Abstract
The antiviral factor CPSF6-358 restricts human immunodeficiency virus type 1 (HIV-1) infection through an interaction with capsid (CA), preventing virus nuclear entry and integration. HIV-1 acquires resistance to CPSF6-358 through an N74D mutation of CA that impairs binding of the antiviral factor. Here we examined the determinants within CPSF6-358 that are necessary for CA-specific interaction. Residues 314 to 322 include amino acids that are essential for CPSF6-358 restriction of HIV-1. Fusion of CPSF6 residues 301 to 358 to rhesus TRIM5α is also sufficient to restrict wild-type but not N74D HIV-1. Restriction is lost if CPSF6 residues in the amino acid 314 to 322 interaction motif are mutated. Examination of the CA targeting motif in CPSF6-358 did not reveal evidence of positive selection. Given the sensitivity of different primate lentiviruses to CPSF6-358 and apparent conservation of this interaction, our data suggest that CPSF6-358-mediated targeting of HIV-1 could provide a broadly effective antiviral strategy.
INTRODUCTION
The capsid (CA) proteins of mature retroviruses form core structures that package the viral nucleic acid and enzymatic proteins required for early steps in replication. CA is initially associated with human immunodeficiency virus type 1 (HIV-1) after entry into cells (8, 10, 19). Notably, retroviral CA is a focal point for restriction by antiviral factors during postentry infection. Early genetic studies identified a relationship between the mouse Fv1 locus and susceptibility to infection by gammaretroviruses (15, 24). Depending on the mouse background, murine leukemia viruses (MLV) were restricted from infection of mouse cells in a CA-dependent manner (31). Restriction to MLV infection by Fv1 was deduced to occur through the interaction of a dominant negative factor (25). Efforts to identify Fv1 were impaired due to low-level expression of the factor. Positional cloning revealed Fv1 to be a retroviral gag-related factor (4). Although genetic studies provided details of the interaction of Fv1 with MLV CA during early infection, attempts to study direct binding were unsuccessful until recently (13). The inability of HIV-1 to replicate in cells from nonhuman primates suggested that similar factors could also regulate lentiviral infection (3, 6, 12, 14, 21, 35, 36). Subsequent studies uncovered products of TRIM5-related genes as restriction factors in primate cells. Rhesus TRIM5α (rhTRIM5α) and owl monkey TRIM-Cyp interact with HIV-1 CA and interfere with postentry replication steps (22, 30, 33, 37). While the mechanism of interaction of TRIM-Cyp with HIV-1 CA was quickly surmised, the interface between rhTRIM5α and HIV-1 CA was first described via genetic studies and more recently elaborated through structural analysis (11).
We identified a carboxyl-terminal (C-terminal) truncation mutant of cleavage and polyadenylation factor 6, CPSF6-358, that restricts HIV-1 infection (18). Truncation of CPSF6 enriches the protein in the cytoplasm and prevents nuclear entry of the HIV-1 preintegration complex (PIC). Primate lentiviruses, including HIV-1, HIV-2, and SIVmac (simian immunodeficiency virus), are restricted by the factor, whereas other lentiviruses, such as feline immunodeficiency virus (FIV) and equine infectious anemia virus (EIAV), are not. HIV-1 interaction with CPSF6-358 is CA dependent. Replacement of HIV-1 CA with that of MLV relieves the restriction to CPSF6-358. Passaging of HIV-1 in cells expressing CPSF6-358 selects for an N74D resistance mutation in CA. Introduction of this single mutation is sufficient to confer CPSF6-358 resistance to different HIV-1 isolates. Recombinant HIV-1 CA with the N74D mutation has reduced binding to CPSF6-358 relative to that of wild-type (WT) CA. The C-terminal 58 residues of CPSF6-358 contribute to antiviral restriction of and binding to WT HIV-1. Primate lentiviruses resistant to TRIM5 or TRIM-Cyp family proteins retain sensitivity to CPSF6-358, indicating a distinct interaction surface in CA.
Here we have mapped a 9-residue stretch of hydrophobic amino acids in CPSF6 that is required for HIV-1 restriction. The interaction domain is modular and can enable CA-specific targeting of HIV-1 by fusion proteins. Depending on the fusion partner, HIV-1 replication can be blocked at different, early replication steps using the CPSF6-358 CA targeting motif. These findings highlight a third mechanism of HIV-1 CA interaction with a cellular protein during early infection (in addition to TRIM5 and CypA mediated) and only the second with a human protein partner.
MATERIALS AND METHODS
Cell culture and antibodies.
The 293T and HeLa cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (HyClone), 100 U/ml penicillin, and 0.1 mg/ml streptomycin. HeLa cell lines transduced with pLPCX-based vectors to stably express CPSF6 mutants were selected and maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 1 μg/ml puromycin (Calbiochem). Antibodies used included monoclonal antibodies to hemagglutinin (HA) tag (Sigma) and tubulin (Sigma) and polyclonal antibody to Aequorea coerulescens green fluorescent protein (AcGFP) (Clontech). Secondary antibodies included horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit antibodies (GE Healthcare).
Virus stocks.
The Moloney MLV-based retroviral vectors pLPCX (Clontech) and pMX (23) were used to generate stable cell lines. The MLV vector stocks were produced by DNA transfections of 293T cells grown in 6-well plates using HilyMax transfection reagent (Dojindo Molecular Technologies, Inc.). Each cell monolayer (well) was cotransfected with pLPCX- or pMX-based expression plasmid and pJK3, pCMV-Tat, and pL-VSV-G plasmids at a 4:2:0.3:1 ratio (2). HIV-1 virus stocks were produced by DNA transfection on monolayer cultures of 293T cells grown in 10-cm culture dishes using HilyMax transfection reagent. Each 10-cm dish was cotransfected with wild-type (WT)- or N74D CA-bearing pHIV-RFP (18) and pL-VSV-G at a 3:1 ratio. Culture supernatants from the 293T cells were collected 48 h posttransfection, clarified by low-speed centrifugation (1,000 × g, 10 min), and filtered through 0.45-μm-pore-size sterile filters. For the HIV-1 vectors, the clarified supernatants were titrated in 293T and HeLa cells to determine the infectious units (IFU) per ml.
CPSF6-358 expression vectors.
pLPC-CPSF6-FL-HA and pLPC-CPSF6-358HA were generated by PCR from full-length human CPSF6 cDNA (GenBank accession number BC000714) and have been described previously (18). The Aequorea coerulescens GFP (AcGFP) gene used in fusions encodes a monomeric form of GFP with spectral properties similar to those of enhanced GFP (Clontech). To generate CPSF6-358 deletion mutants by overlap PCR, forward and reverse primers containing equivalent lengths of sequence on the two sides of the deletion were designed. In the first PCR, two separate reactions were performed. One PCR included the forward primer (complementary to the 5′ end of the CPSF6 gene) containing the EcoRI restriction site and Kozak sequence and the reverse primer for creating the deletion, while the other PCR included the forward primer for creating the deletion and the reverse primer that contained the HA tag coding sequence, stop codon, and a NotI restriction enzyme site, with pLPC-CPSF6-358HA plasmid as the template for both PCRs. In the subsequent second PCR step, the two overlapping PCR products generated in the first PCR step were mixed together and used as a template along with the external forward and reverse primers containing the EcoRI and NotI sequences, respectively, to generate the full-length PCR products containing the desired deletion. Constructs were verified by DNA sequencing. To generate the triple and single alanine mutants, a two-step PCR was performed. In the first PCR step, forward (+) mutant primers that contained base substitutions and the reverse primer that contained the HA tag coding sequence, stop codon, and a NotI restriction enzyme site generated mutant fragments. In the second PCR, these generated fragments were used in combination with a forward primer (complementary to the 5′ end of the CPSF6 gene) containing an EcoRI restriction site and the Kozak sequence (CCATGG). The pLPC-CPSF6-358HA plasmid was used as the template in both PCRs. The presence of a single PCR product of the expected size/length in each of the first set of PCRs was confirmed on 1.0% agarose gels before the second PCR step was begun. The products generated after the second PCR were cut with EcoRI-NotI and cloned into an EcoRI-NotI-digested pLPCX vector. For generating AcGFP fusion proteins, the 301-358HA sequence was amplified from the wild-type or triple alanine mutant CPSF6-358HA construct with forward and reverse primers containing BglII and NotI, respectively, and the generated fragment was cloned into the pAcGFP-N1 multiple cloning site. The AcGFP-301-358HA fusion was subsequently shuttled into the pMX vector. Fusions with rhesus TRIM5α were generated in a similar manner. All fusion and mutant proteins were confirmed by DNA sequencing.
Transductions and virus infections.
For transduction using pLPCX-derived vectors bearing a puromycin resistance gene, HeLa cells were infected with vectors carrying various genes and selected in DMEM containing 1 μg/ml puromycin at 48 to 72 h postinfection. Similarly, for transduction of AcGFP fused proteins using pMX-derived vectors, the HeLa cells were transduced with vectors and cultured at least for 1 week to allow stable expression of introduced proteins. Transduced cells then were sorted twice on the basis of AcGFP expression to obtain stable lines using a fluorescence-activated cell sorter (FACS) (FACSAria II; Becton Dickinson) (see Fig. S1A in the supplemental material). For HIV-1 infection, pretitrated WT HIV-RFP/VSV-G or N74D HIV-RFP/VSV-G virus stocks were used to infect monolayer cultures of HeLa cells, which were plated the previous day at 40,000 cells/well in 12-well tissue culture plates (Corning). At 48 h postinfection, the cells were trypsinized and resuspended in phosphate-buffered saline (PBS) containing 2% FBS. The percentage of red fluorescent protein (RFP)-expressing, infected cells was measured by FACS analysis (FACSCalibur; Becton Dickinson).
Western blot analysis.
Transfected or transduced cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma) (50 mM Tris-HCl, pH 8.0, with 150 mM sodium chloride, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate). Prior to loading, the cell lysates were mixed with a 2× Laemmli sample loading buffer (Bio-Rad) (62.5 mM Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, 0.01% bromophenol blue, and 5% 2-mercaptoethanol) and boiled, and proteins were separated on 10.0% fixed or 4 to 15% gradient polyacrylamide gels containing SDS. Following electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane by electroblotting and incubated for 1 h at room temperature in blocking buffer (5% nonfat dry milk in PBS). The blocked blot was exposed to the appropriate primary antibody in blocking buffer with constant shaking. After extensive washing, bound antibodies were detected by chemiluminescence using horseradish peroxidase-conjugated species-specific secondary antibodies as described by the manufacturer (GE Healthcare).
Real-time qPCR analysis of virus infection.
HIV-1 vectors were treated with DNase I (Roche) at 37°C for 30 min. These DNase I-treated HIV-1 vectors were used to infect HeLa cells at a multiplicity of infection (MOI) of 1 for different times. At each time point, cells were collected and lysed, and genomic DNA was extracted using a QIAmp blood minikit (Qiagen). As a control, an equivalent amount of virus was either heat inactivated at 56°C for 30 min before infecting cells or treated with the reverse transcriptase inhibitor efavirenz (100 nM) at the time of infection. Samples were assayed by quantitative PCR (qPCR) using Platinum qPCR SuperMix-UDG (Invitrogen), primers, and probes as described previously (16). Duplicate samples of serial dilutions of plasmid DNAs containing the target sequences were used to generate a standard curve, which was used for quantification of PCR products.
Primate gene sequences.
TRIM5 and APOBEC3G gene sequences for human, chimpanzee, gorilla, orangutan, white-cheeked gibbon, rhesus macaque, and marmoset species were obtained from GenBank. CPSF6 sequences for the same species were obtained from the UCSC genome database using the BLAT alignment tool (17).
PAML analysis.
Multiple alignments were created with CLUSTAL_X 2.0 (34). Maximum-likelihood analysis was performed with codeml in the PAML4.2 software package (39). To detect selection, multiple alignments were fitted to the M7 (neutral model, codon values of dN/dS [ratio of nonsynonymous to synonymous substitutions] fit to a beta distribution, dN/dS of >1 disallowed) and M8 (positive-selection model, similar to M7 but with an extra class of dN/dS of >1 allowed) models using the f61 model of codon frequencies. Likelihood ratio tests were performed to assess whether permitting codons to evolve under positive selection gives a significantly better fit to the data (M7 versus M8).
RESULTS
Deletion mutagenesis of CPSF6-358.
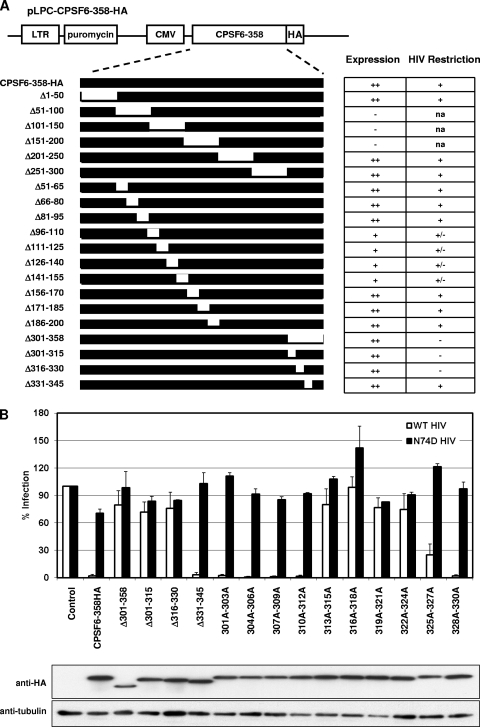
To identify CPSF6-358 determinants responsible for CA-specific inhibition of HIV-1, we first created CPSF6-358 mutants with sequential, nonoverlapping deletions of 50 or more amino acids along the length of the protein: CPSF6-358(Δ1–50), CPSF6-358(Δ51–100), CPSF6-358(Δ101–150), CPSF6-358(Δ151–200), CPSF6-358(Δ201–250), CPSF6-358(Δ251–300), and CPSF6-358(Δ301–358) (Fig. 1A). Each mutant was assayed for stable expression in HeLa cells after vector transduction and antibiotic selection. All proteins contained a C-terminal HA epitope tag to enable comparison of expression levels. Vectors expressing CPSF6-358(Δ51–100), CPSF6-358(Δ101–150), and CPSF6-358(Δ151–200) mutants could not be tested, as loss of these regions destabilized expression. These proteins could not be detected from lysates of cell lines by using antibody to CPSF6 or the HA epitope. In contrast, CPSF6-358(Δ1–50), CPSF6-358(Δ201–250), and CPSF6-358(Δ251–300) were expressed at levels comparable to that of CPSF6-358 and restricted WT HIV-1 but not N74D HIV-1 infection. The mutant CPSF6-358(Δ301–358) was also stably expressed but failed to restrict HIV-1 infection. We previously reported that this mutant had diminished binding to WT CA-NC complexes relative to that seen with CPSF6-358 (18).
Fig 1.
CPSF6-358 residues 313 to 327 contribute to antiviral activity. (A) Summary of CPSF6-358 domain mapping by deletion mutagenesis. Briefly, HA-tagged CPSF6-358 was cloned into a pLPCX vector containing a puromycin gene as a selectable marker, and various CPSF6-358 deletion mutants were generated as described in the text. HeLa cells were transduced with CPSF6-358HA or CPSF6-358HA constructs carrying various deletions and then tested for their HIV-1 restriction by infecting cells with WT HIV-1 and N74D HIV-1. CMV, cytomegalovirus. Expression column: ++, high level; +, detectable; −, none. HIV Restriction column: +, strong; +/−, partial; −, none; na, not attempted. (B) (Top) WT or N74D HIV-RFP/VSV-G infection of HeLa cells expressing control vector, CPSF6-358, or various CPSF6-358 mutants as shown. After quantification of RFP-positive cells via FACS, percent infection relative to that for control vector-expressing cells was calculated. Error bars indicate standard deviations. (Bottom) Western blot analysis of introduced proteins expressed in stable cell lines.
Because 50-amino-acid deletions between residues 51 and 200 interfered with protein expression or stability, we attempted to examine the role of this region in CPSF6-358 restriction by using smaller, 15-amino-acid deletions. These mutant proteins displayed improved expression. For proteins with expression comparable to that of CPSF6-358, similar levels of WT HIV-1 restriction were observed. For mutant proteins that contained deletions between residues 96 and 155, expression was detected but was diminished relative to that of CPSF6-358. Weak inhibition of WT HIV-1 infection by these mutants was also observed, and this effect was specific, as N74D HIV-1 infection was unaffected (data not shown).
We thus focused on further mutagenesis of residues 301 to 358 in CPSF6-358. CPSF6-358(Δ301-315) and CPSF6-358(Δ316-330) mutant proteins did not restrict WT HIV-1 infection, whereas CPSF6-358(Δ331-345) was still inhibitory to WT HIV-1 infection without affecting N74D HIV-1 infection (Fig. 1B). Attempts to derive cell lines expressing CPSF6-345, missing residues 346 to 358 of CPSF6-358, were unsuccessful. A CPSF6-340-AcGFP fusion protein was instead tested and was antiviral to WT HIV-1 infection (data not shown). These experiments suggested that CPSF6 residues 301 to 330 contained the CA-specific determinant. To examine the contribution of these residues more carefully, we replaced every three consecutive residues in the amino acid 301 to 330 stretch of CPSF6-358 with alanines and expressed these proteins in HeLa cells. CPSF6-358 bearing triple alanine substitutions between residues 301 and 312 strongly inhibited WT HIV-1 infection. CPSF6-358 with a triple alanine substitution between residues 328 and 330 also restricted WT HIV-1. In contrast, CPSF6-358(313A-315A), CPSF6-358(316A-318A), CPSF6-358(319A-321A), and CPSF6-358(322A-324A) displayed no restriction of HIV-1 despite stable expression of the mutant proteins. Cells expressing CPSF6-358(325A-327A) also showed reduced susceptibility to HIV-1 infection.
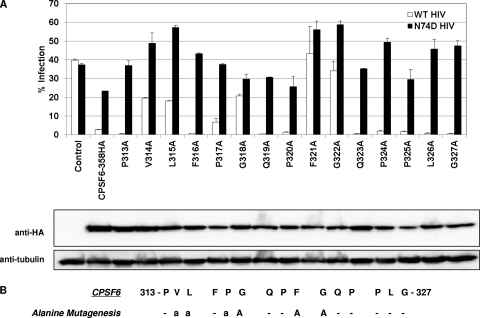
Single alanine substitution of CPSF6 residues 313 to 327.
We next assessed the contribution of individual amino acids in residues 313 to 327 of CPSF6-358 to the inhibition of WT HIV-1 infection. Individual residues were mutated to alanine, mutant proteins were stably expressed in HeLa cells, and cells were infected with WT versus N74D HIV-1 (Fig. 2A). CPSF6(V314A), CPSF6(L315A), and CPSF6(P317A) showed loss of some antiviral activity to WT HIV-1 infection compared to that seen with CPSF6-358-expressing cells. CPSF6(G318A), CPSF6(F321A), and CPSF6(G322A) lost almost all antiviral activity to WT HIV-1 infection. Interpretation of loss of activity was based on comparison to N74D HIV-1 infection of cells plated in parallel to control for differences in plating efficiency or cell proliferation. MLV infections were also conducted in parallel to control for cell number variations (data not shown). In comparison, alanine mutation of CPSF6-358 residue 313, 316, 319, 320, 323, 324, 325, 326, or 327 yielded proteins that maintained strong antiviral activity against WT HIV-1 infection. We also examined effects of single alanine mutations in CPSF6-358 by transient expression in 293T cells and confirmed that alanine mutations at residues 314, 315, 317, 318, 321, and 322 of CPSF6-358 affected the WT HIV-1 infection block (data not shown). To verify the contribution of this region of CPSF6-358 to HIV-1 CA interaction, we made individual tryptophan substitutions to introduce a bulkier hydrophobic side chain in place of residues 316 to 321. These mutations diminished the antiviral activity of CPSF6-358 to various degrees (see Fig. S2 in the supplemental material). While such substitutions helped confirm this region of CPSF6 as a binding interface, in some instances loss of binding could occur through steric hindrance, especially in cases where alanine substitutions were biochemically significant (e.g., P320A) but yielded no phenotype. Consistent with this interpretation, tryptophan mutant proteins that showed residual restrictive activity (e.g., P320W) segregated into this category. This combined analysis supports a role for residues 314, 315, 317, 318, 321, and 322 in CPSF6-358 restriction (Fig. 2B).
Fig 2.
Functional analysis of CPSF6-358 residues 313 to 327 by alanine scanning mutagenesis. (A) (Top) WT and N74D HIV-RFP/VSV-G infection of HeLa cells expressing CPSF6-358 mutants carrying single alanine mutations from residues 313 to 327. Percent infected cells was measured by FACS for RFP expression at 48 h postinfection. Error bars indicate standard deviations. (Bottom) Western blot analysis of stably transduced proteins. (B) Summary of alanine scanning mutagenesis. “a” represents single alanine mutants of CPSF6-358 with reduced antiviral activity. “A” represents alanine mutants of CPSF6-358 with minimal or no restriction of WT HIV-1.
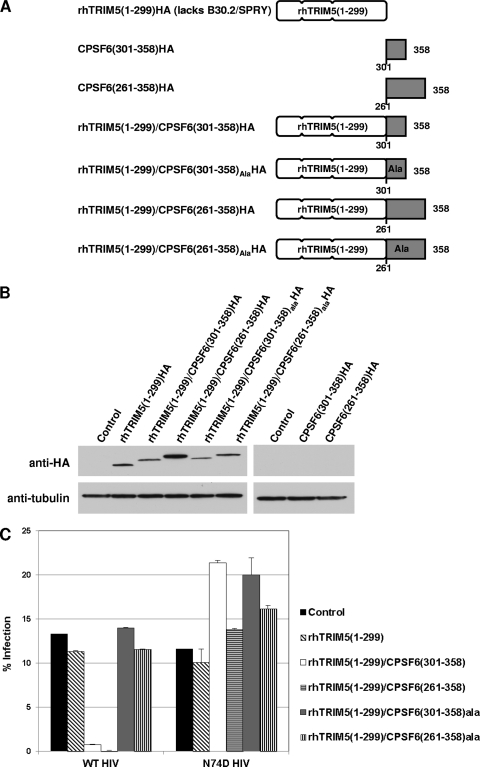
HIV-1 core specificity of the CPSF6-358 C terminus is transferable to rhesus TRIM5α.
The TRIM5α protein is a CA-dependent retroviral restriction factor (33). The protein contains separable domains that provide antiviral function and CA specificity. Gene shuffling events that created TRIM-Cyp proteins in owl monkeys first illustrated these modular properties (22, 30). Such proteins were recapitulated using human or rhesus TRIM5α lacking a CA-interacting B30.2/SPRY domain [TRIM5α ΔB30.2/TRIM5α(1–299)] fused to the HIV-1 CA binding protein cyclophilin A (CypA). The engineered TRIM-Cyp proteins restricted HIV-1 in a manner that was dependent on interaction with CA, with specificity provided by the fused CypA (40). Our characterization of CPSF6-358 indicated residues 314 to 322 to be necessary for CA-dependent HIV-1 inhibition. To determine whether sequences in the CPSF6-358 C-terminal region were sufficient for CA interaction, we fused CPSF6-358(261–358) or CPSF6-358(301–358) to rhTRIM5α(1–299) containing RBCC domains (the tripartite RING motif, B box, and coiled-coil motif) but not the B30.2/SPRY domain (Fig. 3A). As controls, CPSF6-358(261–358)ala and CPSF6-358(301–358)ala containing alanine mutations in residues 319 to 321 were also fused to rhTRIM5(1–299). At the same time, we also wanted to test if CPSF6-358(261–358) or CPSF6-358(301–358) protein fragments without a fusion could be antiviral. The different proteins illustrated in Fig. 3A were transduced into HeLa cells and tested for their expression by Western blot analysis. The unfused rhTRIM5(1–299) and the rhTRIM5(1–299) fused with CPSF6(261–358), CPSF6(301–358), CPSF6(261–358)ala, or CPSF6(301–358)ala coding sequence were efficiently expressed in HeLa cells; however, the unfused CPSF6(261–358) and CPSF6(301–358) proteins were not expressed (Fig. 3B). We tested antiviral function of TRIM5/CPSF6 fusion proteins that were stably expressed by infecting cells with WT HIV-1 or N74D HIV-1. Empty-vector-transduced control cells and rhTRIM5(1–299)-expressing cells were efficiently infected by both WT and N74D HIV-1 (Fig. 3C). WT HIV-1 infection was markedly inhibited in cells expressing rhTRIM5(1–299)/CPSF6(261–358) and rhTRIM5(1–299)/CPSF6(301–358) fusion proteins, while WT HIV-1 infection of CPSF6-358(261–358)ala- and CPSF6-358(301–358)ala-expressing cells was similar to that seen with control cells. Infectivity of N74D HIV-1 was not significantly decreased in any of the rhTRIM5(1–299)/CPSF6 fusion protein-expressing cells. Similar results were obtained through infection of 293T cells transiently transfected with the same expression constructs (see Fig. S3 in the supplemental material).
Fig 3.
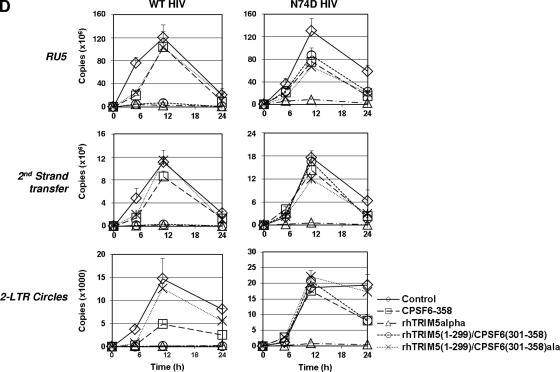
Analysis of TRIM5/CPSF6 fusion proteins. (A) The B30.2/SPRY domain of rhTRIM5α was replaced by CPSF6(261–358), CPSF6(261–358)ala, CPSF6(301–358), or CPSF6(301–358)ala. (B) Lysates of HeLa cells stably expressing TRIM5 and TRIM5/CPSF6 fusion proteins were evaluated for relative protein expression levels by Western blot analysis. (C) HeLa cells expressing control protein and TRIM/CPSF6 fusion proteins were infected with WT HIV-RFP or N74D HIV-RFP. (D) HIV-1 DNA synthesis was measured by real-time qPCR. Genomic DNA extracted from cells infected WT HIV-RFP and N74D HIV-RFP was collected at various time points (0 h, 5 h, 11 h, and 24 h) and assayed by qPCR to detect RU5, second-strand transfer, and 2-LTR circle junction vDNA levels. Error bars indicate standard deviations.
Next, to investigate which HIV-1 replication step was affected by rhTRIM5(1–299)/CPSF6 fusion proteins, the stable lines were infected with either WT or N74D HIV-1 at an MOI of 1 and analyzed by quantitative PCR for minus-strand strong stop (RU5), second-strand transfer, and two long terminal repeat (2-LTR) circle viral DNA (vDNA) products (Fig. 3D). As controls, we included rhTRIM5α (early reverse transcription block to both WT and N74D HIV-1 [unpublished data]) and CPSF6-358 (nuclear entry block to WT HIV-1 only [18]). As expected, RU5 levels of both WT HIV-1 and N74D HIV-1 were greatly diminished in cells expressing full-length rhTRIM5α, implying that WT HIV-1 and N74D HIV-1 were blocked at early reverse transcription. Also, as previously described, cells expressing CPSF6-358 showed normal reverse transcription products for both WT and N74D HIV-1. However, levels of 2-LTR circle junction products were significantly diminished only for WT HIV-1. Notably, the rhTRIM5(1–299)/CPSF6(301–358) cells blocked WT HIV-1 at early reverse transcription as rhTRIM5α but did not affect the N74D HIV-1 as CPSF6-358. Moreover, rhTRIM5(1–299)/CPSF6(301–358)ala, which possesses alanine mutations that prevent CA interaction, did not block WT HIV-1. Taken together, these results confirm that wild-type CA-specific HIV-1 core binding by CPSF6(301–358) was responsible for rhTRIM5(1–299) targeting of WT but not N74D HIV-1.
The last 58 residues of CPSF6-358 are sufficient for HIV-1 restriction.
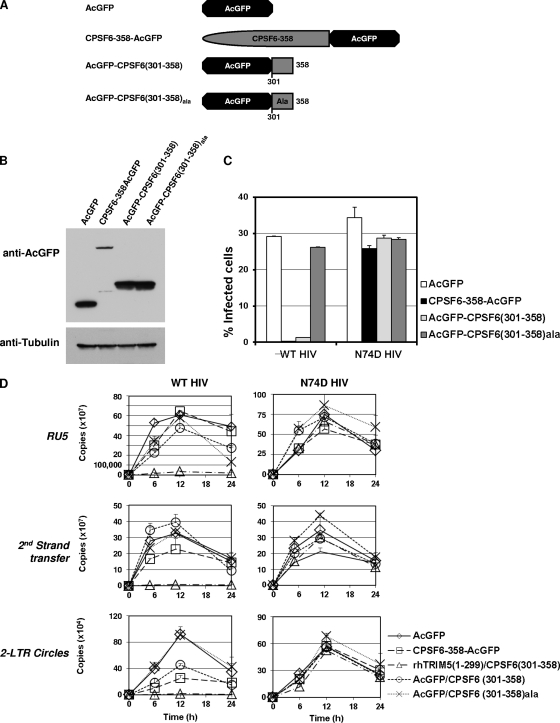
Because the C-terminal 58 residues in CPSF6-358 could substitute for the core binding B30.2/SPRY domain of rhTRIM5α, we inferred that these residues in CPSF6-358 were required for restriction through binding to the HIV-1 core. To further elucidate whether these residues were also sufficient for HIV-1 restriction, we tried to stably express this CPSF6 antiviral domain, but we were not able to express it by itself (Fig. 3B). Therefore, we expressed it as a fusion protein with AcGFP, a monomeric form of GFP (Fig. 4A). We first tested inhibition of WT HIV-RFP/VSV-G in cells transiently transfected with vector AcGFP alone, CPSF6-358-AcGFP, AcGFP-CPSF6(301–358), or AcGFP-CPSF6(301–358)ala. These results indicated potent restriction by CPSF6-358-AcGFP, but cells transfected with AcGFP-CPSF6(301–358) appeared to restrict WT HIV-RFP/VSV-G infection only in cells brightest for GFP fluorescence (data not shown). To eliminate transfection efficiency variability, we sought to make stable lines using the retroviral vectors expressing the different fusion proteins. AcGFP-CPSF6(301–358) stable cells and control cells were FACS sorted for elevated levels of the fusion protein (Fig. 4B; also see Fig. S1A in the supplemental material). Stable cell lines expressing AcGFP alone, CPSF6-358-AcGFP, AcGFP-CPSF6(301–358), or AcGFP-CPSF6(301–358)ala were then tested for susceptibility to HIV-1 infection (Fig. 4C; also see Fig. S1B in the supplemental material). The cells expressing AcGFP-CPSF6(301–358) were resistant to WT HIV-1 infection, as were CPSF6-358-AcGFP-expressing cells, while the control cells or cells expressing AcGFP-CPSF6(301–358)ala remained susceptible to infection with WT HIV-1. Importantly, all HeLa stable cell lines remained susceptible to infection with N74D HIV-1, indicating that the HIV-1 blocks observed for CPSF6-358-AcGFP and AcGFP-CPSF6(301–358) were CA dependent.
Fig 4.
A 58-amino-acid fragment of CPSF6 fused to AcGFP can block HIV-1 infection in a CA-dependent manner. (A) CPSF6-358, CPSF6(301–358), and CPSF6(301–358)ala were fused to AcGFP. (B) Protein expression was detected by AcGFP antibody. (C) HeLa cells stably expressing AcGFP fused CPSF6 proteins were infected with WT HIV-RFP and N74D HIV-RFP. (D) Cells infected with WT HIV and N74D HIV were collected for genomic extraction at various time points (0 h, 5 h, 11 h, and 24 h). Real-time qPCR was performed to detect RU5, second-strand transfer, and 2-LTR circle junction vDNA levels. Error bars indicate standard deviations.
To further characterize the nature of the block conferred by AcGFP-CPSF6(301–358), we examined virus-infected cells by quantitative PCR (Fig. 4D). Both WT and N74D HIV-1 displayed similar reverse transcription profiles in the control AcGFP-expressing cells. The rhTRIM5(1–299)-CPSF6(301–358)-expressing cells were also used as controls, and as described above, these cells blocked WT but not N74D virions during early reverse transcription. However, 2-LTR circle junction products were significantly diminished in cells expressing the CPSF6-358-AcGFP or AcGFP-CPSF6(301–358) fusion protein, although their reverse transcription products of RU5 or second-strand transfer were at levels comparable to those for infected control cells. No effect on levels of WT HIV-1 early reverse transcription, second-strand transfer, or 2-LTR circle products was observed in the AcGFP-CPSF6(301-358)ala cells. Reverse transcription by N74D HIV-1 in both AcGFP-CPSF6(301-358) and AcGFP-CPSF6(301–358)ala cells was similar to control cell levels. These results indicate that residues 301 to 358 of CPSF6-358, when expressed at high levels, are sufficient for HIV-1 restriction.
DISCUSSION
Removal of the C-terminal 58 residues of CPSF6-358 impairs specific binding to HIV-1 CA (18). Here we define the amino acids within this region, CPSF6-358 residues 314 to 322, necessary for the restriction of HIV-1. Similarly to CypA, this region can confer specificity of restriction when introduced as a fusion to primate TRIM5α. The CPSF6-358 HIV-1 interaction domain was not stable when expressed alone. However, fusion to a monomeric form of GFP indicated that it could be sufficient, if expressed at high levels, to mediate a CA-specific HIV-1 restriction. The stage at which virus infection was blocked depended on the fusion partner. Similar observations have been made in the analysis of artificial TRIM-Cyp fusions, in which different TRIM genes were fused to CypA (40). In our study, the presumptive virus targeting domain is altered instead. When CPSF6 residues 301 to 358 are C-terminally fused to rhTRIM5(1–299), this protein potently restricts wild-type but not N74D HIV-1. Unlike CPSF6-358, which blocks HIV-1 replication following reverse transcription, the rhTRIM5(1–299)/CPSF6(301–358) fusion prevents the initiation reverse transcription akin to rhTRIM5α restriction, further indicating that CPSF6-358 interacts with HIV-1 CA.
Because high cellular levels of AcGFP-CPSF6(301–358) were required to impair HIV-1 infection, the CPSF6(301–358) region appears to be inefficient in restriction even if it retains specificity of interaction with CA. Unlike TRIM5α, which has regions that contribute to multimerization and antiviral function that are separable from virus interaction, no other domains in CPSF6-358 emerged in the mutational analysis as being essential for restriction activity. It is possible that determinants present in CPSF6 residues 96 to 155 contribute to restriction, but these were difficult to assess carefully, as deletions in this region impaired expression/stability of the proteins.
Unlike genes encoding other cellular proteins known to interact with lentiviruses, we find no evidence that positive selection has acted on the CPSF6 gene (see Fig. S4 in the supplemental material). Positive-selection analysis has been used to predict virus interaction interfaces with primate Apobec3 and TRIM5α proteins (9, 28, 29). In contrast to these factors, the CPSF6 CA binding domain is perfectly conserved in primate orthologs spanning approximately 35 million years of primate evolution. This strong sequence conservation is consistent with evolutionary constraint acting on this domain, potentially due to another cellular binding partner. However, another intriguing possibility is that the interaction of this factor with primate lentiviruses is relatively new and has not yet affected the sequence of CPSF6 through arms-race dynamics (20). This model is consistent with our observation that two other mammalian lentiviruses, FIV and EIAV, do not interact with CPSF6-358 (18; data not shown).
The role of endogenous CPSF6 in HIV-1 infection has not been determined. CPSF6 is a nuclear protein involved in pre-mRNA processing and the regulation of alternative polyadenylation (26, 27). It localizes to nuclear paraspeckles, which are enriched in SR family proteins (7). Depletion of the protein in transformed cell lines increases HIV-1 infection (18). However, N74D mutation of CA to prevent interaction with CPSF6-358 has profound effects on HIV-1 replication, leading to an apparent loss of interaction with cell factors, such as TNPO3. It is possible that the interaction of CPSF6 with HIV-1, similarly to results seen with initial experiments to elucidate the role of CypA in HIV-1 infection, will turn out to be cell type dependent.
Recent studies of TNPO3 have suggested that HIV-1 CA might enter the nucleus in association with vDNA and that the removal of CA is necessary for infection to proceed (41). In this work, it was observed that N74D CA failed to enter the nucleus under conditions where WT CA was detected. Should this model of CA transport to the nucleus hold, a potential role of nuclear CPSF6 in HIV-1 PIC interactions through CA will have to be considered.
N74D HIV-1 resistance to CPSF6-358 suggests that this residue in helix 4 of the CA N-terminal domain (NTD) is present at a binding interface. Consistent with this interpretation, N74D mutation impairs binding of CPSF6-358 to recombinant CA-NC core complexes in sedimentation assays (18). Other mutations in CA helix 4 have also been observed to impair HIV-1 restriction by CPSF6-358 (18; data not shown). Notably, the antiviral compound PF-3450074 targets the same cavity of the CA NTD (5). PF-3450074 treatment of cells appears to prevent infection by accelerating CA loss from HIV-1 reverse transcription complexes (32). It will be of interest to determine whether potential cross-competitive interactions by CPSF6-358 and PF-3450074 engender viral cross-resistance.
A recent study has provided a structure of CPSF6 (38). Unfortunately, this and another effort have been unsuccessful in synthesizing soluble, recombinant CPSF6 that includes regions that we have identified as being critical for HIV-1 restriction (7, 38). The central 200 residues of CPSF6 are 47% proline (26). The role of this proline-rich hinge is unknown, but it has been suggested to provide an interface for interactions with multiple proteins and possibly contribute to nuclear retention of the protein (7, 26). Because recombinant forms of CPSF6 containing the proline-rich central region have proven difficult to work with, smaller fragments or peptides containing CPSF6 residues 314 to 322, should they retain specificity of CA interaction, may provide insight into the mechanisms and contacts made by CPSF6-358 in the CA NTD pocket.
As we have demonstrated, the CPSF6-358 CA interaction domain can be used to target other antiviral proteins to HIV-1. Given that primate lentiviruses resistant to rhTRIM5α remain sensitive to CPSF6-358, this interaction interface appears to be well conserved. In fact, N74 and adjoining CA residues are invariant among known HIV-1 isolates (18). N74D-mediated HIV-1 resistance to CPSF6-358 has a cost and leads to loss of macrophage tropism (1). This CA interface, if mediating interaction with a host cofactor necessary for replication in macrophages, may be especially susceptible as a therapeutic target. Further elaboration of the CPSF6 interaction with CA is likely to provide an increased structural and functional understanding of the postentry replication of HIV-1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan Rein for manuscript comments.
This work was supported by the National Cancer Institute's intramural Center for Cancer Research, which supports the HIV Drug Resistance Program (V.N.K.). N.R.M. is supported by a National Science Foundation graduate research fellowship.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 1 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ambrose Z, et al. Human immunodeficiency virus type 1 (HIV-1) capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection. J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartz SR, Vodicka MA. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337–342 [DOI] [PubMed] [Google Scholar]
- 3. Besnier C, Takeuchi Y, Towers G. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. U. S. A. 99:11920–11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829 [DOI] [PubMed] [Google Scholar]
- 5. Blair WS, et al. 2010. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS pathogens 6:e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowan S, et al. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. U. S. A. 99:11914–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 279:35788–35797 [DOI] [PubMed] [Google Scholar]
- 8. Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duggal NK, Malik HS, Emerman M. 2011. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J. Virol. 85:11361–11371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ganser-Pornillos BK, et al. 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U. S. A. 108:534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hilditch L, et al. 2011. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc. Natl. Acad. Sci. U. S. A. 108:5771–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hofmann W, et al. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020–10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jolicoeur P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:67–122 [DOI] [PubMed] [Google Scholar]
- 16. Julias JG, Ferris AL, Boyer PL, Hughes SH. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karolchik D, et al. 2003. The UCSC Genome Browser database. Nucleic Acids Res. 31:51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee K, et al. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDonald D, et al. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyerson NR, Sawyer SL. 2011. Two-stepping through time: mammals and viruses. Trends Microbiol. 19:286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Munk C, Brandt SM, Lucero G, Landau NR. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U. S. A. 99:13843–13848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nisole S, Lynch C, Stoye JP, Yap MW. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U. S. A. 101:13324–13328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Onishi M, et al. 1996. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 24:324–329 [PubMed] [Google Scholar]
- 24. Pincus T, Rowe WP, Lilly F. 1971. A major genetic locus affecting resistance to infection with murine leukemia viruses. II. Apparent identity to a major locus described for resistance to friend murine leukemia virus. J. Exp. Med. 133:1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rein A, Kashmiri SV, Bassin RH, Gerwin BL, Duran-Troise G. 1976. Phenotypic mixing between N- and B-tropic murine leukemia viruses: infectious particles with dual sensitivity to Fv-1 restriction. Cell 7:373–379 [DOI] [PubMed] [Google Scholar]
- 26. Ruegsegger U, Blank D, Keller W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243–253 [DOI] [PubMed] [Google Scholar]
- 27. Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. 2008. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol. Reprod. 78:472–482 [DOI] [PubMed] [Google Scholar]
- 28. Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573 [DOI] [PubMed] [Google Scholar]
- 31. Schindler J, Gautsch JW, Lerner RA, Hopkins N. 1981. Biochemical analysis of the p30's of N-, B-, and B leads to NB-tropic murine leukemia viruses of BALB/c origin. J. Virol. 39:703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2011. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 34. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Towers G, et al. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. U. S. A. 97:12295–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Towers GJ, et al. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138–1143 [DOI] [PubMed] [Google Scholar]
- 37. Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. 2006. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U. S. A. 103:7465–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Q, Coseno M, Gilmartin GM, Doublie S. 2011. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555–556 [DOI] [PubMed] [Google Scholar]
- 40. Yap MW, Dodding MP, Stoye JP. 2006. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J. Virol. 80:4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou L, et al. 2011. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 7:e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.