ABSTRACT
Spatial organization within bacteria is fundamental to many cellular processes, although the basic mechanisms underlying localization of proteins to specific sites within bacteria are poorly understood. The study of protein positioning has been limited by a paucity of methods that allow rapid large-scale screening for mutants in which protein positioning is altered. We developed a genetic reporter system for protein localization to the pole within the bacterial cytoplasm that allows saturation screening for mutants in Escherichia coli in which protein localization is altered. Utilizing this system, we identify proteins required for proper positioning of the Shigella autotransporter IcsA. Autotransporters, widely distributed bacterial virulence proteins, are secreted at the bacterial pole. We show that the conserved cell division protein FtsQ is required for localization of IcsA and other autotransporters to the pole. We demonstrate further that this system can be applied to the study of proteins other than autotransporters that display polar positioning within bacterial cells.
IMPORTANCE
Many proteins localize to specific sites within bacterial cells, and localization to these sites is frequently critical to proper protein function. The mechanisms that underlie protein localization are incompletely understood, in part because of the paucity of methods that allow saturation screening for mutants in which protein localization is altered. We developed a genetic reporter assay that enables screening of bacterial populations for changes in localization of proteins to the bacterial pole, and we demonstrate the utility of the system in identifying factors required for proper localization of the polar Shigella autotransporter protein IcsA. Using this method, we identify the conserved cell division protein FtsQ as being required for positioning of IcsA to the bacterial pole. We demonstrate further that the requirement for FtsQ for polar positioning applies to other autotransporters and that the method can be applied to polar proteins other than autotransporters.
Introduction
The proper localization of proteins in bacterial cells is critical to many cellular processes, including virulence, DNA replication, chromosome segregation, cell division, chemotaxis, gene transfer, and motility. Detailed molecular understanding of how proteins localize in bacteria is incomplete. A major challenge in the investigation of the mechanism of protein localization has been the paucity of methods that enable screening of large numbers of mutants for defects in protein localization. While previously published large-scale screens have yielded important insights, they have depended on microscopic visualization of individual mutants (e.g., see references 1, 2), which is tedious and time-consuming.
In rod-shaped bacteria, a subset of proteins localize to the bacterial pole. Among these are autotransporter proteins, the largest group of secreted proteins in Gram-negative bacteria. Autotransporters are outer membrane proteins that are secreted at the bacterial pole, contain a large domain exposed on the bacterial surface (see Fig. S1a in the supplemental material), and commonly play a role in virulence (3–5). The model autotransporter IcsA from Shigella mediates assembly of an actin tail at the bacterial pole that propels the intracellular bacterium through the intestinal epithelium during infection in humans (6). Like other autotransporters, IcsA is secreted across the cytoplasmic membrane via the Sec secretion apparatus (7), is inserted into and translocated across the outer membrane in a process that requires the outer membrane insertase BamA (YaeT, Omp85) (8), and is localized to the bacterial pole (4).
Localization of IcsA to the bacterial pole occurs in the cytoplasm prior to secretion across the cytoplasmic membrane (7, 9). Although IcsA is present only in Shigella spp., the molecular mechanism that localizes IcsA to the pole is conserved among a wide variety of Gram-negative bacteria (9, 10).
Currently, to our knowledge, there are no methods that enable large-scale genetic screening of spatial positioning of proteins in bacteria. Using IcsA, we developed a reporter assay for protein localization within the bacterial cytoplasm that allows saturation screening for mutants in which protein localization is altered. Utilizing this assay, we identify proteins required for proper positioning of IcsA and other autotransporters at the bacterial pole. We demonstrate further that this system can be successfully applied to the study of proteins other than autotransporters that localize to the bacterial pole.
RESULTS
Reporter system for IcsA localization in the bacterial cytoplasm.
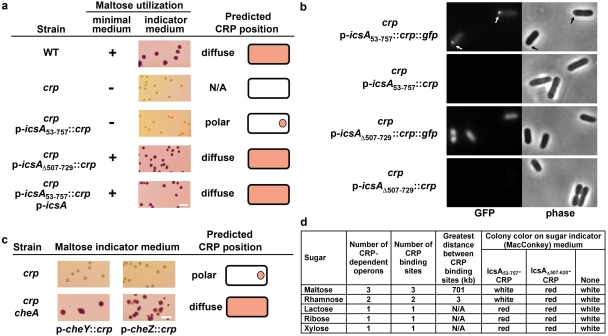
To identify proteins involved in polar localization of IcsA, we developed a reporter system for IcsA localization in the bacterial cytoplasm based on the premise that fusion of a transcription regulator to IcsA in the cytoplasm might sequester the transcription regulator at the pole and disrupt its ability to regulate its cognate operons. CRP (cyclic AMP receptor protein), a transcription factor required for uptake and utilization of maltose (11), was translationally fused to the carboxy terminus of a derivative of IcsA that localizes to the pole and yet remains in the cytoplasm because it lacks a signal peptide (the entire α domain, residues 53 to 757 [see Fig. S1b in the supplemental material]) (9). On maltose utilization indicator (maltose MacConkey) medium, wild-type (WT) Escherichia coli (strain MG1655), which can utilize maltose as a carbon source, formed red colonies and a crp mutant, which cannot utilize maltose, formed white colonies; similarly, on maltose minimal medium, the wild-type strain grew well, but the crp mutant did not grow (Fig. 1a). A crp mutant directing the synthesis of the fusion of IcsA53-757 to CRP (IcsA53-757-CRP) formed white colonies on indicator medium and did not grow on maltose minimal medium, whereas the same mutant directing the synthesis of CRP alone or an IcsA-CRP fusion protein lacking the IcsA polar localization signal (IcsAΔ507-729-CRP) was able to use maltose on both media (Fig. 1a). Because an IcsA53-757-GFP fusion protein localizes to the bacterial pole and an IcsAΔ507-729-GFP fusion protein is diffuse (see Fig. S1c) (9), the inability of cells containing IcsA53-757-CRP to utilize maltose is likely due to sequestration of the fusion protein at the pole, thereby disrupting CRP activity, and the ability of IcsAΔ507-729-CRP to utilize maltose is likely due to delocalization of the fusion protein. To further test the hypothesis that sequestration of the fusion protein at the pole is responsible for the maltose utilization-minus phenotype of cells containing IcsA53-757-CRP, we provided excess full-length untagged IcsA from a multicopy vector, which is capable of displacing localized (nonaggregated) IcsA-green fluorescent protein (GFP) fusion proteins from the pole (9, 12, 13). Cells containing the IcsA53-757-CRP fusion protein together with excess full-length IcsA were once again able to utilize maltose (Fig. 1a), providing strong support for the inference that the IcsA53-757 moiety can sequester CRP at the cell pole.
FIG 1 .
Reporter assay for positioning of proteins at the bacterial pole. (a) Maltose utilization phenotype of indicated E. coli MG1655 strains. icsA53-757, polar cytoplasmic derivative of IcsA; icsAΔ507-729, delocalized derivative of IcsA; p-icsA, full-length icsA. + or −, growth or absence of growth, respectively, on maltose minimal medium. Red colony, maltose utilization; white colony, maltose nonutilization on maltose MacConkey indicator agar. (b) Positions of IcsA53-757-CRP-GFP and IcsAΔ507-729-CRP-GFP in MG1655 crp. Arrows, polar foci of IcsA53-757-CRP-GFP. (c) Maltose utilization phenotype of MG1655 crp or crp cheA, carrying a plasmid carrying cheY::crp or cheZ::crp. Predicted CRP position, putative positioning of fusion based on observed maltose utilization phenotype. (d) Colony phenotype on indicator medium containing various sugars. Note the correlation of the specificity of the phenotype with multiple CRP-dependent operons being required for sugar utilization. N/A, not applicable. Bars, 5 mm.
To examine the position of the CRP reporter fusions in these cells, we placed a GFP tag at the C termini of IcsA53-757-CRP and IcsAΔ507-729-CRP. Cells containing IcsA53-757-CRP-GFP displayed foci that were polarly localized (Fig. 1b, top row, arrows), whereas cells containing IcsAΔ507-729-CRP-GFP displayed a diffuse signal (Fig. 1b, third row). The GFP foci were visualized only upon induction of expression by addition of low concentrations of the inducer anhydrotetracycline (50 ng/ml), which reflects the low level of synthesis of IcsA53-757-CRP and IcsAΔ507-729-CRP from the same vector in the experiments described above, which were performed in the absence of inducer (see comparison to level of synthesis of native IcsA [see Fig. S2a in the supplemental material]). The two GFP fusion proteins were produced at similar levels (see Fig. S2b). These data provide additional evidence for the inference that IcsA53-757-CRP sequesters CRP at the pole and IcsAΔ507-729-CRP does not.
Further validation of this reporter system was provided by the observation that fusion of CRP to each of two other polar proteins, the soluble chemotaxis proteins CheY and CheZ, also disrupted its activity (Fig. 1c). The localization of CheY or CheZ to the pole is dependent on the polar membrane-associated histidine kinase CheA (14), and expression of either the cheY::crp or cheZ::crp fusion from a multicopy plasmid in a cheA crp strain rescued maltose utilization (Fig. 1c), consistent with sequestration of CRP at the pole by the fusion proteins being dependent on CheA. These results demonstrate that IcsA is not unique in its ability to interfere with the function of a fused transcription regulator by sequestering it at the pole and suggest that this capability may be a general feature of polar proteins. Thus, the inability of a CRP-minus strain containing a polar protein fusion to CRP to utilize maltose as a carbon source correlates with polar localization of the protein-CRP fusion.
As CRP is required for the transcriptional activation of operons that encode proteins for the utilization of any of several sugars as carbon sources by the bacterium, we tested whether utilization of sugars other than maltose could serve as a readout for this reporter system. On indicator medium that contained rhamnose, growth of the E. coli crp strain carrying the polar IcsA53-757-CRP reporter construct or the nonpolar IcsAΔ507-729-CRP control construct reproduced the phenotypes observed on maltose indicator medium, with cells carrying IcsA53-757-CRP growing as white colonies and cells carrying IcsAΔ507-729-CRP growing as red colonies (Fig. 1d). However, on indicator medium that contained lactose, ribose, or xylose, both the cells carrying IcsAΔ507-729-CRP and the cells carrying IcsA53-757-CRP grew as red colonies (Fig. 1d). The parent E. coli crp strain was unable to metabolize any of these sugars, since in all cases it grew as white colonies (Fig. 1d), indicating that the red colony color of cells carrying IcsA53-757-CRP upon growth on lactose, ribose, or xylose was due to binding of the CRP moiety of the fusion protein to cognate CRP promoters in these cells. We speculate that these differences reflect the ability of IcsA-CRP molecules that are sequestered at the pole to access the transcriptional units required for utilization of each of these sugars. For the utilization of maltose or rhamnose, CRP regulates multiple operons, and multiple CRP binding sites at a distance from one another must be occupied to activate transcription of these operons (11, 15). In contrast, for utilization of lactose, ribose, or xylose, either sugar utilization itself depends on transcription of only a single operon, or if transcription of multiple operons is required, CRP activates only one of them (16–18). The position of the CRP binding sites relative to the E. coli origin was not sufficient to explain the sugar utilization phenotype on the basis of the origin being generally positioned near the bacterial pole (see Table S1 in the supplemental material). Instead, when CRP-dependent activation of only a single operon is required, we speculate that, within a subpopulation of cells in each colony, the chromosome will at times be positioned such that the cognate CRP binding site will be sufficiently close to the bacterial pole to permit binding and activation, leading to sugar utilization and red colony color (see Fig. S3). Since only 3 kb separates the two sites required for rhamnose utilization, relatively short distances between CRP binding sites may be sufficient to prevent coordinated binding of sequestered CRP; alternatively, the two rhamnose CRP binding sites may not be simultaneously accessible due to other aspects of chromosome positioning or structure. Alternatively, these findings could potentially be explained by the level of production of IcsA53-757-CRP being sufficient to occupy one binding site but insufficient to occupy two or three binding sites or by differences in the affinities of IcsA53-757-CRP for the various binding sites. However, the findings described below, in which elevated levels of RpoS resulting from disruption of rssB lead both to rescue of maltose utilization (Fig. 2a) with no change in the level of production of IcsA53-757-CRP (see Fig. S2a) and to delocalization of IcsA507-620-GFP (Fig. 2b), indicate that this level of production of IcsA53-757-CRP is sufficient to occupy all three CRP binding sites within the maltose operons and that differences in affinities for these sites do not determine maltose utilization. These findings suggest that IcsA-CRP sequestered at the pole retains its ability to activate CRP-dependent promoters and that the limitation on its ability to activate cognate promoters may result from the positioning of the chromosome in the cells.
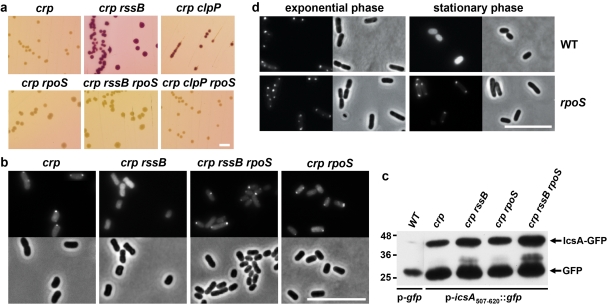
FIG 2 .
High levels of RpoS disrupt localization of IcsA to the bacterial pole. (a) Maltose utilization phenotypes on maltose MacConkey indicator agar of the indicated E. coli MG1655 strains, each carrying the icsA53-757::crp reporter. (b and d) Positions of IcsA507-620-GFP in each of the indicated derivatives of E. coli MG1655. GFP fluorescence and phase micrographs. (c) Levels of IcsA53-757-GFP in indicated derivatives of MG1655, corresponding to panel b. For each construct, breakdown of fusion protein yields some free GFP, which is likely responsible for the diffuse signal in those strains with polar foci and for part of the diffuse signal in the crp rssB strain. Numbers at left are molecular masses in kilodaltons. Bars, 5 mm (a) and 10 µm (b and d).
High levels of RpoS (sigma S) disrupt localization of IcsA to the pole.
To identify nonessential genes involved in targeting IcsA to the pole, we performed a large-scale transposon mutagenesis of crp cells containing IcsA53-757-CRP, first selecting for mutants that displayed rescue of growth on maltose minimal medium and subsequently screening these mutants for red colony color on maltose indicator medium. Utilization of maltose was rescued only by transposon insertions in rssB or in the plasmid carrying icsA53-757::crp. Insertions in the plasmid most likely led to production of nonsequestered CRP and were thus not studied further. A strain with a nonpolar deletion of rssB also formed red colonies, and maltose nonutilization was rescued by expression of rssB in trans (Fig. 2a; Table 1), indicating that the maltose utilization phenotype of the rssB mutant was due to the transposon insertion in rssB. To visualize localization of IcsA in the cytoplasm, we tagged IcsA with GFP, using for these studies IcsA residues 507 to 620 (IcsA507-620-GFP [see Fig. S1d in the supplemental material]), the minimal internal fragment of IcsA53-757 that localizes to the pole, because this fragment is slightly more efficient at localizing the fused GFP moiety to the pole than are residues 53 to 757 (9). The rssB mutation caused IcsA delocalization in the cytoplasm, since IcsA507-620-GFP was diffuse in crp rssB cells and yet polar in crp cells (Fig. 2b and 2c; see also Table S2) while being produced at similar levels under the two conditions.
TABLE 1 .
Maltose utilization and RpoS levels in E. coli MG1655 strains
| Strain | Maltose utilization |
RpoS levela | |
|---|---|---|---|
| Growth on minimal medium | Colony color on indicator medium | ||
| WT, stationary phase | + | Red | 1.0 |
| WT, exponential phase | NDb | ND | BDc |
| crp p-icsA53-757::crp strain | − | White | BD |
| crp rssB p-icsA53-757::crp strain | + | Red | 1.4 |
| crp rssB p-icsA53-757::crp p-rssB strain | ND | White | ND |
| crp rpoS p-icsA53-757::crp strain | − | White | BD |
| crp rssB rpoS p-icsA53-757::crp strain | − | White | BD |
| crp clpP p-icsA53-757::crp strain | ND | Red | 1.3 |
| crp clpP rpoS p-icsA53-757::crp strain | ND | White | BD |
RpoS levels were determined for exponential-phase growth in rich media, except where otherwise indicated, and were normalized to the level in WT stationary-phase cells.
ND, not determined.
BD, below the level of detection.
The protein encoded by rssB, RssB (SprE), targets the sigma factor RpoS (sigma S) to the ClpXP protease complex during the exponential phase of growth, leading to RpoS degradation (19) (see Fig. S2c in the supplemental material). During late exponential phase, RpoS levels in the crp rssB mutant were 10-fold higher than those in crp cells (see Fig. S2e), as seen previously (20, 21). IcsA delocalization was due to the high levels of RpoS, since disruption of rpoS in the crp rssB p-IcsA53-757-CRP strain caused loss of maltose utilization (Fig. 2a; Table 1) and in the crp rssB p-IcsA507-620-GFP strain restored polar GFP foci (Fig. 2b; see also Table S2). Moreover, inactivation of the ClpXP protease complex by disruption of clpP in the crp p-IcsA53-757-CRP strain restored maltose utilization in a manner dependent on rpoS (Fig. 2a; Table 1). The fainter red color of the crp clpP colonies was not due to lower levels of RpoS (see Fig. S2f) but may be due to either the slower growth of Clp strains (22) or the accumulation of another ClpP substrate that also influences maltose utilization. Of note, insertions in clpP and clpX were not isolated in our transposon mutagenesis selection, likely because the slower growth of these strains may cause the mutants to be missed. We did not attempt to overexpress rpoS from an inducible promoter, since it is difficult to increase levels of RpoS by this approach due to its tight posttranslational regulation. Altogether, these data demonstrate that high levels of RpoS disrupt polar localization of IcsA and validate the ability of the CRP reporter system to identify cells in which IcsA is delocalized.
RpoS accumulates in stationary phase. In wild-type cells, IcsA507-620-GFP localized to the pole during exponential phase but was delocalized at high cell density in a manner dependent on rpoS (Fig. 2c; see also Table S2 in the supplemental material), while IcsA507-620-GFP levels were similar. In Shigella flexneri, the disruption of rssB led to undetectable IcsA on the bacterial surface (IcsA as polar on 99% ± 2% of wild-type [WT] cells versus <1% ± 0% of rssB cells, P = 6 × 10−8 [see Fig. S4a and Table S3]) and in cell lysates (see Fig. S4b). The absence of IcsA in this strain could be consistent with the presence of high levels of RpoS interfering with proper targeting of IcsA that in turn causes it to be unstable. We were unable to directly test whether the block in IcsA secretion and/or stability in these cells was due to elevated RpoS levels, since introduction of a mutation in rpoS into the S. flexneri rssB (but not the E. coli rssB) background led to a high rate of cell lysis (see Table S3). We have previously observed a significant decrease in IcsA signal and localization to the pole in stationary-phase S. flexneri serotype 5 strain M90T (23); however, this effect or the levels of RpoS in stationary phase appear to be somewhat strain dependent, as in the current study using serotype 2a strain 2457T, the decreases in IcsA signal and localization in stationary phase were modest (IcsA was polar on 99% ± 2% of exponential-phase cells versus 54% ± 34% of stationary-phase cells, P = 0.08 [see Fig. S4a and Table S3]). This suggests that while markedly increased levels of RpoS seen in rssB strains may be sufficient to interfere with IcsA localization, moderately increased levels seen in stationary phase have strain-dependent effects on IcsA localization.
The conserved cell division protein FtsQ is required for IcsA localization to the bacterial pole.
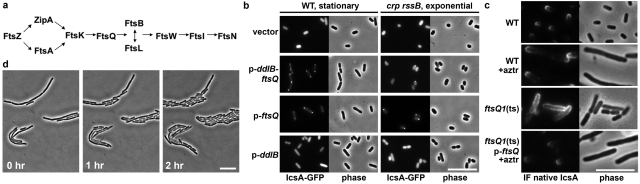
RpoS activates the transcription of a large number of genes (19). We postulated that a component of the IcsA polar localization machinery might be inhibited by a factor synthesized under the control of RpoS, in which case overproduction of this component of the polar localization machinery might titrate out the inhibitory factor, thereby rescuing polar localization of IcsA. From among plasmids of a multicopy E. coli chromosomal library introduced into the crp rssB p-IcsA53-757-CRP strain, the essential cell division gene ftsQ (Fig. 3a) in an operon with the peptidoglycan biosynthesis gene ddlB rescued the white colony phenotype on maltose indicator medium. In the presence of the ddlB-ftsQ region, but not of vector alone, IcsA507-620-GFP localized to the pole in both exponential-phase crp rssB cells and stationary-phase wild-type cells (Fig. 3b; Table 2), without significant alteration of IcsA507-620-GFP or RpoS levels (see Fig. S2d and S2e in the supplemental material). ftsQ alone restored polar localization of IcsA507-620-GFP, whereas ddlB alone did not (Fig. 3b; Table 2), suggesting that the function of FtsQ in this phenotype may be inhibited by elevated levels of RpoS and that FtsQ may be a component of the machinery required to localize IcsA to the pole.
FIG 3 .
Conserved cell division protein FtsQ is required for localization of IcsA to the pole. (a) Hierarchical recruitment of cell division proteins (33). (b) Position of IcsA507-620-GFP in E. coli MG1655 or an MG1655 crp rssB mutant, carrying vector, p-ddlB-ftsQ, p-ddlB, or p-ftsQ, grown to stationary or exponential phase. (c) Distribution of native IcsA on the surface of intact cells of WT S. flexneri 2457T or an ftsQ1(Ts) derivative visualized by immunofluorescence (IF). Growth was at 37°C, the restrictive temperature for ftsQ1(Ts) cells, for 1 h. Aztreonam was used to filament WT (WT + aztr) and complemented ftsQ1(Ts) cells during the 1-h period; addition of aztreonam per se had no effect on distribution of IcsA. (d) Rescue of cell division upon shift of S. flexneri 2457T ftsQ1(Ts) to 30°C following 2 h of growth at 37°C. Bars, 10 µm.
TABLE 2 .
Distribution of GFP- or mCherry-tagged cytoplasmic derivative of IcsA (IcsA507-620-GFP or IcsA507-620-mCherry) in E. colia
| Relevant genotype (growth phase) | Distribution of IcsA-GFP or IcsA-mCherry (% of cells) |
|
|---|---|---|
| Foci at poles | Diffuse | |
| crp rssB p-vector (exponential) | 15 ± 5b,c | 86 ± 5 |
| crp rssB p-ddlB-ftsQ (exponential) | 85 ± 1b | 15 ± 1 |
| crp rssB p-ftsQ (exponential) | 75 ± 8c | 26 ± 8 |
| crp rssB p-ddlB (exponential) | 14 ± 3 | 86 ± 3 |
| WT p-vector (stationary) | 19 ± 8d,e | 81 ± 8 |
| WT p-ddlB-ftsQ (stationary) | 97 ± 1d | 3 ± 1 |
| WT p-ftsQ (stationary) | 91 ± 4e | 9 ± 4 |
| WT p-ddlB (stationary) | 7 ± 1 | 93 ± 1 |
| WT λatt-gfp (stationary) | 16 ± 6f,g | 84 ± 6 |
| WT λatt-gfp::qqq (stationary) | 82 ± 6f | 18 ± 6 |
| WT λatt-gfp::ffq (stationary) | 85 ± 4g | 15 ± 4 |
All strains are MG1655 and its derivatives, except for chromosomal integrants of gfp, gfp::qqq, and gfp::ffq, which are in the KS272 background.
P = 0.03.
P = 0.01.
P = 0.03.
P = 0.003.
P = 0.007.
P = 0.005.
In its native context, IcsA localizes to the poles in the outer membrane of Shigella sp. (4). The ftsQ1(Ts) allele of ftsQ contains an E125K mutation, which leads to dysfunction of FtsQ at elevated temperatures, inhibition of septation, and filamentation (24). Since IcsA expression is optimal at 37°C, this was used as the restrictive temperature in these experiments. In S. flexneri cells in which FtsQ encoded by the ftsQ1(Ts) allele had been inactivated by growth at 37°C, IcsA was distributed circumferentially around the filamented cells (Fig. 3c), whereas in wild-type cells grown at 37°C, similar to previous results (4), IcsA was localized to the poles on 43% ± 6% of the cells, reflective of its localization under these growth conditions to the older of the two bacterial poles of most cells. To test whether filamentation per se influenced IcsA localization, wild-type cells were filamented with aztreonam, which inhibits the cell division protein FtsI; IcsA localized to the poles of these cells (Fig. 3c) [filament poles with IcsA at 37°C, 2% ± 2% of ftsQ1(Ts) cells versus 98% ± 2% of WT cells, P = 5.5 × 10−7]. The presence of IcsA at both poles of most filamented wild-type cells, but only the older pole of dividing wild-type cells, has been noted previously (12) and suggests that the newer pole matures during filamentation in such a way as to enable localization to these sites. Polar localization of IcsA in the ftsQ1 strain grown at 37°C was rescued by the presence of ftsQ in trans (Fig. 3c). Altered IcsA localization was not due to loss of cell viability, since shifting cells to 30°C after growth at 37°C rescued both cell division (Fig. 3d) and polar localization of IcsA (IcsA was polar on 17% ± 7% and 27% ± 1% of cells after shifting to 30°C for 1 and 2 h, respectively). These findings indicate that FtsQ function is required for localization of native IcsA to the poles of S. flexneri.
Cell division proteins other than FtsQ are not required for establishing IcsA polarity.
The cell division proteins FtsB and FtsL form a complex with FtsQ (Fig. 3a) (25). We examined whether these and cell division proteins that lie downstream of FtsQ in the cell division recruitment cascade might also be required for proper IcsA localization. Chromosomal overproduction of FtsB was able to rescue polarity of cytoplasmic IcsA in stationary-phase E. coli (see Fig. S5a in the supplemental material), as did multicopy or chromosomal overproduction of FtsQ (Fig. 3b; see also Fig. S5a), while chromosomal overproduction of all other downstream cell division proteins did not (see Fig. S5a), raising the possibility that FtsB might be required for establishing IcsA polarity. However, on S. flexneri cells that had been depleted of FtsB, IcsA localized to the pole similarly to its localization on wild-type S. flexneri (see Fig. S5b), suggesting that FtsB is not required for establishing IcsA polarity and that overproduction of FtsB under our experimental conditions may increase FtsQ stability or alter FtsQ function.
The periplasmic domain of FtsQ is sufficient for establishing IcsA polarity.
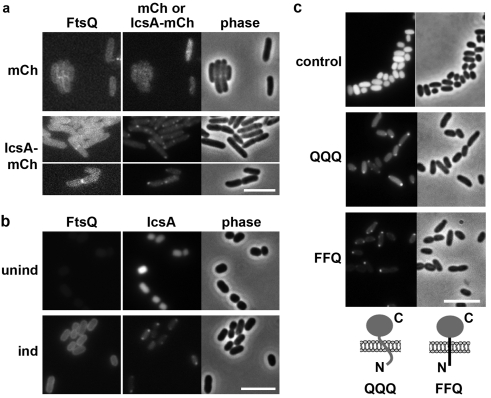
FtsQ is a bitopic membrane protein with a short cytoplasmic tail (residues 1 to 24), a transmembrane segment (residues 25 to 49), and a 227-residue periplasmic domain (residues 50 to 276). During septation, FtsQ localizes to the cytokinetic ring at midcell (Fig. 4a) (26). In stationary-phase cells, GFP-tagged FtsQ localized to the membrane circumferentially around the entire cell, whereas cytoplasmic IcsA507-620-mCherry localized to the pole (Fig. 4b). An FtsQ chimeric protein in which the cytoplasmic and transmembrane segments of MalF have replaced these domains of FtsQ (FFQ) is mildly defective in localization to midcell but recruits downstream cell division proteins (26). The FFQ construct rescued IcsA507-620 localization (Fig. 4c and Table 2), indicating that the periplasmic domain of FtsQ is sufficient to establish IcsA polarity. The ability of the periplasmic domain of FtsQ to rescue polar localization of cytoplasmic IcsA indicates that the role of FtsQ in IcsA polarity is due to an indirect relationship between the two proteins.
FIG 4 .
Periplasmic domain of FtsQ is sufficient to establish polar localization of IcsA. (a and b) Distinct distribution of IcsA507-620-mCherry and GFP-FtsQ. Exponential (a)- or stationary (b)-phase cells expressing functional GFP-FtsQ, synthesized under the control of IPTG (strain JOE226 [exponential phase] or JOE654 [stationary phase]) and carrying episomally encoded IcsA507-620-mCherry (IcsA-mCh) or mCherry (mCh). Unind or ind, uninduced or induced synthesis of GFP-FtsQ, respectively. IcsA507-620-mCherry synthesis was induced under all conditions. (c) Distribution of IcsA507-620-mCherry in stationary-phase E. coli MC4100 carrying gfp (control), gfp-qqq (QQQ), or gfp-ffq (FFQ). Diagram of QQQ and FFQ constructs is below the panels. Gray shading, FtsQ domains; black shading, MalF domains. Bars, 5 µm.
FtsQ is required for establishing polarity of other autotransporter proteins.
Like IcsA, other bacterial autotransporters are secreted at the cell pole (3). We tested whether the Shigella autotransporter SepA required FtsQ for localization to the pole in the bacterial cytoplasm by determining the localization of a GFP-tagged derivative of SepA that remains in the cytoplasm by virtue of lacking the Sec recognition sequence within its signal peptide (SepA1-24/57-1042-GFP) (3). Like IcsA, the GFP-tagged cytoplasmic derivative of SepA localizes GFP to the pole of E. coli during the exponential phase of growth (3). Also like IcsA, during stationary-phase growth, this GFP-tagged SepA was diffuse in wild-type E. coli but, in the presence of multicopy FtsQ, localized to the pole (see Fig. S6a in the supplemental material), indicating that FtsQ is required for the establishment of the polar positional information recognized by both IcsA and SepA in the bacterial cytoplasm.
We tested whether the localization of autotransporters other than IcsA to the pole on the bacterial surface is dependent on FtsQ. SepA is not well suited to this analysis because surface-exposed SepA is efficiently cleaved and is thus difficult to detect (3). BrkA, an autotransporter from the distantly related Gram-negative coccobacillary pathogen Bordetella pertussis, is secreted at the pole of B. pertussis and, upon heterologous expression, at the pole of E. coli as well (3). We introduced a plasmid carrying brkA into an S. flexneri derivative that lacks icsA and sepA by virtue of having been cured of the Shigella large plasmid that carries these genes (strain BS103). On the surface of BS103, BrkA was distributed asymmetrically (see Fig. S6b in the supplemental material). Its distribution was less polarized than that of IcsA, likely because IcsA is maintained in tight polar caps on Shigella in part as a result of its regulated cleavage from the surface by specific proteases (3, 27, 28). In BS103 ftsQ1(Ts) cells in which FtsQ had been depleted by growth at 37°C, BrkA was absent from the poles and localized along the length of the cell (see Fig. S6c). In contrast, in the presence of FtsQ, either by virtue of growing the ftsQ1(Ts) strain at 25°C or by complementing cells grown at the restrictive temperature with a wild-type allele of ftsQ, BrkA localized asymmetrically at and near the poles of the cell (see Fig. S6c). On the other hand, FtsQ had no effect on localization to the pole of the nonautotransporter polar proteins CheY and CheZ. These findings indicate that FtsQ is required for localization to the pole of multiple autotransporters from distantly related organisms, suggesting that FtsQ may be generally required for the localization of autotransporters to the pole.
DISCUSSION
Spatial positioning of proteins is critical to multiple bacterial cellular processes. Bacterial proteins critical to virulence, DNA replication, chromosome segregation, cell division, chemotaxis, and other processes are positioned at one or both poles, at midcell, on the chromosome, to an endospore, or to another specific site within the cell. The proper positioning of many of these proteins is a prerequisite for normal function. Enzymatic assays have existed that indicate whether a specific protein is located within the cytoplasm or in an extracytoplasmic compartment; these assays typically use beta-galactosidase or alkaline phosphatase as a reporter and permit the determination of the subcellular localization of a protein on a large scale within the cells of bacterial colonies (29, 30). However, because their ability to distinguish bacterial compartments is based on the redox potential of the compartment, which is constant throughout the compartment, these enzymatic assays provide no information on protein positioning within the compartments.
Despite the importance of spatial positioning within the cytoplasm, to our knowledge, no methods have existed previously for detecting on a large scale the position of specific proteins within the cytoplasm of bacterial cells. The reporter assay that we describe here provides this functionality. We demonstrate the ability not only to discriminate in individual bacterial strains whether the polar protein IcsA is at the pole or is delocalized from the pole but also to perform large-scale selection for second-site mutations that lead to its delocalization and large-scale screening for loci that rescue its polar positioning in a strain background in which it is delocalized. This reporter system is adaptable to proteins other than IcsA that localize to the pole in the cytoplasm, since the polar chemotaxis proteins CheY and CheZ each functioned similarly to IcsA in the assay (Fig. 1c).
Our findings are consistent with a model in which, in any given cell, when CRP is sequestered at the bacterial pole, it cannot access all of the binding sites necessary to enable sugar utilization (see Fig. S3 in the supplemental material). Our results suggest that the high specificity of the reporter system for maltose (or rhamnose) utilization is due to the requirement for CRP to bind to multiple promoter binding sites for utilization of that particular sugar (Fig. 1d). Consequently, we anticipate that the system will be adaptable to proteins that are not polar and have specific nonpolar positions, since distant binding sites will be equally inaccessible in aggregate to an activator that is sequestered anywhere in the cytoplasm. Similarly, it seems likely that transcriptional reporters other than CRP that must bind multiple chromosomal sites to produce a particular phenotype will also work as reporters. Thus, this reporter system is likely to be applicable to the study of a wide range of bacterial proteins. The ability to discern the distribution of a protein within the bacterial cytoplasm in a high-throughput fashion will likely have broad-reaching applications.
One potential limitation of this system is the requirement that the readout (e.g., maltose utilization) depend on the reporter binding to multiple sites on the chromosome; the requirement for CRP in either maltose utilization or rhamnose utilization meets these requirements. A second limitation of the system is that the reporter must retain its activity upon fusion to the targeting protein. Given that multiple transcription activators are functional when fused to bait proteins in two-hybrid systems, this limitation is unlikely to be a major restriction. Finally, a potential limitation is that because the number of copies of the fusion protein in our cells is very low compared to native IcsA (see Fig. S2a in the supplemental material), it can be difficult to visualize the position of the IcsA-CRP reporter fusion protein in individual cells. We were not able to detect it by immunofluorescence using antibodies to IcsA or CRP; we were able to detect it only after fusion to a GFP tag and induction of expression with low concentrations of anhydrotetracycline (Fig. 1b). However, with different reporters or different antibodies, detection of an untagged reporter by immunofluorescence might be possible.
Our initial selection, designed to identify nonessential genes required for polarity of an IcsA-CRP reporter, resulted in the demonstration that elevated levels of RpoS block IcsA polarity. RpoS could theoretically block IcsA polarity by negatively regulating expression of ftsQ; instead, however, RpoS is known to be an inducer of expression from one of two ftsQ promoters in E. coli (31, 32), indicating that RpoS regulation of ftsQ is not the cause of the FtsQ-dependent phenotype that we observed. RpoS regulates entry into stationary phase, yet levels of IcsA in S. flexneri are diminished during stationary phase (23). Moreover, during infection of mammalian cells, Shigella spp. are continuously dividing and using IcsA to spread into adjacent cells, such that bacterial cell density likely never reaches that of stationary phase.
We demonstrate that the cell division protein FtsQ is required for localization of IcsA and other autotransporters to the bacterial pole. In most bacteria, FtsQ is required for cell division, a process that involves assembly of a cytokinetic ring of FtsZ at midcell, followed by constriction of that ring, leading to invagination of the cell envelope and formation of a septum at midcell. FtsQ functions in the middle of the cell division cascade (Fig. 3a), is dependent on FtsK for its recruitment to midcell, and in turn is required for recruitment of FtsB and FtsL (33). The fact that the periplasmic domain of FtsQ is sufficient for its function in IcsA localization suggests that an extracytoplasmic activity of FtsQ contributes to the establishment of bacterial poles. Other than its role in recruitment of FtsB and FtsL, the molecular function of FtsQ in the cell division process is unknown. We speculate, as others have done previously (34, 35), that the periplasmic domain of FtsQ is critical to the remodeling of peptidoglycan that normally occurs in conjunction with cell division and that may contribute to the establishment of autotransporter polarity. The methodology for assaying protein position described here, which enabled the identification of a role for FtsQ in polar localization of autotransporters, has the potential to be a powerful tool for the study of positional information relevant to a wide range of proteins that localize to the bacterial pole.
MATERIALS AND METHODS
Bacterial strains, plasmids, and libraries.
Strains and plasmids used in this study are listed in Table S4 in the supplemental material. Unless otherwise indicated, E. coli strains were grown in LB at 37°C, and S. flexneri strains were grown in TCS (trypto casein soy broth) at 37°C. ftsQ1(Ts) strains were maintained at 30°C. Maltose MacConkey indicator plates were MacConkey agar base (Difco) supplemented with 0.2% maltose. Maltose minimal medium was M63 medium supplemented with 0.2% maltose. Where appropriate, antibiotics were added at the indicated concentrations: kanamycin, 50 µg/ml; ampicillin 100 µg/ml; chloramphenicol, 25 µg/ml; tetracycline, 12.5 µg/ml; spectinomycin, 100 µg/ml.
Genetic manipulations.
pBAD-IcsA507-620-GFP, pBAD-IcsA1-104-GFP, and pBAD-IcsAΔ507-729-GFP, which encode translational fusions of IcsA polypeptides to GFP or mCherry under the control of the arabinose promoter, as well as pBR322-icsA, which carries icsA under the control of the native promoter, have been described elsewhere (9, 36). pBAD-IcsA507-620-mCherry is pBAD33 carrying a translational fusion of the coding sequence for icsA507-620 to mCherry. To generate p-icsA53-757::crp, sequence encoding IcsA amino acids 53 to 757 with an initiating methionine, fused in frame via a 3-alanine linker (GCG GCC GCA) to amino acid 2 of CRP, was cloned into a derivative of pZE21-MCS-1 (37). The C-terminal CRP fusion to IcsAΔ507-729, which lacks polar targeting sequences and is localized diffusely (9), was constructed in the same manner. Insertion of the GFP tag at the C terminus of IcsA53-757-CRP and IcsAΔ507-729-CRP was by overlap extension PCR using as template for the GFP tag a derivative of gfp in which TTG replaces the ATG (lab stock). Expression of each icsA::crp fusion is under the control of a leaky, tetracycline-inducible promoter and was performed without the addition of exogenous inducer, with the exception of experiments involving microscopic analysis of the position of IcsA53-757-CRP-GFP and IcsAΔ507-729-CRP-GFP, in which case cells were harvested following induction with 50 ng/ml of anhydrotetracycline for 50 min at 37°C.
pAJ61 was created from p-icsA53-757::crp by swapping the origin of replication of p-icsA53-757::crp with that of pZA31-luc (37). pVS1, carrying cheY, and pVS53, carrying cheZ, were the gift of H. Berg (14). p-cheY::crp was created by amplifying the coding sequence of cheY as a BamHI-NotI fragment from pVS1 by PCR. p-cheZ::crp was created by amplifying the coding sequence of cheZ as a BamHI-NotI fragment from pVS53 by PCR. The resulting products were digested with BamHI and NotI and ligated into the BamHI and NotI sites of p-icsA53-757::crp. p-crp was created by amplifying the coding sequence of crp as a BamHI-MluI fragment from the chromosome by PCR. The resulting product was digested with BamHI and MluI and ligated into the BamHI and MluI sites of p-icsA53-757::crp.
A multicopy library of the E. coli MC4100 chromosome was constructed by partially digesting MC4100 chromosomal DNA with Sau3AI, isolating DNA fragments between 3 and 10 kb by agarose gel electrophoresis, and ligating the size-selected DNA into the medium-copy-number vector pACYC184. Within this library, 80% of clones contained inserts larger than 3 kb. The library was transformed into E. coli crp rssB (p-icsA53-757::crp) cells, and transformants were plated on maltose MacConkey agar. A total of 7,500 colonies were screened, 8% of which were white. Plasmid DNA was isolated from 73 randomly selected white colonies, and the 5′ and 3′ ends of the inserts were sequenced to identify the genetic content of the plasmid insert.
To create p-ddlB, the coding sequence of ddlB along with 762 bp upstream of its translation start codon was amplified from p-ddlB-ftsQ as an XbaI-SphI fragment by PCR. The resulting product was digested with XbaI and SphI and ligated into the XbaI and SphI sites of pAYC184. To create p-ftsQ, the coding sequence of ftsQ along with 575 bp upstream of its start codon was amplified from p-ddlB-ftsQ as a HindIII and SphI fragment by PCR. The resulting product was digested with HindIII and SphI and ligated into the HindIII and SphI sites of pAYC184.
Sequence analysis was performed to verify that each construct was correct. Transduction was performed with P1L4 phage by standard procedures. The sequences of primers used in PCR and sequencing are available from the authors upon request.
The ftsQ1(Ts) allele was introduced into S. flexneri wild-type strain 2457T and virulence plasmid-cured strain BS103 by P1 transduction of the allele, which is linked to leu::Tn10, from E. coli strain MDG149. The presence of the ftsQ1(Ts) allele was verified by testing transductants for temperature-sensitive growth and filamentation at elevated temperatures.
Transposon mutagenesis.
A transposon library was generated in E. coli crp (p-icsA53-757::crp) cells using a mini-Tn10 (Cmr) delivered by lambda NK1324, as described elsewhere (38). The selection of transposon insertions that result in utilization of maltose was performed by growth on maltose minimal medium. Approximately 13,000 individual transposon insertion mutants were screened, as estimated by plating in parallel on maltose MacConkey agar. The ability of colonies that grew on maltose minimal medium to utilize maltose was verified by red colony color upon replica plating on maltose MacConkey agar.
Localization of IcsA, FtsQ, and CRP fusions.
The localization of IcsA507-620-GFP or -mCherry was determined by induction of protein synthesis from an arabinose-inducible promoter following growth either to mid-exponential phase or overnight to stationary phase at 37°C. For determination of IcsA507-620-GFP or -mCherry localization in mid-exponential-phase CRP+ cells, l-arabinose was added to 0.2% and growth was continued for an additional 30 min at 30°C. Because CRP is transcriptionally active at arabinose promoters, cells carrying a deletion of crp displayed reduced expression from arabinose-inducible promoters; therefore, induction of arabinose-inducible IcsA fusions from these promoters was for 90 to 120 min at 37°C, which resulted in levels of IcsA507-620-GFP comparable to those in CRP+ cells. To determine IcsA localization in stationary phase, expression of IcsA507-620-GFP or -mCherry was induced by the addition of 0.2% l-arabinose to overnight cultures for 60 to 90 min at 30°C. For stationary-phase colocalization of GFP-FtsQ or expression of FFQ or QQQ, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was included in overnight cultures. SepA1-24/57-1042-GFP expression was induced by the addition of l-arabinose to 0.2% for 40 min at 30°C in stationary-phase MG1655. Depletion of functional FtsQ in ftsQ1 strains was accomplished by shifting to growth at 37°C or 42°C for 60 to 150 min. Depletion of FtsB or FtsL in DJS86 and DJS87 was accomplished by growth in the absence of arabinose for 60 to 180 min. Filamentation with aztreonam was performed by adding it to 1 µg/ml for 50 to 60 min. Fixation and detection of IcsA on the surface of bacteria were performed as described previously (4). Alternatively, cells were labeled with primary and secondary antibodies prior to fixation.
Western blot analysis.
Whole-cell proteins were prepared from bacterial cultures immediately before microscopy. Protein loading was normalized to cell density, and all sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels consisted of 5% stacking and 10% separating layers. Western blot analysis was carried out using standard protocols, using antibodies to IcsA (4) at a 1:10,000 dilution, isocitrate dehydrogenase (ICDH; gift from A. L. Sonenshein) at a 1:10,000 dilution, RpoS (Neoclone) at a 1:5,000 dilution, or GFP (Molecular Probes or Roche) at a 1:1,000 dilution. RpoS levels were determined by band densitometry using IP Laboratory (Scanalytics) software.
Microscopy.
For microscopic observation, cells were mounted onto 1% agarose pads or glass coverslips. Microscopy was performed by using a 100× oil-immersion objective lens on a Nikon TE300 microscope with Nikon or Chroma Technology filters. Images were captured digitally using a black-and-white CoolSnap HQ charge-coupled device camera and IP Laboratory (Scanalytics) software.
Statistical analysis.
Tabulation of protein localization was conducted in a blinded fashion. For each set of experiments, 30 or more cells were analyzed in each of three independent experiments. For GFP fusions, if a focus was present at a cell pole, the cell was scored as polar, and if there were no foci, it was scored as diffuse. We never observed cells with foci that were only at sites other than a pole. The statistical significance of differences between experimental results was determined using a Student t test.
SUPPLEMENTAL MATERIAL
Schematic of the Shigella autotransporter IcsA. (a) Domain organization, which is common among autotransporter proteins: N-terminal signal peptide (SP, blue bar); large extracellular, functional (α) domain (green bar), which is exposed on the bacterial surface and/or proteolytically released into the extracellular space; and C-terminal beta (β) domain, which forms a beta barrel in the outer membrane (OM) (black bar). IcsAFL, full-length IcsA. (b, c, and d) Regions of IcsA in constructs used in this study and their ability to localize a GFP fusion to the pole (9). Numbers indicate amino acid residues of IcsA. Download Figure S1, TIF file, 1.3 MB.
Levels of IcsA-CRP, IcsA-GFP, and RpoS in strains used in this study. (a) Production of IcsA53-757-CRP in E. coli is substantially lower than native levels of IcsA in S. flexneri. Western blot analysis of whole-cell proteins prepared from the indicated strains grown to exponential phase using IcsA antiserum specific to IcsA residues 53 to 757. WT S. flexneri expressed icsA from its native promoter. Load, relative amount of protein preparation loaded into each lane. IcsAFL, full-length mature IcsA, which consists of residues 53 to 1102. *, nonspecific band. (b) Levels of IcsA53-757-CRP-GFP and IcsAΔ507-729-CRP-GFP in E. coli crp. Western blot analysis of whole-cell proteins prepared from the indicated strains, grown to exponential phase and induced with 50 ng/ml anhydrotetracycline for 50 min, using antibody to GFP. (c) The RpoS (σS) degradation pathway in exponential phase and stationary phase. (d, e, and f) The levels of IcsA507-620-GFP or GFP (d) and RpoS (d, e, and f) in the indicated E. coli strains, as determined by Western blot analysis using RpoS antibody. In panel d, cells were harvested in stationary phase. In panels e and f, cells were harvested in exponential phase, except for the first lane of panel f, in which the cells were harvested in stationary phase. The cytoplasmic protein isocitrate dehydrogenase (ICDH) was used as a loading control. Molecular masses are indicated in kDa. Download Figure S2, TIF file, 1.6 MB.
Possible model for why different sugars lead to distinct CRP-dependent sugar utilization phenotypes. Shown are cells that contain a deletion of the chromosomal gene encoding the transcription activator CRP and that carry a plasmid-borne translational fusion of IcsA53-757 (light green oval) to CRP (dark blue arch). (a) CRP-dependent activation of three operons (purple boxes) is required for utilization of the sugar on which these cells are grown. Consequently, the colony formed by these cells is white. (b) CRP-dependent activation of only a single operon (light blue box) is required for utilization of the sugar on which these cells are grown. Within a subpopulation of cells in the colony, the chromosome will be positioned such that the cognate CRP binding site will be sufficiently close to the bacterial pole to permit binding by CRP and transcription activation, leading to sugar utilization. Consequently, the colony formed by these cells is red. Download Figure S3, TIF file, 0.3 MB.
IcsA in S. flexneri rssB and rpoS. (a) Native IcsA on the surface of intact exponential-phase or stationary-phase wild-type S. flexneri strain 2457T or its ΔrssB or rpoS::Tn10 derivatives. Immunofluorescence (IF) with IcsA antibody (left panels) and phase (right panels). Bars, 5 µm. (b) Levels of IcsA and RpoS in the same strains under the same growth conditions as shown in panel a. Pellets contain full-length mature IcsA (IcsA); supernatants (supnt) contain truncated IcsA (IcsA′) that has been proteolytically cleaved and released at the bacterial surface. Western blot analyses using antibody to IcsA or RpoS. Loading was normalized to optical density at 600 nm (OD600). All lanes are from the same blots. Of note, more protein was loaded than that loaded in Fig. S2e and S2f, and the S. flexneri rssB rpoS double mutant displayed considerable cell lysis, as observed by microscopy, and therefore was omitted from these analyses. Download Figure S4, TIF file, 2.6 MB.
IcsA localization to the pole appears to be independent of other cell division proteins. (a) Localization of IcsA507-620-mCherry in derivatives of E. coli MC4100 containing chromosomally integrated and IPTG-inducible gfp (vector) or translational gfp fusions to the indicated cell division proteins, grown to stationary phase in the presence of IPTG. Right panels, IcsA507-620-mCherry; left panels, phase. Synthesis of the indicated cell division proteins was verified by Western blot analysis. (b) Distribution of native IcsA on the surface of intact cells of S. flexneri 2457T and its derivatives by immunofluorescence. Aztreonam was used to filament WT cells (WT + aztr); addition of aztreonam per se had no effect on localization of IcsA. Bars, 10 µm. depl, depleted; IF, immunofluorescence. Download Figure S5, TIF file, 2.5 MB.
FtsQ is required for polar localization of autotransporters other than IcsA. (a) Localization of GFP-tagged cytoplasmic derivative of Shigella autotransporter SepA. Stationary-phase E. coli MG1655 carrying SepA1-24/57-1042-GFP, which lacks the SepA Sec secretion signal, and carrying vector or p-ftsQ. (b) Distribution of B. pertussis autotransporter BrkA on the surface of intact Shigella cells. BS103, an S. flexneri derivative cured of the virulence plasmid, carrying pDO6935 (p-brkA), which carries brkA, or no vector. (c) Distribution of BrkA on the surface of intact cells of S. flexneri BS103 ftsQ1(Ts) derivatives. Growth was at 37°C, the restrictive temperature for ftsQ1(Ts). Aztreonam was used to filament BS103 WT and complemented ftsQ1(Ts) cells; addition of aztreonam per se had no effect on localization of BrkA. Arrows, asymmetrically distributed BrkA; arrowheads, nonpolar BrkA. Bars, 10 µm. Download Figure S6, TIF file, 1 MB.
Chromosome position of CRP binding sites in operons required for utilization of various sugars and of oriC in MG1655.
Distribution of GFP-tagged cytoplasmic derivative of IcsA (IcsA507-620-GFP) in E. coli.
Distribution of native IcsA on the surface of S. flexneri.
Strains and plasmids used in this study.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R01 AI035817 from the National Institutes of Health (to M.B.G.) and American Heart Association fellowship 0425855T (to A.J.).
We thank C.-R. Yi and L. Stamm for careful reading of the manuscript; J. Kahane, D. Raj, and W. Lane for technical assistance; and A. L. Sonenshein, J. Beckwith, and R. Fernandez for reagents.
Footnotes
Citation Fixen KR, et al. 2012. Genetic reporter system for positioning of proteins at the bacterial pole. mBio 3(2):e00251-11. doi:10.1128/mBio.00251-11.
REFERENCES
- 1. Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- 2. Nilsen T, Yan AW, Gale G, Goldberg MB. 2005. Presence of multiple sites containing polar material in spherical Escherichia coli cells that lack MreB. J. Bacteriol. 187:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jain S, et al. 2006. Polar localization of the autotransporter family of large bacterial virulence proteins. J. Bacteriol. 188:4841–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldberg MB, Bârzu O, Parsot C, Sansonetti PJ. 1993. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J. Bacteriol. 175:2189–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldberg MB. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandon LD, et al. 2003. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol. Microbiol. 50:45–60 [DOI] [PubMed] [Google Scholar]
- 8. Jain S, Goldberg MB. 2007. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 189:5393–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charles M, Pérez M, Kobil JH, Goldberg MB. 2001. Polar targeting of Shigella virulence factor IcsA in Enterobacteriaceae and Vibrio. Proc. Natl. Acad. Sci. U. S. A. 98:9871–9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sandlin RC, Maurelli AT. 1999. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect. Immun. 67:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapon C. 1982. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J. Bacteriol. 150:722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janakiraman A, Goldberg MB. 2004. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl. Acad. Sci. U. S. A. 101:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nilsen T, Ghosh AS, Goldberg MB, Young KD. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sourjik V, Berg HC. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740–751 [DOI] [PubMed] [Google Scholar]
- 15. Wickstrum JR, Santangelo TJ, Egan SM. 2005. Cyclic AMP receptor protein and RhaR synergistically activate transcription from the L-rhamnose-responsive rhaSR promoter in Escherichia coli. J. Bacteriol. 187:6708–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lopilato JE, Garwin JL, Emr SD, Silhavy TJ, Beckwith JR. 1984. D-ribose metabolism in Escherichia coli K-12: genetics, regulation, and transport. J. Bacteriol. 158:665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spassky A, Busby S, Buc H. 1984. On the action of the cyclic AMP–cyclic AMP receptor protein complex at the Escherichia coli lactose and galactose promoter regions. EMBO J. 3:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song S, Park C. 1997. Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 179:7025–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. 1996. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333–1339 [PMC free article] [PubMed] [Google Scholar]
- 21. Pratt LA, Silhavy TJ. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U. S. A. 93:2488–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Damerau K, St John AC. 1993. Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J. Bacteriol. 175:53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldberg MB, Theriot JA, Sansonetti PJ. 1994. Regulation of surface presentation of IcsA, a Shigella protein essential to intracellular movement and spread, is growth phase dependent. Infect. Immun. 62:5664–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen JC, Minev M, Beckwith J. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 184:695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buddelmeijer N, Beckwith J. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315–1327 [DOI] [PubMed] [Google Scholar]
- 26. Chen JC, Weiss DS, Ghigo JM, Beckwith J. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181:521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shere KD, Sallustio S, Manessis A, D’Aversa TG, Goldberg MB. 1997. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol. Microbiol. 25:451–462 [DOI] [PubMed] [Google Scholar]
- 28. Egile C, d’Hauteville H, Parsot C, Sansonetti PJ. 1997. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol. Microbiol. 23:1063–1073 [DOI] [PubMed] [Google Scholar]
- 29. Broome-Smith JK, Spratt BG. 1986. A vector for the construction of translational fusions to TEM beta-lactamase and the analysis of protein export signals and membrane protein topology. Gene 49:341–349 [DOI] [PubMed] [Google Scholar]
- 30. Manoil C, Beckwith J. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403–1408 [DOI] [PubMed] [Google Scholar]
- 31. Sitnikov DM, Schineller JB, Baldwin TO. 1996. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. U. S. A. 93:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ballesteros M, Kusano S, Ishihama A, Vicente M. 1998. The ftsQ1p gearbox promoter of Escherichia coli is a major sigma S-dependent promoter in the ddlB-ftsA region. Mol. Microbiol. 30:419–430 [DOI] [PubMed] [Google Scholar]
- 33. Buddelmeijer N, Beckwith J. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553–557 [DOI] [PubMed] [Google Scholar]
- 34. Mengin-Lecreulx D, van Heijenoort J, Park JT. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate: L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uehara T, Park JT. 2002. Role of the murein precursor UDP-N-acetylmuramyl-L-Ala-gamma-D-Glu-meso-diaminopimelic acid-D-Ala-D-Ala in repression of beta-lactamase induction in cell division mutants. J. Bacteriol. 184:4233–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Magdalena J, Goldberg MB. 2002. Quantification of Shigella IcsA required for bacterial actin polymerization. Cell Motil. Cytoskeleton 51:187–196 [DOI] [PubMed] [Google Scholar]
- 37. Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kleckner N, Bender J, Gottesman S. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139–180 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of the Shigella autotransporter IcsA. (a) Domain organization, which is common among autotransporter proteins: N-terminal signal peptide (SP, blue bar); large extracellular, functional (α) domain (green bar), which is exposed on the bacterial surface and/or proteolytically released into the extracellular space; and C-terminal beta (β) domain, which forms a beta barrel in the outer membrane (OM) (black bar). IcsAFL, full-length IcsA. (b, c, and d) Regions of IcsA in constructs used in this study and their ability to localize a GFP fusion to the pole (9). Numbers indicate amino acid residues of IcsA. Download Figure S1, TIF file, 1.3 MB.
Levels of IcsA-CRP, IcsA-GFP, and RpoS in strains used in this study. (a) Production of IcsA53-757-CRP in E. coli is substantially lower than native levels of IcsA in S. flexneri. Western blot analysis of whole-cell proteins prepared from the indicated strains grown to exponential phase using IcsA antiserum specific to IcsA residues 53 to 757. WT S. flexneri expressed icsA from its native promoter. Load, relative amount of protein preparation loaded into each lane. IcsAFL, full-length mature IcsA, which consists of residues 53 to 1102. *, nonspecific band. (b) Levels of IcsA53-757-CRP-GFP and IcsAΔ507-729-CRP-GFP in E. coli crp. Western blot analysis of whole-cell proteins prepared from the indicated strains, grown to exponential phase and induced with 50 ng/ml anhydrotetracycline for 50 min, using antibody to GFP. (c) The RpoS (σS) degradation pathway in exponential phase and stationary phase. (d, e, and f) The levels of IcsA507-620-GFP or GFP (d) and RpoS (d, e, and f) in the indicated E. coli strains, as determined by Western blot analysis using RpoS antibody. In panel d, cells were harvested in stationary phase. In panels e and f, cells were harvested in exponential phase, except for the first lane of panel f, in which the cells were harvested in stationary phase. The cytoplasmic protein isocitrate dehydrogenase (ICDH) was used as a loading control. Molecular masses are indicated in kDa. Download Figure S2, TIF file, 1.6 MB.
Possible model for why different sugars lead to distinct CRP-dependent sugar utilization phenotypes. Shown are cells that contain a deletion of the chromosomal gene encoding the transcription activator CRP and that carry a plasmid-borne translational fusion of IcsA53-757 (light green oval) to CRP (dark blue arch). (a) CRP-dependent activation of three operons (purple boxes) is required for utilization of the sugar on which these cells are grown. Consequently, the colony formed by these cells is white. (b) CRP-dependent activation of only a single operon (light blue box) is required for utilization of the sugar on which these cells are grown. Within a subpopulation of cells in the colony, the chromosome will be positioned such that the cognate CRP binding site will be sufficiently close to the bacterial pole to permit binding by CRP and transcription activation, leading to sugar utilization. Consequently, the colony formed by these cells is red. Download Figure S3, TIF file, 0.3 MB.
IcsA in S. flexneri rssB and rpoS. (a) Native IcsA on the surface of intact exponential-phase or stationary-phase wild-type S. flexneri strain 2457T or its ΔrssB or rpoS::Tn10 derivatives. Immunofluorescence (IF) with IcsA antibody (left panels) and phase (right panels). Bars, 5 µm. (b) Levels of IcsA and RpoS in the same strains under the same growth conditions as shown in panel a. Pellets contain full-length mature IcsA (IcsA); supernatants (supnt) contain truncated IcsA (IcsA′) that has been proteolytically cleaved and released at the bacterial surface. Western blot analyses using antibody to IcsA or RpoS. Loading was normalized to optical density at 600 nm (OD600). All lanes are from the same blots. Of note, more protein was loaded than that loaded in Fig. S2e and S2f, and the S. flexneri rssB rpoS double mutant displayed considerable cell lysis, as observed by microscopy, and therefore was omitted from these analyses. Download Figure S4, TIF file, 2.6 MB.
IcsA localization to the pole appears to be independent of other cell division proteins. (a) Localization of IcsA507-620-mCherry in derivatives of E. coli MC4100 containing chromosomally integrated and IPTG-inducible gfp (vector) or translational gfp fusions to the indicated cell division proteins, grown to stationary phase in the presence of IPTG. Right panels, IcsA507-620-mCherry; left panels, phase. Synthesis of the indicated cell division proteins was verified by Western blot analysis. (b) Distribution of native IcsA on the surface of intact cells of S. flexneri 2457T and its derivatives by immunofluorescence. Aztreonam was used to filament WT cells (WT + aztr); addition of aztreonam per se had no effect on localization of IcsA. Bars, 10 µm. depl, depleted; IF, immunofluorescence. Download Figure S5, TIF file, 2.5 MB.
FtsQ is required for polar localization of autotransporters other than IcsA. (a) Localization of GFP-tagged cytoplasmic derivative of Shigella autotransporter SepA. Stationary-phase E. coli MG1655 carrying SepA1-24/57-1042-GFP, which lacks the SepA Sec secretion signal, and carrying vector or p-ftsQ. (b) Distribution of B. pertussis autotransporter BrkA on the surface of intact Shigella cells. BS103, an S. flexneri derivative cured of the virulence plasmid, carrying pDO6935 (p-brkA), which carries brkA, or no vector. (c) Distribution of BrkA on the surface of intact cells of S. flexneri BS103 ftsQ1(Ts) derivatives. Growth was at 37°C, the restrictive temperature for ftsQ1(Ts). Aztreonam was used to filament BS103 WT and complemented ftsQ1(Ts) cells; addition of aztreonam per se had no effect on localization of BrkA. Arrows, asymmetrically distributed BrkA; arrowheads, nonpolar BrkA. Bars, 10 µm. Download Figure S6, TIF file, 1 MB.
Chromosome position of CRP binding sites in operons required for utilization of various sugars and of oriC in MG1655.
Distribution of GFP-tagged cytoplasmic derivative of IcsA (IcsA507-620-GFP) in E. coli.
Distribution of native IcsA on the surface of S. flexneri.
Strains and plasmids used in this study.