et al. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650 [DOI] [PubMed] [Google Scholar]
- 55. Hehl EA, Joshi P, Kalpana GV, Prasad VR. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hindmarsh P, et al. 1999. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 73:2994–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ikeda T, et al. 2004. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 78:11563–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ishikawa T, et al. 1999. Monoclonal antibodies against the minimal DNA-binding domain in the carboxyl-terminal region of human immunodeficiency virus type 1 integrase. J. Virol. 73:4475–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jayappa KD, Ao Z, Yang M, Wang J, Yao X. 2011. Identification of critical motifs within HIV-1 integrase required for importin α3 interaction and viral cDNA nuclear import. J. Mol. Biol. 410:847–862 [DOI] [PubMed] [Google Scholar]
- 60. Jenkins TM, Engelman A, Ghirlando R, Craigie R. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and the C-terminal domains in multimerization. J. Biol. Chem. 271:7712–7718 [DOI] [PubMed] [Google Scholar]
- 61. Jenkins TM, Esposito D, Engelman A, Craigie R. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. U. S. A. 99:9515–9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalpana GV, et al. 1999. Isolation and characterization of an oligomerization-negative mutant of HIV-1 integrase. Virology 259:274–285 [DOI] [PubMed] [Google Scholar]
- 64. Krishnan L, et al. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 107:15010–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kulkosky J, Katz RA, Merkel G, Skalka AM. 1995. Activities and substrate specificity of the evolutionarily conserved central domain of retroviral integrase. Virology 206:448–456 [DOI] [PubMed] [Google Scholar]
- 66. LaFemina R, et al. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Leavitt AD, Robles G, Alesandro N, Varmus HE. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leavitt AD, Shiue L, Varmus HE. 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 268:2113–2119 [PubMed] [Google Scholar]
- 69. Li L, et al. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li M, Craigie R. 2005. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 280:29334–29339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li M, Mizuuchi M, Burke TR, Craigie R. 2006. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li X, Krishnan L, Cherepanov P, Engelman A. 2011. Strucutral biology of retroviral DNA integration. Virology 411:194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liao W-H, Wang C-T. 2004. Characterization of human immunodeficiency virus type 1 Pr160 gag-pol mutants with truncations downstream of the protease domain. Virology 329:180–188 [DOI] [PubMed] [Google Scholar]
- 74. Lide DR. 1998. Handbook of chemistry and physics, 79th ed CRC Press, Boca Raton, FL [Google Scholar]
- 75. Limón A, et al. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lloyd AG, Ng YS, Muesing MA, Simon V, Mulder LCF. 2007. Characterization of HIV-1 integrase N-terminal mutant viruses. Virology 360:129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lu R, Ghory HZ, Engelman A. 2005. Genetic analyses of conserved residues in the carboxyl terminal domain of human immunodeficiency virus type 1 integrase. J. Virol. 79:10356–10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lu R, et al. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a post-nuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lu R, Limon A, Ghory HZ, Engelman A. 2005. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. J. Virol. 79:2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lu R, Vandegraaff N, Cherepanov P, Engelman A. 2005. Lys-34, dispensable for integrase catalysis, is required for preintegration complex function and human immunodeficiency virus type 1 replication. J. Virol. 79:12584–12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Maertens G, et al. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528–33539 [DOI] [PubMed] [Google Scholar]
- 82. Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration through X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mandal D, Prasad VR. 2009. Analysis of 2-LTR circle junctions of viral DNA in infected cells. Methods Mol. Biol. 485:73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Masuda T, Planelles V, Krogstad P, Chen IS. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 85:7818–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. McKee CJ, et al. 2008. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J. Biol. Chem. 283:31802–31812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. McWilliams MJ, Julias JG, Hughes SH. 2008. Mutations in the human immunodeficiency virus type 1 polypurine tract (PPT) reduce the rate of PPT cleavage and plus-strand DNA synthesis. J. Virol. 82:5104–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miller M, Farnet C, Bushman F. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nakamura T, et al. 1997. Lack of infectivity of HIV-1 integrase zinc finger-like domain mutant with morphologically normal maturation. Biochem. Biophys. Res. Commun. 239:715–722 [DOI] [PubMed] [Google Scholar]
- 90. Nishitsuji H, et al. 2009. Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 is critical for HIV-1 infection. PLoS One 4:e7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Oude Essink BB, Back NKT, Berkhout B. 1997. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 25:3212–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pandey KK, et al. 2007. Inhibition of human immunodeficiency virus type 1 concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J. Virol. 81:12189–12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pandey KK, Sinha S, Grandgenett DP. 2007. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J. Virol. 81:3969–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pauza C. 1990. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology 179:886–889 [DOI] [PubMed] [Google Scholar]
- 95. Petit C, Schwartz O, Mammano F. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Petit C, Schwartz O, Mammano F. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Poeschla EM. 2008. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 65:1403–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Puras Lutzke RA, Plasterk RHA. 1998. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J. Virol. 72:4841–4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Puras Lutzke RA, Vink C, Plasterk RHA. 1994. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 22:4125–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Quillent C, Borman AM, Paulous S, Dauguet C, Clavel F. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 219:29–36 [DOI] [PubMed] [Google Scholar]
- 101. Raghavendra NK, Engelman A. 2007. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology 360:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rahman S, Lu R, Vandegraaff N, Cherepanov P, Engelman A. 2007. Structure-based mutagenesis of the integrase-LEDGF/p75 interface uncouples a strict correlation between in vitro protein binding and HIV-1 fitness. Virology 357:79–90 [DOI] [PubMed] [Google Scholar]
- 103. Rakotobe D, et al. 2007. Human Polycomb group EED protein negatively affects HIV-1 assembly and release. Retrovirology 4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Riviere L, Darlix J-L, Cimarelli A. 2010. Analysis of the viral elements required in the nuclear import of HIV-1 DNA. J. Virol. 84:729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Saenz DT, et al. 2004. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J. Virol. 78:2906–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sakai H, et al. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shin C, Taddeo B, Haseltine W, Farnet C. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sinha S, Grandgenett DP. 2005. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 79:8208–8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Stevenson M, et al. 1990. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J. Virol. 64:2421–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Svarovskaia ES, et al. 2004. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat circle junctions. J. Virol. 78:3210–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Taddeo B, Haseltine W, Farnet C. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J. Virol. 68:8401–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tsiang M, et al. 2009. Affinities between the binding partners of the HIV-1 integrase dimer–lens epithelium-derived growth factor (IN dimer-LEDGF) complex. J. Biol. Chem. 284:33580–33599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tsurutani N, et al. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Turlure F, Devroe E, Silver PA, Engelman A. 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187–3208 [DOI] [PubMed] [Google Scholar]
- 115. Valkov E, et al. 2009. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 37:243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. 2006. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology 346:415–426 [DOI] [PubMed] [Google Scholar]
- 117. van Gent DC, Groeneger AA, Plasterk RHA. 1992. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl. Acad. Sci. U. S. A. 89:9598–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. 2006. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 31:98–105 [DOI] [PubMed] [Google Scholar]
- 119. Vatakis DN, Kim S, Kim N, Chow SA, Zack JA. 2009. Human immunodeficiency virus integration efficiency and site selection in quiescent CD4+ T cells. J. Virol. 83:6222–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Vink C, Oude Groeneger AM, Plasterk RHA. 1993. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 21:1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Vink C, Yeheskiely E, van der Marel GA, Van Boom JH, Plasterk RHA. 1991. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 19:6691–6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Violot S, et al. 2003. The human Polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J. Virol. 77:12507–12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wilkinson TA, et al. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 284:7931–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wiskerchen M, Muesing MA. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Woerner AM, Marcus-Sekura CJ. 1993. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 21:3507–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wu X, et al. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wu X, et al. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Yung E, et al. 2004. Specificity of interaction of INI1/hSNF5 with retroviral integrases and its functional significance. J. Virol. 78:2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhao Z, et al. 2008. Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 integrase during viral DNA binding. J. Biol. Chem. 283:5632–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhu K, Dobard C, Chow SA. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]