Abstract
Human noroviruses are genetically and antigenically highly divergent. Monoclonal antibodies raised in mice against one kind of norovirus virus-like particle (VLP), however, were found to have broad recognition. In this study, we present the crystal structure of the antigen-binding fragment (Fab) for one of these broadly reactive monoclonal antibodies, 5B18, in complex with the capsid-protruding domain from a genogroup II genotype 10 (GII.10) norovirus at 3.3-Å resolution and, also, the cryo-electron microscopy structure of the GII.10 VLP at ∼10-Å resolution. The GII.10 VLP structure was more similar in overall architecture to the GV.1 murine norovirus virion than to the prototype GI.1 human norovirus VLP, with the GII.10 protruding domain raised ∼15 Å off the shell domain and rotated ∼40° relative to the GI.1 protruding domain. In the crystal structure, the 5B18 Fab bound to a highly conserved region of the protruding domain. Based on the VLP structure, this region is involved in interactions with other regions of the capsid and is buried in the virus particle. Despite the occluded nature of the recognized epitope in the VLP structure, enzyme-linked immunosorbent assay (ELISA) binding suggested that the 5B18 antibody was able to capture intact VLPs. Together, the results provide evidence that the norovirus particle is capable of extreme conformational flexibility, which may allow for antibody recognition of conserved surfaces that would otherwise be buried on intact particles.
INTRODUCTION
The family Caliciviridae contains four genera, Norovirus, Sapovirus, Lagovirus, and Vesivirus, which include norovirus, sapovirus, rabbit hemorrhagic disease virus, and feline calicivirus strains, respectively. Human noroviruses are the dominant cause of outbreaks of gastroenteritis and are genetically and antigenically distinct (21). Two main genogroups (GI and -II) of human noroviruses are mostly responsible for causing human infections, and these two genogroups are further subdivided into numerous genotypes (GI.1 to -8 and GII.1 to -17) (72). The human norovirus genome has three open reading frames (ORF1 to -3), where ORF1 encodes the nonstructural proteins, ORF2 encodes the capsid protein, and ORF3 encodes a small structural protein. Human noroviruses cannot be grown in cell culture, but the expression of the capsid protein in a baculovirus expression system leads to the self-assembly of nucleic acid-free virus-like particles (VLPs) that are believed to be morphologically and antigenically similar to the native virion (27).
The cryo-electron microscopy (cryo-EM) and X-ray crystal structures of the prototype norovirus VLP (GI.1, Norwalk virus) show that the VLP form a T=3 icosahedral structure (53, 54). The VLP can be divided into two domains, the shell (S) and the protruding (P) domains. The S domain forms a scaffold surrounding the RNA, whereas the P domain, which is further subdivided into P1 and P2 subdomains, is thought to contain the determinants of cell attachment and strain diversity (27, 53, 60). The P domain forms 90 dimer subunits, termed A/B and C/C. Interestingly, the P domains alone can be expressed in Escherichia coli, and these form P domain dimers that are biologically relevant (60). The X-ray crystal structures of several human norovirus P domains (GI.1, GII.4, GII.10, and GII.12) indicate that their overall structures are similar and resemble the P domain on the VLPs, with a single α-helix in the P1 subdomain and six antiparallel β-strands in the P2 subdomain (7, 10, 13, 22). The X-ray crystal structure of a GV.1 murine norovirus P domain reveals a structure that is similar overall to the human norovirus P domains (63). However, cryo-electron microscopy (cryo-EM) studies show that in the GV.1 murine norovirus virion, the P domain is raised off the S domain by ∼16 Å (30), while in the GI.1 and GII.4 (Grimsby virus strain) human norovirus VLPs, the P domain rests directly on the S domain (12, 53, 54). In addition, the GV.1 murine norovirus P domain is rotated 40° clockwise with respect to the GI.1 and GII.4 human norovirus P domains.
Human noroviruses are generally detected using reverse transcription-PCR (RT-PCR) with degenerate primers or enzyme-linked immunosorbent assay (ELISA) with norovirus-specific antibodies. Many of the polyclonal and monoclonal antibodies (MAbs) used in the ELISA kits were developed in mice immunized with norovirus VLPs (15, 28, 55, 57), and most have broad recognition (21, 39, 40, 50, 59, 70). Several antibodies are found to bind to the S domain (39, 70), while others bound to the P domain (50, 59). However, structural details of the antibody mode of binding to the VLPs, S domain or P domain, have been lacking. In the studies presented here, we used X-ray crystallography to define the recognition site of the broadly reactive antibody 5B18 on the norovirus GII.10 P domain, cryo-EM to determine the structure of the intact GII.10 VLP, and ELISA to determine whether this antibody recognizes intact or disassembled virus particles. Overall, the findings have implications related to the dynamic nature of the P domain in the context of the norovirus VLP and to its recognition by antibody.
MATERIALS AND METHODS
Norovirus VLP expression.
The GII.4 Saga (GenBank accession number BAG70515), GII.10 Vietnam026 (GenBank accession number AF504671) (23), and GII.12 Hiro (GenBank accession number AB044366) (23) VLPs were expressed as previously described (23). The VLPs were purified using CsCl equilibrium gradient ultracentrifugation at 35,000 rpm for 24 h at 4°C (Beckman SW55 rotor). A distinct viral band was removed from the side of the tube with a syringe and the VLPs were stored in phosphate-buffered saline (PBS) (pH 7.3) at 4°C. The integrity of the VLPs was confirmed by negative-stain EM. Briefly, the VLP samples were applied to a carbon-coated 300-mesh EM grid and stained with 2% uranyl acetate (pH 4). Grids were examined using a Jeol JEM-1220 transmission electron microscope operated at 80 kV.
Antibody ELISA binding to GII.4, GII.10, and GII.12 VLPs.
An antibody ELISA was used to compare the cross-reactivities of VLPs from the three different GII norovirus genotypes (GII.4, GII.10, and GII.12) with 5B18 IgG. Wells of 96-well microtiter plates (MaxiSorp; Nunc, Denmark) were each coated with 100 μl of ∼100 ng of purified VLPs (in PBS, pH 7.3) and incubated overnight at 4°C. The wells were washed four times with PBS containing 0.1% (vol/vol) Tween 20 (PBS-T) and then were blocked with PBS containing 5% (wt/vol) skim milk for 1 h at room temperature. After the wells were washed four times with PBS-T, 100 μl of 2-fold serially diluted horseradish peroxidase (HRP)-conjugated labeled 5B18 IgG, from a 1:5,000 starting dilution in PBS containing 0.05% (wt/vol) skim milk, was added to each well, and the plates were incubated for 1 h at 37°C. The wells were washed four times with PBS-T, and then 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate was added to each well. The reaction was stopped by the addition of 100 μl of 2 N H2SO4 to each well, and the absorbance was measured at 450 nm (A450). The titer was expressed as the reciprocal of the highest dilution of antiserum giving a value of A450 of >0.2, as previously described (21).
Western blotting.
The VLPs and P domains were separated by SDS-PAGE and electrotransferred to polyvinylidene difluoride (PVDF) membranes by using iBLOT, following the manufacturer's protocol. Proteins were detected with 5B18 IgG at a dilution of 1:5,000, and then, following the manufacturer's instructions, the blots were developed by chemiluminescence using enhanced chemiluminescence (ECL) detection reagent (Amersham Biosciences, England).
Protein expression and purification of GII.10 P domain.
The GII.10 P domain was expressed in E. coli and purified as previously described (22). Briefly, the P domain was optimized for E. coli expression, cloned in a modified pMal-c2x vector at the BamHI and NotI restriction sites (New England BioLabs), and transformed into BL21(DE3) cells (Invitrogen). Expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM) for 18 h at 22°C. After a series of purification steps and protease cleavage, the P domain was concentrated to 2 to 10 mg/ml and stored in gel filtration buffer (0.35 M NaCl, 2.5 mM Tris, pH 7.0, 0.02% NaN3).
Preparation of 5B18 Fab fragment.
The 5B18 IgG monoclonal antibody was produced from a mouse immunized with GII.4 norovirus-445 VLPs (GenBank accession number DQ093064) (Denkaseiken, Japan). The 5B18 IgG is currently used as a GII broad-range capture antibody in a commercially available ELISA kit (Denkaseiken, Japan). The 5B18 Fab was prepared using a modified method (34). Approximately 60 mg of purified 5B18 IgG was used for Fab preparation. IgG was reduced in 100 mM dithiothreitol (DTT) (pH 7.6) for 1 h at 37°C. The reduced IgG was added to a dialysis cassette, and the DTT was removed by placing the cassette in GFB (0.35 M NaCl, 2.5 mM Tris, pH 7.0, 0.02% NaN3) supplemented with 20 mM HEPES (pH 7.7) for 1 h at 4°C. The IgG was alkylated in the same buffer supplemented with 2 mM iodoacetamide for 48 h at 4°C, and then the cassette was transferred to a fresh solution without the iodoacetamide for 1 h at 4°C. The IgG was concentrated to 5 mg/ml and then digested with papain using a commercial kit (Pierce, Rockford, United States). The Fab was separated from the Fc in a protein A column, and the resulting Fab was further purified by size exclusion chromatography with a Superdex 200 column (GE), concentrated to 5 mg/ml, and stored in GFB. The purified GII.10 P domain and Fab were mixed 1.4:1 for 1 h at 25°C, and finally, the GII.10 P domain-Fab complex was purified by size exclusion chromatography.
Preparation and cocrystallization of GII.10 P domain-Fab complex for X-ray crystallography.
Crystals of the GII.10 P domain-Fab complex were grown by the hanging drop vapor diffusion method, mixing the protein and reservoir solution (40% [vol/vol] polyethylene glycol [PEG] 400, 5% [wt/vol] PEG 3350, and 0.1 M acetic acid, pH 5.5) (42) in a 1:1 ratio. Crystals grew over 1 week at a temperature of 20°C. Prior to data collection, crystals were transferred to 50% (vol/vol) PEG 400.
X-ray crystallography data collection, structure solution, and refinement.
X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-BM at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, and processed with HKL2000 (49). Despite the large size of the crystals (perfectly formed pyramids of up to 0.3 mm per edge), the diffraction data were poor due to split reflections, high background, and most diffraction extending to less than 4 Å. These resulted in Chi2 values of 0 for several wedges of data. Despite these difficulties, relatively complete data (90%) was obtained from 180 degrees of oscillation, though with lower than expected redundancy (2.7-fold), and the overall quality of data which passed the Chi2 tests appeared fine. Structures were solved by molecular replacement in PHASER (44), using the structure with Protein Data Bank identifier (PDB ID) 3ONU for the GII.10 P domain and the structure with PDB ID 1WEJ for the Fab as a search model. Manual model building was performed in COOT (18), and positional refinement together with translation/liberation/screw (TLS) refinement were performed using REFMAC (14) and PHENIX (1).
Cryo-EM data collection and refinement.
VLPs at a concentration of 1.0 mg/ml were applied to a glow-discharged Quantifoil R1.2/1.3 Mo 200-mesh holey carbon grid with a thin layer of carbon over the holes. The sample was rapidly plunged into liquid ethane after automatic blotting for 7 to 8 s at 8°C and 100% humidity using an FEI MarkIV Vitrobot. Grids were examined with a JEOL JEM-2200FFC microscope equipped with a field emission gun and an in-column (omega-type) energy filter operated at an acceleration voltage of 200 kV. Images of the frozen VLPs were collected at a nominal magnification of 40,000 using a 4k × 4k (4,096- by 4,096-pixel) Tietz charge-coupled-device (CCD) camera, resulting in a pixel size of 2.4 Å. Particles were extracted from raw micrographs using the swarm semiautomatic particle-picking algorithm in EMAN2 into boxes of 250 by 250 square pixels (61). The raw particles were normalized, phase flipped, and high-pass filtered before single-particle analysis. Approximately 8,000 particle images were subjected to two-dimensional (2-D) reference-free alignment and classification through several rounds of multireference alignment and multivariate statistical analysis in IMAGIC to a total of 50 classes (65). Approximately 80% of the data were used for the reconstruction. Particles with cross-correlation values 1.5 σ below the mean were not used (∼20%). Representative class averages were used as the basis for initial model generation in EMAN2. The initial model with the best match between reprojections and class averages was chosen as the starting model for several rounds of projection-matching refinement with increasing angular sampling. The final reconstruction, at ∼10-Å resolution (0.5 Fourier shell correlation [FSC] criterion), was used for atomic coordinate fitting and structure comparison in Chimera (version 1.5.3) (52).
Structure analysis, sequence analysis, and figures.
Figures were rendered using PyMOL (version 1.2r3; Schroedinger LLC) and Chimera (version 1.5.3) (52). The N-terminal amino acid sequences for the Fab heavy and kappa (κ) chains were determined at the Columbia University Medical Center, Protein Core Facility. Degenerate primers for the Fab sequence were designed from constant regions and from the N-terminal amino acid sequence based on closely matching murine antibody N-terminal nucleotide sequences in GenBank. Norovirus complete-capsid amino acid sequences were aligned and analyzed with Genetyx-Mac software (version 16.0.0).
Antibody ELISA binding to GII.10 VLPs at different pHs.
An antibody ELISA was used to determine the ability of 5B18 IgG to bind to GII.10 VLPs. The GII.10 VLPs were diluted in PBS at pHs 5.3, 6.3, 7.3, 8.3, and 9.3 to a final concentration of 7.5 μg/ml. At high PBS pHs, the VLPs were found to be partially broken, while at low pHs, most of the VLPs were intact. Wells of 96-well microtiter plates (MaxiSorp; Nunc, Denmark) were each coated with 100 μl of 7.5 μg/ml of VLPs at the different pHs and incubated overnight at 4°C. The VLPs were detected as described above, except that 5B18 IgG and goat HRP–anti-mouse IgG (used as a secondary antibody) replaced the HRP-labeled 5B18 IgG. After the addition of the secondary antibody, the wells were washed four times with PBS-T, and then 100 μl of substrate o-phenylenediamine and H2O2 was added to each well and the plates left in the dark for 30 min at room temperature. The reaction was stopped by the addition of 50 μl of 2 N H2SO4 to each well, and the absorbance was measured at 492 nm (A492).
Sequence conservation on the VLP and P domain.
To show the sequence conservation on the GII VLP, a model of the GII.10 VLP was built as described previously (22) using the unbound GII.10 P domain structure (PDB ID 3ONU) and the S domain from the Norwalk virus capsid structure (PDB ID 1IHM). Amino acid sequence conservation was analyzed as previously described (22). Briefly, an alignment of a representative set of GII sequences was used to compute residue conservation scores using the AL2CO server (51) and mapped (using a color range for highly variable to highly conserved residues) onto the surface of the GII.10 VLP model and unbound GII.10 P domain structure.
Accession numbers.
Atomic coordinate and structure factors of the X-ray crystal structure were deposited in the Protein Data Bank under accession number 3V7A. The 3-D cryo-EM map was deposited in the EMDataBank with accession number EMD-5374.
RESULTS
5B18 binds to several GII genotypes.
To confirm the ability of the 5B18 IgG to bind diverse GII norovirus genotypes, we expressed VLPs from three GII genotypes (GII.4, GII.10, and GII.12) and examined the binding using ELISA and Western blotting. The EM results showed that GII.4 VLPs were a mixture of small and native-size particles, while the GII.10 and GII.12 VLPs were mostly of native size (data not shown). The ELISA results showed that 5B18 IgG was capable of cross-reacting with all three GII genotypes having an equal titer of 320,000 (see Fig. S1A in the supplemental material). The Western blotting results also showed that the 5B18 IgG could detect the three GII genotypes (see Fig. S1B).
X-ray crystal structure of GII.10 P domain-Fab complex.
The GII.10 P domain was used to determine the precise binding location of the 5B18 antibody on the norovirus capsid using X-ray crystallography. The GII.10 P domain and Fab proteins were mixed together (1.4:1 molar ratio) for 1 h at room temperature. The GII.10 P domain-Fab complex was purified using size exclusion chromatography. Two main peaks were observed, corresponding to the complex and free P domain (data not shown). The fractions containing the GII.10 P domain-Fab complex were pooled and concentrated to ∼4 mg/ml, and the complex was crystallized by the hanging-drop vapor diffusion method. A single GII.10 P domain-Fab complex crystal diffracted X rays to ∼3.3 Å. The structure was solved using molecular replacement with a GII.10 P domain monomer (PDB ID 3ONU) and a mouse Fab (PDB ID 1WEJ) as search models. Molecular replacement indicated that two P domain monomers and two 5B18 Fabs each containing a kappa (κ) chain and heavy chain were in the asymmetrical unit and these were related by a noncrystallographic 2-fold. Water molecules were not added to the structure since the resolution was at 3.3 Å. The 5B18 Fab was shown to bind to the wall of the P1 subdomain and involved a monomeric interaction with the P1 subdomain (Fig. 1A). The electron densities of the P domains and 5B18 Fabs were generally well defined, and refinement led to an Rvalue of 0.230 (Rfree of 0.283) (Table 1; also see Fig. S2 in the supplemental material). The P domain dimer had a single helix in each P1 subdomain and six antiparallel β-strands in each P2 subdomain as previously described (22).
Fig 1.
The X-ray crystal structure of the GII.10 P domain-Fab complex shows that the Fab bound to the lower side of the P1 subdomain. The GII.10 P domain dimer is colored according to monomers and subdomains, i.e., chain A: P1 (blue), chain A: P2 (light blue), chain B: P1 (violet), and chain B: P2 (salmon), whereas the Fab is colored according to chain, i.e., κ chain (yellow) and heavy chain (green). (A) The Fab bound to the wall of the P1 subdomain (considered to be inside the virus particle) and involved a monomeric interaction. (B) A close-up stereoview of the interacting P domain residues for chain A (Val433, Glu496, Asn530, Tyr533, Thr534, and Leu535) and Fab residues (κ chain, Tyr32, Tyr92, Gly93, Ser94, and Trp97, and heavy chain, Asn52). The hydrogen bond interactions included both side chain and main chain interactions (also see Table S1 in the supplemental material).
Table 1.
Data collection and refinement statistics for the GII.10 norovirus P domain-Fab complex structure
| Parameter | Valuea |
|---|---|
| Data collection | |
| Space group | P 4322 |
| Cell dimensions | |
| a, b, c (Å) | 145.48, 145.48, 216.33 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50-3.30 (3.42-3.30)b |
| Rsym | 15.0 (55.1) |
| I /σI | 8.3 (1.8) |
| Completeness (%) | 87.6 (89.0) |
| Redundancy | 2.7 (2.7) |
| No. of unique reflections | 31,300 |
| Refinement | |
| Resolution (Å) | 31.54–3.30 |
| No. of reflections | 30,833 |
| Rwork/Rfree | 0.227/0.283 |
| No. of atoms | 11,459 |
| Average B factors (Å2) | |
| Overall | 84.7 |
| P domain | 82.4 |
| Fab | 86.3 |
| Ramachandran (%) | |
| Outliers | 0.00 |
| Favored | 93.31 |
| RMS deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.795 |
The data set was collected from a single crystal of the 026_P domain-Fab complex (PDB ID 3V7A).
Values in parentheses are for the highest-resolution shell.
GII.10 P domain interaction with the 5B18 Fab.
The total interface area of the GII.10 P domain and 5B18 Fab was 1,500 Å2 (770 Å2 on the P domain and 730 Å2 on the 5B18 Fab), as calculated using PISA software (33). The GII.10 P domain and 5B18 Fab interaction included nine hydrogen bonds, eight of which were formed between the P1 subdomain and κ chain and one between the P1 subdomain and heavy chain (Fig. 1B; also see Table S1 in the supplemental material). Six P1 subdomain amino acids interacted with the 5B18 Fab, Tyr533 formed a single hydrogen bond with Tyr92κ, Thr534 formed three hydrogen bonds with Gly93κ and one hydrogen bond with Trp97κ, Leu535 formed a hydrogen bond with Tyr32κ, Glu496 formed a hydrogen bond with Tyr92κ, Asn530 formed a hydrogen bond with Ser94κ, and Val433 formed a hydrogen bond with Asn52 heavy chain. Superposition of the apo GII.10 P domain dimer and the Fab-bound GII.10 P domain showed that each of the P1 subdomains shifted slightly (∼1 to 2 Å) toward the center of the dimer, while the P2 subdomain showed little conformational change (see Fig. S3 in the supplemental material). The electrostatic potential of the Fab was calculated (16), and the interacting residues on the P domain bound at two negatively charged pockets on the Fab at the variable regions (Fig. 2).
Fig 2.
The binding site on the Fab was coordinated by negative charge regions on the Fab. (A) The GII.10 P domain is colored as in Fig. 1. The 5B18 Fab surface is color-coded according to electrostatic potential from red (negative charge) to blue (positive charge). (B) A close-up view of the electrostatic potential on the Fab, showing the four P domain side chain residues on chain A that interacted with the Fab. The P domain side chains bound near the negative charge (red) regions on the Fab.
Conservation of the 5B18 Fab-binding site on GII P domains.
The 5B18 Fab formed hydrogen bonds with residues at three different sites on the P1 subdomain, termed A, B, and C (Fig. 3). An amino acid alignment of representatives from 10 GII norovirus genotypes indicated that Val433 (site A) was the most variable, with other genotypes having threonine, serine, asparagine, leucine, or methionine at this position. Thr534 (site C) was mostly conserved, as the only other amino acid at this position was a serine. Glu496 (site B), Asn530 (site C), Tyr533 (site C), and Leu535 (site C) were all highly conserved among the representative GII genotypes. Superposition of other known GII norovirus P domains (GV.1, GII.4-TCH05, GII.4-VA387, GII.12, GII.9-VA207, and GI.1) showed that the equivalent GII.10 interacting side chains were mostly in the same conformation (see Fig. S4 in the supplemental material). GI norovirus side chains also appeared to be similar to the GII.10 interacting side chains (see Fig. S4).
Fig 3.
Amino acid alignment of GII capsid sequences indicated that four GII.10 P domain residues involved in binding 5B18 Fab were highly conserved among other GII genotypes. Ten different GII genotypes' capsid sequences were aligned (strain name is followed by genotype), and the GII.10 capsid sequence was used as the consensus (21). The GII.10 P domain residues that interacted with the 5B18 Fab involved three different sites on the P domain, termed A, B, and C. The blue shading shows the six GII.10 P domain residues that interacted with the 5B18 Fab, and from the alignment, four of six residues were highly conserved among other GII genotypes. The red circles show the suspected binding site of MAb14-1 monoclonal antibody (59).
GII.10 VLP cryo-EM structure.
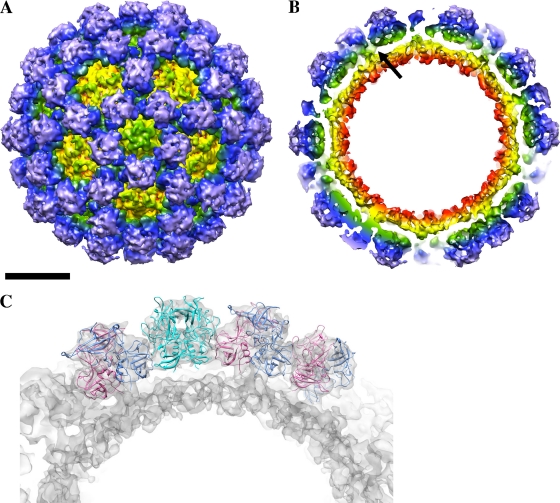
From the general location of the epitope (Fig. 1) and the known structures of other caliciviruses (7, 10, 13, 22), it was not clear how the monoclonal antibody, raised against intact VLPs, could bind at this occluded site on intact particles. To this end, the cryo-EM structure of the GII.10 VLP (in an unbound state) was determined to define the arrangement of the GII.10 P domains with respect to the entire capsid. The GII.10 VLPs appeared as homogeneous, monodisperse particles in ice (see Fig. S5A in the supplemental material). Reference-free class averages of the VLPs showed icosahedral particles with spike-like structures extending from the vertices (see Fig. S5). The cryo-EM reconstruction of the GII.10 VLP at ∼10-Å resolution (0.5 FSC criterion) showed several striking features (Fig. 4). The GII.10 S domain was noticeably surface exposed at the 3- and 5-fold axes (Fig. 4A). The GII.10 P domain appeared as a second outer shell, and a central section through the VLP revealed that the P domain was raised off the S domain by ∼15 Å (Fig. 4B). The electron density at the tip of the P domain (the P2 subdomain) was significantly weaker than at the base of the P1 domain, suggesting that there was marked flexibility in the P2 subdomains. This was consistent with what has been observed with several other reconstructions of calicivirus particles (4, 5) and suggests that there is a great deal of conformational heterogeneity in the P domains.
Fig 4.
The cryo-EM structure of the GII.10 VLP consisted of an S domain surrounded by 90 P domain dimers. (A) The GII.10 S domain was surface exposed (yellow and yellow to green). The P domain dimers (green to blue and purple) were raised off the S domain by ∼15 Å. (B) The inner surface of the S domain is colored red. The S domain was connected to the P domain monomer by a narrow hinge region (single arrow; the other hinges were not labeled for clarity). The scale bar for panels A and B indicates 100 Å. (C) Fitting and modeling of the GII.10 P domain (apo P domain structure) into the A/B dimer subunit (light blue and pink, respectively) and C/C dimer subunit (cyan).
Fitting of the GII.10 P domain and P domain-Fab complex into the GII.10 VLP cryo-EM structure.
At ∼10-Å resolution, the GII.10 P domain monomers on the VLP were easily distinguished. Fitting of the crystal structures of the GII.10 P domain and P domain-Fab complex into the GII.10 VLP cryo-EM map was performed manually and guided by previous fitting results of GV.1 P domain dimers into the GV.1 cryo-EM map (63). This approximate alignment was adjusted computationally using the Fit-in-Map function in UCSF Chimera (52) to a cross-correlation coefficient of 0.94 (Fig. 4). Using this method, the X-ray structure of the GII.10 P domain dimer (PDB ID 3ONU) was unambiguously fitted into the corresponding density in the cryo-EM map, except for several loops on the P2 subdomain (Fig. 4C). This is probably due to flexibility in these domains, as the electron density of the P2 subdomain loops was weak and the tips of the P2 domains were less ordered than the S domain and P1 domains in the cryo-EM reconstruction (data not shown). The P domain dimers appeared to be connected to adjacent, icosahedrally related P1 subdomains in the VLP, whereas the P2 subdomains had no such connections (Fig. 4C). When the P domain from the X-ray structure of the P domain-Fab complex was fitted into the A/B dimer subunit of the reconstruction, the 5B18 Fab was located under the neighboring P domain dimer and rested on top of the S domain at the space at the 2-fold axes (Fig. 5A). When the P domain from the X-ray structure of the P domain-Fab complex was instead fitted into the C/C dimer subunit, the 5B18 Fab made contact with a neighboring P domain dimer and clashed with a star-like protrusion on the S domain at the space at the 5-fold axes (Fig. 5B). Essentially, the epitope of 5B18 overlapped the region of the P1 subdomain that made interactions with icosahedrally related, adjacent P domains in the VLPs when in this “floating P domain” conformation. Based on this modeling, it appeared that the VLP probably could not be saturated with 5B18 antibodies, as this would create a highly unstable structure, as well as additional IgG-IgG steric clashes at the axis spaces. Two possibilities are likely, (i) that 5B18 recognition of intact norovirus particles occurs at select, transiently exposed P domains or (ii) that 5B18 recognition occurs at places where the particle has a defect, where the P domain is exposed because the particle is not appropriately formed.
Fig 5.
The X-ray crystal structure of the GII.10 P domain-Fab complex fitted into the cryo-EM structure of the GII.10 VLP and the X-ray crystal structure of the GI.1 VLP (PDB ID 1IHM). (A) The P domain (light blue and pink) from the P domain-Fab was fitted into the A/B dimer subunit on the VLP. The boxed region shows a close-up stereoview of the interaction. The Fab appeared to make slight contact with the S domain at the space at the 6-fold axes and was under a neighboring domain. (B) The P domain (cyan) from the P domain-Fab was fitted into the C/C dimer subunit on the VLP. The boxed region shows a close-up stereoview of the interaction. The Fab appeared to make contact with a raised S domain structure at the space at the 5-fold axes and was for the most part hidden under a neighboring domain. (C) The GII.10 P domain (colored as in Fig. 1 and rotated 90° from the views in Fig. 5A and 5B) was highly similar to the GI.1 P domain (light gray), but the Fabs clashed with the GI.1 S domain (orange).
The 5B18 IgG bound equally well with intact and partially broken GII.10 VLPs.
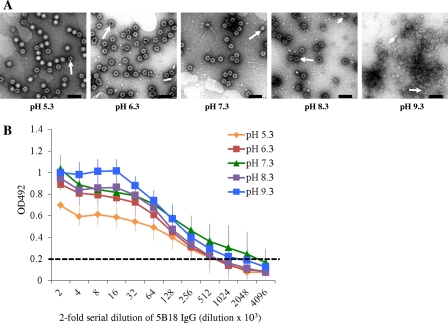
To test whether 5B18 recognition occurs with intact or with broken particles, we assessed the pH behavior of 5B18 recognition, as norovirus VLPs become less stable and appear broken at high pH values (2). We observed that at low and neutral pHs (5.3, 6.3, and 7.3), the GII.10 VLPs were mostly homogenous in size and unbroken, whereas at higher pHs (8.3 and 9.3), the GII.10 VLPs appeared less homogenous in size and partially broken (Fig. 6A). The 5B18 IgG detected GII.10 VLPs at different pH values with nearly identical efficacy, regardless of the fraction of damaged particles (Fig. 6B). At pH 5.3, 6.3, and 8.3, the titer was 512,000, at pH 9.3, the titer was 1,024,000, and at pH 7.3, the titer was 2,048,000 (optical density [OD] cutoff of >0.2) (21). Together, these results suggest that 5B18 appears capable of detecting nominally intact GII.10 VLPs.
Fig 6.
An antibody ELISA was used to determine the binding ability of IgG to GII.10 VLPs. (A) The morphology of the VLPs was examined using EM. At low pHs (from pH 5.3 to 7.3), the majority of the VLPs appeared intact, while above pH 7.3, many of the VLPs appeared broken (long arrows). Small VLPs were also found (short arrows). Scale bar, 100 nm. (B) The same VLPs shown in panel A were used in an ELISA to compare the binding ability of 5B18 IgG. The OD values represent the means of the results for 4 wells; error bars are shown. The OD at 492 nm (OD492) was determined; the dashed line shows the OD492 cutoff of 0.2 (21). The 5B18 IgG detected GII.10 VLPs at different pH values. At pH 7.3, the titer was 2,048,000, while the titers of the other pH values were 2- or 4-fold lower, indicating similar cross-reactivities.
Norovirus variability and 5B18 detection.
As stated previously, norovirus displays considerable genetic variation. Mapping this variation onto the VLP structure shows that this variation is concentrated on the outer surface of the virion, with buried portions of the P and S domains being much more conserved (Fig. 7). If one were only able to access the outer surface of the VLP, this genetic variation would make pan-recognition extremely difficult. The 5B18 mode of recognition thus provides a mechanism to achieve broad VLP detection by recognizing a conserved surface that is transiently exposed in a dynamic manner. Indeed, the Fab footprint was highly conserved on the GII P domains (Fig. 7C).
Fig 7.
Surface representations of GII amino acid conservation. Noroviruses are genetically and antigenically distinct, with the S domain being more conserved than the P domain. (A) The GII amino acid variability was mapped onto the model of the GII.10 VLP (GI.1 S domain and GII.10 P domain). Amino acid conservation ranges are color-coded from deep purple (highly conserved) to white (highly variable). (B) The amino acid variability was mapped onto the GII.10 P domain apo structure (3ONU) with the 5B18 Fab bound. The top of the P domain was highly variable (left side), while the bottom half of the P domain was more conserved (right side). (C) The Fab binding footprint was mapped onto the P domain (yellow line). The footprint was at a highly conserved area on the wall of the P domain (inside the particle).
DISCUSSION
Human noroviruses are genetically and antigenically distinct, but broad-range monoclonal antibodies capable of detecting multiple norovirus genogroups and genotypes have been described (21, 39, 40, 50, 59, 70). One such antibody, 5B18, is currently in use in a commercial norovirus ELISA detection kit (Denka Seiken, Japan) and was found to bind to numerous GII genotypes but not GI noroviruses (unpublished data). To describe the precise binding location of 5B18, we determined the X-ray crystal structure of the GII.10 P domain-Fab complex. We also determined the cryo-EM structure of the GII.10 VLPs in an attempt to understand the 5B18 Fab binding interaction in the context of the entire virus particle.
The 5B18 Fab binds to a face of the GII.10 P1 subdomain close to the S domain and not openly exposed at the VLP surface. Six amino acid residues on the P1 subdomain make main chain and side chain interactions with the Fab. Four of these residues are highly conserved among numerous GII norovirus genotypes (Fig. 3). Variation at these residues appears to be tolerated, as the 5B18 antibody detects both GII.4 VLPs, which had Thr433 (instead of Val433), and GII.12 VLPs, which had Thr433 and Ser534 (instead of Val433 and Thr534, respectively). Surprisingly, the 5B18 Fab contact residues are almost identical to those of another broad-range monoclonal antibody, MAb14-1 (Fig. 3) (59). Furthermore, the epitopes of two other broad-range monoclonal antibodies, NV3901 and NV3912, are in this general region (50). The MAb14-1 antibody was shown to bind VLPs from many GII genotypes and several GI genotypes, including a GI.1 genotype, whereas the NV3901 and NV3912 antibodies were found to only bind GI genotypes. Interestingly, the 5B18, MAb14-1, and NV3901/NV3912 antibodies were raised in different mice immunized with different VLPs and their binding sites were all in close proximity on the P1 subdomain (50). Although the precise structural binding details of MAb14-1, NV3901, and NV3912 antibodies have not been described, it suggests that the P1 subdomain was an important antigenic site for GI and GII noroviruses. Moreover, the P1 subdomain likely contained GI and GII cross-reactive epitopes. Superpositioning of published X-ray crystal structures of norovirus P domain (GI.1, GII.4, GII.9, GII.12, and GV.1) onto the GII.10 P domain-Fab complex structure showed that three of six amino acids involved in the 5B18 Fab binding were highly conserved for three norovirus genogroups and that the conformation of their side chains closely resembles those of GII.10 (see Table S2 and Fig. S4 in the supplemental material). Taken together, the results indicate that the 5B18 binding epitope represents an important site for antibody recognition (Fig. 7).
Initially, the X-ray crystal structure of the GI.1 VLP (53) was used for fitting the GII.10 P domain-Fab complex and to describe the binding interaction in the context of the entire particle. The P domains of GI.1 and GII.10 matched well (root mean square deviation [RMSD], 1.3 Å), but the 5B18 Fab clashed with the GI.1 S domain (Fig. 5C). Indeed, the P domains in GI.1 VLPs rest on the S domains, and this necessarily placed most of the Fab structure into a position that overlapped the S domain (Fig. 5C). In an attempt to understand the 5B18 antibody interaction in the context of a GII VLP, the cryo-EM structure of the GII.10 VLP was determined to an ∼10-Å resolution. Recent cryo-EM studies have shown that GI.1 and GV.1 norovirus capsid structures are strikingly different (30, 67), whereas another study indicated that GI.1 and GII.4 (Grimsby virus) capsids are highly similar (12). The cryo-EM structure of the GII.10 VLPs showed several structural similarities to the GV.1 virion, including a raised P domain, P1-P1 subdomain contacts, and an extended hinge region (see Fig. S6 in the supplemental material). In addition, the GII.10 and GV.1 P domain dimers were rotated ∼40° clockwise compared to the orientation of the GI.1 P domain dimer (data not shown). Fitting of the X-ray crystal structure of the GII.10 P domain-Fab complex into the GII.10 VLP structure showed that the P domain could be positioned unambiguously into the P domain density of the EM map; however, this placement resulted in significant overlap between Fab and neighboring P and S domains in the virus particle (Fig. 5). One potential explanation for this result is that the VLPs flexibly expose the P domain to the 5B18 antibody by rotating the P domains out of the conformation observed in the cryo-EM reconstruction and breaking the P1-P1 domain contacts seen in the VLP. This may be possible since the S domain-P1 subdomain connection in GII noroviruses is particularly long and flexible.
The structural differences between the GI.1 and GII.10 norovirus VLPs do not appear to be a consequence of sequence diversity, since the GI.1 and GII.4 VLP structures are similar and distinct from the GV.1 virion and GII.10 VLP structures. Moreover, the VLP preparation and cryo-EM techniques appear to be essentially the same (54). Two factors that may have affected the particle structures were the insect cell type and the pH of the VLPs. The GI.1 VLPs were expressed in Spodoptera frugiperda (Sf9) cells, purified by CsCl ultracentrifugation, and then resuspended in water (pH not described in text) (53, 54), and the GII.10 VLPs were expressed in Trichopulsia ni (H5) cells, purified by CsCl ultracentrifugation, and then resuspended in PBS (pH 7.3). We note parenthetically that the cryo-EM structures of hepatitis E virus VLPs expressed in Sf9 and H5 cells are similar, although the processing of the viral protein appeared different (38). Our EM results showed that GII.10 VLPs were intact particles at pH 5.3, 6.3, and 7.3, while another study found that the diameter of norovirus VLPs remained virtually unchanged at pH 3 to 7 but appeared smaller at pH 8 (2). This suggests that the insect cell line and water/PBS (neutral pH) did not affect the overall structure of the VLPs. However, another study has shown that a pH change from 7.6 to 5.0 could cause large structural changes in Nudaurelia capensis ω virus VLPs (43, 62). It is possible that these varied conformations do not represent different, stable norovirus structures but are rather all part of a wide spectrum of conformations afforded by the flexible tether between the P and S domains. From previous studies (30), it is clear that this “floating P domain” conformation is independent of whether the sample is a VLP or infectious virion. Since this extended conformation is now observed in rabbit hemorrhagic disease virus (also a calicivirus, genus Lagovirus), it also cannot be dependent upon calicivirus genera. It is possible that the energy differences between the conformations represented by these viruses is relatively small and that subtle protein-protein interaction differences favor one conformation under particular conditions. It would be particularly interesting to examine the conformations of these viruses under a broad range of conditions that mimic the expected environments during the viral life cycle. Such changes in virion structure have been observed with numerous other viruses (3, 9, 46, 64, 71). In the case of GV.1 norovirus, where there is an animal model (69) and infectious clone (66), it would also be important to determine what role this flexible tether region has in the replication of the virions and pathology of the disease.
It is important to note that the observed ELISA binding of 5B18 IgG may not occur with intact VLPs. It is possible that denatured or partially broken VLPs or the presence of contaminating GII.10 VP1 was responsible for the binding observed in the ELISA (19, 20, 24). However, it is known that high pH (8.3 or above), partially breaks or denatures norovirus VLPs (2). Despite this pH dependence, the titer remained almost identical, especially in the comparison between pHs 7.3 to 9.3 (Fig. 6), suggesting that only intact or structurally stable virions are being detected. Moreover, the 5B18 antibody could detect GII.10 VLPs that were bound to the plates via histo-blood group antigens, which required a dimeric interaction (22; also unpublished data). Finally three other antibodies, MAb14-1 and NV3901/NV3912, which bound in close proximity to the 5B18 were all shown to detect VLPs (50, 59). These data therefore favor a model in which apparently intact norovirus capsids can indeed bind the 5B18 antibody (and other antibodies) despite significant steric clashes with the VLP structure.
Viruses often use remarkable conformational changes in their envelope or capsid structures to protect their genetic material by waiting for the proper cellular trigger to release their genome into the host cell. For example, the hemagglutinin spike in influenza undergoes a drastic pH-dependent conformational change in the endosome that initiates membrane fusion (8, 68). Similarly large pH-dependent changes have been observed with the enveloped flaviviruses (31, 32, 47) and alphaviruses (36, 45). Such changes due to environmental cues can expose or hide antigenic sites (e.g., see references 41, 45, and 47). Viruses can also receive cues via interactions with cellular receptors, as is the case with human rhinovirus (25, 26, 48). Viruses also undergo small, dynamic structural changes, “breathing” (6, 35, 37, 56), that are probably a prelude to the far larger conformational changes that occur during uncoating. These dynamic motions can transiently expose more-conserved antigenic sites that can be leveraged in designing vaccines (29, 37). However, the fact that these norovirus antibodies are recognizing deeply occluded portions of the P1 domain in apparently intact virions represents a different kind of viral dynamic: for this recognition to occur, the P domains must be capable of extremely large conformational changes without any obvious environmental cue. Such recognition would probably involve just one or a few P domains of a VLP being recognized by antibody 5B18; indeed, images of VLPs after incubation with an excess of antibody 5B18 for 1 h at 37°C (the same incubation used in the ELISA) shows them to be intact, with bound IgG difficult to detect (see Fig. S7 in the supplemental material).
Other antibodies have recently been described that bind to occluded sites on virions. With West Nile virus, the fusion loop-specific antibody E53 recognizes an epitope that should be inaccessible on mature virions. However, this antibody could neutralize mature West Nile virus in a time- and temperature-dependent manner, indicating a role of virus “breathing” or conformational dynamics in antibody recognition (17). With HIV-1, broadly neutralizing antibodies against the membrane-proximal external region of the virus do not appear to recognize the native viral spike (11, 58), again implicating conformational rearrangements to permit antibody recognition. These studies, along with the present study on norovirus recognition by 5B18, suggest substantial flexibility in certain virus particles as being important biologically for antibody-mediated recognition.
In summary, we have shown that a broadly reactive monoclonal antibody binds to an occluded site on the GII.10 P1 subdomain. The binding site was in close proximity to other monoclonal antibody binding sites, suggesting that the site contained an immunodominant region. We also found that the GII.10 VLP structure was more closely related to a GV.1 virion structure than to a GI.1 VLP structure and has marked flexibility in the P domains. These studies suggest that the P domain of noroviruses is capable of adopting variable conformations with respect to the S domain. Despite the vaunted diversity of noroviruses, especially on the exposed outer surface of the virion, one mechanism to achieve near pan-recognition by antibody may be to target a highly conserved domain interface that is dynamically exposed to the environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Stuckey for assistance with figures and members of the Structural Biology Section at the NIH Vaccine Research Center for help with Fab preparation and comments on the manuscript, K. Nagayama for generous help and insightful discussions, and M. Kataoka for assistance with electron microscopy.
G.S.H. and P.D.K. conceived the project; G.S.H. performed X-ray crystallography and biochemical assays with assistance from J.S.M., S.-Y.P., and P.D.K.; D.W.T. determined the cryo-EM structure with assistance from K.M.; M.Y., F.G., M.M., and K.K. provided the 5B18 IgG; I.G. mapped sequence conservation onto the GII.10 VLP; and G.S.H., D.W.T., J.S.M., T.J.S., I.G., J.R.H.T., K.M., and P.D.K. analyzed the data and wrote the paper, on which all authors commented.
Support for this work was provided by the Intramural Research Program of the National Institutes of Health (NIAID [P.D.K.]), USA, and by a Grant-in-Aid for Scientific Research, grants from the Ministry of Health, Labor, and Welfare of Japan, and a grant from the National Institute of Natural Sciences (NINS), Japan (K.M.). D.W.T is an NSF Graduate Research Fellow and performed this work in Japan as a JSPS/NSF East Asia and Pacific Summer Institute Fellow. Use of Sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
Footnotes
Published ahead of print 25 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adams PD, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66(Pt 2):213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ausar SF, Foubert TR, Hudson MH, Vedvick TS, Middaugh CR. 2006. Conformational stability and disassembly of Norwalk virus-like particles. Effect of pH and temperature. J. Biol. Chem. 281:19478–19488 [DOI] [PubMed] [Google Scholar]
- 3. Belnap DM, et al. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhella D, Gatherer D, Chaudhry Y, Pink R, Goodfellow IG. 2008. Structural insights into calicivirus attachment and uncoating. J. Virol. 82:8051–8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhella D, Goodfellow IG. 2011. The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J. Virol. 85:11381–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. 1998. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J. Biol. Chem. 273:673–676 [DOI] [PubMed] [Google Scholar]
- 7. Bu W, et al. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43 [DOI] [PubMed] [Google Scholar]
- 9. Canady MA, Tihova M, Hanzlik TN, Johnson JE, Yeager M. 2000. Large conformational changes in the maturation of a simple RNA virus, nudaurelia capensis omega virus (NomegaV). J. Mol. Biol. 299:573–584 [DOI] [PubMed] [Google Scholar]
- 10. Cao S, et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chakrabarti BK, et al. 2011. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J. Virol. 85:8217–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen R, et al. 2004. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 78:6469–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi JM, Hutson AM, Estes MK, Prasad BV. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175–9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collaborative Computational Project N 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 15. de Bruin E, Duizer E, Vennema H, Koopmans MP. 2006. Diagnosis of Norovirus outbreaks by commercial ELISA or RT-PCR. J. Virol. Methods 137:259–264 [DOI] [PubMed] [Google Scholar]
- 16. Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. 2011. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7:e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of COOT. Acta Crystallogr. D. Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graham DY, et al. 1994. Norwalk virus infection of volunteers: new insights based on improved assays. J. Infect. Dis. 170:34–43 [DOI] [PubMed] [Google Scholar]
- 20. Greenberg HB, et al. 1981. Proteins of Norwalk virus. J. Virol. 37:994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansman GS, et al. 2006. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 87:909–919 [DOI] [PubMed] [Google Scholar]
- 22. Hansman GS, et al. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J. Virol. 85:6687–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansman GS, et al. 2004. Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch. Virol. 149:1673–1688 [DOI] [PubMed] [Google Scholar]
- 24. Hardy ME, White LJ, Ball JM, Estes MK. 1995. Specific proteolytic cleavage of recombinant Norwalk virus capsid protein. J. Virol. 69:1693–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hewat EA, Blaas D. 2004. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J. Virol. 78:2935–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoover-Litty H, Greve JM. 1993. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J. Virol. 67:390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang X, Wang M, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamata K, et al. 2005. Expression and antigenicity of virus-like particles of norovirus and their application for detection of noroviruses in stool samples. J. Med. Virol. 76:129–136 [DOI] [PubMed] [Google Scholar]
- 29. Katpally U, Fu T, Freed DC, Casimiro DR, Smith TJ. 2009. Antibodies to the buried N-terminus of rhinovirus VP4 exhibit cross-serotypic neutralization. J. Virol. 83:7040–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Katpally U, et al. 2010. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J. Virol. 84:5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaufmann B, et al. 2009. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 5:e1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaufmann B, et al. 2006. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 103:12400–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 34. Kwong PD, et al. 1999. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274:4115–4123 [DOI] [PubMed] [Google Scholar]
- 35. Lewis JK, Bothner B, Smith TJ, Siuzdak G. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. U. S. A. 95:6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Q, Yafal AG, Lee YMH, Hogle J, Chow M. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of the sequences at physiological temperatures. J. Virol. 68:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li TC, et al. 2005. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 79:12999–13006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X, Zhou R, Tian X, Li H, Zhou Z. 2010. Characterization of a cross-reactive monoclonal antibody against Norovirus genogroups I, II, III and V. Virus Res. 151:142–147 [DOI] [PubMed] [Google Scholar]
- 40. Lochridge VP, Jutila KL, Graff JW, Hardy ME. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799–2806 [DOI] [PubMed] [Google Scholar]
- 41. Lok S-M, et al. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312–317 [DOI] [PubMed] [Google Scholar]
- 42. Majeed S, et al. 2003. Enhancing protein crystallization through precipitant synergy. Structure 11:1061–1070 [DOI] [PubMed] [Google Scholar]
- 43. Matsui T, Lander G, Johnson JE. 2009. Characterization of large conformational changes and autoproteolysis in the maturation of a T=4 virus capsid. J. Virol. 83:1126–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCoy AJ, et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meyer WJ, Gidwitz S, Ayers VK, Schoepp RJ, Johnston RE. 1992. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J. Virol. 66:3504–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miao Y, Johnson JE, Ortoleva PJ. 2010. All-atom multiscale simulation of cowpea chlorotic mottle virus capsid swelling. J. Phys. Chem. B. 114:11181–11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 48. Olson NH, et al. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. U. S. A. 90:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 50. Parker TD, Kitamoto N, Tanaka T, Hutson AM, Estes MK. 2005. Identification of genogroup I and genogroup II broadly reactive epitopes on the norovirus capsid. J. Virol. 79:7402–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pei J, Grishin NV. 2001. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics 17:700–712 [DOI] [PubMed] [Google Scholar]
- 52. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 53. Prasad BV, et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 54. Prasad BV, Rothnagel R, Jiang X, Estes MK. 1994. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 68:5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rabenau HF, et al. 2003. Laboratory diagnosis of norovirus: which method is the best? Intervirology 46:232–238 [DOI] [PubMed] [Google Scholar]
- 56. Reisdorph N, et al. 2003. Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virology 314:34–44 [DOI] [PubMed] [Google Scholar]
- 57. Richards AF, et al. 2003. Evaluation of a commercial ELISA for detecting Norwalk-like virus antigen in faeces. J. Clin. Virol. 26:109–115 [DOI] [PubMed] [Google Scholar]
- 58. Ruprecht CR, et al. 2011. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J. Exp. Med. 208:439–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shiota T, et al. 2007. Characterization of a broadly reactive monoclonal antibody against norovirus genogroups I and II: recognition of a novel conformational epitope. J. Virol. 81:12298–12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan M, Hegde RS, Jiang X. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 78:6233–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang G, et al. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46 [DOI] [PubMed] [Google Scholar]
- 62. Tang J, et al. 2009. Dynamics and stability in maturation of a T=4 virus. J. Mol. Biol. 392:803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Taube S, et al. 2010. High-resolution X-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J. Virol. 84:5695–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Trus BL, et al. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447–462 [DOI] [PubMed] [Google Scholar]
- 65. van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17–24 [DOI] [PubMed] [Google Scholar]
- 66. Ward VK, et al. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U. S. A. 104:11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Widdowson M-A, et al. 2005. Detection of serum antibodies to bovine norovirus in veterinarians and the general population in the Netherlands. J. Med. Virol. 76:119–128 [DOI] [PubMed] [Google Scholar]
- 68. Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature 289:366–373 [DOI] [PubMed] [Google Scholar]
- 69. Wobus CE, Thackray LB, Virgin HW. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yoda T, et al. 2003. Precise characterization of norovirus (Norwalk-like virus)-specific monoclonal antibodies with broad reactivity. J. Clin. Microbiol. 41:2367–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yu IM, et al. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837 [DOI] [PubMed] [Google Scholar]
- 72. Zheng DP, et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.