Abstract
The incidence of malignant melanoma is rising faster than that of any other cancer in the United States. Due to its high expression on the surface of melanomas, MC1R has been investigated as a target for selective imaging and therapeutic agents against melanoma. Eight ligands were screened against cell lines engineered to over-express MC1R, MC4R or MC5R. Of these, compound 1 (4-phenylbutyryl-His-Dphe-Arg-Trp-NH2) exhibited high (0.2 nM) binding affinity for MC1R, and low (high nM) affinities for MC4R and MC5R. Subsequently functionalization of the ligand at the C-terminus with an alkyne for use in Cu-catalyzed click chemistry was shown not to affect the binding affinity. Finally, formation of the targeted-polymer, as well as the targeted micelle formulation, also resulted in constructs with low nM binding affinity.
Keywords: Click reaction, MC1R, micelle, melanoma, melanocortin receptor, solid-phase synthesis, targeted therapies
Introduction
The incidence of malignant melanoma is rising faster than that of any other cancer in the United States, with diagnoses having doubled from 1986 to 20011. Melanoma progression is associated with altered expression of cell surface proteins, including adhesion proteins and receptors2. Over 80% of malignant melanomas express high levels of isoform 1 of the melanocyte stimulating hormone (αMSH) receptor (melanocortin 1 receptor, or MC1R).3 Thus, MC1R has been investigated as a target for selective imaging and therapeutic agents. MC1R belongs to a family of five G protein-coupled melanocortin receptors (MC1R–MC5R). Melanocortin receptors have been discovered in a wide range of tissues and organs throughout the body, ranging from the hair/skin (MC1R)4, kidneys (MC5R)5, adrenal glands (MC2R)6 and hypothalamus (MC3R/MC4R)7 and are known to play a role in skin pigmentation, hair coloration, obesity, metabolism, diabetes, sexual behavior, erectile dysfunction, stress response and mood.4–8 Endogenously, agonists for the melanocortins are the α-, β-, γ-melanocyte stimulating hormones (MSH) and adrenocorticotropic hormone (ACTH, MC2R specific), all of which contain the same central sequence of His-Phe-Arg-Trp.9 This high degree of pharmacophore homology makes it difficult to design selective ligands that are highly specific for receptor subtypes.
Due to its high expression on the surface of melanomas, MC1R has been investigated as a target for selective imaging and therapeutic agents, and a number of selective ligands have been developed.10 The most well known of these, [Nle4,D-Phe7]-α-MSH (NDP-α-MSH)11, has been investigated extensively by Chen who showed that 99αTc-CGCG labeled NDP-α-MSH bound to melanomas with very high avidity (6.5% ID/g)12. However, it is not selective, as NDP -α-MSH has strong nanomolar binding affinities to MC3R, MC4R and MC5R as well13. Such off-target binding is undesirable given the presence of these receptors in sensitive organs such as the kidney and brain. A co-injection of lysine has been reported to diminish off-target binding in the kidneys12, 14, and presumably most agents will not be able to cross the blood-brain barrier. Nonetheless, the need for the development of highly specific and selective ligands against MC1R for melanomas is of critical importance.
The development of ligands that can be attached to micelles and/or liposomes to target cancer cells relative to healthy organs is a major hurdle in current research. Stabilized micelles and liposomes are emerging as important platforms for delivery of lipophilic therapies to target tissues. Many such attempts fail either from (1) a loss of affinity resulting from the attachment of small peptides to large micelles or liposomes; (2) an inherent instability that results in collapse before entering the vicinity of the tumor; or (3) a nanoparticle size that is too large to escape the vasculature. In order to effectively design targeted nanoparticles, each of these issues must be addressed.
In the current work, we screened 8 putatively MC1R-specific compounds (Table 1) against cell lines that were engineered to overexpress MC1R, MC4R or MC5R, respectively. Compounds were tested for their ability to compete with Eu-NDP-α-MSH using a readout of time resolved fluorescence (TRF) 13b, 15. Of these, compound 1 exhibited high binding affinity for MC1R, and low affinities for MC4R and MC5R. Two analogs (compounds 2 and 3) have been synthesized to allow attachment via their N-termini; however they exhibited a reduced (2) or complete loss (3) of binding affinity. Therefore, 1 was modified at the C-terminus (compound 4) to allow attachment to the micelle and this modification was shown to not affect the binding affinity. Attachment to the stabilized polymer-based micelles was accomplished with Cu-catalyzed click chemistry, and the resulting construct was also shown to retain low nM binding affinity.
Table 1.
Structures of ligands screened for MC1R selectivity.
| Compound | Structure |
|---|---|
| 1 | 4-phenylbutyryl-His-DPhe-Arg-Trp-NH2 |
| 2 | Ac-Homophenylalanine-His-DPhe-Arg-Trp-NH2 |
| 3 | 4-hydroxycinnamoyl-His-DPhe-Arg-Trp-NH2 |
| 4 | 4-phenylbutyryl-His-DPhe-Arg-Trp-Gly-Lys(hex-5-ynoyl)-NH2 |
| 5 | H-Tyr-Val-Nle-Gly-His-DNal(2′)-Arg-DTrp-Asp-Arg-Phe-Gly-NH2 |
| 6 | H-Lys(hex-5-ynoyl)-Tyr-Val-Nle-Gly-His-DNal(2′)-Arg-DTrp-Asp-Arg-Phe-Gly-NH2 |
| 7 | H-Tyr-Val-Nle-Gly-His-DNal(2′)-Arg-DPhe-Asp-Arg-Phe-Gly-NH2 |
| 8 | H- Lys(hex-5-ynoyl)Tyr-Val-Nle-Gly-His-DNal(2′)-Arg-DPhe-Asp-Arg-Phe-Gly-NH2 |
Results
Competitive Binding Assays
Competitive binding assays were performed using HCT116/hMC1R, HEK293/hMC4R and HEK293/hMC5R cells with ligands 1–8. Seven of these were observed to bind to MC1R with low nM affinity. Of these, only three related compounds (compounds 1, 2, and 4) were found to bind selectively to the hMC1R receptor, vis-à-vis MC4R or MC5R (Table 2). Compound 1 displayed slightly higher affinities for hMC1R as compared to 2 (Ki is 0.17 nM vs 1.77 nM for 1 and 2, respectively), as well as higher selectivity, especially compared to MC5R (selectivity ratios of 160 and 33, respectively). Ligands 5–8 demonstrated a high affinity for hMC1R as well; however, they were also shown to have an even stronger affinity for hMC4R and hMC5R (Table 2). Ligand 3 demonstrated no affinity for the hMC1R.
Table 2.
Affinity and selectivity of ligands assayed in this publication.
| Ki (nM) | |||||
|---|---|---|---|---|---|
| Ligand | MC1R | MC4R | MC5R | 1R/4R | 1R/5R |
| 1 | 0.17 | 160 | 27 | 950 | 160 |
| 2 | 1.8 | 988 | 58 | 560 | 33 |
| 3 | NB⫞ | NB | NB | - | - |
| 4 | 0.24 | 254 | 46 | 1058 | 192 |
| 5 | 2.0 | 0.75 | 0.76 | 0.38 | 0.38 |
| 6 | 2.6 | 1.7 | 1.4 | 0.67 | 0.57 |
| 7 | 5.6 | 0.77 | 0.71 | 0.13 | 0.13 |
| 8 | 4.2 | 4.4 | 3.9 | 1.0 | 0.93 |
| 4-targeted polymer | 26 | NB | NB | - | - |
| 4-targeted micelles | 2.9 | NB | NB | - | - |
| NDP-a-MSH | 1.8 | 19 | 9.9 | 10 | 5.5 |
NB = Non-binding.
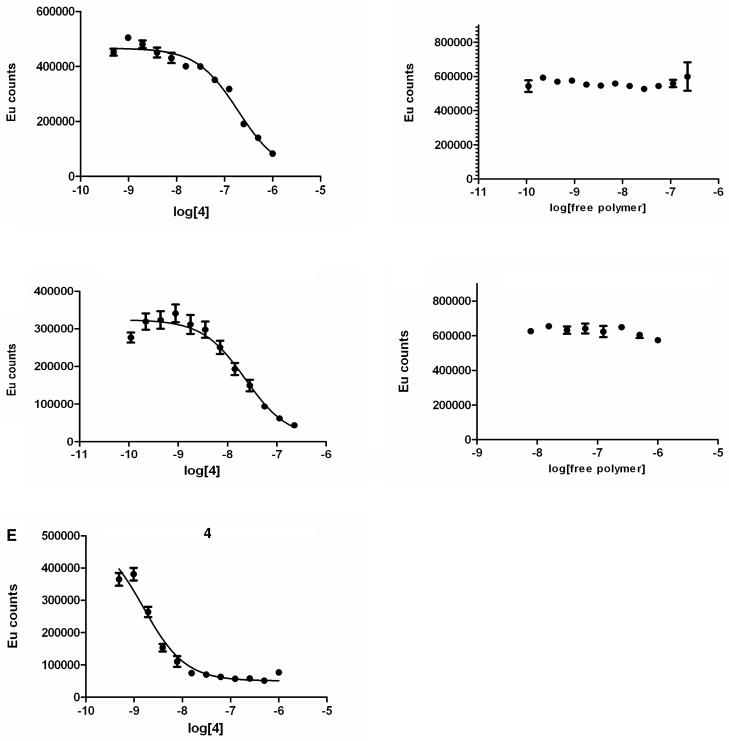
Competitive binding assays were also performed using 4-targeted triblock polymers, 4-targeted stabilized triblock polymer micelles, as well as untargeted polymer and untargeted, stabilized micelles as controls (Figure 1, Table 2). The 4-targeted micelle exhibits an increased binding avidity to the hMC1R receptor compared to the targeted polymer, and both are less avid than the native ligand (Ki are 2.9 nM, 25.7 nM and 0.24 nM for the micelle, polymer and ligand, respectively). No binding is observed with the untargeted polymer or untargeted micelles, indicating that the tri-block polymer does not interact non-specifically with the cell surface. Most notably, there is no measurable interaction (≤10−5) between the 4-targeted micelle and hMC4R or hMC5R, indicating that the conjugation of the ligand to a micelle results in an increased selectivity.
Figure 1.
Representative competitive binding assays for (A) 4-targeted polymer; (B) untargeted polymer; (C) 4-targeted micelles; (D) untargeted micelles and (E) 4 against HCT116/hMC1R cells. X-axis concentrations for (A), (C) and (E) were normalized to the targeting ligand. Concentrations for the ligand in (A), (C) and (E) and polymer and micelle in (B) and (D) were deliberately chosen to be the same for all assays.
Micelle physical properties
Dynamic light scattering (DLS) and zeta potential measurements showed the size and surface charge of the 4-targeted micelles to be 91 +/− 2 nm and −10.6 +/− 0.9 mV, respectively.
Discussion
Historically, ligands which are known to interact with the hMC1R receptor also demonstrate cross-reactivity with other melanocortin receptors, including hMC4R and hMC5R. While MC1R is known to be expressed almost exclusively in melanoma cells and melanocytes, hMC4R and hMC5R have high expression levels in normal tissues, including kidney and brain, thus non-specific ligand binding is not ideal. To combat this problem and minimize off-target effects, we chose several ligands from the literature that have previously been reported to possess nanomolar binding affinities for hMC1R.
Ligand 1 was reported to have a high affinity and selectivity for MC1R (1R/4R selectivity ratio of 1200)10c, 16 and was consequently chosen as a template for the design of the novel ligands 2, 3, which contain the same parent amino acid sequence, as well as 4, which possesses a terminal alkyne. Ligands 2 and 3 were intially designed for potential attachment to a micelle through the N-terminus. Compound 2 represents the α-amino analog of 1 and compound 3 possesses a more rigid C2 (cinnamate) aromatic linker compared to the 4-phenylbutyryl, 1, which has free rotation about the C3 chain. The 4-hydroxy group in compound 3 was intended as an attachment point to the micelles via O-alkylation. As mentioned, the rigid C2 linker in 3 reduces the free rotation of compound 1 and thus we reasoned might potentially result in improved interaction with the hydrophobic cleft of the MC1R. Unfortunately, compound 3 exhibited no affinity to the MC1R receptor, and compound 2 exhibited 10 times lower binding affinity as compared to 1. Therefore, we chose to proceed with a C-terminal modification, (i.e. compound 4) as the binding affinity was not changed compared to the parent compound 1. Additionally, ligands 5 and 7 were reported to have moderate hMC1R selectivities over hMC4R and hMC5R (1R/4R = 20.00 and 12.50 nM for 5 and 7, respectively; 1R/5R = 11.67 and 2.06 for 5 and 7, respectively) 13a; thus, each was functionalized with a terminal alkyne for attachment to a nanoparticle scaffold via click chemistry and screened for retained selectivity to MC1R.
The most specific of these ligands, 1, was determined by us to have an hMC1R/hMC4R selectivity of 950, which is in good agreement with the reported selectivity of 1200. Likewise, ligand 2, based on the same parent amino acid sequence as 1, was also found to have high 1R/4R selectivity; however, its 1R/5R selectivity was substantially lower. Unfortunately, ligands 3 and 5–8 were found to be not at all selective for MC1R, with ligand 3 possessing no affinity for any of the receptors tested. Our results for ligands 5 and 7 deviate from that which has been previously reported; however, this discrepancy may be due to differences in the binding assays used to derive the affinity constants. As detailed in the experimental section, our lab derives Ki values based on europium time-resolved fluorescence assays; however, previously determined EC50 values for these ligands were derived via 125I-labeled competitive binding assays.
As 1 was determined to be the ligand with the highest hMC1R affinity and selectivity, it was chosen for modification with a terminal alkyne for attachment of a triblock polymer micelle. Compound 4 did not demonstrate a loss of affinity of MC1R following alkyne functionalization.
As predicted, 1 and 2 have similar binding profiles given the similarity in their structures; however, it was surprising to see a complete loss of affinity in 3. The differences in affinity among these three ligands arise from the structural differences at the N-terminal end of the peptide, given that they all share the same R-HfRW-NH2 parent scaffold. However, whereas 1 and 2 contain Ph-(CH2)3-CO-and Ac-Hpe groups, both of which are non-polar, at the N-terminus, 3 contains a 4-hydroxyPh-CH=CH-CO-, which is more polar due the incorporation of the hydroxyl. Conversely, several analogues of 3 reported in the literature possess low nanomolar affinities against MC1R with varying selectivities. Thus, it seems reasonable to conclude that the loss in affinity experienced by 3 results from the incorporation of the alkene, rather than the increased polarity that arises from the addition of the hydroxyl group. While the exact reasons behind the affinity of 3, or lack thereof, remain unclear, it is plausible that incorporation of the alkene in this ligand causes the peptide to adopt too rigid a structure, thereby reducing its ability to conform to the receptor binding pocket.
Ligands 5–8 are about twice as large and display binding affinities one-to-two orders of magnitude higher with hMC1R as compared to those described above. Ligands 6 and 8 were synthesized as analogues of ligands 5 and 7, respectively, to be used for potential attachment to nanoparticles. The similarity in binding affinities of 5 versus 6 and 7 versus 8 further demonstrates that the C-terminal end of these peptides is a suitable location for the placement of an attachment of a scaffold, as it does not seem to impact the binding ability of the ligand.
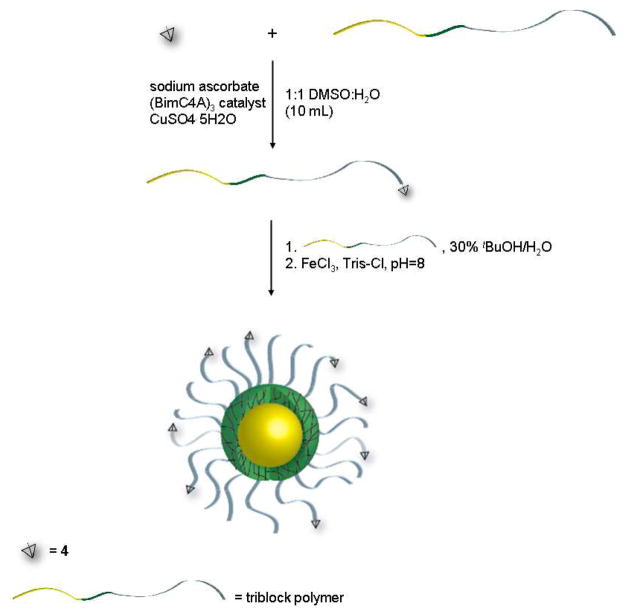
A targeted, stabilized triblock polymer micelle was prepared by combining 10% 4-targeted polymer with 90% untargeted polymer (Scheme 2). As a control, competitive binding assays were performed with targeted and untargeted polymer, as well as untargeted stabilized micelles. For consistency, all binding assays were normalized to the concentration of the ligand. As previously stated, the 4-targeted micelle exhibits an increased binding avidity to the hMC1R receptor as compared to the targeted-polymer and a slightly weaker avidity than the native ligand. The increase in binding avidity for the targeted micelles as compared to the targeted-polymer is noteworthy in that it (1) demonstrates the in-vitro stability of this micelle system; and (2) it indicates that the binding avidity of the 4-targeted micelles may be benefiting from multivalent interactions. Additionally, the 4-targeted polymer and 4-targeted micelle demonstrated no measurable interactions with either MC4R or MC5R, thereby indicating that the targeted micelle is itself more specific than the ligand alone.
Scheme 2.
Synthesis of targeted, stabilized micelle system.
The decrease in binding affinity of the targeted polymer relative to the native ligand can be explained by the conjugation of a large, flexible PEG group to a relatively small ligand. In addition to adding to the entropy of the ligand system through increased flexibility and size, PEG chains are known to have at least moderate interaction with non-polar hydrophobic groups18. Consequently, it is possible that the PEG moiety on the end of the triblock polymer is weakly interacting with the hydrophobic amino acids of the targeting group, thereby decreasing its affinity for MC1R.
However tempting it may be to ascribe the modest increase in avidity of the targeted micelles relative to targeted polymer solely to multivalent binding, such interactions are of high complexity and can be attributed to the sum of multiple factors. These variables include the enthalpies of individual binding events, the entropic consequences experienced by the micelle polymer upon binding, the topography of ligand positioning on the micelle, the statistical proximity effect and differences in rates of cellular uptake.
Improved avidity through multivalent interactions is more readily observed with ligands with relatively low affinity. 19. In the case of 4, it is possible that multivalent interactions would not greatly enhance binding avidity given the high affinity of the targeting ligand for binding the MC1R receptor. Also, assembled polymer micelles have decreased entropy relative to free polymer, via the hydrophobic effect, thereby potentially leading to slower ligand-receptor off rates.20 Additionally, ligand proximity and steric repulsions between ligands and polymer chains are known to be important factors that influence the degree, if any, to which multivalency is experienced in micellar systems19b.
The targeted micelles herein are calculated to have approximately 10 targeting groups each. This may seem to be a large number; however, since these stabilized micelles are relatively inflexible, ligands binding to the surface of a cell positioned on one side of the micelle could prohibit binding of ligands positioned on the opposite side. Since total ligand concentration was considered when calculating avidities, having a large fraction of ligands inaccessible to receptor at any given time would effectively reduce the calculated avidity. However, even if multivalent binding is completely inhibited in this manner, increased binding avidity can be still observed due to the statistical effect where proximal ligands can readily bind following release of an existing interaction.21
Lastly, the rate of cellular uptake is likely different for the targeted micelle relative to the targeted polymer or free ligand. Constructs with faster rates of uptake can be calculated to have higher avidities due to the loss of “off-rate” once internalized. The sum of these variables and others not mentioned may contribute to the observed binding avidity of these supramolecular systems.
Conclusion
An hMC1R ligand was modified for attachment to a stabilized, triblock polymer micelle. Functionalization and subsequent attachment of the ligand to a ~100 nm polymer micelle resulted in a slight decrease in affinity, but increase in specificity, to MC1R. Presumably, this decrease results from the thermodynamic hurdles encountered in appending a small peptide to a large nanoparticle, as well as an inherent handicap in the assay design. As mentioned in the introduction, three hurdles must be overcome in the design of an effective targeted nanoparticle delivery system: (1) it must be insured that there is no loss of ligand affinity resulting from the attachment of a small peptide to a large nanoparticles, or any such loss in affinity must be compensated by multivalent binding interactions; (2) nanoparticles must be inherently stable; and (3) nanoparticles must be sufficiently small to escape the vasculature and enter the tumor. We believe the current publication addresses the first two of these concerns. We have shown that our ligand remains selective after attachment and the increased binding affinity observed between the 4-targeted polymer and 4-targeted stabilized micelle has demonstrated the in-vitro stability of the system. Based on our DLS data, we are confident that our micelles are of sufficient size to escape the vasculature and in-vivo studies to evaluate the selectivity and stability of this targeted micellar system in mice are currently underway.
Experimental Section
Ligand Synthesis
N-α-Fmoc-protected amino acids, HBTU, HCTU, HOCt and HOBt were purchased from Anaspec (San Jose, CA) or Novabiochem (San Diego, CA). Rink amide Tentagel S and R resins were acquired from Rapp Polymere (Tubingen, Germany). Rink amide 1%-DVB PS resin was acquired from Novabiochem (San Diego, CA). For the N-α -Fmoc-protected amino acids, the following side chain protecting groups were used: Arg(Ng-Pbf); Asp(O-tBu); His(Nim-Trt); Trp(Ni-Boc); Tyr(tBu), and Lys(Nε-Aloc). Reagent grade solvents, reagents, and acetonitrile (ACN) for HPLC were acquired from VWR (West Chester, PA) or Aldrich-Sigma (Milwaukee, WI), and were used without further purification unless otherwise noted. N-terminal heterocyclic acids, NMI, and scavengers were obtained from Sigma-Aldrich or TCI. The solid-phase synthesis was performed in fritted syringes using a Domino manual synthesizer obtained from Torviq (Niles, MI). The C-18 Sep-Pak™ RC cartdridges for solid phase extraction were purchased from Waters (Milford, MA).
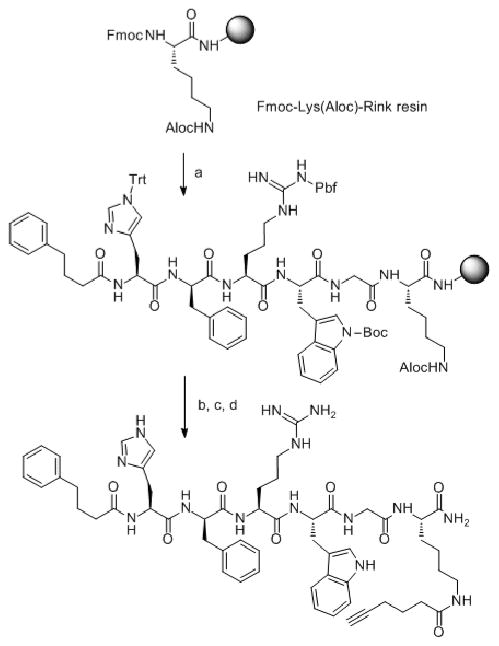
Ligands 1–8 were prepared as previously published by solid-phase synthesis as summarized in Scheme 1 on Rink Amide Tentagel resin (0.23 mmol/g) using a Fmoc/tBu synthetic strategy and standard activations.15, 22 After final deprotection of the Fmoc group, the resin was coupled with HOBt ester of 4-phenylbutyric acid (compounds 1, 4), acetylated with acetic anhydride/pyridine (compound 2) or left unreacted as a free amino group (compounds 5–8). 4-hydroxycinnamoyl-His-DPhe-Arg-Trp-NH NH-resin was treated with 50% piperidine in DMF to remove 4-hydroxycinnamoyl oligomers. The ligands were cleaved off the resins with TFA-scavenger cocktail (91% TFA, 3% water, 3% thioanisole, 3% ethanedithiol), extracted with cold diethylether, then dissolved in 1.0 M aqueous acetic acid. The crude ligands were purified by SEC and HPLC. All final compounds were >95% pure by HPLC analysis. The pure compounds were dissolved in DI water or DMSO at approximately 1.0 mM concentrations and concentration was determined by Trp-HPLC measurement23. Ligand purification methods, mass spectra and HPLC characterization data are provided in Supplemental Information
Scheme 1.
Synthetic route for compounds 1–8. a) (i) Fmoc-AA-OH (3eq), HOCt or HOBt (3eq), and DIC (3eq) in DMF/DCM (10 mL/1g of resin) for amino acid couplings; (iii) Piperidine/DMF (1:10, 2 + 20 minutes); (iv) 4-phenylbutyric acid (6eq), and DIC (3eq) in DMF/DCM; b) (i) Pd(0)tetrakistriphenylphosphine (0.01eq), N,N′-dimethylbarbituric acid (5eq) in degassed DCM (2 × 30 minutes) (ii) 5-hexynoic acid (5eq) and DIC (3eq) in DMF/DCM for compound 1; S-Trt-3-propanoic acid (5eq) and DIC (3eq) in DMF/DCM for compound 2; c) (i) TFA-scavengers cocktail (91% TFA, 3% water, 3% thioanisole, 3% ethanedithiol); (ii) ether extraction; d) purification.
Cell Culture
HCT116 cells overexpressing hMC1R and HEK293 cells overexpressing hMC4R13b or hMC5R were used in all studies. The parental human colorectal carcinoma cell line, HCT116 (American Type Culture Collection, CCL 247) was also used. Cells were maintained under standard conditions (37°C and 5% CO2) and were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS and 5% penicillin/streptomycin. For HCT116/hMC1R cells, geneticin (G418S, 0.4mg/mL) was added to the media to ensure proper selection.
Europium Binding Assays
Competitive binding assays were performed using HCT116/hMC1R cells and HEK293/hMC4R or hMC5R as previously described, with slight modifications13b. HCT116/hMC1R cells were plated in black PerkinElmer 96-well plates and HEK293/hMC4R and HEK293/hMC5R cells were plated on SigmaScreen Poly-D-Lysine Coated Plates (Sigma-Aldrich), all at a density of 10,000–30,000 cells/well. Poly-D-Lysine Coated Plates contain a PDL polymer coating that creates a uniform positive charge at the surface of the plastic, thereby facilitating cell attachment, growth and differentiation. Both PerkinElmer and SigmaScreen plates were evaluated for non-specific binding. Cells were grown in the 96-well plates for 2–3 days. On the day of the experiment, the media was aspirated and 50 μL of non-labeled competing ligand was added to each well in a series of decreasing concentrations (ranging from ~1 μM to 0.1 nM), followed by 50 μL of Eu-NDP-α-MSH at 10 nM. Both labeled and non-labeled ligands were diluted in binding media (DMEM, 1 mM 1,10-phenanthroline, 200 mg/L bacitracin, 0.5 mg/L leupeptin, 0.3% BSA). In the case of the triblock polymer micelle solutions, micelles were allowed to equilibrate in solution for a period of 30 min prior to cell addition. Cells were incubated with labeled and non-labeled ligands for 1 hour at 37°C. Following incubation, cells were washed three times with wash buffer (50 mM Tris–HCl, 0.2% BSA, 30 mM NaCl) and 100 μL of enhancement solution (PerkinElmer) was added to each well. Cells were incubated for an additional 30 min at 37°C prior to reading. The plates were read on a PerkinElmer VICTORx4 2030 multilabel reader using the standard Eu time-resolved fluorescence (TRF) measurement (340 nm excitation, 400 μs delay, and emission collection for 400 μs at 615 nm). Competition curves were analyzed with GraphPad Prism software using the sigmoidal dose–response (variable slope) classical equation for nonlinear regression analysis.
Synthesis of Targeted Triblock Polymers
Triblock polymer, azido-Poly(ethylene glycol)-block-poly(aspartic acid)-block-poly(leucine-co-tyrosine), was obtained from Intezyne Inc. (Tampa, FL). Alkyne functionalized ligand 4 was conjugated to the terminal azide of the polymer by copper assisted click chemistry (CuAAC)24 (Scheme 2) using the following method. To a solution of 1:1 DMSO:H2O (10 mL) was added 4 (16 mmol, 1.2 equiv), triblock polymer (13.3 mmol, 1 equiv), sodium ascorbate (334.15 mmol, 25 equiv), (BimC4A)3 catalyst25 (13.42 mmol, 0.2 equiv) and CuSO4·5H2O (13.3 mmol, 1 equiv). The solution was heated to 50°C and stirred for 2 days. The mixture was then cooled and placed in a 3500 MW dialysis bag (Spectra Por) and dialyzed against EDTA/H2O (x3) and H2O (x3). Following purification by dialysis, the solution was lyophilized. Successful click coupling was verified through visualization of the triazole-H in 1H NMR (8.02 ppm).
Micelle Formulation
Triblock polymers were dissolved at 20 mg/mL in 30% tert-butanol/H2O at room temperature, stirred for 4 hours and then lyophilized (Scheme 2).26 Micelles were stabilized with an Fe(III) crosslinking (Scheme 2) by dissolving the micelle (20 mg/mL) in FeCl3 (1.35 mg/mL) in TRIS buffer, adjusting to pH = 8 and stirring for 12 hours. For the targeted micelle system, 10% targeted polymer and 90% untargeted polymer were used in the formulation mixture. Micelle size was determined by dynamic light scattering (DLS, Wyatt Technology, DynaPro) and surface charge was determined by zeta measurement (Malvern, Zetasizer).
Supplementary Material
Acknowledgments
Funding Sources
Research was funded by the Bankhead Coley Melanoma Pre-SPORE Program (Grant 02-15066-10-03) to D.L.M., NIH/NCI (Grant R01 CA 097360) to R.J. G. and D.L.M., and Intezyne Inc. (Award #84-16301-01-01) to R.J.G. for a portion of N.M.B.’s salary and polymer materials.
The authors wish to acknowledge Intezyne Inc. for their support and scientific contributions.
ABBREVIATIONS
Abbreviations used for amino acids and designation of peptides follow the rules of the IUPAC-IUB Commission of Biochemical Nomenclature in J. Biol. Chem. 1972, 247, 977–983. The following additional abbreviations are used:
- Aloc
allyloxycarbonyl
- (BimC4A)3
potassium 5,5′,5″-(2,2′,2″-(nitrilotris(methylene))tris(1H-benzimidazole-2,1-diyl)) tripentanoate
- Boc
tert-butyloxycarbonyl
- tBu
tert-butyl
- DMSO
dimethylsulfoxide
- DVB
divinylbenzene
- Fmoc
(9H-fluoren-9-ylmethoxy)carbonyl
- HBTU
2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluoro-phosphate
- HOBt
N-hydroxybenzotriazole
- HOCt
6-chloro-1H-hydroxybenzotriazole
- NMI
N-methylimidazole
- Pbf
2,2,4,6,7-pentamethyl-dihydrobenzofuran-5-sulfonyl
- PS
polystyrene
- RP-HPLC
reverse-phase high performance liquid chromatography
- TFA
trifluoroacetic acid
- and Trt
triphenylmethyl (trityl)
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. Complete information on general methods and synthesis, solid phase synthesis QC and purification, and mass spectroscopy are included in the supplemental information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Welch HG, Woloshin S, Schwartz LM. Skin biopsy rates and incidence of melanoma: population based ecological study. BMJ. 2005;331:481–489. doi: 10.1136/bmj.38516.649537.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Lehmann JM, Holzmann B, Breitbart EW, Schmiegelow P, Riethmuller G, Johnson JP. Discrimination between benign and malignant cells of melanocytic lineage by two novel antigens, a glycoprotein with a molecular weight of 113,000 and a protein with a molecular weight of 76,000. Cancer Res. 1987;47(3):841–845. [PubMed] [Google Scholar]; (b) Wang R, Kobayashi R, Bishop JM. Cellular adherence elicits ligand-independent activation of the Met cell-surface receptor. Proc Natl Acad Sci U S A. 1996;93(16):8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wang Y, Rao U, Mascari R, Richards TJ, Panson AJ, Edington HD, Shipe-Spotloe JM, Donnelly SS, Kirkwood JM, Becker D. Molecular analysis of melanoma precursor lesions. Cell Growth Differ. 1996;7(12):1733–1740. [PubMed] [Google Scholar]; (d) Wang Z, Margulies L, Hicklin DJ, Ferrone S. Molecular and functional phenotypes of melanoma cells with abnormalities in HLA class I antigen expression. Tissue Antigens. 1996;47(5):382–390. doi: 10.1111/j.1399-0039.1996.tb02573.x. [DOI] [PubMed] [Google Scholar]; (e) Yang P, Farkas DL, Kirkwood JM, Abernethy JL, Edington HD, Becker D. Macroscopic spectral imaging and gene expression analysis of the early stages of melanoma. Mol Med. 1999;5(12):785–794. [PMC free article] [PubMed] [Google Scholar]; (f) Hsu M, Andl T, Li G, Meinkoth JL, Herlyn M. Cadherin repertoire determines partner-specific gap junctional communication during melanoma progression. J Cell Sci. 2000;113(Pt 9):1535–1542. doi: 10.1242/jcs.113.9.1535. [DOI] [PubMed] [Google Scholar]
- 3.Siegrist W, Solca F, Stutz S, Giuffre L, Carrel S, Girard J, Eberle AN. Characterization of receptors for alpha-melanocyte-stimulating hormone on human melanoma cells. Cancer Res. 1989;49(22):6352–6358. [PubMed] [Google Scholar]
- 4.Garcia-Borron JC, Sanchez-Laorden BL, Jimenez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues AR, Pignatelli D, Almeida H, Gouveiaa AM. Melanocortin 5 receptor activates ERK1/2 through a PI3K-regulated signaling mechanism. Molecular and Cellular Endocrinology. 2009;303:74–81. doi: 10.1016/j.mce.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Webb TR, Clark AJL. Minireview: The Melanocortin 2 Receptor Accessory Proteins. Molecular Endocrinology. 2009;24(3):475–484. doi: 10.1210/me.2009-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Ploeg LHTVd, Martin WJ, Howard AD, Nargund RP, Austin CP, Guan X, Drisko J, Cashen D, Sebhat I, Patchett AA, Figueroa DJ, DiLella AG, Connolly BM, Weinberg DH, Tan CP, Palyha OC, Pong S-S, MacNeil T, Rosenblum C, Vongs A, Tang R, Yu H, Sailer AW, Fong TM, Huang C, Tota MR, Chang RS, Stearns R, Tamvakopoulos C, Christ G, Drazen DL, Spar BD, Nelson RJ, MacIntyre DE. A role for the melanocortin 4 receptor in sexual function. PNAS. 2002;99(17):11381–11386. doi: 10.1073/pnas.172378699. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Begriche K, Sutton GM, Fang J, Butler AA. The role of melanocortin neuronal pathways in circadian biology: a new homeostatic output involving melanocortin-3 receptors? obesity reviews. 2009;10(Suppl 2):14–24. doi: 10.1111/j.1467-789X.2009.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Hall JE, Silva AAd, do Carmo Jussara M, Dubinion J, Hamza S, Munusamy S, Smith G, Stec DE. Obesity-induced Hypertension: Role of Sympathetic Nervous System, Leptin, and Melanocortins. J Bio Chem. 2010;28(23):17271–17276. doi: 10.1074/jbc.R110.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jun D-J, Na K-Y, Kim W, Kwak D, Kwon Eun-Jeong, Yoon JH, Yea K, Lee H, Kim J, Suh P-G, an SHR, Kim K-T. Melanocortins induce interleukin 6 gene expression and secretion through melanocortin receptors 2 and 5 in 3T3-L1 adipocytes. Journal of Molecular Endocrinology. 2010;44:225–236. doi: 10.1677/JME-09-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Fridmanis D, Petrovskaa R, Kalnina I, Peculisa MS, Schiöth HB, Klovins J. Identification of domains responsible for specific membrane transport and ligand specificity of the ACTH receptor (MC2R) Molecular and Cellular Endocrinology. 2010;321:175–183. doi: 10.1016/j.mce.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 9.Corander MP, Fenech M, Coll AP. The science of self preservation: how melanocortin action in the brain modulates body weight, blood pressure and ischaemic damage. Circulation. 2010;120(22):2260–2268. doi: 10.1161/CIRCULATIONAHA.109.854612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Cai M, Varga EV, Stankova M, Mayorov A, Perry JW, Yamamura HI, Trivedi D, Hruby VJ. Cell signaling and trafficking of human melanocortin receptors in real time using two-photon fluorescence and confocal laser microscopy: differentiation of agonists and antagonists. Chem Biol Drug Des. 2006;68(4):183–193. doi: 10.1111/j.1747-0285.2006.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mayorov AV, Han SY, Cai M, Hammer MR, Trivedi D, Hruby VJ. Effects of macrocycle size and rigidity on melanocortin receptor-1 and -5 selectivity in cyclic lactam alpha-melanocyte-stimulating hormone analogs. Chem Biol Drug Des. 2006;67(5):329–335. doi: 10.1111/j.1747-0285.2006.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Koikov LN, Ebetino FH, Solinsky MG, Cross-Doersen D, Knittel JJ. Sub-nanomolar hMC1R agonists by end-capping of the melanocortin tetrapeptide His-D-Phe-Arg-Trp-NH(2) Bioorg Med Chem Lett. 2003;13(16):2647–2650. doi: 10.1016/s0960-894x(03)00552-3. [DOI] [PubMed] [Google Scholar]
- 11.Sawyer TK, Sanfilippo PJ, Hruby VJ, Engel MH, Heward CB, Burnett JB, Hadley ME. 4-Norleucine, 7-D-phenylalanine-a-melanocyte-stimulating hormone: A highly potent a-melanotropin with ultralong biological activity. Proc Natl Acad Sci USA. 1980;77(10):5754–5758. doi: 10.1073/pnas.77.10.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Giblin MF, Wang N, Jurisson SS, Quinn TP. In vivo evaluation of 99mTc/188Re-labeled linear alpha-melanocyte stimulating hormone analogs for specific melanoma targeting. Nucl Med Biol. 1999;26(6):687–693. doi: 10.1016/s0969-8051(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 13.(a) Cai M, Mayorov AV, Cabello C, Stankova M, Trivedi D, Hruby VJ. Novel 3D Pharmacophore of r-MSH/α-MSH Hybrids Leads to Selective Human MC1R and MC3R Analogues. J Med Chem. 2005;48(6):1839–1848. doi: 10.1021/jm049579s. [DOI] [PubMed] [Google Scholar]; (b) Handl HL, Vagner J, Yamamura HI, Hruby VJ, Gillies RJ. Lanthanide-based time-resolved fluorescence of in cyto ligand-receptor interactions. Anal Biochem. 2004;330(2):242–250. doi: 10.1016/j.ab.2004.04.012. [DOI] [PubMed] [Google Scholar]; (c) Yang Y, Hruby VJ, Chen M, Crasto C, Cai M, Harmon CM. Novel Binding Motif of ACTH Analogues at the Melanocortin Receptors. Biochemistry. 2009;48:9775–9784. doi: 10.1021/bi900634e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Wei L, Butcher C, Miao Y, Gallazzi F, Quinn TP, Welch MJ, Lewis JS. Synthesis and Biologic Evaluation of 64Cu-Labeled Rhenium-Cyclized a-MSH Peptide Analog Using a Cross-Bridged Cyclam Chelator. The Journal Of Nuclear Medicine. 2007;48(1):64–72. [PubMed] [Google Scholar]; (b) Chen J, Cheng Z, Hoffman TJ, Jurisson SS, Quinn TP. Melanoma-targeting properties of (99m)technetium-labeled cyclic alpha-melanocyte-stimulating hormone peptide analogues. Cancer Res. 2000;60(20):5649–5658. [PubMed] [Google Scholar]
- 15.Handl HL, Sankaranarayanan R, Josan J, Vagner J, Mash EA, Gillies RJ, Hruby VJ. Bioconj Chem. 2007;18:1101–1109. doi: 10.1021/bc0603642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koikov LN, Ebetino FH, Solinsky MG, Cross-Doersenb D, Knittela JJ. Analogs of sub-nanomolar hMC1R agonist LK-184 [Ph(CH2)3CO-His-D-Phe-Arg-Trp-NH2]. An additional binding site within the human melanocortin receptor 1? Bioorganic& Medicinal Chemistry Letters. 2004;14:3997–4000. doi: 10.1016/j.bmcl.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 17.Koikov LN, Ebetino FH, Hayes JC, Cross-Doersen D, Knittel JJ. End-capping of the modified melanocortin tetrapeptide (p-Cl)Phe-D-Phe-Arg-Trp-NH2 as a route to hMC1R agonists. Bioorg Med Chem Lett. 2004;14:4389–4842. doi: 10.1016/j.bmcl.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 18.Sheth SR, Efremova N, Leckband DE. Interactions of poly(ethylene oxide) brushes with chemically selective surfaces. J Phys Chem B. 2000;104:7652–7662. [Google Scholar]
- 19.(a) Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective Tumor Cell Targeting Using Low-Affinity, Multivalent Interactions. ACS Chem Bio. 2007;2(2):119–127. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]; (b) Martin AL, Li B, Gillies ER. Surface Functionalization of Nanomaterials with Dendritic Groups: Toward Enhanced Binding to Biological Targets. J Am Chem Soc. 2008;131:734–741. doi: 10.1021/ja807220u. [DOI] [PubMed] [Google Scholar]
- 20.Tanford C. The hydrophobic effect and the organization of living matter. Science. 1977;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 21.Handl H, Vagner J, Han H, Mash E, Hruby V, Gillies R. Hitting Multiple Targets with Multimeric Ligands. Expert Opinion on Therapeutic Targets. 2004;8(6):565–586. doi: 10.1517/14728222.8.6.565. [DOI] [PubMed] [Google Scholar]
- 22.Vagner J, Handl HL, Gillies RJ, Hruby VJ. Bioorg Med Chem Lett. 2004;14:211–215. doi: 10.1016/j.bmcl.2003.09.079. [DOI] [PubMed] [Google Scholar]
- 23.(a) Vagner J, Xu L, Handl HL, Josan JS, Morse DL, Mash EA, Gillies RJ, Hruby VJ. Heterobivalent Ligands Crosslink Multiple Cell-Surface Receptors: The Human Melanocortin-4 and δ-Opioid Receptors. Angew Chem Int Ed. 2008;47:1685–1688. doi: 10.1002/anie.200702770. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Josan JS, Vagner J, Handl HL, Sankaranarayanan R, Gillies RJ, Hruby VJ. Solid-Phase Synthesis of Heterobivalent Ligands Targeted to Melanocortin and Cholecystokinin Receptors. Int J Pept Res Ther. 2008;14:293–300. doi: 10.1007/s10989-008-9150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis WG, Magallon FG, Fokin VV, Finn MG. Discovery and Characterization of Catalysts for Azide-Alkyne Cycloaddition by Fluorescence Quenching. J Am Chem Soc. 2004;126:9152–9153. doi: 10.1021/ja048425z. [DOI] [PubMed] [Google Scholar]
- 25.Rodionov VO, Presolski SI, Gardinier S, Lim YH, Finn MG. Benzimidazole and Related Ligands for Cu-Catalyzed Azide-Alkyne Cycloaddition. J Am Chem Soc. 2007;129(42):12696–12704. doi: 10.1021/ja072678l. [DOI] [PubMed] [Google Scholar]
- 26.Fournier E, Dufresne MH, Smith DC, Ranger M, Leroux JC. A Novel OneStep Drug-Loading Procedure for Water-Soluble Amphiphilic Nanocarriers. Pharmaceutical Research. 2004;21(6):962–968. doi: 10.1023/b:pham.0000029284.40637.69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.