ABSTRACT
Pseudomonas aeruginosa is a common cause of infection in the lungs of patients with cystic fibrosis (CF). In addition, biofilm formation and antibiotic resistance of Pseudomonas are major problems that can complicate antibiotic therapy. We evaluated the efficacy of using bacteriophages to kill the pathogen in both biofilms and in the murine lung. We isolated and characterized two phages from a local wastewater treatment plant, a myovirus (ϕNH-4) and a podovirus (ϕMR299-2). Both phages were active against clinical isolates of P. aeruginosa. Together, the two phages killed all 9 clinical isolate strains tested, including both mucoid and nonmucoid strains. An equal mixture of the two phages was effective in killing P. aeruginosa NH57388A (mucoid) and P. aeruginosa MR299 (nonmucoid) strains when growing as a biofilm on a cystic fibrosis bronchial epithelial CFBE41o- cell line. Phage titers increased almost 100-fold over a 24-h period, confirming replication of the phage. Furthermore, the phage mix was also effective in killing the pathogen in murine lungs containing 1 × 107 to 2 × 107 P. aeruginosa. Pseudomonas was effectively cleared (reduced by a magnitude of at least 3 to 4 log units) from murine lungs in 6 h. Our study demonstrates the efficacy of these two phages in killing clinical Pseudomonas isolates in the murine lung or as a biofilm on a pulmonary cell line and supports the growing interest in using phage therapy for the control and treatment of multidrug-resistant Pseudomonas lung infections in CF patients.
IMPORTANCE
Given the rise in antibiotic resistance, nonantibiotic therapies are required for the treatment of infection. This is particularly true for the treatment of Pseudomonas infection in patients with cystic fibrosis. We have identified two bacterial viruses (bacteriophages) that can kill Pseudomonas growing on human lung cells and in an animal model of lung infection. The use of bacteriophages is particularly appropriate because the killing agent can replicate on the target cell, generating fresh copies of the bacteriophage. Thus, in the presence of a target, the killing agent multiplies. By using two bacteriophages we can reduce the risk of resistant colonies developing at the site of infection. Bacteriophage therapy is an exciting field, and this study represents an important demonstration of efficacy in validated infection models.
Introduction
Cystic fibrosis (CF) is an inherited genetic disorder that chronically affects the lungs and digestive system (pancreas and intestine) of children and adults worldwide. CF also affects the mucus and glands of the liver, sinuses, and sex organs causing progressive disability due to multiorgan system failure. CF results from mutations in the transmembrane conductance regulator gene (1–3). The defective enzyme leads to the production of unusually thick and sticky mucus and high levels of chloride containing secretions into ducts and body cavities. These secretions clog the lungs, leading to life-threatening infections (4, 5).
The lungs of CF patients are often colonized at infancy or in early childhood with Pseudomonas aeruginosa that may damage the epithelial surface, resulting in altered airway physiology and impairment of mucocillary clearance. This chronic infection is one of the main causes of lung function decline and mortality in CF patients (6, 7). Indeed, 80 to 95% of patients with CF succumb to respiratory failure brought about by chronic bacterial infection and concomitant airway inflammation (7). P. aeruginosa is particularly persistent in the lungs due to its aerobic nature and its ability to form biofilms in the lungs of CF patients. Another significant factor is the inherent resistance of P. aeruginosa to many antibiotics due to membrane impermeability (8–12). Other acquired mechanisms of resistance include production of β-lactamases and carbapenemases (13) and multidrug efflux pumps (14). It has also been noted that the most prevalent and severe chronic lung infections in CF patients are caused by mucoid P. aeruginosa strains (4, 11, 15).
Reports have shown that organisms in biofilms are able to tolerate 10- to 1,000-fold-higher levels of antibiotics than planktonic bacteria (16, 17), and this can mean that the antibiotic concentration needed to eradicate a biofilm is higher than the peak serum concentration (18), rendering it ineffective. The continued emergence and reemergence of biofilm-forming Pseudomonas resistant to one or more antibiotics pose a continuous challenge in the treatment of lung infections in CF patients. As a result, there is a need for alternative, nonantibiotic approaches such as phage therapy (19, 20). Although phage therapy has been practiced in Eastern European countries for decades, it has been neglected by the Western world for many years. However, there is now a growing interest in the use of phage therapy for the control and treatment of multidrug-resistant bacterial infections in general, and for Pseudomonas lung infections in CF patients in particular. Studies on phage efficacy in clearing of biofilms formed on abiotic surfaces (catheters and microtiter plates) by P. aeruginosa confirmed that phage can reduce the bacterial load in these biofilms by 50 to 99% (21, 22). A recent in vitro study showed that Pseudomonas phage PT-6 was able to reduce the viscosity of alginate polymers extracted from P. aeruginosa by almost 65%, a mechanism used by phages to attack the exopolysaccharide matrix of mucoid Pseudomonas to gain access to the host cell (23). Recent reports show the promising role phage therapy could play in the treatment of acute lung infections in an in vivo murine lung model with P. aeruginosa (24, 25) and Burkholderia cenocepacia (26). The effectiveness of phage therapy in rescuing larvae (an invertebrate infection model) infected with Burkholderia cepacia complex from death was reported by Seed and Dennis (27).
Currently, there is little information concerning the effect of bacteriophage on biofilms growing on a lung tissue model. In this report, we describe how a mixture of two newly isolated phages can kill and clear lux-tagged Pseudomonas from the lungs of infected mice and in biofilms growing on the surface of a cystic fibrosis bronchial epithelial (CFBE41o-) monolayer. In this study, we used bioluminescence imaging—a powerful tool for studying bacterial infections in small-animal models, since it allows accurate real-time in vivo temporal and spatial tracking of tagged bacteria in living animals (25, 28). Monitoring the light emitted by tagged Pseudomonas cells in vivo was a valuable tool in verifying the effectiveness of the phage mix to kill the pathogen. Pseudomonas cell numbers were reduced by a magnitude of 3 to 4 log units when the phage mix was tested in both in vivo and in vitro systems.
RESULTS
Isolation of phage from a sewage treatment plant.
The overall aim of this study was to assess the potential of bacteriophage therapy to treat Pseudomonas lung infections. We initially isolated two phages, ϕNH-4 and ϕMR299-2, from sewage obtained from a water treatment plant. Both phages were demonstrated to be virulent to P. aeruginosa. Scanning electron microscopy revealed that phage ϕMR299-2 virions have isometric capsids of 40 to 60 nm in diameter and very short tails measuring 10 to 20 nm (Fig. 1A). Morphologically, phage ϕMR299-2 shows similarity to Pseudomonas ϕPap3 (29), a podovirus that has an isometric capsid and short tail. We have assigned ϕMR299-2 to type species coliphage T7 and to the family Podoviridae (Report of the International Committee on Taxonomy of Viruses [30, 31]). Phage ϕNH-4 possessed an isometric capsid of 50 to 60 nm in diameter and a contractile nonflexible tail with cross striations (noncontracted tail length of 150 nm and contracted tail length of 85 nm; tail diameter of 20 nm when contracted). In addition, a putative DNA injecting structure of 70 nm in length (narrower than the contracted tail sheath) and tail fibers were observed (Fig. 1B). Morphologically, ϕNH-4 shows similarity to ϕPB1, ϕLBL3, and ϕSN that are classified into the T4 morphological group of the Myoviridae (32). On the basis of structural characteristics obtained from the microscopic analysis, we assign ϕNH-4 as a member of the Myoviridae family according to the International Committee on Taxonomy of Viruses (30, 31).
FIG 1 .
Scanning electron microscopy images of phage ϕ229-2, a podophage (A) and phage ϕNH-4, a myophage (B), stained with 0.2% phosphotungstic acid. The arrows point to the short tail (10 to 20 nm long) of the podophage in panel A and to the contracted tail sheath of the myovirus in panel B.
An equal ratio of ϕNH-4 and ϕMR299-2 was used in all experiments to assess their ability to kill Pseudomonas. The host range of ϕNH-4 and ϕMR299-2 was determined by exposing different CF Pseudomonas isolates to each individual phage using a plaque assay. Of the ten Pseudomonas isolates tested, eight were sensitive to both phages, while all were sensitive to at least one phage (Table 1). When the ϕ299-2 and ϕNH-4 phages were added to Pseudomonas in LB broth either separately or in combination, the individual phage resulted in reduction of 3 to 4 log units while the combination of the two resulted in a reduction of about 4.5 log units (see Table S1 in the supplemental material).
TABLE 1 .
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Sourceb or reference |
Sensitivityc of strain to the following phage: |
|
|---|---|---|---|---|
| ϕNH-4 | ϕMR299-2 | |||
| Pseudomonas aeruginosa strains | ||||
| MR299 | Human CF sputum isolate | CUH | +++ | +++ |
| MR299::p16Slux | lux-tagged MR299 | This study | +++ | +++ |
| NH57388A | Stable mucoid CF mouse sputum isolate | 20 | +++ | ++ |
| NH57388A::p16Slux | lux-tagged NH57388A | This study | +++ | ++ |
| MR300 | Human CF sputum isolate | CUH | ++ | − |
| MR325 | Human CF sputum isolate | CUH | − | + |
| MR326 | Human CF sputum isolate | CUH | +++ | ++ |
| MR327 | Human CF sputum isolate | CUH | ++ | ++ |
| MR330 | Human CF sputum isolate | CUH | +++ | ++ |
| MR331 | Human CF sputum isolate | CUH | ++ | + |
| CH001 | Human CF sputum isolate | AH | +++ | +++ |
| POA1 | UCC culture collection | UCC | +++ | ++ |
| Plasmid p16Slux | lux-tagged plasmid vector | 19 | ||
UCC, University College Cork.
CUH, Cork University Hospital (Cork, Ireland); AH, Alimentary Health Ltd. (Cork, Ireland).
Symbols: +++, very strong lysis; ++, strong lysis; +, moderate lysis; −, no lysis.
Phage genome overview.
Before phage can be used in human therapy, it is important to assess any potential risk associated with the phage genomes such as the presence of genetic determinants for toxins or other virulence factors or for the capability to integrate into the host genome. The complete sequences of phages ϕ299-2 and ϕNH-4 were determined using 454 pyrosequencing. Phage ϕ299-2 consists of a double-stranded DNA (dsDNA) molecule of 44,789 bp with a GC content of 52%, significantly lower than the 66.6% of P. aeruginosa. Both the genome size and GC content of this phage are similar to the closely related Pseudomonas phage ϕPaP3 (29). A total of 68 open reading frames (ORFs) are predicted, 21 of which have a leftward orientation and 47 of which are transcribed to the right (Fig. 2). Three tRNA genes (tRNAAsn, tRNAAsp, and tRNAPro) were also identified in phage ϕ299-2, clustered at the 5’ end of the genome. While ϕ299-2 is a lytic phage, comparative genomic analysis revealed that its closest homolog is the temperate Pseudomonas phage, ϕPaP3 (29), isolated from hospital sewage. Interestingly, a recent study calls into question the temperate nature of ϕPaP3, since no site-specific recombinase is encoded up- or downstream of the ϕPaP3 attP site, and immunity or reactivation of the integrated ϕPaP3 DNA was not demonstrated (33). Of the 68 predicted protein products of ϕ299-2, 56 display the highest amino acid identity to proteins from ϕPaP3 (see Table S3 in the supplemental material). Among these are proteins involved in particle formation (ORF03, ORF04, ORF06, and ORF07), genome replication (ORF29, ORF33/40, encoding DNA polymerase I subunits and ORF41, a putative primase/helicase), a putative lysozyme-like endolysin (ORF02) and 47 proteins of hypothetical function. A conserved genomic organization is also evident on comparing ϕ299-2 to ϕPaP3. The remaining 12 ORFs show significant identity to proteins from the lytic Pseudomonas phage LUZ24 (33). According to Ceyssens et al., ϕPaP3 and ϕLUZ24 represent a new genus within the Podoviridae family (33). Considering the close relationship between ϕ299-2 and ϕPaP3 and ϕLUZ24, it is likely that ϕ299-2 also belongs to this genus.
FIG 2 .
Genome organization of phage ϕMR299-2 (top) and ϕNH-4 (bottom). The predicted open reading frames are indicated by the thick arrows, which are shaded to show the level of protein identity to the corresponding regions of the closest P. aeruginosa (phage PaP3 for ϕMR299-2) or phage LMA2 (for ϕNH-4).
Phage ϕNH-4 is a member of the widespread and conserved PB1-like viruses, with a genome of 66,116 bp and a GC content of 55.5%, also significantly lower than that of its host. A total of 94 ORFs were identified in the sequence; the predicted protein products of 56 of these ORFs have the highest amino acid identity to ϕLMA2 (ST4), isolated from a river in Maastricht, Holland, in 2007 (32). Other ϕNH-4 proteins show significant identity to predicted proteins from PB1-like viruses such as ϕ14-1, ϕSN, ϕLBL3, ϕJG024, and ϕPB1 itself (32). As is the case with other PB1-like viruses, the genes in ϕNH-4 are arranged in a compact manner and appear to be organized into at least 7 transcriptional blocks, alternating on both strands (Fig. 2). Based primarily on sequence similarity to ORFs in the genomes of phages LMA2 and JG024, genomic regions encoding phage particle formation (ORF19 to -47) and phage DNA replication (ORF55 to -70) could be identified. ORF48 encodes a putative endolysin with 100% identity to the endolysin of phage ϕLMA2 and belongs to a lysozyme-like superfamily.
Biofilm growth by lux-tagged Pseudomonas on CFBE41o- cell monolayer.
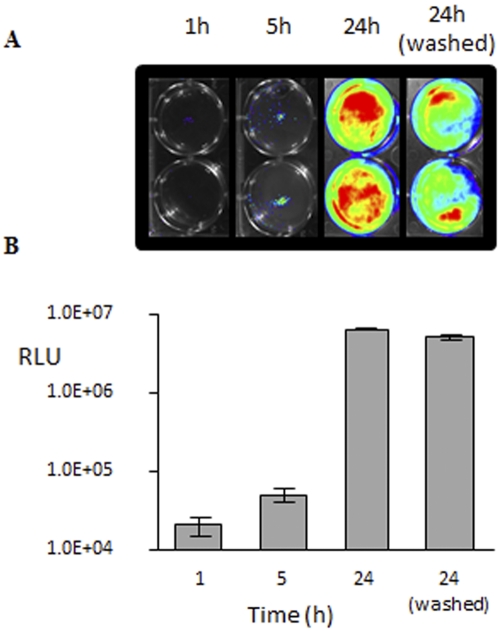
We studied the ability of phage to penetrate and attack Pseudomonas cells growing as biofilms on the surface of a layer of human lung epithelial cells. CFBE41o- cells (CFBE stands for cystic fibrosis bronchial epithelial) were grown in standard 6-well tissue culture plates, and a confluent culture with a tight junction between cells was achieved after 8 to 10 days of incubation (12). The confluent culture was inoculated with lux-tagged Pseudomonas strains NH57388A::p16Slux or MR299::p16Slux, and growth was monitored in this static system for 24 h (Fig. 3A). The addition of arginine in the minimal essential medium (MEM) enhanced the formation of biofilms and helped preserve the integrity of the CFBE41o- cells, as suggested by Anderson et al. (12). The amount of luminescence recorded for the growing biofilms increased by 2 log units during the 24-h incubation (Fig. 3B). Washing the biofilm monolayer culture twice with MEM removed all planktonic Pseudomonas cells. The absence of motile cells and the presence of only adhered clusters of microcolonies of various sizes scattered across the epithelial cell monolayer were confirmed by phase-contrast microscopy (see Fig. S2 in the supplemental material). The amount of bioluminescence recorded after removing planktonic cells was only 25% lower than that obtained before washing (Fig. 3A), which confirmed that the majority of Pseudomonas cells were adhered to the epithelial monolayer. We determined the number of CFU after washing the monolayer at 2.6 × 107 to 3.8 × 107 CFU/well and 4.2 to 5.4 × 107 CFU/ well (well area of 9.5 cm2) for strains NH57388A and MR299, respectively.
FIG 3 .
(A) Growth of lux-tagged Pseudomonas biofilms on the surface of the CFBE410- cell monolayer. Light was measured 1, 5, and 24 h (before and after the monolayer was washed with MEM). (B) Readings from 6 wells are shown. Values are shown as means ± standard deviations (SD) (error bars).
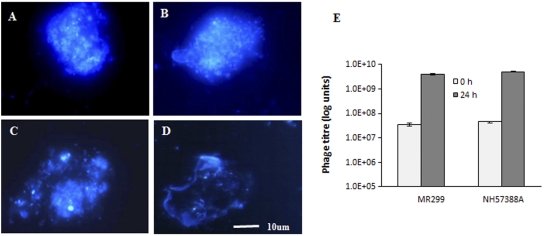
Calcofluor white, a fluorescence enhancer that binds to the β(1–3) and β(1–4) polysaccharide linkages found in biofilm matrices produced by exopolysaccharide-producing organisms (12, 34, 35) was used to stain biofilms. The staining revealed that the Pseudomonas cells were contained within a polysaccharide matrix (Fig. 4A and B). The structures formed by the two Pseudomonas strains NH57388A and MR299 measured on average 20 to 30 µm by 30 to 40 µm in diameter. The presence of abundant numbers of Pseudomonas cells packed in polysaccharide matrices and attached to the cell line led us to conclude that the microcolony structures formed on the monolayer fit the definition of biofilms.
FIG 4 .
Fluorescent image of 24-h-old culture of P. aeruginosa cells grown on a CFBE41o- cell monolayer after Calcofluor white (fluorescent enhancer) staining. Staining confirms that P. aeruginosa NH57388A (A) and MR299 (B) are embedded in an exopolysaccharide structure prior to phage exposure. After 24-h incubation in the presence of mixed phages, staining indicates open and weak matrices with reduced numbers of cells for both NH57388A (C) and MR299 (D). An increase in phage titer was observed over the 24-h incubation period for both MR299 and NH57388A strains (E).
Clearing of biofilms by phage.
We examined the ability of the mixture of phages to clear 24-h-old biofilms of P. aeruginosa NH57388A or MR299. Calcofluor white staining revealed a change to open and weak matrices in the presence of the ϕNH-4 and ϕMR299-2 phages, indicating considerable destruction to the biofilm structure (Fig. 4C and D). At the same time, phage titers increased almost 2 log units during the 24-h period, confirming significant phage replication (Fig. 4E). The amount of luminescence recorded for both Pseudomonas biofilms also decreased by 2 log units over a 24-h period (Fig. 5). In contrast, the light level remained high and unchanged in the control biofilms (with no phage added) over the same period. Direct plating results also confirm that the significant reduction in light was a direct result of the destruction of the Pseudomonas cells by the phage. The numbers of CFU estimated for the biofilms before phage added were 2.6 × 107 to 3.8 × 107 CFU/well and 4.2 × 107 to 5.4 × 107 CFU/well (well area of 9.5 cm2) for strains NH57388A and MR299, respectively, and the amount of phage added was 0.5 × 108 to 1.0 × 108/well (multiplicity of infection [MOI] of 2 to 5). During the 24-h incubation, the number of Pseudomonas cells in the biofilms was reduced by 3 to 4 log units in the presence of phage.
FIG 5 .
(A) Light emitted from nonmucoid P. aeruginosa MR299 strain and mucoid NH57388A strain grown on a CFBE410- cell monolayer for 24 h in the presence (+) and absence (−) of phage mix. (B) The RLU values are mean ± SD readings from 3 wells.
Clearing of Pseudomonas from murine lungs.
The ability of phage to kill Pseudomonas in situ in the lungs of infected 8-week-old female BALB/c mice (n = 16) was also examined (Fig. 6). In this regard, lux tagging of Pseudomonas cells was very useful in monitoring the fate of these cells in the lungs of infected mice. The presence of Pseudomonas was evident in the lungs of both test and control mice 2 h after infection. The amount of light recorded in the control mice (without phage) increased 3-fold and reached its maximum level after 6 h. The amount of light recorded in mice treated with phage decreased significantly during the same period.
FIG 6 .
Mice (n = 8) were infected with nonmucoid P. aeruginosa MR299 (A) and mucoid NH57388A (mucoid strain) (B). Test mice (+) were treated with the phage mix (ϕMR299-2 and ϕNH-4B). Phage was given 2 h after the mice were infected with Pseudomonas. Control mice (−) did not receive the phage mix.
DISCUSSION
There has been increased interest in phage therapy as a means of combating bacterial lung infections. However, there is little information about the efficacy of bacteriophage in clearing Pseudomonas growing on CF lung tissue or in an animal model. It has been well documented that the CF lung environment causes normally motile, planktonic P. aeruginosa to form mucoid biofilms (36–38). Our data show that 1 h after the addition of lux-tagged Pseudomonas cells to a CF epithelial monolayer, only a very low level of localized light was detected. The signal then increased over 100-fold in 24 h (Fig. 3). Washing the 24-h-old biofilm with MEM medium resulted in 25% reduction in the amount of light, indicating that 75% (2.6 × 107 to 5.4 × 107 CFU/well) of the Pseudomonas adhered as microcolonies over the entire surface of the CF cell monolayer. It is well documented in the literature that microcolony dispersal in Pseudomonas is a feature of biofilm maturation (39–42).
To be effective, phage must be able to penetrate the biofilm exopolysaccharide. This may account for the fact that it took more time (22 to 24 h) for phage (at similar MOI) to clear Pseudomonas growing on CFBE41o- cells than the 5 to 6 h required to clear recently introduced planktonic cells from the lungs of infected mice. We observed proliferation of Pseudomonas cells in the lungs of mice in the absence of phage, whereas phage treatment prevented growth and reduced the bacterial load to a nondetectable level during the 6-h period. A recent study reported that the amount of light measured in mice infected with a lux-tagged Pseudomonas PAK strain also decreased over 6 h when phage PAK-P1 was administered 2 h after infection (24, 25). The amount of light increased in the control mice and in the mice that received a delayed phage treatment. The authors concluded that administering phage 2 h after infection was critical to resolving infection in the mouse model system and suggested that the efficacy of phage needed to be tested against biofilms.
One advantage of using phage to control bacterial infection is that they can replicate at the infection site (24). In addition to being a very effective way of clearing Pseudomonas, the use of a phage mix has another advantage over the use of a single phage in that it reduces the likelihood of a phage resistance population emerging. Different phages often use different bacterial receptors and therefore will require independent mutations to generate resistance to each phage (43). Consequently, unless mutants with generalized resistance mechanism evolve, a mix of different phages for which there is no cross-resistance should be able to prevent multiple phage resistance and provide indefinite control of bacterial population.
In conclusion, this study demonstrates that Pseudomonas growing on a CF airway tissue monolayer could be killed by phage. Since chronic lung infections in CF patients are associated with Pseudomonas biofilms rather than planktonic cells, we anticipate that biofilm clearing from lungs of CF patients by bacteriophage might take place in a similar manner to that observed with the exopolysaccharide-producing microcolonies growing on an epithelial cell monolayer. Moreover, we subsequently demonstrate that the same phage mix was effective in killing the pathogen in the lungs of infected mice. Our study reinforces the growing interest in using phage therapy as a means of attacking multidrug-resistant CF infections.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Pseudomonas strains isolated from CF patients and used in this study are shown in Table 1. LB medium was used throughout this study to culture Pseudomonas strains. A double-strength LB medium was made for phage isolation by doubling the weight of dry ingredients required to prepare single-strength LB broth. Cultures were grown at 37°C under aerobic conditions and shaking at 180 rpm. Solid media and soft agar overlays contained 1.5% and 0.7% agar (BD Difco, Oxford, United Kingdom), respectively.
Transformation of Pseudomonas with p16lux plasmid.
P. aeruginosa strains NH57388A (mucoid) and MR299 (nonmucoid) were transformed with p16Slux plasmid (44) by the method of Shen et al. (45). In brief, Pseudomonas strains were grown in LB medium at 37°C until an optical density (OD) at 600 nm of 0.8 is reached. To facilitate electroporation, Pseudomonas exopolysaccharide was digested by adding alginate-lyase (catalog no. A1603; Sigma, Japan) to a final concentration of 2 U ml−1. The cell enzyme mixture was incubated at 37°C for 30 min. The cells were centrifuged at 10,000 × g (4°C) for 10 min. The resultant pellet was washed twice with chilled electroporation buffer (containing 300 mM glucose, 5 mm CaCl2, and 25 mM HEPES in distilled water [pH 7.0]) and resuspended in 0.1 ml of buffer (1 × 109 to 1 × 1010 CFU/ml). These electrocompetent cells were mixed with 10 µl of p16Slux-tagged plasmid DNA (10 µg) and incubated on ice for 10 to 15 min. The mixture was immediately transferred to a chilled electroporation cuvette (0.2-cm electrode gap Gene-pulser cuvette; Bio-Rad, Hercules, CA) and subjected to a single voltage shock by applying a pulse with an ECM 630 BTX Harvard precision pulse apparatus (Holliston, MA) at the following settings: capacitor, 25 µF; resistor, 200 Ω; and voltage, 2.5 kV. Immediately after the electric shock, 900 µl of chilled SOC medium (catalog no. S1797; Sigma-Aldrich) was added to the mixture and incubated on ice for 10 min, followed by incubation at 30°C for 2 to 3 h. The cells were concentrated by centrifugation at 10,000 × g (room temperature), and transformants were obtained by plating cells on LB agar containing erythromycin (800 µg/ml) and incubating at a permissive temperature (30°C) for 24 to 48 h. Eryr colonies were checked for light emission using a VivoVision IVIS100 imaging system (Xenogen, Alameda, CA), luminescence was measured in relative light units (RLU) (in photons second−1), and the presence of p16Slux was confirmed by miniprep and restriction analyses. A standard curve was generated to determine the relationship between light measurements and CFU (see Fig. S3 and Table S2 in the supplemental material).
Isolation of phage from sewage.
Pseudomonas phages were isolated from fresh sewage obtained from a local sewage treatment plant as described previously (46) with some modifications. Sewage samples were centrifuged at 3,200 × g value (Heraeus Labofuge 400 centrifuge; Thermo Fisher Scientific, Inc., MA) for 12 min, and the supernatant was filtered using a 0.45-µm-pore-size filter (Sarstedt, Actiengeselischaft and Co., Germany). The sewage filtrate (5 ml) was mixed with an equal volume of double-strength LB broth, supplemented with 10 mM CaCl2, and inoculated with a mixture of ten CF P. aeruginosa cultures (50 µl each of 1 × 107 to 2 × 107/ml). The samples were incubated overnight aerobically (37°C) with slow shaking (20 to 30 rpm). Following overnight incubation, the cultures were centrifuged at 3,200 × g value for 12 min to remove bacterial cells and debris, and the supernatant was filtered through a sterile 0.45-µm-pore-size filter. A double-layer LB agar plate containing a lawn of individual host culture supplemented with 10 mM CaCl2 was prepared, and 10 µl of cell-free filtrate containing phage was applied to the plate. The plates were examined for the presence of plaques after incubating aerobically for 18 to 24 h at 37°C.
Plaque purification and bacteriophage titers.
Phages were purified by successive single plaque isolation and propagation. In general, a single plaque was picked from a plate using a sterile capillary tube and added to a mid-log-phase Pseudomonas culture (108 CFU/ml) supplemented with 10 mM CaCl2. The culture mixture and phage mixture were incubated at 37°C overnight. The lysate was filtered through a 0.45-µm-pore-size sterile filter, serial dilutions were made, and plaques were allowed to form on a lawn of the same host culture. Single plaques were purified through 3 successive rounds of plaquing and repeated three additional times after which purified phages were obtained. Phage titer was determined as the number of PFU/ml by plaque assay as previously described (47).
DNA extraction and restriction digestion analysis.
High-titer-purified phage suspension was prepared by concentration of phage particles from 400 ml cell lysate in LB medium to a final volume of 1 ml in sterile ice-cold ammonium acetate (0.1 M, pH 7.2) as described before (46), and DNA was extracted from the high-titer-purified phage as previously described (46, 48, 49). Phage DNA was digested with restriction endonuclease EcoRI (New England Biolabs, MA) according to the supplier’s recommendation, and digested samples were analyzed by gel electrophoresis using agarose gel (0.7%) containing ethidium bromide (SF1).
Genome sequencing and annotation.
The genomes of phages ϕNH-4 and ϕ299-2 were sequenced by Beckman Coulter Genomics (Sanger sequencing services; Beckman Coulter, Takeley, United Kingdom) on a 454 GS-FLX sequencer, and sequences were assembled into contigs using the Newbler program (Roche Applied Sciences). The quality of the sequence was assessed using Hawkeye (Amos) (50). To confirm phage genome structure, primers were designed at contig ends for PCR amplification using Platinum PCR SuperMix (Invitrogen) followed by direct sequencing of the PCR products, and full phage genome assembly was performed using the Phred-Phrap-Consed package (51, 52). ORFs were predicted using GLIMMER 3.02 (53). The resulting gene models were fed into GAMOLA (54) for annotation. Complementary annotation was provided using the RAST annotation server (55). Data were manually curated using Artemis version 11 (56) where additional programs were then used, including BLASTp (57), GATU (58) and RBS finder (53). Comparative genomics with reference phages (ϕPaP3 and ϕLMA2) were analyzed using the Artemis comparison tool (ACT) (7) and the Mauve alignment tool (59).
Electron microscopy and phage characterization.
Electron microscopy image of phages was obtained by applying a drop of high-titer phage suspension (1 × 109 to 2 × 109 PFU/ml) deposited on carbon-coated copper grids, negatively stained with 2% (wt/vol) potassium phosphotungstate (pH 7.2) and examined with Zeiss Supra 40VP scanning electron microscope (Carl Zeiss SMT Ltd., Cambridge, United Kingdom) fitted with a scanning transmission electron microscope detector (STEM) operating in bright-field mode at an accelerating voltage of 25 kV (Moorepark National Food Imaging Centre, TEAGASC Food Research Centre [TFRC], and Advanced Microscope Research Facility, University College Cork, Cork, Ireland).
Cell culture and Pseudomonas biofilm formation on the surface of a monolayer of human bronchial epithelial cells (CFBE41o- cells).
Cystic fibrosis bronchial epithelial (CFBE41o-) cell (60, 61) cultures were grown by the method of Anderson et al. (12). In general, CFBE41o- cells were seeded in sterile 6-well, flat-bottom tissue culture plates (Sarstedt, Newton, NC) at a concentration of 106 cells/well and maintained in minimal essential medium (MEM) containing 10% fetal bovine serum, 2 mM l-glutamate, 100 µg/ml penicillin, and 100 µg/ml streptomycin (all from Invitrogen GIBCO, United Kingdom). The cells were grown at 37°C and 5% CO2 using a Jouan IGO150 cell life incubator (Jouan, St. Herblain, France) for 8 to 10 days until cells form a confluent monolayer and tight junctions. The medium (MEM) was changed every 2 or 3 days until confluent growth was achieved.
For biofilm formation, lux-tagged P. aeruginosa cells were grown on the confluent CFBE41o- cell monolayer using a coculture model system (12). Once the monolayer growth was achieved (between 8 and 10 days), the medium was replaced with 1.5 ml fresh MEM (without fetal bovine serum, penicillin, and streptomycin), and lux-tagged Pseudomonas cells were inoculated (1 × 107 to 2 × 107 CFU/well). The plates were incubated at 37°C and 5% CO2 for 1 h. The medium containing the unattached (planktonic) Pseudomonas cells was then removed using a sterile serological pipette and replaced with fresh MEM supplemented with 0.4% arginine and incubated for 24 h. Planktonic Pseudomonas cells were removed, and the biofilm culture was washed twice using MEM supplemented with 0.4% arginine. The integrity of the epithelial cell monolayer and the presence of growing Pseudomonas microcolonies were assessed by phase-contrast microscopy (Olympus IX50 inverted system microscope; Olympus Co., Tokyo, Japan). Luminescence from biofilms was monitored by Vivo Vision IVIS100 imaging system (Xenogen, Alameda, CA). Biofilm CFU was estimated by the method of Wirtanen et al. (62) with some modifications. A 24-h biofilm growing on an epithelial cell monolayer was washed twice with MEM and then scraped off using a tissue culture scrapper and transferred to a 2-ml Eppendorf tube containing 1 ml phosphate buffer. The tube was then vortexed thoroughly for 1 to 2 min to release the cells. After this, the samples were then serially diluted, and a plate count was made on a LB plate incubated at 37°C overnight.
Applying phages to biofilms and lung infections in mice.
Fifty microliters of the phage mixture (1 × 109 to 2 × 109 PFU/ml of defined phage [containing ϕNH-4 and ϕMR299-2] in a 1:1 mixture) was applied to wells containing 24-h-old biofilms on a CFBE41o- cell monolayer, and the plates were incubated at 37°C and 5% CO2 for 24 h. Biofilm clearing was monitored by measuring light and images taken for times indicated, using the IVIS100 imaging system. The lungs of 6- to 8-week-old conventional female BALB/c mice (n = 16) were infected with Pseudomonas by the method of Riedel et al. (28). The mice were infected intranasally with 50 µl lux-tagged Pseudomonas NH57388A or MR299 in phosphate buffer (2 × 108 to 5 × 108 CFU/ml). Two hours following infection, 50 µl of a phage mix suspension in phosphate buffer containing Pseudomonas phages ϕNH-4 and ϕMR299-2 (2 × 109 to 5 × 109 PFU/ml for a multiplicity of infection [MOI] of 10) were given intranasally. Fifty microliters of phosphate buffer was given to the control groups. Animals were anesthetized with isoflurane, light was monitored, and images were taken using the IVIS 100 system at the time points indicated. Animals were kept in an animal colony, and all experiments were approved by the animal ethics committee of University College Cork.
Biofilm staining.
Pseudomonas biofilms were stained with the fluorescent enhancer Calcofluor white (fluorescent brightener 28, catalog no. F3543; Sigma Aldrich, China). In brief, the monolayers of CFBE41o- cells containing biofilms were removed from wells using a sterile cell scraper (Sarstedt, Actiengeselischaft and Co., Germany) and mixed with a drop of Calcofluor white (0.1%) on a microscope slide. A coverslip was applied to the slide; the edges were sealed with paraffin oil, and the microscope slide was incubated for 1 h (37°C). Stained biofilm preparations were assessed by using an Olympus BX51 fluorescence microscope fitted with a U-RFL-T fluorescent power supply unit, and images were taken with a DP50 integrated camera (all from Olympus Optical Co., Japan).
Nucleotide sequence accession numbers.
The completed phage genome sequences of phages NH-4 and MR299-2 were deposited in GenBank database and assigned accession numbers JN254800 and JN254801, respectively.
SUPPLEMENTAL MATERIAL
EcoRI restriction digestion profile of phage DNA. Lane 1, hyperladder I markers; lanes 2 and 3, DNA restriction profiles for phages ϕMR299-2 and ϕNH-4, respectively. Download Figure S1, PDF file, 0.1 MB.
(A) Confluent CFBE41o- (cystic fibrosis bronchial epithelial) cell monolayer (8 to 10 days old). (B) Pseudomonas microcolonies (biofilms) (24 h old) adhered to the surface of the CFBE41o- cell monolayer. Download Figure S2, PDF file, 0.1 MB.
Standard curve showing the relationship between light measurements of lux-tagged P. aeruginosa and CFU. The graph shows linearity to a cell count of l × 108 CFU and the light saturate afterward. Cell number (CFU) estimates were made within the linear range of the standard curve. Data points are means ± standard errors of the means of six separate readings taken from ST2. Download Figure S3, PDF file, 0.1 MB.
Pseudomonas cell lyses in vitro (LB broth) in the presence of phage ϕ299-2 and ϕNH4 separately and phage mix containing the combination of the two phages
CFU of lux-tagged Pseudomonas cells and corresponding luminescence light measured by IVIS100
Features of phage MR299-2 putative gene assignments based on protein PSI-BLAST comparisons
Features of phage ϕNH-4 putative gene assignments based on PSI-BLAST comparisons
ACKNOWLEDGMENTS
The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan. This work was supported by SFI grants 02/CEB124 and 07/CE/B1368.
We thank Dieter C. Gruenert (California Pacific Medical Center, University of California, San Francisco, CA) for providing CFBE41o- cells and Nadine Hoffmann (Institute of Medical Microbiology and Immunology, University of Copenhagen, Copenhagen, Denmark) for providing the mucoid P. aeruginosa NH57388A strain. We also thank Mark Auty (Moorepark Imaging Centre, TFRC) and Don O’Leary (Advanced Microscope Research Facility, University College Cork) for electron microscropy (EM) imaging.
Footnotes
Citation Alemayehu D, et al. 2012. Bacteriophages ϕMR299-2 and ϕNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. mBio 3(2):e00029-12. doi:10.1128/mBio.00029-12.
REFERENCES
- 1. Kerem B, et al. 1998. Identification of the cystic fibrosis gene: genetic analysis. Science 245:1073–1080 [DOI] [PubMed] [Google Scholar]
- 2. Riordan JR, et al. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073 [DOI] [PubMed] [Google Scholar]
- 3. Rommens JM, et al. 1998. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 245:1059–1065 [DOI] [PubMed] [Google Scholar]
- 4. Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951 [DOI] [PubMed] [Google Scholar]
- 5. Saiman L, Siegel J. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burns JL, et al. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444–452 [DOI] [PubMed] [Google Scholar]
- 7. Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Høiby N, Frederiksen B, Pressler T. 2005. Eradication of early Pseudomonas aeruginosa infection. J. Cyst. Fibros. 4(Suppl. 2):49–54 [DOI] [PubMed] [Google Scholar]
- 10. Lam J, Chan R, Lam K, Costerton JW. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pedersen SS. 1992. Lung infection with alginate-producing, mucoid P. aeruginosa in cystic fibrosis. APMIS 28(Suppl.):1–79 [PubMed] [Google Scholar]
- 12. Anderson GG, Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 76:1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zavascki AP, Gaspareto PB, Martins AF, Gonçalves AL, Barth AL. 2005. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing SPM-1 metallo-{beta}-lactamase in a teaching hospital in southern Brazil. J. Antimicrob. Chemother. 56:1148–1151 [DOI] [PubMed] [Google Scholar]
- 14. Pasca MR, et al. 2012. Evaluation of fluoroquinolone resistance mechanisms in Pseudomonas aeruginosa multidrug resistance clinical isolates. Microb. Drug Resist. 18:23–32 [DOI] [PubMed] [Google Scholar]
- 15. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2(9):1051–1060 [DOI] [PubMed] [Google Scholar]
- 16. Azeredo A, Southerland WI. 2008. The use of phages for the removal of infectious biofilms. Curr. Pharm. Biotechnol. 9:261–266 [DOI] [PubMed] [Google Scholar]
- 17. Cerca N, et al. 2005. Comparative assessment of antibiotic susceptibility of coagulase-negative staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J. Antimicrob. Chemother. 56(2):331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monzón M, Oteiza C, Leiva J, Lamata M, Amorena B. 2002. Biofilm testing of Staphylococcus epidermidis clinical isolates: low performance of vancomycin in relation to other antibiotics. Diagn. Microbiol. Infect. Dis. 44(4):319–324 [DOI] [PubMed] [Google Scholar]
- 19. Travis J. 2000. Viruses that slay bacteria drew new interest. Science News 157:356–360 [Google Scholar]
- 20. Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ. 2008. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. MicroBiol. 104(1):1–13 [DOI] [PubMed] [Google Scholar]
- 21. Fu W, et al. 2010. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob. Agents Chemother. 54:397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knezevic P, Petrovic O. 2008. A colorimetric microtiter plate method for assessment of phage effect on Pseudomonas aeruginosa biofilm. J. Microbiol. Methods 74(2-3):114–118 [DOI] [PubMed] [Google Scholar]
- 23. Glonti T, Chanishvili N, Taylor PW. 2010. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. MicroBiol. 108(2):695–702 [DOI] [PubMed] [Google Scholar]
- 24. Morello E, et al. 2011. Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: first steps towards treatment and prevention. PLoS One 6(2):e16963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Debarbieux L, et al. 2010. Bacteriophages can treat and prevent P. aeruginosa lung infections. Infect. Dis. 201(7):1096–1104 [DOI] [PubMed] [Google Scholar]
- 26. Carmody LA, et al. 2010. Efficacy of bacteriophage therapy in a model of Burkholderia cenocepacia pulmonary infection. J. Infect. Dis. 201(2):264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seed KD, Dennis JJ. 2009. Experimental bacteriophage therapy increases survival of Galleria mellonella larvae infected with clinically relevant strains of the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 53(5):2205–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riedel CU, et al. 2007. Construction of p16Slux, a novel vector for improved bioluminescent labeling of Gram-negative bacteria. Appl. Environ. MicroBiol. 73(21):7092–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan Y, et al. 2007. Whole genome sequencing of a novel temperate bacteriophage of P. aeruginosa: evidence of tRNA gene mediating integration of the phage genome into the host bacterial chromosome. Cell. MicroBiol. 9(2):479–491 [DOI] [PubMed] [Google Scholar]
- 30. Van Regenmortel MHV, et al. 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the international committee on taxonomy of viruses. Academic Press, San Diego, CA. [Google Scholar]
- 31. Murphy FA, et al. 1995. Virus taxonomy: classification and nomenclature of viruses. Sixth Report of the International Committee on Taxonomy of Viruses, p. 51–63 Springer-Verlag, Vienna, Austria: [Google Scholar]
- 32. Ceyssens PJ, et al. 2009. Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ. MicroBiol. 11(11):2874–2883 [DOI] [PubMed] [Google Scholar]
- 33. Ceyssens PJ, et al. 2008. The intron-containing genome of the lytic Pseudomonas phage LUZ24 resembles the temperate phage PaP3. Virology 377(2):233–238 [DOI] [PubMed] [Google Scholar]
- 34. Ross P, Mayer R, Benziman M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:35–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. MicroBiol. 39(6):1452–1463 [DOI] [PubMed] [Google Scholar]
- 36. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 37. O’May CY, Reid DW, Kirov SM. 2006. Anaerobic culture conditions favour biofilm-like phenotypes in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. FEMS Immunol. Med. MicroBiol. 48(3):373–380 [DOI] [PubMed] [Google Scholar]
- 38. Worlitzsch D, et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109(3):317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirov SM, et al. 2007. Biofilm differentiation and dispersal in mucoid P. aeruginosa isolates from patients with cystic fibrosis. Microbiology 153:3264–3274 [DOI] [PubMed] [Google Scholar]
- 40. Shrout JD, et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. MicroBiol. 62(5):1264–1277 [DOI] [PubMed] [Google Scholar]
- 41. Purevdorj-Gage B, Costerton WJ, Stoodley P. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151(5):1569–1576 [DOI] [PubMed] [Google Scholar]
- 42. Webb JS, et al. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Levin BR, Bull JJ. 2004. Population and evolutionary dynamics of phage therapy. Nat. Rev. MicroBiol. 2:166–173 [DOI] [PubMed] [Google Scholar]
- 44. Hoffmann N, et al. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect. Immun. 73:2504–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen H, Han F, Yuzi Lin Y, Yu W. 2006. A high efficient electroporation of Pseudomonas sp. QDA pretreated with alginate lyase. Enzyme Microb. Technol. 39(4):677–682 [Google Scholar]
- 46. Alemayehu D, et al. 2009. Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 448(1):29–39 [DOI] [PubMed] [Google Scholar]
- 47. O’Sullivan D, et al. 2001. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. MicroBiol. 67:929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Capra ML, et al. 2006. Characterization of a new virulent phage (MLC-A) of Lactobacillus paracasei. J. Dairy Sci. 89(7):2414–2423 [DOI] [PubMed] [Google Scholar]
- 49. Moineau S, Pandian S, Klaenhammer TR. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. MicroBiol. 60:1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schatz MC, Phillippy AM, Shneiderman B, Salzberg SL. 2007. Hawkeye: an interactive visual analytics tool for genome assemblies. Genome Biol. 8(3):R34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gordon D. 2003. Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinformatics Chapter 11, Unit 11.2 [DOI] [PubMed] [Google Scholar]
- 52. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8(3):186–194 [PubMed] [Google Scholar]
- 53. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27(23):4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altermann E, Klaenhammer TR. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7(2):161–169 [DOI] [PubMed] [Google Scholar]
- 55. Aziz RK, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carver T, et al. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24(23):2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 58. Tcherepanov V, Ehlers A, Upton C. 2006. Genome annotation transfer utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Darling AE, Treangen TJ, Messeguer X, Perna NT. 2007. Analyzing patterns of microbial evolution using the mauve genome alignment system. Methods Mol. Biol. 396:135–152 [DOI] [PubMed] [Google Scholar]
- 60. Bruscia E, et al. 2002. Isolation of CF cell lines corrected at ΔF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 9(11):683–685 [DOI] [PubMed] [Google Scholar]
- 61. Cozens AL, et al. 1994. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 10(1):38–47 [DOI] [PubMed] [Google Scholar]
- 62. Wirtanen G, Salo S, Helander IM, Mattila-Sandholm T. 2001. Microbiological methods for testing disinfectant efficiency on Pseudomonas biofilm. Colloids Surf. B Biointerfaces 20:37–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EcoRI restriction digestion profile of phage DNA. Lane 1, hyperladder I markers; lanes 2 and 3, DNA restriction profiles for phages ϕMR299-2 and ϕNH-4, respectively. Download Figure S1, PDF file, 0.1 MB.
(A) Confluent CFBE41o- (cystic fibrosis bronchial epithelial) cell monolayer (8 to 10 days old). (B) Pseudomonas microcolonies (biofilms) (24 h old) adhered to the surface of the CFBE41o- cell monolayer. Download Figure S2, PDF file, 0.1 MB.
Standard curve showing the relationship between light measurements of lux-tagged P. aeruginosa and CFU. The graph shows linearity to a cell count of l × 108 CFU and the light saturate afterward. Cell number (CFU) estimates were made within the linear range of the standard curve. Data points are means ± standard errors of the means of six separate readings taken from ST2. Download Figure S3, PDF file, 0.1 MB.
Pseudomonas cell lyses in vitro (LB broth) in the presence of phage ϕ299-2 and ϕNH4 separately and phage mix containing the combination of the two phages
CFU of lux-tagged Pseudomonas cells and corresponding luminescence light measured by IVIS100
Features of phage MR299-2 putative gene assignments based on protein PSI-BLAST comparisons
Features of phage ϕNH-4 putative gene assignments based on PSI-BLAST comparisons