Abstract
The photolyase/cryptochrome family of proteins are FAD-containing flavoproteins which carry out blue-light dependent functions including DNA repair, plant growth and development, and regulation of the circadian clock. In addition to FAD, many members of the family contain a second chromophore which functions as a photoantenna, harvesting light and transferring the excitation energy to FAD and thus increasing the efficiency of the system. The second chromophore is methenyltetrahydrofolate (MTHF) in most photolyases characterized to date and FAD, FMN, or 5-deazariboflavin in others. To date no second chromophore has been identified in cryptochromes. Drosophila contains 3 members of the cryptochrome/photolyase family: cyclobutane pyrimidine dimer (CPD) photolyase, (6-4) photoproduct photolyase, and cryptochrome. We developed an expression system capable of incorporating all known second chromophores into the cognate cryptochrome/photolyase family members. Using this system we demonstrate that Drosophila CPD photolyase and (6-4) photolyase employ 5-deazariboflavin as their second chromophore but Drosophila cryptochrome, which is evolutionarily closer to (6-4) photolyase than the CPD photolyase, lacks a second chromophore.
The photolyase/cryptochrome family encompasses a large group of flavoproteins involved in blue-light dependent and independent functions including DNA repair, regulation of plant growth and development, and regulation of the circadian clock in animals (1-9). Phylogenetic analyses have revealed numerous classes of the family. Functionally, there are 3 classes, cyclobutane pyrimidine dimer photolyase (CPD photolyase or simply photolyase), pyrimidone-pyrimidine (6-4) photoproduct (10) photolyase [(6-4) photolyase], and cryptochrome (CRY). Some organisms such as humans have only cryptochrome, some others such as opossum have cryptochrome and photolyase but no (6-4) photolyase and some such as Drosophila melanogaster and Arabidopsis thaliana have photolyase, (6-4) photolyase, and cryptochrome (1,4).
The photolyase/cryptochrome family members contain reduced FAD (FADH-) as the catalytic cofactor. All CPD photolyases characterized to date also contain a second chromophore which, because its extinction coefficient is several fold higher than FADH-, and because it has an absorption peak at a wavelength longer than FADH-, is more efficient at absorbing environmental radiation than FADH- (4). The second chromophore transfers the excitation energy to FADH- by Förster resonance energy transfer and thus increases the efficiency of the incident light in repair. In most cases the second chromophore is methenyltetrahydrofolate (MTHF). However, FAD (11), FMN (12), and 7,8-didemethyl-8-hydroxy-5-deazariboflavin (F0) (4,13) have also been found in several photolyases. So far there is no definitive evidence that CRYs contain a second chromophore. Significantly, it is unknown whether an organism that contains both folate and Fo and all 3 members of the photolyase/CRY family utilize the same second chromophore in all 3 family members. We developed a baculovirus/insect cell expression system and examined the 3 members of the Drosophila photolyase/CRY family to address this question. We found that Drosophila photolyase and (6-4) photolyase which are evolutionarily rather distant both contain Fo as the second chromophore but Dm CRY, which is phylogenetically closer to (6-4) photolyase than the CPD photolyase, does not carry a second chromophore. Finally, to determine if the lack of a second chromophore is a general property of cryptochrome, we tested A. thaliana CRY2 (AtCRY2) and zebrafish CRY4 (ZfCRY4) in this system. Both proteins contained full complement of FAD catalytic cofactor but no folate or Fo chromophore. Thus we conclude that both of the Drosophila photolyases contain Fo as the second chromophore and that within the limitation of our assay neither plant nor animal cryptochromes seem to carry a second chromophore.
MATERIALS AND METHODS
Expression Vectors
The fbiC gene from Mycobacterium smegmatis was amplified by PCR from the pMsfbiC construct (14) kindly provided by Dr. Robert White, Virginia Tech, Blacksburg, Virginia) and inserted into the pRSFDuet-1 vector (Novagen) to give pfbiRS which encodes fbiC with a polyhistidine tag at the amino terminus when expressed in bacteria.
Other proteins were expressed using the Gibco BRL Bac to Bac Baculovirus Expression System. Viruses for expressing the Drosophila photolyases were made by first amplifying by PCR the respective genes encoding (6-4) photolyase and CPD photolyase from a library provided by Dr. Dongping Zhong (Ohio State University). Each gene was inserted into the pFastBac1 vector with a flag-tag-encoding sequence at the amino terminus. These constructs, p64PLA5 and pFBCPDL6 were each transformed into E. coli DH10 Bac cells, from which bacmids were isolated. Bacmids were transfected into Sf21 cells, and the virus-containing culture media was harvested after 3 days. Next, high titer virus for each photolyase was generated by successively amplifying the virus two times. The virus for expressing A. nidulans photolyase was made in a similar manner. The photolyase gene was amplified from pUNC1993 (15) and inserted into pFastBac1 to create pAnPLF2. The viruses used to express DmCry protein (16), ZfCRY4 (17) and AtCRY2 (18) proteins have been described.
The sequence of each plasmid construct generated in this study was verified.
Deazariboflavin Biosynthesis and Analysis
E. coli BL21 was transformed with pfbiRS and grown at 37°C in 1-liter volumes of Luria Broth with 50 ug/ml kanamycin to an OD of approximately 0.5 at 595 nm. Cells were then cooled on ice to room temperature, IPTG was added to 0.3 mM, and growth continued overnight at room temperature. Cells were then pelleted, and the supernatants containing Fo were sterilized by filtration, and stored at 4°C for up to several months. Mass spectroscopy was unsuccessful in identification of pure Fo or the Fo associated with the enzymes. Its identity was determined by absorption and fluorescence spectroscopy and by paper chromatography and solid phase electrophoresis.
Protein Purification
500 ml volumes of SF21 cells were grown in spinner culture at 27°C in Grace’s medium with 10 % FBS to a density of 1 to 2 × 106 cells/ml. Cells were then pelleted and resuspended in 500 ml of fresh Grace’s medium with 10% FBS and 5 ml of the high titer virus. In tests of F0 incorporation, 50 ml of LB supernatant containing F0 was included along with 5 ml of the high titer virus and the final concentration of FBS was adjusted to 10 %. Infection continued for two days. Cells were harvested by centrifugation, washed one time with PBS, and pellets were frozen in dry ice-ethanol and stored at -80°C.
To purify protein, pellets were thawed and resuspended in 25 to 30 ml of TBS200 (50 mM Tris-HCl pH 7.5; 200 mM NaCl) containing 0.5% NP40 and a protease inhibitor (Roche 11836153001, one tablet per 50 ml). Suspensions were passed several times through a 19 gauge needle, sonicated 10 × 10 sec, passed several times through a 22 gauge needle, and pelleted in a Sorvall centrifuge at 15,000 rpm for 1 hour to obtain cell-free extracts.
Anti-flag affinity resin was used to purify A. nidulans photolyase, Dm CRY, Dm (6-4) photolyase, Dm CPD photolyase, and ZfCRY4. 300 ul of resin (Sigma A2220) was equilibrated with TBS200 and then added to each extract. Resin and the cell extract were mixed by rotation at 4°C for about 3 hours. Resin was then washed three times with TBS200 and then eluted by mixing for about 1 hour with 300 ul of TBS200 containing Flag peptide (0.2 mg/ml) at 4°C. Resin was pelleted and supernatants containing eluted protein were collected.
Nickel affinity resin was used to purify AtCRY2. Pellets were resuspended in pH 7.4 Wash buffer (6.7 mM KCl; 3.7 mM KH2PO4; 345 mM NaCl; 20 mM Na2HPO4; 20 mM imidazole; 10% glycerol) containing 0.5 % NP40. Extracts were prepared as described above. 200 ul of Ni-NTA Agarose (Quiagen), washed with Wash buffer, was mixed with the cell-free extract by rotating at 4°C for about 3 hours. Resin was then washed 3 times with Wash buffer and then the bound protein was eluted by mixing for about 1 hour with Wash buffer in which imidazole was adjusted to 250 mM. Resin was pelleted and the supernatant containing eluted protein was collected.
Absorption and Fluorescence Spectroscopy
The absorbance was analysed using a Shimadzu UV-1601 UV-visible spectrophotometer. Values of absorbance at 0.5 nm intervals were transferred from the spectrophotometer into Microsoft Excel which was used to generate absorption curves. Smoothed curves for A. nidulans photolyase (−F0) and for Drosophila CPD photolyase (−F0) were obtained using Graph Pad Prism Software. Fluorescence was measured using a Shimadzu RF5000U Spectrofluorophotometer. Values for fluorescence were interpolated from printouts from the fluorimeter and were entered into the Excel program to generate the spectra shown
RESULTS
An In Vivo System for Introducing Deazariboflavin into Recombinant Proteins
So far, 4 cofactor/chromophores have been identified in photolyases and cryptochromes: FAD (in two-electron reduced or anion radical form) is the universal active site cofactor, and MTHF, FMN (oxidized), FAD (oxidized), or F0 function as photoantenna/second chromophores (8). Recombinant photolyases/CRYs are commonly expressed in E. coli, insect cells, or mammalian cells using plasmid or viral vectors and are purified by affinity chromatography. All these hosts synthesize FAD, FMN, and MTHF which can be incorporated into the cognate enzymes. However, none of the hosts (E. coli, insect cells, human cells) synthesize F0 (4,19) and hence an enzyme which in its natural host contains both the FAD cofactor and the F0 chromophore will incorporate only FAD when expressed in these hosts and it would not be possible to tell whether the enzyme might have Fo as the second chromophore. In some cases this problem is addressed by in vitro reconstitution experiments with purified F0 and enzyme in vitro (15,20). However, F0 is not commercially available and the in vitro reconstitution experiments do not always work (21-23). Therefore, we developed a strategy for incorporating F0 into its cognate enzymes in vivo using the baculovirus/insect cell expression system. 8-Hydroxy-5-deazariboflavin is synthesized by F0 synthase from an intermediate of riboflavin biosynthesis and 4-hydroxyphenylpyruvate which is a precursor in tyrosine biosynthesis (14). F0 synthase is not widely distributed in nature having been found in some archaea, mycobacteria and some photosynthetic eukaryotes (23) but not in eubacteria, and not in model organisms such as budding yeast, Drosophila, mice humans, or Arabidopsis. F0 synthase, however, has been cloned from several sources and the cloned gene expresses the enzyme in E. coli (14) and conceptually an E. coli strain expressing both F0 synthase and a photolyase should incorporate F0 into photolyase. However, some of the photolyases and CRYs we used in this study are not properly folded in E. coli. In contrast most of these proteins are expressed at high levels and can be purified in good quantities from SF21 insect cells. Hence we decided to supplement Sf21 growth media with F0 made in E. coli from recombinant Fo synthase: E. coli expressing F0 synthase and secreting F0 into the growth medium was grown to stationary phase in Luria Broth and then the cells were removed from the medium by centrifugation and the supernatant was sterilized by filtration. Figure 1 shows that medium prepared in this manner contains F0 as indicated by its spectroscopic properties (13,14). Then, the medium containing F0 was added at 1/10 ratio to Sf21 cultures expressing the appropriate photolyase/CRY protein which was eventually purified and characterized.
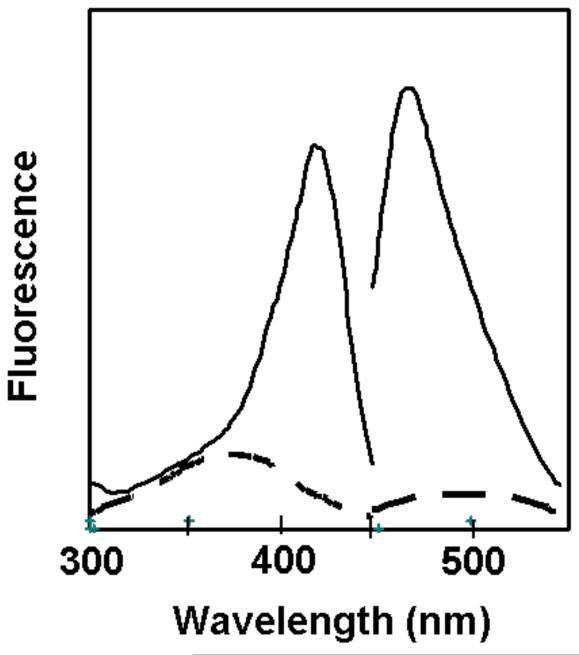
Figure 1.
Production of F0 in E. coli. BL21 cells were transformed with pfbiRS and grown in the presence (solid line) or absence (dashed line) of IPTG. Cells were pelleted, and then the supernatants were filtered and fluorescence was measured. The left side (300-450 nm) shows excitation spectra with emission at 480 nm and the right side shows emission spectra with excitation at 420 nm. Excitation and emission peaks were exhibited at 419 and 469 nm, respectively.
Analysis of Drosophila Photolyase/CRY Family Chromophores
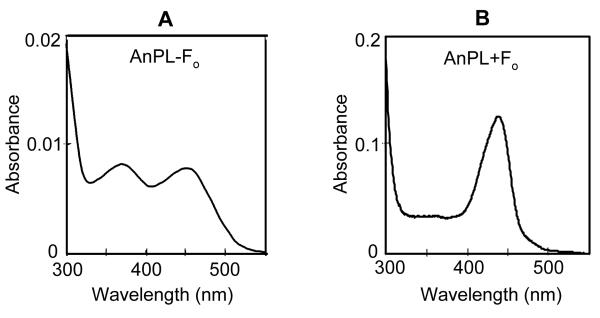
We infected Sf21 cells grown in standard or F0-supplemented media with baculoviruses expressing the 3 members of the Drosophila cryptochrome/photolyase family, photolyase, (6-4) photolyase, and cryptochrome, as well as with a baculovirus expressing the deazaflavin class Anacystis nidulans photolyase (which is known to contain F0 in its native host; 13) as a positive control. The proteins were isolated to a high degree of purity using affinity resin (Figure 2) and then analyzed spectroscopically for cofactor content. As seen in Figure 3A, the chromophore of A. nidulans photolyase purified from cells grown in standard medium exhibits an absorption spectrum typical of FAD, in agreement with previous reports (15) and with the fact that Sf21 cells do not contain F0 synthase or the necessary precursor to F0 biosynthesis and therefore cannot synthesize deazaflavin. In contrast, as seen in Figure 3B, when these cells are grown in F0-supplemented medium, they make A. nidulans photolyase with an absorption spectrum typical of F0- (and FAD-) containing enzyme with an absorption peak at approximately 440 nm. Denaturation of the photolyase releases the chromophore which displays an absorption peak at approximately 420 nm, which is characteristic of free F0 (data not shown).
Figure 2.
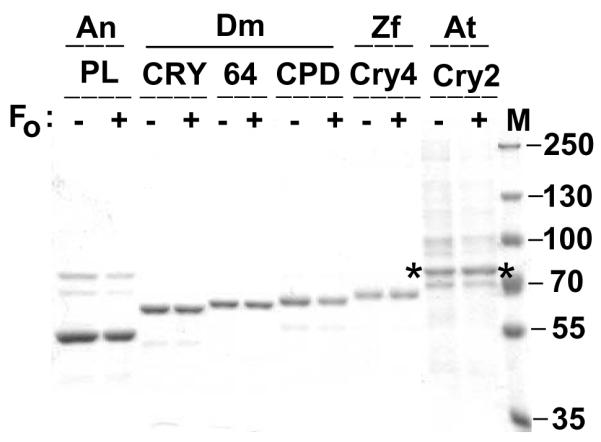
SDS-PAGE analysis of photolyase/cryptochrome family proteins used in this study. Approximately 5 ug of each protein, expressed in Sf21 cells in the presence (+) or absence (−) of Fo, were resolved with 8% PAGE. The gel was stained with Coomassie Blue. The AtCRY2 protein with the polyhistidine tag has an expected molecular weight of 73,301 Da and corresponds to the bands indicated by asterisks in the last two lanes. M, molecular weight markers.
Figure 3.
Chromophores in Anacystis photolyase. Sf21 cells expressing Anacystis photolyase were grown either in the absence (A) or presence (B) of F0 in the culture medium. Spectrum (A) is the spectrum of chromophore liberated from the purified enzyme, while spectrum B is of the holoenzyme. The spectrum characteristic of oxidized, free FAD is seen in (A) while that of enzyme-bound F0 is seen in (B).
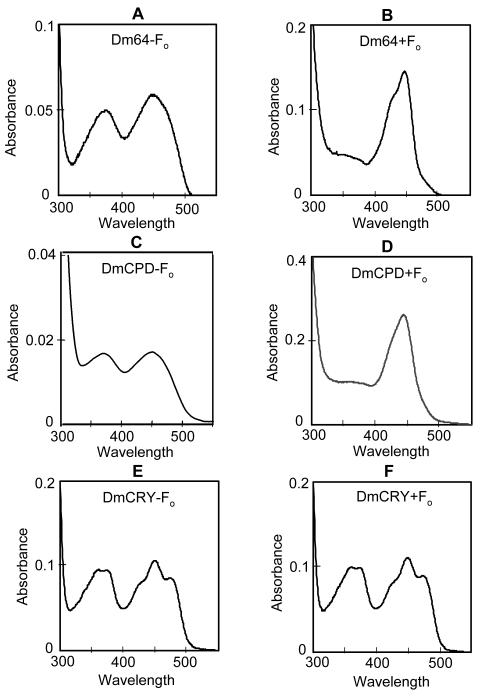
Having thus established that this system is appropriate for incorporating F0 into cognate enzymes, we proceeded to analyze the incorporation of F0 into the photolyase/CRY family members of Drosophila, as shown in Figure 4. As expected from a recent in vitro reconstitution experiment (24), Drosophila (6-4) photolyase expressed in Sf21 cells grown in standard medium contains FAD but lacks a second chromophore (Fig. 4A). However, when the enzyme is purified from cells grown in F0-containing medium it exhibits the characteristic enzyme-bound deazaflavin absorption spectrum (Fig. 4B). which also is in agreement with the in vitro reconstitution study (24). Significantly, and rather unexpectedly, when the Drosophila CPD photolyase, which is only distantly related to Drosophila (6-4) photolyase, was expressed in F0-supplemented medium, it also contained F0 in addition to FAD based on spectroscopic properties (Fig. 4C,D) and analysis by paper and electrophoretic chromatography (data not shown). In contrast, Drosophila CRY purified from Sf21 cells grown in media with or without F0 exhibited only the spectra of enzyme-bound FAD (Fig. 4E,F) with no indication of any F0 incorporation. This was somewhat surprising because phylogenetically, DmCRY is closer to (6-4) photolyase than to CPD photolyase. Regardless of the evolutionary implications, based on these findings we conclude that of the 3 members of the photolyase/cryptochrome family expressed in Drosophila, photolyases for both CPD and the (6-4) photoproduct contain the catalytic FAD cofactor and are capable of binding F0 which may function as a photoantenna whereas cryptochrome contains only the FAD cofactor which functions both as the photosensory and the effector chromophore/cofactor.
Figure 4.
Chromophores of CRY/photolyase family members from D. melanogaster. D. melanogaster (6-4) photolyase (A,B), CPD-photolyase (C,D), and CRY (E,F) were expressed in Sf 21 cells in the absence (A,C,E) or presence (B,D,F) of F0 in the growth medium. Spectra for the photolyases were recorded as in Figure 3, with panels A and C showing spectra of chromophores released by denaturation and the remaining panels showing spectra of native, purified enzymes. The spectra in B and D are dominated by the absorbance of F0, and the presence of FAD is indicated by its much weaker absorbance at approximately 366 nm. Protein concentrations before dilution were 1.3 mg/ml in B, 1.2 mg/ml in D, 0.9 mg/ml in E, and 0.9 mg/ml in F, as determined by comparing intensity of full-length proteins with known amounts of BSA on SDS-PAGE gels stained with Coomassie Blue. The sample in B was diluted ¼ before measuring; all other samples were measured undiluted.
Analysis of Plant and Vertebrate Cryptochromes for Second Chromophores
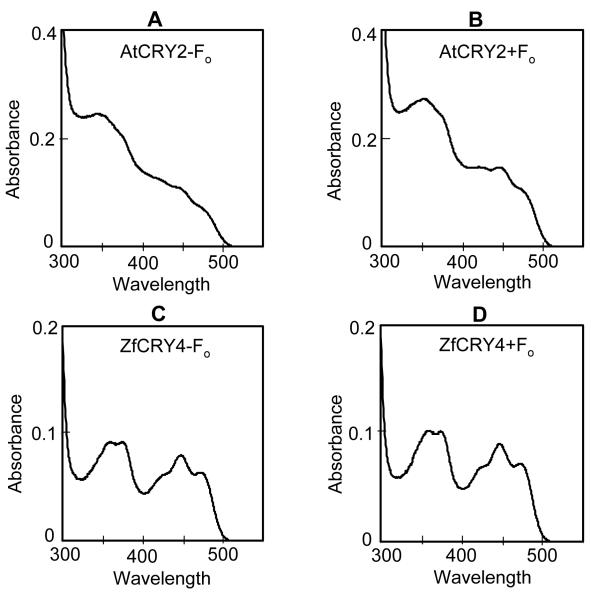
Most of the plant and vertebrate animal cryptochromes and insect Type 2 CRYs (which are phylogenetically related to vertebrate CRYs; 25) when expressed in heterologous systems contain grossly substoichiometric amounts of FAD making biochemical characterization of these CRYs rather challenging and raising some doubt about proper folding of these proteins in heterologous systems (26,27). Consequently, determination of cofactor composition of these proteins has been inconclusive. Rare exceptions to this generalization are Arabidopsis CRY2 (26) and the zebrafish CRY4 (17) which can be purified with stoichiometric FAD content and hence appear to be properly folded. Hence, we decided to test these CRYs for F0 incorporation in our in vivo system. The proteins were expressed in Sf21 cells, purified by affinity chromatography (Fig. 1) and analyzed for cofactor composition by spectroscopy. As seen in Figure 5, both AtCRY2 (A,B) and ZfCRY4 (C,D), expressed with and without F0, contained FAD as previously reported. Importantly, neither photoreceptor exhibited any indication of the presence of F0. Formally, it is possible that AtCRY2 and ZfCRY4 (as well as DmCRY) did in fact bind to F0 in vivo, but dissociated from the cofactor during purification. However, this seems unlikely and with this caveat, in summary, we find that our data is consistent with other findings in that all known photolyases characterized to date have two chromophores whereas, as of now, there is no evidence that CRYs from any source contain a second chromophore.
Figure 5.
Analysis of chromophores of AtCRY2 (A,B) and ZfCRY4 (C,D). Spectra of native proteins expressed in the absence (A,C) and presence (B,D) of F0 are shown.
DISCUSSION
The photolyase/cryptochrome family is an ancient family of flavoproteins, most of which employ blue light as a co-substrate/cofactor to carry out various cellular functions including DNA repair, plant growth and morphogenesis, circadian photoreception and core circadian clock control (1,3,4,8). In addition to the catalytic cofactor FAD, all CPD photolyases have been shown to contain a second chromophore in the form of MTHF, FAD, FMN, or F0 (8). However, there is no convincing evidence that the so-called Type 2 photolyases from animal and plant sources contain a second chromophore. In fact, based on an action spectrum for DNA repair with cell-free extracts prepared from Drosophila ovaries, one report suggested that Drosophila CPD photolyase contained deazaflavin as the second chromophore (28) while another report, again based mainly on an action spectra of recombinant enzyme purified from E. coli, claimed MTHF as the second chromophore (29). Interestingly, Drosophila is unique among the commonly used model organisms in having all 3 members of the PHR/CRY family, CPD photolyase, (6-4) photolyase, and cryptochrome. Equally interesting is the fact that while the two photolyases, which are phylogenetically quite distant (1,3,5), contain the same second chromophore, the CRY, which phylogenetically is closer to (6-4) photolyase than the CPD photolyase, does not bind F0. In fact, presently there is no convincing evidence that any bona fide cryptochrome contains a second chromophore. There are a few reports suggesting the presence of a second chromophore in some CRYs (18, 30-32). However, these reports relied mainly on fluorescence spectroscopy of proteins expressed in heterologous systems which revealed folate-like excitation and emission spectra. However, as folate is a relatively abundant cofactor the possibility that the detected fluorescence was due to low-level impurities in the affinity-purified CRY preparations was not eliminated. In addition, in all previous work on CRYs, the search for a second chromophore emphasized folate because deazaflavin was considered a rare cofactor synthesized only by some Archaea and a few photosynthetic bacteria (19). However, the recent appreciation that the F0 cofactor is widespread in nature (14) and that even if it is not made by a given organism it might be supplied by a symbiont, such as occurs in Drosophila (24), makes it a realistic candidate for a second chromophore in any member of the photolyase/CRY family (24). Thus, while our currently available data fail to reveal binding of CRY to either MTHF or F0 as a second chromophore, both of these, as well as FMN and FAD, remain as viable candidates for future studies on CRYs from a variety of sources. It is also possible that eventually, some novel compound(s) may be found to function as a second chromophore for a cryptochrome.
ACKNOWLEDGEMENTS
We thank Dr. R.H. White (Virginia Tech.) for the F0 synthase (fbiC) clone and Dr. Rui Ye for useful comments on the manuscript.
Abbreviations
- FAD
flavin adenine dinucleotide
- FADH2
reduced FAD
- FMN
flavin mononucleotide
- MTHF
methenyltetrahydrofolate
- CRY
cryptochrome
- CPD
cyclobutane pyrimidine dimer
- F0
7,8-didemethyl-8-hydroxy-5-deazariboflavin
- Dm
Drosophila melanogaster
- (6-4) PL
(6-4) phtotolyase
- AnPL
Anacystis nidulans photolyase
- ZfCRY4
zebrafish CRY4
- AtCRY2
Arabidopsis thaliana CRY2
- Dm64
Drosophila melanogaster (6-4) photolyase
- DmCPD
Drosophila melanogaster CPD photolyase
Footnotes
This work was supported by NIH grant GM31082
REFERENCES
- 1.Todo T. Functional diversity of the DNA photolyase/blue light receptor family. Res. 1999;434:89–97.1. doi: 10.1016/s0921-8777(99)00013-0. [DOI] [PubMed] [Google Scholar]
- 2.Cashmore AR. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 3.Lin C, Shalitin D. Cryptochrome structure and signal transduction. Ann. Rev. Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. Stucture and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 5.Partch CL, Sancar A. Photochemistry and photobiology of cryptochrome blue-light photopigments: The search for a photocycle. Photochem. Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 6.Essen LD, Klar T. Light-driven DNA repair by photolyases. Cell Mol. Life Sci. 2006;63:1266–1277. doi: 10.1007/s00018-005-5447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kao YT, Saxena C, Wang L, Sancar A, Zhong D. Femtochemistry in enzyme catalysis: DNA photolyase. Cell Biochem. Biophys. 2007;48:32–44. doi: 10.1007/s12013-007-0034-5. [DOI] [PubMed] [Google Scholar]
- 8.Sancar A. Structure and function of photolyase and in vivo enzymology. J.Biol. Chem. 2008;283:32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen L-O, van der Horst GTJ, Batschauer A, Ahmed M. The cryptochromes: blue light photoreceptors in plants and animals. Ann. Rev. Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 10.Taylor JS. Unraveling the molecular pathway from sunlight to skin cancer. Acc. Chem. Res. 1994;27:76–82. [Google Scholar]
- 11.Fujihashi M, Numoto N, Kobayashi Y, Mizushima A, Tsujimura M, Nakamura A, Kawarabayasi Y, Miki K. Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J. Molec. Biol. 2007;365:903–910. doi: 10.1016/j.jmb.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Ueda T, Kato A, Kuramitsu S, Terasawa H, Shimada I. Identification and characterization of a second chromophore of DNA photolyase from Thermus thermophilus HB27. J. Biol. Chem. 2005;280:36237–36243. doi: 10.1074/jbc.M507972200. [DOI] [PubMed] [Google Scholar]
- 13.Eker AP, Kooiman P, Hessels JKC, Yasui A. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J. Biol. Chem. 1990;265:8009–8015. [PubMed] [Google Scholar]
- 14.Graham DE, Xu H, White RH. Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F420 biosynthesis. Arch. Microbiol. 2003;180:455–464. doi: 10.1007/s00203-003-0614-8. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra K, Kim S-T, Walsh C, Sancar A. Roles of FAD and 8-hydroxy-5-deazaflavin chromophores in photoreactivation by Anacystis nidulans DNA photolyase. J. Biol. Chem. 1992;267:15406–15411. [PubMed] [Google Scholar]
- 16.Ozturk N, Song S-H, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 17.Ozturk N, Selby CP, Song S-H, Ye R, Tan C, Kao Y-T, Zhong D, Sancar A. Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry. 2009;48:8585–8593. doi: 10.1021/bi901043s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozgur S, Sancar A. Purification and properties of human blue-light photoreceptor cryptochrome 2. Biochemistry. 2003;42:2926–2932. doi: 10.1021/bi026963n. [DOI] [PubMed] [Google Scholar]
- 19.Walsh CT. Naturally occurring 5-deazaflavin coenzymes-Biological redox roles. Acc. Chem. Res. 1984;19:216–221. [Google Scholar]
- 20.Tamada T, Kitadokoro K, Higuchi Y, Inaka K, Yasui A, de Ruiter PE, Eker APM, Miki K. Crystal structure of DNA photolyase from Anacystis nidulans. Nature Structural Biol. 1997;4:887–891. doi: 10.1038/nsb1197-887. [DOI] [PubMed] [Google Scholar]
- 21.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klar T, Kaiser G, Hennecke U, Carell T, Batschauer A, Essen L-O. Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. Chem. Bio. Chem. 2006;7:1798–1806. doi: 10.1002/cbic.200600206. [DOI] [PubMed] [Google Scholar]
- 23.Petersen JL, Ronan PJ. Critical role of 7,8-didemethyl-8-hydroxy-5-deazariboflavin for photoreactivation in Chlamydomonas reinhardtii. J. Biol. Chem. 2010;285:32467–32475. doi: 10.1074/jbc.M110.146050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glas AF, Maul MJ, Cryle M, Barends TRM, Schneider S, Kaya E, Schlichting I, Carell T. The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes. Proc. Natl. Acad. Sci. USA. 2009;106:11540–11545. doi: 10.1073/pnas.0812665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Q, Metterville D, Briscoe AC, Reppert SM. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 26.Ozgur S, Sancar A. Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry. 2006;45:13369–13374. doi: 10.1021/bi061556n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozturk N, Song SH, Ozgur S, Selby CP, Morrison L, Partch C, Zhong D, Sancar A. Structure and function of animal cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 2007;72:119–131. doi: 10.1101/sqb.2007.72.015. [DOI] [PubMed] [Google Scholar]
- 28.Todo T, Ryo H, Takemori H, Toh H, Nomura T, Kondo S. High-level expression of the photorepair gene in Drosophila ovary and its evolutionary implications. Mutat. Res. 1994;315:213–228. doi: 10.1016/0921-8777(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim ST, Malhotra K, Ryo H, Sancar A, Todo T. Purification and characterization of Drosophila melanogaster photolyase. Mutat. Res. 1996;363:97–104. doi: 10.1016/0921-8777(96)00003-1. [DOI] [PubMed] [Google Scholar]
- 30.Malhotra K, Kim S-T, Batschauer A, Dawut L, Sancar A. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 31.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science. 1995;269:968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 32.Selby CP, Sancar A. A third member of the photolyase/blue-light photoreceptor family in Drosophila: a putative circadian photoreceptor. Photochem. Photobiol. 1999;69:105–107. [PubMed] [Google Scholar]