Abstract
Background
Cigarette smoking is a major risk factor for COPD and COPD severity. Previous genome-wide association studies (GWAS) have identified numerous single nucleotide polymorphisms (SNPs) associated with the number of cigarettes smoked per day (CPD) and a Dopamine Beta-Hydroxylase (DBH) locus associated with smoking cessation in multiple populations.
Objective
To identify SNPs associated with lifetime average and current CPD, age at smoking initiation, and smoking cessation in COPD subjects.
Methods
GWAS were conducted in 4 independent cohorts encompassing 3,441 ever-smoking COPD subjects (GOLD stage II or higher). Untyped SNPs were imputed using HapMap (phase II) panel. Results from all cohorts were meta-analyzed.
Results
Several SNPs near the HLA region on chromosome 6p21 and in an intergenic region on chromosome 2q21 showed associations with age at smoking initiation, both with the lowest p=2×10−7. No SNPs were associated with lifetime average CPD, current CPD or smoking cessation with p<10−6. Nominally significant associations with candidate SNPs within alpha-nicotinic acetylcholine receptors 3/5 (CHRNA3/CHRNA5; e.g. p=0.00011 for SNP rs1051730) and Cytochrome P450 2A6 (CYP2A6; e.g. p=2.78×10−5 for a nonsynonymous SNP rs1801272) regions were observed for lifetime average CPD, however only CYP2A6 showed evidence of significant association with current CPD. A candidate SNP (rs3025343) in the DBH was significantly (p=0.015) associated with smoking cessation.
Conclusion
We identified two candidate regions associated with age at smoking initiation in COPD subjects. Associations of CHRNA3/CHRNA5 and CYP2A6 loci with CPD and DBH with smoking cessation are also likely of importance in the smoking behaviors of COPD patients.
Keywords: Chronic Obstructive Pulmonary Disease (COPD), Genome Wide Association study (GWAS), smoking behaviors, Single Nucleotide Polymorphism (SNP)
INTRODUCTION
Chronic Obstructive Pulmonary Disease (COPD) is a common, genetically complex disease caused, and accelerated in its progression, predominantly by tobacco smoking. High smoking intensity, likely related at least in part to nicotine addiction, increases the risk of developing COPD. Although many patients quit smoking after they are diagnosed with COPD, some continue to smoke, placing them at high risk for continued disease progression.
Smoking behaviors, such as age at smoking initiation, smoking cessation, and number of cigarettes smoked per day (CPD), are partially genetically determined and have substantial heritability.1–4 Numerous loci and candidate genes have been suggested to contain genetic markers affecting smoking behaviors using genome-wide linkage scans 5 and genome wide association study approaches.6,7 Recent Genome Wide Association Studies (GWAS) have identified loci associated with smoking cessation (Dopamine Beta-Hydroxylase on chromosome 9q34) and CPD (e.g. nicotinic acetylcholine receptor locus on chromosome 15q25 and Cytochrome P2A6 (CYP2A6) locus on chromosome 19q13) in multiple populations,8–11 although GWAS of smoking behaviors specifically within COPD subjects have not been reported. Interestingly, the same single nucleotide polymorphisms (SNPs) in the 15q25 locus were previously associated with development of COPD,12 but the role of this locus in smoking behaviors in COPD patients and whether the sole effect of this locus on COPD susceptibility relates to smoking behavior remain unclear.
Due to their typically heavy lifetime smoking exposures, potentially related to (at least in part) enrichment in genetic variants responsible for nicotine addiction, COPD subjects can be considered as a unique population for studying smoking behaviors. However, diagnosis and further progression of COPD are likely to modify smoking status (i.e. increased efforts to quit smoking) and smoking intensity (e.g. reduction of CPD). Identifying genetic factors involved in smoking cessation is of special importance in clinical practice, since quitting smoking may reduce subsequent loss of lung function in COPD patients.13,14 Smoking cessation results in an improvement of respiratory symptoms in COPD patients, and is associated with reduced mortality due to COPD.13–16 Another smoking-related phenotype, age at smoking onset, correlates with nicotine dependence in adulthood 17 and mortality due to COPD.15 Taken together, it is of special interest to search for markers associated with smoking behaviors uniquely among COPD patients. Likewise, it is of importance to assess whether SNPs, regarded as established genetic determinants of smoking cessation and CPD in other populations, associate with these traits in COPD patients as well. Identification and description of such genetic variations may have significant consequences on future, targeted therapy aiming to reduce smoking among COPD patients.
The aim of the current study was to identify SNPs associated with age at smoking initiation, smoking cessation, current and lifetime average CPD among COPD subjects, using GWAS in four independent cohorts: National Emphysema Treatment Trial (NETT), Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE), GenKOLS cohort from Bergen, Norway, and COPDGene. We hypothesized that a subset of smoking behavior genetic determinants in other population samples would influence smoking behavior in COPD subjects.
METHODS
Subjects and phenotypes
Ever-smoking, Caucasian subjects with at least moderate COPD (GOLD stage II or higher) from four independent cohorts (NETT,18 ECLIPSE,19 GenKOLS cohort from Bergen, Norway,20 and COPDGene (first 1000 subjects)21) were studied (Table 1). In total 3,441 subjects had at least 1 (out of 4) non-missing, smoking-related phenotype (Table 1). All phenotypes were self-reported using either Case Report Form (smoking status and the lifetime average CPD in the ECLIPSE cohort) or modified versions of the American Thoracic Society/Division of Lung Diseases Respiratory Disease Questionnaire.22 Since current smokers were not eligible for the NETT study, this cohort did not contribute to the analyses on smoking cessation and current CPD.
Table 1.
Characteristics of COPD subjects and smoking-related phenotypes studied
| Cohort | ||||||
|---|---|---|---|---|---|---|
| NETT n=362* |
Norway n=851* |
ECLIPSE n=1,734* |
COPDGene n=494* |
|||
| Characteristics | Males* | N (%) | 234 (64.6) | 511 (60.0) | 1,160 (66.9) | 242 (49.0) |
| Age in years* | Mean (SD) | 67.4 (5.8) | 65.5 (10.1) | 63.7 (7.1) | 64.7 (8.1) | |
| Pack-years smoked* | Mean (SD) | 66.1 (30.9) | 32.1 (18.6) | 50.4 (27.4) | 54.8 (26.8) | |
| Post-bronchodilator FEV1 (% pred.)* | Mean (SD) | 29.1 (7.8) | 50.7 (17.5) | 44.8 (14.7) | 48.7 (18.4) | |
| Post-bronchodilator FEV1/FVC* | Mean (SD) | 0.32 (0.06) | 0.51 (0.13) | 0.45 (0.12) | 0.48 (0.13) | |
| Enrollment | Years(s) | 1998–2002 | 2003–2005 | 2005–2007 | 2008–2009 | |
| Genotyping technology | Chip | Illumina Quad 610 | Illumina HumanHap 550 V1, V3, and Duo | Illumina HumanHap 550 V3 | Illumina Human Omni1-Quad | |
| Phenotype studied | Age at smoking initiation | Mean (SD) | 16.6 (3.6) | 18.7 (5.1) | 16.9 (4.4) | 16.8 (4.4) |
| Number of subjects with non-missing phenotype | 362 | 851 | 1,690 | 494 | ||
| Lambda inflation factor | 0.986 | 0.989 | 1.019 | 0.997 | ||
| Lifetime average CPD | Mean (SD) | 32.4 (13.5) | 15.7 (7.8) | 25.5 (12.4) | 27.6 (11.8) | |
| Number of subjects with non-missing phenotype | 361 | 851 | 1,734 | 494 | ||
| Lambda inflation factor | 1.002 | 0.996 | 1.018 | 0.996 | ||
| Smoking cessation | Current smokers, n (%) | 0 (0) | 404 (47.5) | 610 (35.3) | 150 (30.6) | |
| Former Smokers, n (%) | 362 (100) | 447 (52.5) | 1120 (64.7) | 340 (69.4) | ||
| Lambda inflation factor | - | 1.000 | 0.998 | 0.995 | ||
| Current CPD† | Mean (SD) | - | 13.1 (6.9) | 15.6 (10.8) | 18.4 (12.4) | |
| Number of subjects with non-missing phenotype | - | 398 | 565 | 150 | ||
| Lambda inflation factor | - | 0.995 | 1.005 | 0.997 | ||
calculated for subjects with at least 1 non-missing phenotype
for subjects with reported current number of cigarettes smoked per day > 0
SD=Standard Deviation; FEV1=Forced Expiratory Volume in 1 second; FVC=Forced Vital Capacity; CPD=number of cigarettes smoked per day
Genotyping and quality control (QC)
Different genotyping chips from Illumina (Illumina Inc., San Diego, CA) were used (table 1). QC steps were previously described in detail for the NETT, Norwegian and ECLIPSE cohorts,23 and were applied to the COPDGene study as well. Briefly, QC steps consisted of filtering SNPs based on missing call rates (>5%) and Hardy-Weinberg Equilibrium deviation (p<10−8), and filtering subjects based on genotyping call rate (<95%), sex discrepancy, unexpected relatedness (PLINK pi-hat cutoff of 0.125), and ethnicity. Removal of cases that were outliers for genetic ancestry was performed based on principal components (PCs) analysis in cases only. In the primary analysis, untyped markers were imputed using 120 founder Caucasian (Centre d’Etude du Polymorphisme Humain (Utah residents with ancestry from northern and western Europe) (CEU)) haplotypes from HapMap reference panel (phase II) and, as a secondary analysis, using 1000 Genomes Project data.24,25 We limited our analysis to SNPs genotyped/imputed in at least 2 cohorts, with imputation r2 coefficient ≥0.3 (for imputed SNPs only). Overall, approximately 2.5 and 6.3 million SNPs per phenotype were analyzed using the reference HapMap Project and 1000 Genomes Project panels, respectively.
Association analysis of candidate SNPs
We extracted candidate SNPs achieving genome-wide significance in the previous studies on smoking cessation and CPD.8–11,26 Following previous GWAS,8,9 we additionally extracted two SNPs from CYP2A6 (rs1801272 (Leu160His)) and CHRNA5 (rs588765) based on their biological function. Since a candidate SNP from the DBH locus and most of the candidate SNPs from the CYP2A6 locus were imputed in all cohorts, we searched for the best proxy SNP genotyped in at least 3 of the cohorts.
Statistics
According to Box-Cox transformation 27 that identified approximately the best transformation of dependent variables, which could be applied to all cohorts, we studied log2-transformed age at smoking initiation and lifetime average CPD, and the square root of the current CPD. Regression models were run under an additive model coded SNPs, while adjusting for potential confounders (see online supplementary material for details). A fixed effect model was used for all meta-analyses. Effect allele was defined as the one associated with later age at smoking initiation, higher CPD or higher odds for smoking cessation. Meta-analytic p<5×10−8 was considered as genome-wide significant;28 5×10−8 ≤p<5×10−7 was interpreted as a suggestive association in the genome-wide panel with p<0.05 a suggestive association for candidate SNPs.
Software
Box-Cox transformations were performed with MASS package 27, while lambda inflation factors were calculated with GenABEL package 29 in R (ver. 2.10.1).30 SNP imputation was performed with MACH (ver. 1.0).31 PCs, reflecting genetic structure of each study population, were calculated and analyzed with the EIGENSOFT package (ver. 2.0).32 Genetic association analyses and meta-analyses were run with PLINK (ver. 1.0.7).33,34 SNAP (ver. 2.2) 35 was used to search for proxy SNPs (r2≥0.8) using the Caucasian HapMap phase II panel and to assess linkage disequilibrium (LD) coefficients in 1000 Genomes Project data. SNP/gene annotations and regional association plots were created with LocusZoom (ver. 1.1; human genome build hg18).36
RESULTS
Age at smoking initiation
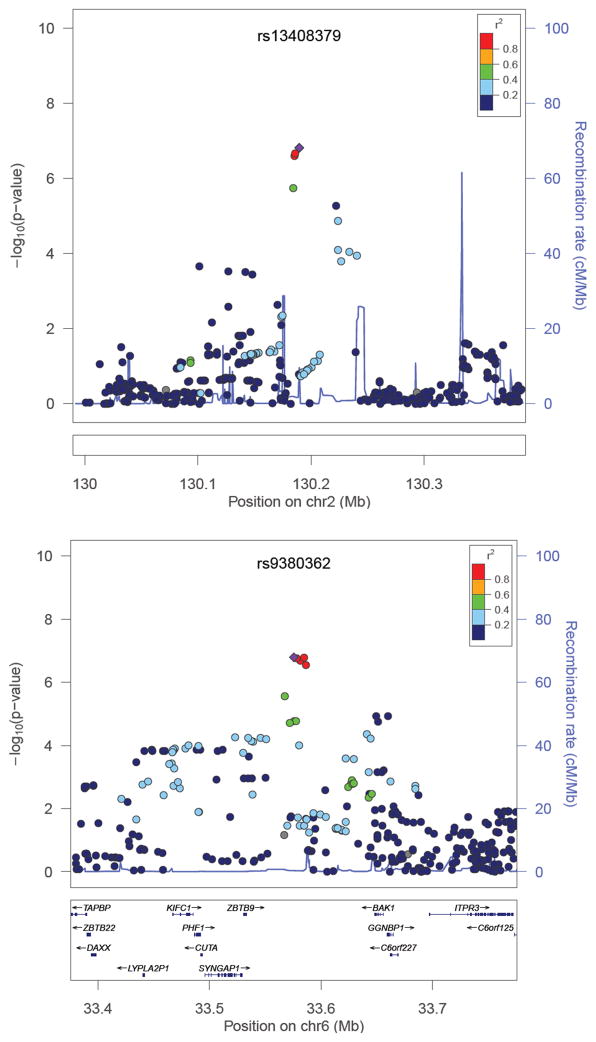
Analyses in the individual cohorts were adjusted for sex and principal components for genetic ancestry. In total, meta-analysis included 3,397 subjects for the age at smoking initiation phenotype. Lambda inflation factors were between 0.986 and 1.019 for individual cohorts (table 1), and 1.002 for meta-analytic p values (see online supplementary figure 1 for a Q-Q plot of meta-analysis). No SNPs showed meta-analytic association p values below the genome-wide significance level. Numerous SNPs in the intergenic region on chromosome 2q21 and in the region between BCL2-antagonist/killer 1 (BAK1) and Zinc finger and BTB domain containing 9 (ZBTB9) on chromosome 6p21 were associated with age at smoking initiation at a suggestive significance level (figure 1 and online supplementary table 1). In total 24 SNPs were associated with age at smoking initiation with p<10−5 in the meta-analysis (online supplementary table 1).
Figure 1. Regional association plots of two loci including SNPs associated with age at smoking initiation with meta-analytic p<10−6.
Dots correspond to meta-analyzed SNPs. Color of dots corresponds to r2 (linkage disequilibrium (LD) coefficient in the HapMap II CEU population; grey dots correspond to SNPs with missing LD information) relative to the most significantly associated SNP (depicted with diamond) in the region i.e. rs13408379 (top figure) and rs9380362 (bottom figure).
Lifetime average CPD
Analyses in single cohorts were adjusted for sex, age and principal components for genetic ancestry. In total, meta-analysis included 3,440 subjects for the lifetime average CPD phenotype. Lambda inflation factors were between 0.996 and 1.018 for individual cohorts (table 1), and 1.019 for meta-analytic p values (see online supplementary figure 2 for a Q-Q plot of meta-analysis). Thirteen SNPs were associated with lifetime average CPD with meta-analytic p<10−5, however, no SNPs showed association with meta-analytic p<10−6 (table 2).
Table 2.
SNPs associated with the lifetime average CPD (log2-transformed) with meta-analytic p<10−5, and candidate SNPs from CHRNA3/CHRNA5 and CYP2A6 loci identified by previous studies
| Chr. | Top SNP | Nearest gene within 50kb (location) | Effect/Non- Effect allele | B | p | I2 | Q stat. | N/Nimp | Freq. | Effect direction consistent with previous studies | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Top SNPs | 4 | rs13104971 | SCFD2 (intron) | G/A | 0.113 | 1.18×10−6 | 0 | 0.92 | 4/3 | 0.13 | - |
| 9 | rs943306 | ASTN2 (intron) | T/C | 0.073 | 1.25×10−6 | 25 | 0.26 | 4/0 | 0.40 | - | |
| 7 | rs10237067 | - | A/G | 0.072 | 2.98×10−6 | 0 | 0.63 | 4/3 | 0.63 | - | |
| 7 | rs12699125 | CALN1 (intron) | A/G | 0.072 | 3.17×10−6 | 42 | 0.16 | 4/3 | 0.66 | - | |
| 7 | rs4718827 | - | T/C | 0.072 | 3.36×10−6 | 0 | 0.61 | 4/4 | 0.63 | - | |
| 7 | rs4717631 | CALN1 (intron) | A/G | 0.072 | 3.60×10−6 | 43 | 0.15 | 4/4 | 0.66 | - | |
| 7 | rs17170849 | ELMO1 (intron) | C/T | 0.265 | 4.07×10−6 | 44 | 0.15 | 4/3 | 0.02 | - | |
| 7 | rs4577845 | CALN1 (intron) | G/A | 0.071 | 4.57×10−6 | 44 | 0.15 | 4/1 | 0.66 | - | |
| 7 | rs1859485 | CALN1 (intron) | T/C | 0.069 | 6.95×10−6 | 40 | 0.17 | 4/1 | 0.65 | - | |
| 12 | rs11044734 | - | G/C | 0.112 | 7.32×10−6 | 0 | 0.48 | 4/4 | 0.90 | - | |
| 12 | rs11044737 | - | G/A | 0.112 | 7.70×10−6 | 0 | 0.47 | 4/0 | 0.90 | - | |
| 16 | rs190369 | - | C/T | 0.161 | 8.89×10−6 | 47 | 0.13 | 4/1 | 0.05 | - | |
| 20 | rs2869961 | CEBPB (5’ region) | A/G | 0.104 | 9.65×10−6 | 0 | 0.59 | 4/1 | 0.12 | - | |
| Candidate SNPs | 15 | rs1051730 | CHRNA3 (exon, Tyr215Tyr) | A/G | 0.060 | 0.00011 | 37 | 0.19 | 4/0 | 0.41 | Yes |
| 15 | rs16969968 | CHRNA5 (exon, Asp398Asn) | A/G | 0.059 | 0.00015 | 30 | 0.23 | 4/3 | 0.41 | Yes | |
| 15 | rs8034191 | AGPHD1 (intron) | C/T | 0.055 | 0.00036 | 40 | 0.17 | 4/0 | 0.41 | Yes | |
| 15 | rs684513 | CHRNA5 (intron) | C/G | 0.029 | 0.163 | 0 | 0.98 | 4/4 | 0.80 | Yes | |
| 15 | rs578776 | CHRNA3 (3’ UTR) | G/A | 0.041 | 0.020 | 0 | 0.94 | 4/0 | 0.76 | Yes | |
| 15 | rs588765 | CHRNA5 (intron) | C/T | 0.031 | 0.046 | 47 | 0.13 | 4/4 | 0.60 | Yes | |
| 19 | rs3733829 | EGLN2 (intron) | G/A | 0.019 | 0.223 | 0 | 0.48 | 4/3 | 0.39 | Yes | |
| 19 | rs7937 | RAB4B (3’ UTR) | T/C | 0.022 | 0.153 | 0 | 0.96 | 4/0 | 0.60 | Yes | |
| 19 | rs1801272 | CYP2A6 (exon, Leu160His) | A/T | 0.266 | 2.78×10−5 | 0 | 0.72 | 4/4 | 0.96 | Yes | |
| 19 | rs4105144 | CYP2A6 (5’ region) | C/T | 0.073 | 3.92×10−5 | 0 | 0.44 | 4/4 | 0.73 | Yes | |
| 19 | rs7260329 | CYP2B6 (intron) | G/A | 0.014 | 0.404 | 0 | 0.55 | 4/1 | 0.68 | Yes | |
| 19 | rs7251570 | CYP2A6 (3’ region) | G/A | 0.057 | 0.00073 | 0 | 0.84 | 4/4 | 0.71 | Yes | |
| 19 | rs12461383 | CYP2A7 (3’ region) | G/C | 0.063 | 0.00019 | 0 | 0.61 | 4/4 | 0.60 | Yes |
Analyses were adjusted for sex, age and principal components for genetic ancestry.
N/Nimp = Number of studies contributing to meta-analysis / number of studies where SNP was imputed; I2=heterogeneity index; Q stat.=p value for Q statistic; p=p value from the fixed effect meta-analysis; SNP=Single Nucleotide Polymorphism; CPD=number of cigarettes smoked per day; Freq. = Effect allele frequency in 3,441 subjects with at least 1 non-missing phenotype from 4 cohorts studied; Chr.=Chromosome; B=regression coefficient; UTR=Untranslated Region; CHRNA3/CHRNA5=alpha-nicotinic acetylcholine receptor 3/5; CYP2A6=Cytochrome P450 2A6
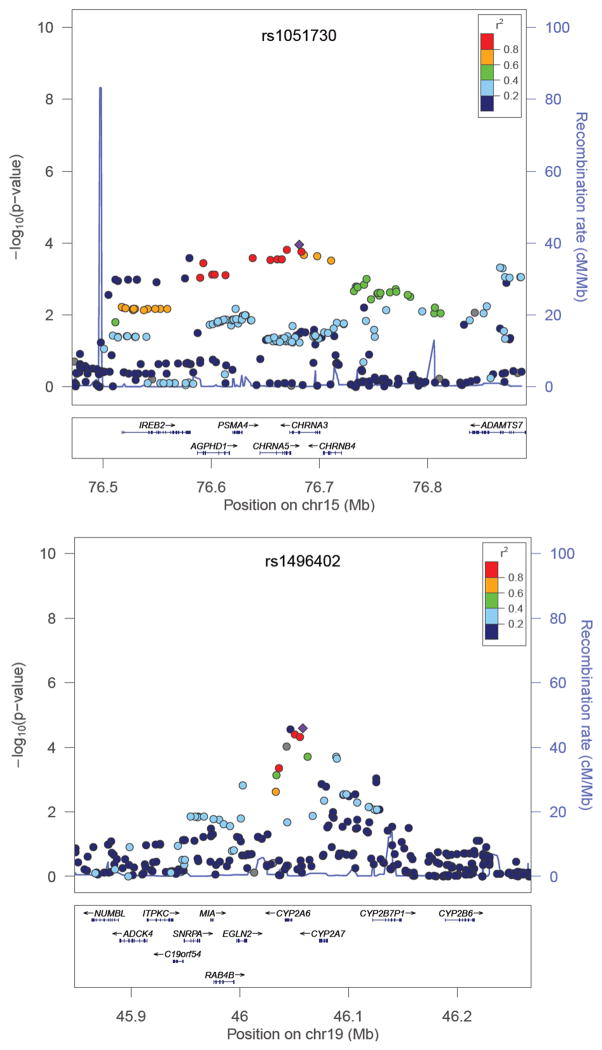
Candidate SNPs from the nicotinic acetylcholine receptor locus on chromosome 15q25 (rs578776, rs588765, rs8034191, rs1051730 and rs16969968) were significantly (p<0.05) associated with lifetime average CPD with the same direction of effect as seen in previous GWAS on CPD (table 2). The synonymous rs1051730 SNP was the top associated SNP in this locus (figure 2).
Figure 2. Regional association plots of the candidate loci centered on CHRNA3 (top) and CYP2A6 (bottom) and their associations with lifetime average number of cigarettes smoked per day.
Dots correspond to meta-analyzed SNPs. Color of dots correspond to r2 (linkage disequilibrium (LD) coefficient in the HapMap II CEU population; grey dots correspond to SNPs with missing LD information) relative to the most significantly associated SNP in the region (depicted with diamond) i.e. rs1051730 in the CHRNA3 (top figure) and rs1496402 (bottom figure; the second top SNP in this region is a nonsynonymous (Leu160His) rs1801272 SNP in the CYP2A6).
CHRNA3=alpha-nicotinic acetylcholine receptor 3; CYP2A6=Cytochrome P450 2A6
Four candidate SNPs (rs7251570, rs4105144, rs1801272 and rs12461383) in the CYP2A6 locus on chromosome 19q13 were significantly (p<0.05) associated with lifetime average CPD with the same direction of effect as seen in previous GWAS on CPD (table 2). A non-synonymous SNP (rs1801272) was the second most significantly associated SNP in this locus (figure 2, table 2). There is, at most, a moderate level of LD between these 4 SNPs according to the HapMap phase II panel (r2≤0.6 and D’≥0.84, see online supplementary figure 3). Analysis of proxy SNPs that were genotyped in at least three cohorts was performed. Analysis of rs8102683 (genotyped in the NETT, ECLIPSE and Norwegian cohorts, and imputed in COPDGene) that is a proxy for rs4105144 (r2=0.87), and rs7251418 (genotyped in all cohorts) that is a proxy for rs7251570 (r2=0.81) confirmed the associations observed (p= 4.65×10−5 (B=0.074 for the “C” effect allele; I2=0) and p=0.0024 (B=0.054 for the “G” effect allele; I2=0), respectively). No genotyped proxy SNPs could be found for rs12461383 and rs1801272 SNPs. We did not replicate, with a nominal p<0.05, associations between SNPs in 7p14, 8p11 and 10q23 loci, reported in previous GWAS on CPD in our analysis on lifetime average CPD (online supplementary table 2).
Smoking cessation
Analyses in single cohorts were adjusted for age, % of predicted FEV1 and principal components summarizing genetic ancestry. In total, meta-analysis included 1,164 current smokers (defined as “No” for smoking cessation) and 1,907 former smokers (defined as “Yes” for smoking cessation). Lambda inflation factors were between 0.995 and 1.000 for individual cohorts (table 1), and 0.998 for meta-analytic p values (see online supplementary figure 4 for a Q-Q plot of meta-analysis). No SNPs showed association with meta-analytic p<10−6 (table 3). Candidate SNP rs3025343 in the DBH locus showed nominally significant association with smoking cessation with the same direction of effect as seen in recent GWAS, yet with substantial heterogeneity between studies (table 3). SNP rs3025316, a proxy for rs3025343 (r2=0.94), was genotyped in all cohorts and confirmed this association (p= 0.002 (OR= 1.32 for the “T” effect allele; I2=18).
Table 3.
SNPs associated with smoking cessation with meta-analytic p<10−5, and a candidate SNP (last row) fromDopamine Beta-Hydroxylase (DBH) locus identified by previous Genome Wide Association studies (GWAS) on smoking cessation
| Chr. | Top SNP | Nearest gene within 50kb | Effect/Non- Effect allele | OR | p | I2 | Q stat | N/Nimp | Freq. | Effect direction consistent with previous GWAS |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | rs10794613 | FLJ46361 (5’ region) | G/C | 1.43 | 3.41×10−6 | 0 | 0.51 | 3/3 | 0.21 | - |
| 3 | rs13064954 | CCNL1 (3' region) | A/G | 1.94 | 5.28×10−6 | 0 | 0.60 | 3/2 | 0.05 | - |
| 10 | rs1896376 | CPMX2 (intron) | C/T | 1.83 | 5.71×10−6 | 57 | 0.10 | 3/2 | 0.95 | - |
| 13 | rs9506942 | - | C/G | 1.29 | 5.96×10−6 | 0 | 0.72 | 3/3 | 0.57 | - |
| 13 | rs9552733 | - | G/A | 1.29 | 5.99×10−6 | 0 | 0.69 | 3/0 | 0.57 | - |
| 3 | rs9866141 | VEPH1 (3'region) | T/C | 1.88 | 7.99×10−6 | 0 | 0.66 | 3/3 | 0.06 | - |
| 3 | rs1165640 | - | C/T | 1.56 | 8.98×10−6 | 0 | 0.57 | 3/0 | 0.10 | - |
| 12 | rs10861185 | TXNRD1 (intron) | C/A | 1.29 | 9.57×10−6 | 16 | 0.31 | 3/2 | 0.57 | - |
| 10 | rs727417 | CPXM2 (intron) | C/G | 1.71 | 9.67×10−6 | 38 | 0.20 | 3/3 | 0.94 | - |
| 9 | rs3025343 | DBH (5’ region) | G/A | 1.24 | 0.015 | 46 | 0.16 | 3/3 | 0.87 | Yes |
Analyses were adjusted for age, % of predicted FEV1 and principal components for genetic ancestry. Current smokers were considered as “controls”, while former smokers were considered as “cases”.
N/Nimp = Number of studies contributing to meta-analysis / number of studies where SNP was imputed; I2=heterogeneity index; Q stat.=p value for Q statistic; p=p value from the fixed effect meta-analysis; SNP=Single Nucleotide Polymorphism; Freq. = Effect allele frequency in 3,441 subjects with at least 1 non-missing phenotype from 4 cohorts studied; Chr.=Chromosome; OR=Odds Ratio
Current CPD
Analyses were adjusted for sex, age, % of predicted FEV1, and principal components for genetic ancestry. In total, meta-analysis included 1,113 current smokers for the current CPD phenotype. Lambda inflation factors were between 0.995 and 1.005 for individual cohorts (table 1), and 1.010 for meta-analytic p values (see online supplementary figure 5 for a Q-Q plot of meta-analysis). No SNPs showed meta-analytic association with p<10−6 (table 4). Among candidate SNPs, rs12461383 from the CYP2A6 locus was significantly associated with current CPD with the same effect direction as compared to previous GWAS on CPD (table 4). Candidate SNPs rs1801272 and rs7251570 showed borderline significant association with the same effect direction as compared to previous GWAS on CPD (table 4). We did not replicate, at a nominal p< 0.05, associations between SNPs in 7p14, 8p11 and 10q23 loci reported in previous GWAS on CPD in our analysis on current CPD (online supplementary table 3).
Table 4.
SNPs associated with the current CPD (square-root transformed) with meta-analytic p<10-5, and candidate SNPs from CHRNA3/CHRNA5 and CYP2A6 loci identified by previous studies
| Chr. | Top SNP | Nearest gene within 50kb | Effect/Non- Effect allele | B | p | I2 | Q stat. | N/Nimp | Freq. | Effect direction consistent with previous studies | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Top SNPs | 3 | rs1881681 | - | C/A | 0.363 | 1.22×10−6 | 33 | 0.22 | 3/0 | 0.88 | - |
| 8 | rs11984631 | - | T/G | 1.351 | 1.56×10−6 | 37 | 0.21 | 2/0 | 0.99 | - | |
| 2 | rs12615264 | SOCS5 (3’ region) | T/G | 0.644 | 3.46×10−6 | 0 | 0.38 | 3/3 | 0.03 | - | |
| 2 | rs11682595 | SOCS5 (3’ region) | T/G | 0.642 | 3.49×10−6 | 0 | 0.41 | 3/3 | 0.03 | - | |
| 2 | rs11125090 | SOCS5 (3’ region) | A/G | 0.641 | 3.58×10−6 | 0 | 0.42 | 3/2 | 0.03 | - | |
| 20 | rs297755 | - | G/A | 0.291 | 4.86×10−6 | 49 | 0.14 | 3/3 | 0.17 | - | |
| 4 | rs3893377 | BST1 (3’ region) | C/T | 0.289 | 4.90×10−6 | 51 | 0.13 | 3/3 | 0.80 | - | |
| 4 | rs10019008 | BST1 (3’ region) | C/T | 0.288 | 4.99×10−6 | 53 | 0.12 | 3/0 | 0.80 | - | |
| 4 | rs11947310 | BST1 (3’ region) | A/C | 0.289 | 5.21×10−6 | 53 | 0.12 | 3/3 | 0.80 | - | |
| 4 | rs10018756 | BST1 (3’ region) | A/T | 0.275 | 7.71×10−6 | 48 | 0.15 | 3/3 | 0.77 | - | |
| Candidate SNPs | 15 | rs1051730 | CHRNA3 (exon, Tyr215Tyr) | G/A | 0.010 | 0.851 | 32 | 0.23 | 3/0 | 0.59 | No |
| 15 | rs16969968 | CHRNA5 (exon, Asp398Asn) | G/A | 0.009 | 0.863 | 30 | 0.24 | 3/2 | 0.59 | No | |
| 15 | rs8034191 | AGPHD1 (intron) | T/C | 0.006 | 0.914 | 5 | 0.35 | 3/0 | 0.59 | No | |
| 15 | rs684513 | CHRNA5 (intron) | C/G | 0.100 | 0.153 | 0 | 0.78 | 3/3 | 0.80 | Yes | |
| 15 | rs578776 | CHRNA3 (3’ UTR) | G/A | 0.052 | 0.388 | 0 | 0.98 | 3/0 | 0.76 | Yes | |
| 15 | rs588765 | CHRNA5 (intron) | T/C | 0.048 | 0.352 | 23 | 0.27 | 3/3 | 0.40 | No | |
| 19 | rs3733829 | EGLN2 (intron) | G/A | 0.005 | 0.930 | 40 | 0.19 | 3/2 | 0.39 | Yes | |
| 19 | rs7937 | RAB4B (3’ UTR) | T/C | 0.027 | 0.590 | 0 | 0.37 | 3/0 | 0.60 | Yes | |
| 19 | rs1801272 | CYP2A6 (exon, Leu160His) | A/T | 0.362 | 0.063 | 0 | 1.00 | 3/3 | 0.96 | Yes | |
| 19 | rs4105144 | CYP2A6 (5’ region) | C/T | 0.069 | 0.217 | 0 | 0.55 | 3/3 | 0.73 | Yes | |
| 19 | rs7260329 | CYP2B6 (intron) | G/A | 0.005 | 0.922 | 0 | 0.76 | 3/0 | 0.68 | Yes | |
| 19 | rs7251570 | CYP2A6 (3’ region) | G/A | 0.105 | 0.052 | 0 | 0.87 | 3/3 | 0.71 | Yes | |
| 19 | rs12461383 | CYP2A7 (3’ region) | G/C | 0.143 | 0.008 | 0 | 0.62 | 3/3 | 0.60 | Yes |
Analyses were adjusted for sex, age, % of predicted FEV1 and principal components for genetic ancestry. N/Nimp =Number of studies contributing to meta-analysis / number of studies where SNP was imputed; I2=heterogeneity index; Q stat.=p value for Q statistic; p=p value from the fixed effect meta-analysis; SNP=Single Nucleotide Polymorphism; CPD=number of cigarettes smoked per day; Freq. = Effect allele frequency in 3,441 subjects with at least 1 non-missing phenotype from 4 cohorts studied; Chr.=Chromosome; B=regression coefficient; UTR=Untranslated Region;CHRNA3/CHRNA5=alpha-nicotinic acetylcholine receptor 3/5;CYP2A6=Cytochrome P450 2A6
Analyses of SNPs Imputed with 1000 Genomes Project Data
Analysis of all phenotypes using SNP genotypes imputed using the 1000 Genomes Project revealed 20 additional associations below the suggestive genome-wide significance level (supplementary table 4). SNP rs9394152 in the 6p21 locus was the most significantly associated SNP with the age at smoking initiation (meta-analytic p=6.55×10−8) and, similarly to the majority of SNPs associated with this phenotype below suggestive genome-wide significance level, was characterized by high (≥0.96), cohort-specific imputation r2 coefficients (supplementary table 4). We observed novel associations of 3 SNPs with the lifetime average CPD; the most significantly associated SNP rs28675338 (r2=0.07 and D’=1.00 to rs1051730 according to 1000 Genomes Project Data) maps to the 15q25 locus. We also found one SNP associated with smoking cessation, with meta-analytic p<5×10−7 (supplementary table 4). Since cohort-specific, imputation r2 coefficients of the 4 SNPs, associated with lifetime average CPD and smoking cessation below suggestive genome-wide significance level, were rather modest (range 0.35–0.64; supplementary table 4), the observed associations certainly require additional confirmation and should be interpreted with caution. The three SNPs associated with current CPD with meta-analytic p<5×10−7 map to regions on chromosomes 2 and 3 (supplementary table 4), which were already ranked as top loci using the HapMap II reference panel (table 4).
DISCUSSION
Cigarette smoking is the most important environmental risk factor for COPD, and smoking cessation is the most important therapeutic intervention to prevent its progression. Our current study identified two loci on chromosomes 2q21 and 6p21 as candidates for containing genes influencing age at smoking initiation in COPD patients, both showing suggestive levels of genome-wide significance. Furthermore, this study confirmed that certain SNPs in the nicotinic acetylcholine receptor locus on chromosome 15q25 and SNPs in the CYP2A6 locus on chromosome 19q13 were associated with CPD in COPD patients. Of importance, we additionally confirmed in COPD subjects the previously reported association of a marker (rs3025343) in the DBH locus with smoking cessation.
The two loci mapping to chromosomes 2q21and 6p21showed evidence of association with age at smoking initiation at a suggestive genome-wide significance level, using both HapMap II and 1000 Genomes projects reference imputation panels. The association peaks found here do not localize within any known genes, suggesting distant regulation of yet unidentified target genes may be involved. SNPs from chromosome 6p21 are located close to the Human Leukocyte Antigen locus, and ZBTB9 and BAK1 are the closest genes to the association peak. The BAK1 gene, encoding a proapoptotic protein, has been previously associated by GWAS with testicular germ cell tumors 37 and platelet number.38 Yet SNPs identified in these studies do not correlate, in terms of LD, with top SNPs from our age at smoking initiation GWAS.
Analysis of candidate SNPs, with respect to the lifetime average CPD, revealed that the majority of those SNPs in the CHRNA3/CHRNA5 (15q25) and CYP2A6 (19q13) regions significantly associated with this trait showed the same direction of effect described previously.8–11,26 However, SNPs from the 7p14, 8p11 (CHRNB3/CHRNA6 locus) and 10q23, as well as some SNPs from the CYP2A6 and CHRNA3/CHRNA5 loci, were not replicated in the current study with a nominal significance threshold. This may be caused by effect sizes that are too small to be detected in our populations or potentially to different genetic determinants of smoking behaviors within COPD subjects.
Besides the “neutral” or reference haplotype, it has been suggested there are at least 2 other haplotypes with independent effects on CPD in this 15q25 region.26,39 Our study confirms previous observations that rs1051730, and tagging SNPs such as rs16969968, exhibit relatively strong effects in this region, and SNPs rs588765 and rs578776, determine independent haplotypes associated with lifetime CPD in COPD patients as well. However, we could not replicate association between lifetime average CPD and rs684513, i.e. the top SNP in the analysis on CPD when conditioning for rs1051730 in the Tobacco and Genetics Consortium.10 Importantly, the effect directions of all candidate SNPs in this 15q25 locus agree with those previously reported, suggesting larger COPD cohorts are required to establish significance of independent SNPs/haplotypes in this region.
Interestingly, effect sizes of replicated SNPs from the CHRNA3/CHRNA5 locus are similar to most of those from the CYP2A6 locus in our study, which contrasts with recent GWAS where bigger effect sizes were seen for the former locus.8 Additionally, variations in the CHRNA3/CHRNA5, but not CYP2A6, locus showed substantial heterogeneity in genetic effects in our study, and as observed in previous GWAS as well.8,10 We speculate that this might be due to between-study differences in general (e.g. educational status or peer smoking40) or COPD-specific (e.g. reporting lifetime average CPD which may be influenced by severity of disease) characteristics.
Analysis of current CPD, which was much less powered than lifetime CPD due to a lower number of subjects, was still able to detect some evidence of association with markers near CYP2A6, yet not for those located in the CHRNA3/CHRNA5 region. CYP2A6 is an enzyme primarily responsible for conversion of nicotine to cotinine in the liver, and rs1801272 (Leu160His) codes for the CYP2A6*2 allele, which inactivates the enzyme.41 This SNP showed the largest effect size on both current and lifetime average CPD among all analyzed candidate SNPs in the region. Leu160His is in LD (D’=1.0) with other SNPs that were replicated in the CYP2A6 locus. This agrees with previous GWAS and suggests that the rs1801272 SNP may be a true causative variant, while the other associations observed may be due to partial tagging by this SNP.8 Importantly, we show that the genotyped proxy SNPs in the CYP2A6 locus confirmed our analysis on imputed SNPs with respect to lifetime average CPD. The lack of convincing association for the proxy SNP rs7251418 with current CPD may be explained by the relatively smaller sample size in this analysis, and incomplete LD between the target and the proxy SNPs. Previous GWAS suggested that other genes may be associated with CPD in the 19q13 region, yet we observed no nominally significant associations for EGLN2, RAB4B and CY2B6.
Analysis of smoking cessation did not reveal any loci associated below the suggestive significance level, and it showed that the previously reported 10 association between rs3025343 in the DBH locus can be replicated in COPD subjects with a nominal significance threshold. DBH is a plausible candidate gene for smoking cessation since it participates in the metabolism of dopamine. This neurotransmitter is released from neurons in response to nicotine,3 and is an important mediator of addiction behaviors such as smoking. The rs3025343 SNP that we extracted showed a substantial heterogeneity in this effect, which may reflect the between-study differences in factors such as use of nicotine replacement therapy or socio-economic status. Interestingly, the best proxy SNP rs3025316 (genotyped in all cohorts) showed a more pronounced effect with somewhat smaller heterogeneity index compared to the rs3025343.
Our study possesses several limitations, and some of those are typical for many GWAS. For example, the size of the current study may have been too small to detect associations at a genome wide significance level. Phenotypes studied are genetically complex and are likely determined by many genes of modest effects, which makes them difficult to detect with genome-wide significance for the currently studied sample size. It is worth noting that the much larger Tobacco and Genetics Consortium did not identify any genome-wide significant associations for age at smoking initiation in a meta-analysis encompassing over 20,000 individuals.10 This emphasizes the need for even larger studies to study smoking behaviors in order to detect variants with presumably low effect sizes. Despite the modest effect sizes of the genetic variants implicated by GWAS, key biological pathways may be identified using such approaches. Secondly, the genotype imputation accuracy could have had an impact on our results. Many top SNPs from the analyses of age at smoking initiation were imputed in all 4 cohorts; however, the association peaks of these regions also contained SNPs genotyped in the majority (or even all) of the cohorts, which likely makes these findings more reliable. Given the fact that different, and not fully overlapping, genotyping chips were used, this imputation was crucial to obtain a comprehensive overview of many genetic associations. In our study, this is of special importance for the nonsynonymous SNP rs1801272 in CYP2A6, which had to be imputed in all cohorts and has no known proxy SNPs. Assuming that the association of this SNP with CPD is a true positive and possibly causal, imputation was the only one way to detect it. Lastly, we must acknowledge that COPD diagnosis has a significant impact on smoking behaviors studied, and especially on current CPD and smoking cessation. We took into account the severity of COPD, reflected in the level of lung function, as a potential confounder when analyzing these two phenotypes. However, we hypothesize that additional factors such as frequency of exacerbations may also affect smoking behaviors in the COPD subjects that were studied here. Additionally, it is plausible that social factors, such as smoking trends changing over time, are important environmental determinants of smoking initiation and intensity, and they might have potentially confounded genetic associations found.
In summary we identified two candidate loci associated with age at smoking initiation in COPD patients. We show that variation in the CHRNA3/CHRNA5 locus on chromosome 15q25 locus and the CYP2A6 locus on chromosome 19q13 were associated with lifetime average CPD among COPD patients. The latter gene may play a significant role in the current smoking intensity among COPD patients. Future studies in larger populations of COPD subjects will be required to determine the overlap between genetic determinants of smoking behavior in the general population and in COPD subjects.
Supplementary Material
Acknowledgments
ECLIPSE Steering Committee: Harvey Coxson (Canada), Lisa Edwards (GlaxoSmithKline, USA), Katharine Knobil (Co-chair, GlaxoSmithKline, UK), David Lomas (UK), William MacNee (UK), Edwin Silverman (USA), Ruth Tal-Singer (GlaxoSmithKline, USA), Jørgen Vestbo (Co-chair, Denmark), Julie Yates (GlaxoSmithKline, USA).
ECLIPSE Scientific Committee: Alvar Agusti (Spain), Peter Calverley (UK), Bartolome Celli (USA), Courtney Crim (GlaxoSmithKline, USA), Bruce Miller (GlaxoSmithKline, UK), William MacNee (Chair, UK), Stephen Rennard (USA), Ruth Tal-Singer (GlaxoSmithKline, USA), Emiel Wouters (The Netherlands), Julie Yates (GlaxoSmithKline, USA).
ECLIPSE Investigators: Bulgaria: Yavor Ivanov, Pleven; Kosta Kostov, Sofia. Canada: Jean Bourbeau, Montreal, Que Mark Fitzgerald, Vancouver, BC; Paul Hernandez, Halifax, NS; Kieran Killian, Hamilton, On; Robert Levy, Vancouver, BC; Francois Maltais, Montreal, Que; Denis O’Donnell, Kingston, On. Czech Republic: Jan Krepelka, Praha. Denmark: Jørgen Vestbo, Hvidovre. Netherlands: Emiel Wouters, Horn-Maastricht. New Zealand: Dean Quinn, Wellington. Norway: Per Bakke, Bergen. Slovenia: Mitja Kosnik, Golnik. Spain: Alvar Agusti, Jaume Sauleda, Palma de Mallorca. Ukraine: Yuri Feschenko, Kiev; Vladamir Gavrisyuk, Kiev; Lyudmila Yashina, Kiev; Nadezhda Monogarova, Donetsk. United Kingdom: Peter Calverley, Liverpool; David Lomas, Cambridge; William MacNee, Edinburgh; David Singh, Manchester; Jadwiga Wedzicha, London. United States of America: Antonio Anzueto, San Antonio, TX; Sidney Braman, Providence, RI; Richard Casaburi, Torrance CA; Bart Celli, Boston, MA; Glenn Giessel, Richmond, VA; Mark Gotfried, Phoenix, AZ; Gary Greenwald, Rancho Mirage, CA; Nicola Hanania, Houston, TX; Don Mahler, Lebanon, NH; Barry Make, Denver, CO; Stephen Rennard, Omaha, NE; Carolyn Rochester, New Haven, CT; Paul Scanlon, Rochester, MN; Dan Schuller, Omaha, NE; Frank Sciurba, Pittsburgh, PA; Amir Sharafkhaneh, Houston, TX; Thomas Siler, St. Charles, MO, Edwin Silverman, Boston, MA; Adam Wanner, Miami, FL; Robert Wise, Baltimore, MD; Richard ZuWallack, Hartford, CT.
We acknowledge the co-investigators in the NETT Genetics Ancillary Study including Joshua Benditt, Gerard Criner, Malcolm DeCamp, Philip Diaz, Mark Ginsburg, Larry Kaiser, Marcia Katz, Mark Krasna, Neil MacIntyre, Barry Make, Rob McKenna, Fernando Martinez, Zab Mosenifar, John Reilly, Andrew Ries, Paul Scanlon, Frank Sciurba, and James Utz.
FUNDING: This work was supported by U.S. National Institutes of Health (NIH) grants R01 HL075478, R01 HL084323, P01 HL083069, U01 HL089856 (Silverman), K12HL089990 (Cho), and U01 HL089897 (Crapo). The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute contracts (N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119), the Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality. The Norway cohort and the ECLIPSE study (Clinicaltrials.gov identifier NCT00292552; GSK Code SCO104960) are funded by GlaxoSmithKline. The COPDGene project was supported by Award Number U01HL089897 and Award Number U01HL089856 from the National Heart, Lung, And Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, And Blood Institute or the National Institutes of Health. The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Pfizer, Boehringer Ingelheim, Novartis and Sepracor.
This article is based on research that is funded in part by grants from the National Institute of Health (NIH) and is therefore subject to the mandatory NIH Public Access Policy. The final, peer-reviewed manuscript must be deposited with the PubMed Central (PMC) database upon acceptance for publication and be made publicly accessible no later than 12 months after publication.
M. Siedlinski is a recipient of a postdoctoral fellowship from the Niels Stensen
Foundation.
COPDGene Investigators
The members of the COPDGene study group as of June 2010
Ann Arbor VA: Jeffrey Curtis, MD (PI), Ella Kazerooni, MD (RAD)
Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, MS (PI), Philip Alapat, MD, Venkata Bandi, MD, Kalpalatha Guntupalli, MD, Elizabeth Guy, MD, Antara Mallampalli, MD, Charles Trinh, MD (RAD), Mustafa Atik, MD
Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, MD, MPH (Co-PI), Craig Hersh, MD, MPH (Co-PI), George Washko, MD, Francine Jacobson, MD, MPH (RAD)
Columbia University, New York, NY: R. Graham Barr, MD, DrPH (PI), Byron Thomashow, MD, John Austin, MD (RAD)
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., MD (PI), Lacey Washington, MD (RAD), H Page McAdams, MD (RAD)
Fallon Clinic, Worcester, MA: Richard Rosiello, MD (PI), Timothy Bresnahan, MD (RAD)
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH (PI), Joseph Tashjian, MD (RAD)
Johns Hopkins University, Baltimore, MD: Robert Wise, MD (PI), Nadia Hansel, MD, MPH, Robert Brown, MD (RAD), Gregory Diette, MD
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, MD (PI), Janos Porszasz, MD, PhD, Hans Fischer, MD, PhD (RAD), Matt Budoff, MD
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, MD (PI), Charles Trinh, MD (RAD), Hirani Kamal, MD, Roham Darvishi, MD
Minneapolis VA: Dennis Niewoehner, MD (PI), Tadashi Allen, MD (RAD), Quentin Anderson, MD (RAD), Kathryn Rice, MD
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS (PI), Gloria Westney, MD, MS, Eugene Berkowitz, MD, PhD (RAD)
National Jewish Health, Denver, CO: Russell Bowler, MD, PhD (PI), Adam Friedlander, MD, David Lynch, MB (RAD), Joyce Schroeder, MD (RAD), John Newell, Jr., MD (RAD)
Temple University, Philadelphia, PA: Gerard Criner, MD (PI), Victor Kim, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, A. James Mamary, MD, Robert Steiner, MD (RAD), Chandra Dass, MD (RAD)
University of Alabama, Birmingham, AL: William Bailey, MD (PI), Mark Dransfield, MD (Co-PI), Hrudaya Nath, MD (RAD)
University of California, San Diego, CA: Joe Ramsdell, MD (PI), Paul Friedman, MD (RAD)
University of Iowa, Iowa City, IA: Geoffrey McLennan, MD, PhD (PI), Edwin JR van Beek, MD, PhD (RAD), Brad Thompson, MD (RAD), Dwight Look, MD
University of Michigan, Ann Arbor, MI: Fernando Martinez, MD (PI), MeiLan Han, MD, Ella Kazerooni, MD (RAD)
University of Minnesota, Minneapolis, MN: Christine Wendt, MD (PI), Tadashi Allen, MD (RAD)
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD (PI), Joel Weissfeld, MD, MPH, Carl Fuhrman, MD (RAD), Jessica Bon, MD
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD (PI), Sandra Adams, MD, Carlos Orozco, MD, Mario Ruiz, MD (RAD)
Administrative Core: James Crapo, MD (PI), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth Regan, MD, Sarah Moyle, MS, Douglas Stinson
Genetic Analysis Core: Terri Beaty, PhD, Barbara Klanderman, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Jacqueline Hetmanski, MS, Tanda Murray
Imaging Core: David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr., MD, John Reilly, MD, Harvey Coxson, PhD, Philip Judy, PhD, Eric Hoffman, PhD, George Washko, MD, Raul San Jose Estepar, PhD, James Ross, MSc, Rebecca Leek, Jordan Zach, Alex Kluiber, Jered Sieren, Heather Baumhauer, Verity McArthur, Dzimitry Kazlouski, Andrew Allen, Tanya Mann, Anastasia Rodionova
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, PhD
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, PhD, Stacey Meyerer, Shivam Chandan, Samantha Bragan
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: James Murphy, PhD, Douglas Everett, PhD, Carla Wilson, MS, Ruthie Knowles, Amber Powell, Joe Piccoli, Maura Robinson, Margaret Forbes, Martina Wamboldt
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, MPH, PhD, Marci Sontag, PhD, Jennifer Black-Shinn, MPH, Gregory Kinney, MPH
Footnotes
COMPETING INTERESTS
No competing interests: MS, AG
GlaxoSmithKline employee: WA, XK
NHLBI grant: JEH, THB
NIH grant: MHC, EKS, JDC
GlaxoSmithKline grant: EKS, DAL, PB
COPD Foundation grant: EKS, JDC
GlaxoSmithKline consulting fee or honorarium and support for travel to meetings for the study or other purposes: EKS, DAL
COPD Foundation support for travel to meetings for the study or other purposes: EKS, JDC
AstraZeneca and GlaxoSmithKline - payment for lectures including service on speakers bureaus and consultancy (outside the submitted work): EKS, PB
Pfizer - payment for lectures including service on speakers bureaus and consultancy (outside the submitted work): PB
GlaxoSmithKline - board membership, consultancy, payment for lectures including service on speakers bureaus and consultancy (outside the submitted work): DAL
Boehringer Ingelheim consultancy (outside the submitted work): DAL
GlaxoSmithKline fees for participation in review activities such as data monitoring boards, statistical analysis, end point committees, and the like: DAL
Consulting fee or honorarium from AstraZeneca, Novartis, Otsuka: SIR
Grant from AstraZeneca, Biomarck, Centocor, Mpex, Nabi, Novartis, Otsuka: SIR
Almirall, Novartis, Nycomed, Pfizer board membership (outside the submitted work): SIR
Almirall, AstraZeneca, Boehringer Ingelheim, California Allergy Society, Creative Educational Concept, France Foundation, GlaxoSmithKline, Information TV, Network for Continuing Education, Novartis, Nycomed, Pfizer - travel/accommodations expenses covered or reimbursed (outside the submitted work): SIR
Able Associates, Adelphi Research, APT Pharma/Britnall, Aradigm, AstraZeneca, Boehringer Ingelheim, Chiesi, CommonHealth, Consult Complete, COPDForum, Data Monitor, Decision Resource, Defined Health, Dey, Dunn Group, Easton Associates, Equinox, Gerson, GlaxoSmithKline, Infomed, KOL Connection, M. Pankove, MedaCorp, MDRx Financial, Mpex, Oriel Therapeutics, Otsuka, Pennside, PharmaVentures, Pharmaxis, Price Waterhouse, Propagate, Pulmatrix, Reckner Associates, Recruiting Resources, Roche, Schlesinger Medical, Scimed, Sudler and Hennessey, TargeGen, Theravance, UBC, Uptake Medical, VantagePoint Mangement - consultancy (outside the submitted work): SIR
References
- 1.Vink JM, Posthuma D, Neale MC, et al. Genome-wide linkage scan to identify Loci for age at first cigarette in Dutch sibling pairs. Behav Genet. 2006;36:100–11. doi: 10.1007/s10519-005-9012-0. [DOI] [PubMed] [Google Scholar]
- 2.Li MD, Cheng R, Ma JZ, et al. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 3.Batra V, Patkar AA, Berrettini WH, et al. The genetic determinants of smoking. Chest. 2003;123:1730–9. doi: 10.1378/chest.123.5.1730. [DOI] [PubMed] [Google Scholar]
- 4.Schnoll RA, Johnson TA, Lerman C. Genetics and smoking behavior. Curr Psychiatry Rep. 2007;9:349–57. doi: 10.1007/s11920-007-0045-3. [DOI] [PubMed] [Google Scholar]
- 5.Han S, Gelernter J, Luo X, et al. Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biol Psychiatry. 67:12–9. doi: 10.1016/j.biopsych.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YZ, Pei YF, Guo YF, et al. Genome-wide association analyses suggested a novel mechanism for smoking behavior regulated by IL15. Mol Psychiatry. 2009;14:668–80. doi: 10.1038/mp.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 42:436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godtfredsen NS, Lam TH, Hansel TT, et al. COPD-related morbidity and mortality after smoking cessation: status of the evidence. Eur Respir J. 2008;32:844–53. doi: 10.1183/09031936.00160007. [DOI] [PubMed] [Google Scholar]
- 14.Andreas S, Hering T, Muhlig S, et al. Smoking cessation in chronic obstructive pulmonary disease: an effective medical intervention. Dtsch Arztebl Int. 2009;106:276–82. doi: 10.3238/arztebl.2009.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (COPD) Lancet. 2004;364:613–20. doi: 10.1016/S0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- 16.Kanner RE, Connett JE, Williams DE, et al. Effects of randomized assignment to a smoking cessation intervention and changes in smoking habits on respiratory symptoms in smokers with early chronic obstructive pulmonary disease: the Lung Health Study. Am J Med. 1999;106:410–6. doi: 10.1016/s0002-9343(99)00056-x. [DOI] [PubMed] [Google Scholar]
- 17.Hu MC, Davies M, Kandel DB. Epidemiology and correlates of daily smoking and nicotine dependence among young adults in the United States. Am J Public Health. 2006;96:299–308. doi: 10.2105/AJPH.2004.057232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishman A, Martinez F, Naunheim K, et al. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348:2059–73. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 19.Vestbo J, Anderson W, Coxson HO, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31:869–73. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 20.Zhu G, Warren L, Aponte J, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176:167–73. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 21.Regan EA, Hokanson JE, Murphy JR, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferris BG. Epidemiology Standardization Project (American Thoracic Society) Am Rev Respir Dis. 1978;118:1–120. [PubMed] [Google Scholar]
- 23.Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 42:200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 6 doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venables WN, Ripley BD. Modern Applied Statistics with S. 4. New York: Springer; 2002. [Google Scholar]
- 28.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aulchenko YS, Ripke S, Isaacs A, et al. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 30.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 31.Li Y, Willer C, Sanna S, et al. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S. PLINK (version 1.0.7) http://pngu.mgh.harvard.edu/purcell/plink/
- 35.Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rapley EA, Turnbull C, Al Olama AA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–10. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soranzo N, Spector TD, Mangino M, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–90. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss RB, Baker TB, Cannon DS, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson EO, Chen LS, Breslau N, et al. Peer smoking and the nicotinic receptor genes: an examination of genetic and environmental risks for nicotine dependence. Addiction. doi: 10.1111/j.1360-0443.2010.03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J Neurogenet. 2009;23:252–61. doi: 10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.