Abstract
Foods with health-promoting effects beyond nutritional values have been gaining increasing research focus in recent years, although not much has been published on this subject in relation to bacterial infections. With respect to treatment, a novel antimicrobial strategy, which is expected to transcend problems with selective pressures for antibiotic resistance, is to interrupt bacterial communication, also known as quorum sensing (QS), by means of signal antagonists, the so-called QS inhibitors (QSIs). Furthermore, QSI agents offer a potential solution to the deficiencies associated with use of traditional antibiotics to treat infections caused by bacterial biofilms and multidrug-resistant bacteria. Several QSIs of natural origin have been identified, and in this study, several common food products and plants were extracted and screened for QSI activity in an attempt to isolate and characterize previously unknown QSI compounds active against the common opportunistic pathogen Pseudomonas aeruginosa. Several extracts displayed activity, but horseradish exhibited the highest activity. Chromatographic separation led to the isolation of a potent QSI compound that was identified by liquid chromatography-diode array detector-mass spectrometry (LC-DAD-MS) and nuclear magnetic resonance (NMR) spectroscopy as iberin—an isothiocyanate produced by many members of the Brassicaceae family. Real-time PCR (RT-PCR) and DNA microarray studies showed that iberin specifically blocks expression of QS-regulated genes in P. aeruginosa.
INTRODUCTION
The emergence of multidrug-resistant bacteria and problems with the use of conventional antibiotics to treat infections caused by bacterial biofilms have prompted scientists to ask whether we are approaching a postantibiotic era (1). Harsh selection pressures in combination with sheer overuse promote the evolution of multidrug-resistant bacteria that pose an increasing threat to human health (45, 60). Another major drawback of the use of conventional antibiotics is their failure to effectively clear chronic infections caused by biofilms, and new measures are required (15, 16).
Since the mid-1990s, novel approaches to the development of antimicrobial drugs have been the subject of extensive experimental work, primarily in academia. One such approach involves interrupting bacterial communication instead of killing the bacteria. Many bacteria regulate phenotypes that are essential to pathogenicity and/or symbiosis through chemical signaling, known as quorum sensing (QS) (57). A common theme in QS is that this takes place in a cell-density-dependent manner in which individual cells release small signal molecules to the surroundings to make their presence known. As more bacteria gather, the concentration of signal molecules gradually increases until a threshold concentration is reached, at which point a cellular response is generated (57). Research by our laboratory has offered compelling evidence that blocking this signaling network reinstates proper action of the immune system, which leads to the removal of the infecting bacteria (7, 23). Several potent QS inhibitors (QSIs) have been identified to date, many of which have been isolated from nature and modified by subsequent organic chemistry (23, 32, 54). This is to be expected, since QS and its countermeasures have presumably existed for millions of years.
Pseudomonas aeruginosa is a Gram-negative, motile aerobic bacterium listed among 6 pathogens identified by the Infectious Disease Society of America (IDSA) as primary targets for antimicrobial research (58). This opportunistic pathogen makes use of QS as a means to coordinate release of an arsenal of virulence factors that contribute greatly to its pathogenicity (12, 59). It is found in nosocomial and life-threatening infections of immunocompromised patients, such as AIDS patients and burn victims (59), and is the predominant cause of chronic lung infections in cystic fibrosis (CF) patients (18, 24, 34). Furthermore, recent work by us has revealed its presence in chronic wounds (17, 31). The P. aeruginosa QS system consist of two sensor systems based on the LuxRI homologues from Vibrio fisheri (19) denoted lasRI and rhlRI. lasI and rhlI encode the signal synthetase, which catalyzes the formation of the required signal molecules, and lasR and rhlR encode the receptor protein that binds the signal molecule followed by a transcription of QS genes. The LasIR system makes use of N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) as a cognate signal molecule, while the RhlIR system utilizes N-butanoyl homoserine lactone (C4-HSL) (37, 46, 65). The two systems work in a hierarchical manner, as the las-encoded system affects the expression of the rhl-encoded system (36, 48, 50). A third sensor system denoted the Pseudomonas quinolone signal (PQS) is present and operates between the two other systems (49). Several investigations have indicated that the maximal production of the PQS signal molecule (2-heptyl-3-hydroxy-4-quinolone) is initiated at the onset of stationary phase and is not regulated by cell population density (14, 39, 41).
Among QS-controlled factors in P. aeruginosa is the production of rhamnolipids, which leads to killing of polymorphonuclear (PMN) leukocytes of the host organism, paving the way for a successful infection by eliminating one of the host's first lines of defense (30). Rhamnolipids are glycolipids with strong surfactant abilities; in particular, one of the most abundant, rhamnolipid B, has been shown to cause necrosis of PMNs (30). The production of rhamnolipid is encoded by the rhlA, rhlB, and rhlC genes (44, 52). In a recent study (61), we showed that a much higher clearance of a rhlA mutant was observed in two different infectious animal models, which led us to hypothesize that a great deal of the tolerance of P. aeruginosa biofilm to agents of the immune system can be attributed to a protective “rhamnolipid shield.”
A field within medical science dealing with food with documented health-promoting or disease-preventing properties has been gaining increasing popularity and media exposure in recent years. It is commonly acknowledged today that a diet rich in fish and a moderate consumption of alcohol reduce the relative risk of coronary heart disease and that dairy products containing probiotics improve gastrointestinal health (27, 38, 55). It has been put forward that, combined with preexisting knowledge (such as family history), functional, tailor-made diets can be made for an individual according to which risk groups he or she falls within. In our experience, it is unlikely that any natural food source has biologically relevant amounts of QSIs sufficient to be considered for therapeutic purposes. However, food sources may offer preventative effects. This might especially be true in the case of CF patients and other groups prone to bacterial infections, for whom a diet enriched in QSI activity might show prophylactic properties.
In this study, a number of common natural food products and plants were tested for QSI activity, using three types of bacterial screens. Column chromatography was used for isolation of activity from crude samples and liquid chromatography-diode array detector-mass spectrometry (LC-DAD-MS) and nuclear magnetic resonance (NMR) spectroscopy for identification of active compounds. Armoracia rusticana (horseradish) stood out from a group of active crude extracts as highly active with respect to QSI activity against P. aeruginosa. Bioassay-guided fractionation and purification led to identification of 1-isothiocyanato-3-(methylsulfinyl) propane, commonly known as iberin, as an active QSI compound in horseradish. Real-time PCR (RT-PCR) and DNA microarray analysis of global gene expression revealed that iberin specifically and extremely effectively targets two of the major QS networks in P. aeruginosa, the LasIR and the RhlIR systems, and was found to downregulate QS-controlled rhamnolipid production in P. aeruginosa wild-type batch cultures.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli and P. aeruginosa strains used in this study are listed in Table 1. Cultures of QSIS1 (E. coli strain), lasB-gfp (P. aeruginosa), rhlA-gfp (P. aeruginosa), lasR and lasB-gfp (E. coli), luxR and luxI-gfp (E. coli), and rhlR and rhlA-lacZ (E. coli) strains used as reporters in QSI screens were grown in BT minimal medium (B medium [11] plus 2.5 mg of thiamine liter−1) supplemented with 10% A10 (11), 0.5% (wt/vol) glucose, and 0.5% (wt/vol) Casamino Acids for 16 h at 30°C and 180 rpm. Overnight cultures of wild-type P. aeruginosa strain PAO1, used for RNA purification for DNA microarray and RT-PCR analyses, were grown in BT media supplemented with 10% A10 and 0.5% (wt/vol) Casamino Acids at 37°C and 180 rpm. Animal experiments were performed with the wild-type P. aeruginosa strain (PAO1) obtained from Barbara Iglewski (University of Rochester Medical Center, Rochester, NY).
Table 1.
Strains used in the study
| Species | Name/genotype | Descriptiona | Reference |
|---|---|---|---|
| P. aeruginosa | PAO1 | Wild type | 25 |
| rhlA-gfp | Gmr-30γ; PAO1-ATCC, PrhlA-gfp(ASV)-Plac-lasR-mini-Tn5 derivative of pMHLAS | 66 | |
| lasB-gfp | Gmr-30γ; PAO1-ATCC, PlasB-gfp(ASV)-Plac-lasR-mini-Tn5 derivative of pMHLAS | 22 | |
| E. coli | QSIS1 | pUC18Not-luxR-PluxI-RBSII-phlA T0-T1, Apr-100γ maintained in CSH37 | 53 |
| rhlA-lacZ | rhlA-lacZ from pECP60 on pJPP8, tacP-rhlR | 47 | |
| lasB-gfp | MT102, PlasB-gfp(ASV)-Plac-lasR-mini-Tn5 derivative of pMHLAS | 22 | |
| luxI-gfp | pJBA132, Tcr; pME6031-luxR-PluxI-RBSII-gfp(ASV)-T0-T1 | 5 |
Gm, gentamicin; Ap, ampicillin; Tc, tetracycline.
Extraction of material from natural sources.
All crude extracts produced in this study were processed through a standard extraction procedure unless otherwise stated. The standard extraction procedure was as follows. Samples were pureed in a standard kitchen blender with approximately 200 ml of ethylacetate (EtOAc) followed by filtration through Whatman no. 4 filter paper. The remaining dry mass was reextracted with approximately 200 ml of methanol (MeOH) and filtered, with the solid residue extracted a third time with approximately 200 ml of sterile water (H2O). Volatile organic solvents were removed in a vacuum using a rotary evaporator, and residual water from the samples was removed by freeze-drying. Samples were finally redissolved in a minimal volume of an appropriate solvent for biological testing.
Dietary supplement tablets were crushed in a mortar and extracted with the set of solvents described above.
Column chromatography.
All chromatographic columns used in the study are listed in Table 2, while further parameters are detailed below.
Table 2.
All chromatographic columns used in the study
| Column type | Instrument | Specifications | Manufacturer |
|---|---|---|---|
| C18 | Isolera | SNAP cartridge (60 g, 81 mm by 39 mm) | Biotage |
| C18 | Semiprep 1 | Luna II (250 by 10 mm, 5 μm pore size) | Phenomenex |
| C18 | LC-DAD-MS | Luna II (50 by 2 mm, 3 μm pore size) | Phenomenex |
C18 columns were processed using a Biotage Isolera flash purification system (Biotage, Uppsala, Sweden) at a flow rate 30 ml/min, with the sample added as a dry load. Samples were collected as 100-ml fractions; the first sample was eluted with 10% MeOH–H2O, the following 10 samples were eluted with a 10% to 100% MeOH gradient (1,000 ml), and all subsequent samples were eluted with 100% MeOH (typically 500 ml).
C18 columns on a semipreparative instrument were run with a 322 pump and a 172 diode array detector attached to a G2250A fraction collector, all from Gilson (Semiprep 1; Gilson, Middleton, WI). The flow rate was set at 5 ml/min with various solvent gradients and time spans (detailed where appropriate in Results).
LC-DAD-MS.
Samples were processed through a C18 column with a high-performance LC (HPLC) and DAD system from the Agilent Technologies 1100 series, run at a flow rate of 0.3 ml/min with an acetonitrile (MeCN) gradient from 15% to 100% in water over a period of 20 min. Formic acid (20 mM) was added to both solvents. The analytes were subsequently positively ionized with the Micromass LockSpray feature and sent to a Micromass LCT mass spectrometer along with leucine-enkephalin as the reference. All MS data were analyzed with MassLynx version 4.1 from Waters' Laboratory Informatics Solutions (Waters Corporation, Milford, MA).
NMR.
Magnetic resonance spectra were recorded at 25°C on a Varian Mercury NMR spectrometer operating at 300 MHz for 1H and 75 MHz for 13C and a Varian Unity Inova spectrometer operating at 500 MHz for heteronuclear multiple-bond coherence (HMBC) and heteronuclear single-quantum coherence (HSQC), using a solvent signal as a reference (CD3OD at 3.3 ppm for 1H and at 49.1 ppm for 13C).
The general QSI screen.
Preparation of the QSIS1 diffusion screen was performed as described by Rasmussen et al. (53). A total of 20 μl of test substance was typically added directly to each well, but in the case of intense sample color, samples were administered via antibiotic disks. The plates were left at room temperature for 1 h before being incubated overnight at 30°C. For a detailed description of the method used, see reference 8.
Specific P. aeruginosa QSI screens.
Growth medium (150 μl of BT with 10% A10, 0.5% [wt/vol] Casamino Acids, and 0.5% [wt/vol] glucose) was added to each well in a 96-well microtiter dish (Black Isoplate; Perkin Elmer, Waltham, MA). Test samples (15 μl) mixed with 135 μl of growth medium were subsequently added into the first well of each row, and a 2-fold serial dilution along the rows was made, leaving the two last wells in each row for reference and all wells with 150 μl. An overnight culture (grown for 16 h at 30°C in BT with 10% A10, 0.5% [wt/vol] Casamino Acids, and 0.5% [wt/vol] glucose) of the lasB-gfp or rhlA-gfp monitor strain was added to a total volume of 300 μl to give a final optical density at 450 nm (OD450) of 0.1 (22, 66). Growth and green fluorescent protein (GFP) expression were monitored using a Victor X4 multilabel plate reader (Perkin Elmer, Waltham, MA) set at a constant temperature of 34°C and measuring every 15 min over a time course of 14 h. Growth was typically monitored at OD450 but in the case of highly colored samples was monitored at OD600. GFP expression was measured as fluorescence at an excitation wavelength of 485 nm and an emission wavelength of 535 nm. For a detailed description of the method used, see reference 8.
Competition of iberin and acyl-homoserine lactone (AHL) molecules.
The following three E. coli reporter strains were used: lasR lasB-gfp, luxR luxI-gfp, and rhlR rhlA-lacZ strains. The methods used for the Las and Lux bioassays were similar to the method described above, with the addition of the cognate signal molecules 3-oxo-C12-HSL and 3-oxo-C6-HSL, respectively, to the wells of the microtiter dish. For the Rhl bioassay, an overnight culture of the rhlR rhlA-lacZ strain was diluted to an OD600 of 0.05 and grown to an OD600 of 0.3 at 37°C with shaking (180 rpm). The culture was thereafter divided in 2-ml aliquots, different concentrations of iberin and C4-HSL were added, and the aliquots were then incubated for 90 min at 37°C. Cells were lysed with lysozyme (USB, Cleveland, OH) (1 mg/ml), and β-galactosidase was monitored using a β-galactosidase enzyme assay system with a reporter lysis buffer kit (Promega, Madison, WI).
Effect of serum albumin on QSI activity.
Neutralizing effects of serum albumin on the QSI activity of iberin were assessed by using the P. aeruginosa lasB-gfp monitor. Bovine serum albumin (BSA) was dissolved in the BT media (11) supplemented with 10% A10 (11), 0.5% (wt/vol) glucose, and 0.5% (wt/vol) Casamino Acids to a concentration of 100 mg/ml. A 300-μl volume was added to the first row of a 96-well microtiter dish (Black Isoplate; Perkin Elmer, Waltham, MA). A 150-μl volume of medium was added to the rest of the rows without serum albumin. A 2-fold serial dilution of serum albumin was made, and iberin was added to achieve the following final concentrations: 100 μM and 200 μM. Finally, 150 μl of the lasB-gfp monitor strain was added to all of the wells to achieve a final OD450 of 0.1. Measurements of GFP and growth were made as described above.
DNA microarray analysis of bacterial transcriptome.
Exponentially growing (OD600 of 0.5) cultures of P. aeruginosa PAO1 maintained at 37°C and 180 rpm in B medium supplemented with 10% A10 and 0.5% Casamino Acids were diluted to an OD600 of 0.05. After the culture reached an OD600 of 0.5, it was divided into 5 cultures of 50 ml and the following four concentrations of iberin were added: 8 μg/ml, 16 μg/ml, 32 μg/ml, and 64 μg/ml (no iberin was added to one culture). When the cultures reached an OD600 of 2.0, samples were retrieved and two volumes of RNAlater (Ambion, Austin, TX) were added. Isolation of RNA was performed using an RNeasy Mini purification kit (Qiagen) with on-column DNase treatment. Synthesis of cDNA and hybridization were performed by the microarray core unit at Rigshospitalet, Denmark, as follows. The gene expression levels were analyzed by the use of ArrayStar version 3.0 software (DNAstar, Madison, WI). DNA microarray analysis of global gene expression was performed according to guidelines provided by Affymetrix and was repeated three times with RNA from three individual growth experiments.
Real-time PCR.
The purified RNA used for DNA microarray was also used for RT-PCR. cDNA was made using 1 μg of RNA and High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). For quantitative real-time PCR, amplification was performed with Power SYBR green Master Mix in a Step-One-Plus Thermal Cycler (Applied Biosystems). The primers were designed using Primer Express 3.0 (Applied Biosystems). Forty cycles were run with denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 60°C for 45 s. The rpoD gene was used as a control for standardization.
The primer sequences were as follows: for rhlA forward, 5′-GGCGATCGGCCATCT-3′; for rhlA reverse, 5′-AGCGAAGCCATGTGCTGAT-3′; for lasB forward, 5′-CGACAACGCGTCGCAGTA-3′; for lasB reverse, 5′-AGGTAGAACGCACGGTTGTACA-3′; for rpoD forward, 5′-ACAAGATCCGCAAGGTACTGAAG-3′; and for rpoD reverse, 5′-CGCCCAGGTGCGAATC-3′.
Rhamnolipid measurements.
Samples for measurements of total rhamnolipid concentrations were retrieved from the cultures grown for DNA microarray and RT-PCR analysis at OD600s of 1.5 and 2.0 and kept at −80°C until further examinations. HPLC performed with high-resolution electrospray ionization mass spectrum (HRESIMS) detection (43) was used to quantify rhamnolipids as their [M + NH4]+ (peak area) based on external-standard quantification of an NMR-validated rhamnolipid B standard. A series of diluted standards was analyzed before and after the samples in order to minimize potential differences in the ionization levels of rhamnolipid between the samples. Other rhamnolipids were assumed to give the same ionization efficiency as rhamnolipid B. The total rhamnolipid concentration was derived from the six major rhamnolipids with the following masses [M + NH4]+: 668.4 (rhamnolipid B; C10-C10-rha-rha), 694.4 (C10-C12D-rha-rha), 696.4 (C10-C12-rha-rha), 522.4 (C10-C10-rha), 548.4 (C10-C12D-rha), and 550.4 (C10-C12-rha).
Animals.
Female BALB/c mice were purchased from Taconic M&B A/S (Ry, Denmark) at 8 to 9 weeks of age and were maintained on standard mouse chow and water ad libitum for a minimum period of 1 week before the challenge. All experiments were authorized by the National Animal Ethics Committee, Denmark.
Foreign-body infection model.
Silicone implants were prepared as described previously by Christensen et al. (10) with modifications. Silicone tube implants with a size of 4 mm (inner diameter, 4 mm; outer diameter, 6 mm) were used instead of square implants and were inserted in the intraperitoneal cavity. A bacterial pellet from a centrifuged overnight culture was resuspended in 0.9% NaCl to an optical density at 600 nm of 0.1. Animals were challenged according to the method of Christensen et al. (10). Mice were anesthetized by subcutaneous (sc) injections in the groin area with hypnorm/midazolam (Roche) (one part hypnorm [0.315 mg/ml fentanyl citrate and 10 mg/ml fluanisone], one part midazolam [5 mg/ml], two parts sterile water) at 10.0 ml/kg of body weight. Pentobarbital (200 mg/ml) with lidocainhydrochloride (DAK) (20 mg/ml) was injected intraperitoneally (ip) to euthanize the mice at the termination of the experiments.
Bacteriology.
After removal from the interperitoneal cavity of the mice, silicone implants were placed in centrifuge tubes containing 2 ml of 0.9% NaCl and keep on ice until the tubes were placed in a ultrasound bath (Bransonic model 2510; Branson Ultrasonic Corporation) for 10 min (5 min of degas followed by 5 min of sonic treatment). After the ultrasound treatment, 100 ml of the solution consisting of NaCl and bacteria was serially diluted and plated on blue agar plates (State Serum Institute, Denmark) for bacterial visualization. Blue agar plates are selective for Gram-negative bacilli (24). The plates were incubated at room temperature for 2 days before determination of CFU per implant.
Preparation of iberin solution for mice experiments.
Iberin purchased from LKT Laboratories (St. Paul, MN) was diluted in 96% ethanol to a concentration of 32 mg/ml followed by a 40× dilution in 0.9% NaCl. The mice were injected with 0.2 ml of the final solution, corresponding to 8 μg/g of body weight. The placebo group was injected with a 2.4% ethanol solution (96% ethanol–0.9% NaCl) corresponding to the amount of ethanol that the iberin-treated group received. Mice were treated every 12 h from day 2 preinsertion to day 2 postinsertion, and treatment was continued until 12 h before the mice were euthanized.
Statistical analysis.
For analyzing the transcriptomic data, the Mann-Whitney U test was used for calculating P values with ArrayStar version 3.0 software (DNAstar, Madison, WI). P values ≤ 0.05 were considered significant.
To compare the bacterial counts (CFU) between two groups of mice, the Mann-Whitney U-test (analysis of nonparametric data) was used for calculating P values in the statistical program GraphPad Prism, version 5.0 (GraphPad Software Inc., San Diego, CA).
RESULTS
Screening for QSIs.
The general QSIS1 screen rules out growth-inhibitory activity in the extracts and was applied as an initial high-throughput screen. Only samples that displayed positive activity in the screen were taken further to the specific P. aeruginosa screen (based on a lasB-gfp fusion or a rhlA-gfp fusion), with the exception of ginkgo biloba (MeOH extraction), kan jang, and vanilla, which made interpretations in the QSIS1 screen difficult due to intense color. The following extracts from natural sources were subjected to the QSIS1 screen: allspice, asparagus, banana, basil (dried), blueberry, broccoli, Camembert cheese, cardamom, cauliflower, cayenne pepper, celery, celery salt, chervil, cinnamon, coriander, cumin, dill, eggplant, elderberry, estragon, fennel, garden cress, ginger, ginkgo biloba (EtOH plus MeOH extraction), goji berries, green tea, hops, horseradish, Iceland moss, juniper berries, kan jang, kiwifruit, kumquat, laurel leaves, lemongrass, lime, liquorice root candy, Lucerne sprouts, marjoram, mint, mustard powder, natural yoghurt, nutmeg, oregano, oyster mushroom, Parmesan cheese, parsley, parsnip, pepper corns, pineapple, pomegranate, Port Salut cheese, potatoes, Pu-erh tea, radish, red onions, rosemary, shiitake mushrooms, shrimp, star anise, strawberry, sugar peas, Tasmanian blue gum, thyme, tomato, turmeric, vanilla, water cress, and white poppy seeds. Table 3 lists extracts giving a positive result with the QSIS1 screen. A number of candidates previously reported to exhibit QSI activity gave negative test results in the present study. These included vanilla (which was shown to inhibit QS in Chromobacterium violaceum), blueberry, cinnamon, oregano, basil, turmeric, and strawberry, which were previously shown to inhibit swarming motility of P. aeruginosa as well as QS in C. violaceum (9, 62). Of all the samples tested for QSI activity, horseradish, Tasmanian blue gum, and ginkgo biloba (MeOH extraction) extracts showed the clearest activity in both screening systems, and of those three, horseradish showed remarkably better inhibitory activity than the other two extracts and was therefore chosen for further testing.
Table 3.
All extracts of natural sources that tested positive in the QSIS1 screen and gave various results in the lasB-gfp and rhlA-gfp screena
| Sample | QSI activity in indicated screen |
|
|---|---|---|
| QSIS1 | lasB-gfprhlA-gfp | |
| Cauliflowerb | + | − |
| Celery saltc | + | − |
| Chervilb | + | − |
| Garden cressb | + | − |
| Ginkgo bilobad (MeOH) | + | ++ |
| Horseradishb | + | +++ |
| Lemongrassb | + | − |
| Parmesan cheeseb | + | − |
| Radishb | + | − |
| Rosemaryb | + | + |
| Tasmanian blue gumc | + | ++ |
| Thymeb | + | − |
| Water cressb | + | − |
+, low QSI activity; ++, medium QSI activity; +++, strong QSI activity; −, no QSI activity.
Fresh products purchased at local grocery stores.
Dried material bought in a herbal shop.
Fresh leaves obtained from the environment.
Purification of iberin from horseradish.
Horseradish is a well-known producer of allyl isothiocyanate; however, this compound proved growth inhibitory at all concentrations tested, suggesting that the observed QSI activity was caused by other compounds.
Horseradish roots were exhaustively extracted by macerating with EtOAc and stirring overnight. After filtration, the solids were extracted a second time with MeOH. The solvents were combined and removed under vacuum; the solid was then redissolved in a minimal volume of EtOAc and partitioned using water. The resulting organic extract was highly oily and not suitable for processing in a water-based C18 matrix. Consequently, it was adsorbed to 50 g of C18 and eluted in two steps, first with MeOH and subsequently with CH2Cl2. Activity was observed only in the clear, yellowish MeOH fraction, which was adsorbed to 5 g of C18 and chromatographed using a flash reverse-phase column and a sharp gradient from 10% to 100% MeOH in H2O, with collection performed every 100 ml. QSI activity was detected in fractions 4 and 5 (40% to 60% MeOH). These fractions were combined and further purified by semipreparative HPLC using a Phenomenex C18 column (5 μm pore size; 250 by 10 mm) at 5 ml/min, starting at 10% MeOH and increasing to 40% over 10 min, with fractions collected every 30 s. Activity was seen in the four fractions collected between 7 and 9 min. LCMS analysis showed the third active fraction, eluting between 8 and 8.5 min, to primarily represent one compound.
The compound was obtained as a pale, yellow oil with a pungent odor. A single UV maximum at 242 nm was observed in the UV spectrum, suggesting very little conjugation. High-resolution electrospray ionization mass spectra (HRESIMS) showed the presence of at least one nitrogen and two sulfur atoms and gave a molecular formula of C5H9NOS2 [HRESI(+)MS m/z calculated for C5H9NOS2] ([M+ + H], 164.0204; found, 164.0202). 1H and 13C NMR data (Table 4) were in very good agreement with published data (35); predicted chemical shifts (ACD Labs) and data from authentic samples (LKT Laboratories, St. Paul, MN) confirmed the compound to be the known isothiocyanate iberin (Fig. 1).
Table 4.
1H and 13C NMR data for iberina
| Position | δC (ppm) | δH mult (J in Hz) |
|---|---|---|
| 1 | 45.0 | 3.77 t (7) |
| 2 | 24.5 | 2.14 ddt (7, 8, 8) |
| 3α | 51.4 | 2.96 dt (8, 13.5) |
| 3β | 51.4 | 2.84 dt (8, 13.5) |
| 4 | 38.2 | 2.68 s |
| 5 | 132.8 |
Recorded in CDCl3. δC, carbon chemical shift value; δH, proton chemical shift value; mult, multiplicity of proton NMR peaks; J, coupling constant(s) in hertz (Hz).
Fig 1.
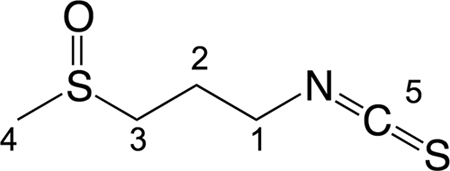
Chemical structure of iberin. The numbers refer to the positions of 13C obtained by NMR data (see Table 4).
QSI activity of iberin.
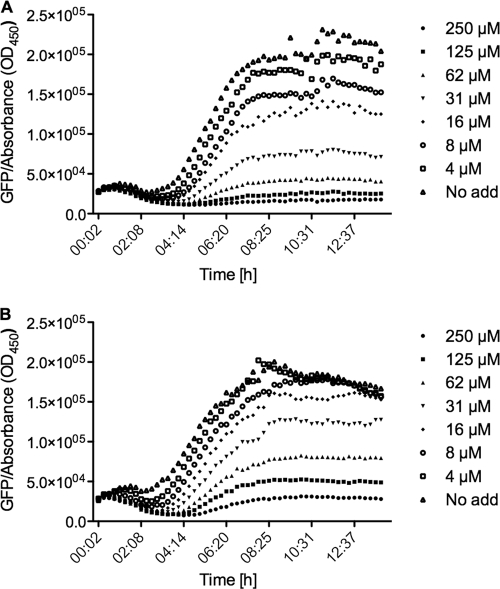
Pure iberin, purchased from LKT Laboratories (St. Paul, MN), displayed activity in the QSIS1 screen (data not shown) and inhibited expression of the lasB-gfp and rhlA-gfp genes in the P. aeruginosa screens. The results of the P. aeruginosa screens illustrated in Fig. 2 show that iberin is capable of completely inhibiting expression of the lasB-gfp fusion at approximately 100 μM (16 μg/ml) without affecting growth. A slightly weaker inhibition was observed in the rhlA-gfp fusion, where approximately 50% of gfp expression was inhibited at 100 μM. Concentrations above 1.0 mM were found to cause significant growth inhibition of P. aeruginosa.
Fig 2.
Dose-response curves (specific fluorescence) of iberin incubated with the QS monitor lasB-gfp (A) and rhlA-gfp (B) strains. No add, no addition of iberin.
Binding of iberin to regulator proteins.
To investigate whether iberin specifically targets one of the QS regulatory R proteins, competition experiments were done with the E. coli lasR lasB-gfp, luxR luxI-gfp, and rhlR rhlA-lacZ reporter strains and their cognate signal molecules. A number of concentrations of iberin and AHL were tested against each other. The results showed no decrease in expression of the Las reporter with non-growth-inhibitory concentrations of iberin, whereas a very clear concentration-dependent decrease in expression could be obtained with both the Rhl and Lux reporters and an almost complete inhibition at a concentration of 31 μM iberin (Fig. 3). The concentrations of the cognate signal molecules used were selected in relation to the lowest concentration where no decrease in AHL-mediated induction could be measured. The concentrations of 3-oxo-C12-HSL, 3-oxo-C6-HSL, and C4-HSL were 25 nm, 25 nM, and 20 μM, respectively.
Fig 3.

Competition between iberin and AHL signal molecules. The reporter lasR lasB-gfp (E. coli) (light gray bars), rhlR rhlA-lacZ (E. coli) (medium gray bars), and luxR-PluxI-gfp (E. coli) (dark gray bars) strains were treated with different concentrations of iberin and with the addition of the following concentrations of their cognate signal molecules: 25 nM, 20 μM, and 25 nM, respectively. Cultures with no addition of iberin were defined as 100%. No add, no addition of iberin. The results are based on three independent experiments. Error bars represent means ± standard deviations (SD).
Interference with rhamnolipid production.
Batch cultures of wild-type PA01 were grown with iberin concentrations of 8 μg/ml (50 μM), 16 μg/ml (100 μM), 32 μg/ml (200 μM), and 64 μg/ml (400 μM), all of which had no effect on the growth of the bacteria compared to a nontreated control according to optical density (OD) measurements. Samples were withdrawn from the batches at OD600s of 1.5 and 2.0 for rhamnolipid measurements. The OD values were obtained after 220 min (OD = 1.5) and 300 min (OD = 2.0) of growth with an exponentially growing culture.
The HPLC measurements showed a steadily decreasing amount of rhamnolipid with increasing iberin concentration in the growth medium, suggesting a concentration-dependent downregulation of rhamnolipid production (Fig. 4). The production of rhamnolipid by P. aeruginosa was completely abolished with a treatment of iberin at 32 μg/ml, and there was no rhamnolipid production at an OD600 of 1.5 in a culture treated at 8 μg/ml. There was an approximately 10-fold-lower concentration of rhamnolipid in the cultures treated with 8 μg/ml compared to the concentration seen with the untreated culture at OD600s of both 1.5 and 2.0. Furthermore, the measurements showed a boost in the concentration of rhamnolipids when the cultures grew from an OD600 of 1.5 to 2.0, which is coherent with the generally acknowledged model of QS-regulated production of rhamnolipids.
Fig 4.
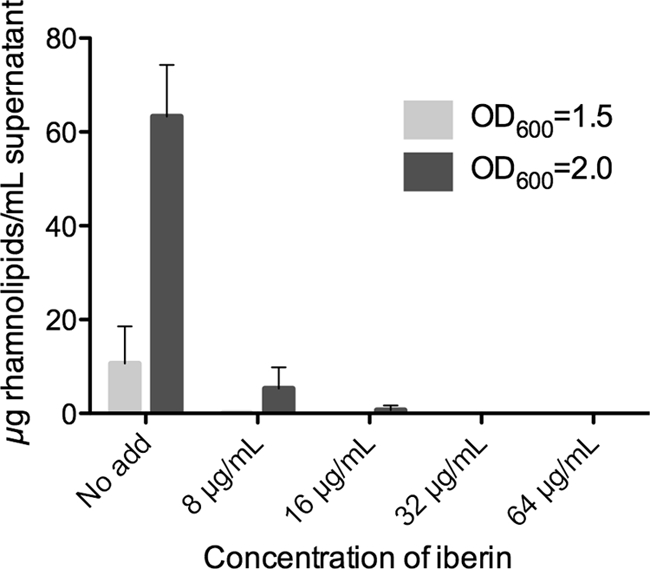
Measurements of rhamnolipid concentrations in batch cultures of P. aeruginosa grown with different concentrations of iberin. Samples were taken at OD600s of 1.5 and 2.0, and measurements were performed with HPLC. No add, no addition of iberin. The results are based on three independent experiments. Error bars represent means ± SD.
RT-PCR and DNA microarray.
RT-PCR and transcriptomic analyses were performed on purified RNA from the same samples taken at an OD600 of 2.0. We choose rhlA and lasB as target genes for RT-PCR because of their central role in virulence. The results, shown in Fig. 5, demonstrate substantial concentration-dependent downregulation of both genes. Expression of lasB appeared to be inhibited substantially more strongly than that of rhlA. The data are based on the results of three independent experiments. It is not possible to establish a significant difference from the standard deviation of the results regarding expression of the lasB transcript determined with the iberin treatments at 32 μg/ml and 64 μg/ml. However, the three independent experiments all demonstrated a substantial downregulation, with 533-, 61-, and 54-fold changes in response to treatment with 32 μg/ml and 928-, 520-, and 133-fold changes in response to treatment with 64 μg/ml.
Fig 5.

Fold change in gene expression of lasB (dark gray bars) and rhlA (light gray bars) in P. aeruginosa grown with various concentrations of iberin measured by real-time PCR. The results are based on three independent experiments. Error bars represent means ± SD.
The cutoff for genes included in the analysis of the microarray data was represented by genes significantly (P < 0.05, Student's t test) downregulated more than 5-fold (Table 5).
Table 5.
Genes significantly downregulated (more than 5-fold) by iberina
| Probe set | Gene | QS | Description of product | Downregulation by iberin at indicated concn (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 |
16 |
32 |
64 |
||||||||
| Fold change | P value | Fold change | P value | Fold change | P value | Fold change | P value | ||||
| PA0059 | osmC | X | Osmotically inducible protein OsmC | −1.3 | 1 | −3.5 | <0.01 | −5.1 | <0.01 | −4.9 | <0.01 |
| PA0122 | X | Conserved hypothetical protein | −2.5 | 0.55 | −9.8 | 0.02 | −27.0 | <0.01 | −46.2 | <0.01 | |
| PA0200 | Hypothetical protein | −1.8 | 0.93 | −2.2 | 0.08 | −3.3 | 0.02 | −6.2 | <0.01 | ||
| PA0852 | cbpD | X | Chitin-binding protein CbpD precursor | −5.8 | 0.43 | −17.8 | <0.01 | −24.7 | <0.01 | −31.4 | <0.01 |
| PA1130 | rhlC | X | Rhamnosyltransferase 2 | −1.4 | 1 | −2.1 | 0.06 | −3.6 | <0.01 | −5.1 | <0.01 |
| PA1323 | X | Hypothetical protein | −1.3 | 1 | −4.2 | <0.01 | −6.0 | <0.01 | −6.4 | <0.01 | |
| PA1324 | X | Hypothetical protein | −1.1 | 1 | −3.5 | 0.03 | −5.3 | <0.01 | −5.0 | <0.01 | |
| PA1708 | popB | Translocator protein PopB | −4.5 | 0.11 | −5.3 | <0.01 | −4.6 | <0.01 | −6.7 | <0.01 | |
| PA1711 | exsE | ExsE | −3.2 | 0.36 | −3.7 | <0.01 | −4.0 | 0.01 | −5.8 | <0.01 | |
| PA1869 | X | Probable acyl carrier protein | −3.7 | 0.14 | −6.1 | <0.01 | −11.4 | <0.01 | −14.0 | <0.01 | |
| PA1871 | lasA | X | LasA protease precursor | −3.9 | 0.43 | −10.1 | <0.01 | −39.6 | <0.01 | −68.4 | <0.01 |
| PA1901 | phzC1, phzC2 | X | Phenazine biosynthesis protein PhzC | −2.5 | 0.93 | −7.0 | 0.03 | −14.5 | <0.01 | −14.7 | <0.01 |
| PA1902 | phzD1, phzD2 | X | Phenazine biosynthesis protein PhzD | −2.8 | 0.96 | −6.4 | 0.02 | −11.7 | <0.01 | −10.8 | <0.01 |
| PA1903 | phzE1, phzE2 | X | Phenazine biosynthesis protein PhzE | −2.5 | 0.86 | −7.8 | 0.02 | −18.7 | <0.01 | −19.2 | <0.01 |
| PA1904 | phzF1, phzF2 | X | Probable phenazine biosynthesis protein | −2.8 | 0.66 | −6.6 | 0.02 | −13.0 | <0.01 | −15.4 | <0.01 |
| PA1905 | phzG1, phzG2 | X | Probable pyridoxamine 5′-phosphate oxidase | −3.1 | 0.41 | −9.9 | 0.01 | −24.4 | <0.01 | −29.0 | <0.01 |
| PA1999 | dhcA | DhcA, dehydrocarnitine CoA transferase, subunit A | −1.8 | 1 | −3.4 | 0.06 | −5.1 | <0.01 | −4.9 | <0.01 | |
| PA2000 | dhcB | DhcB, dehydrocarnitine CoA transferase, subunit B | −2.0 | 1 | −3.4 | 0.06 | −5.6 | <0.01 | −6.1 | <0.01 | |
| PA2030 | X | Hypothetical protein | −3.5 | 0.08 | −5.2 | <0.01 | −5.6 | <0.01 | −5.3 | <0.01 | |
| PA2031 | X | Hypothetical protein | −3.3 | 0.03 | −5.6 | <0.01 | −7.6 | <0.01 | −7.6 | <0.01 | |
| PA2067 | Probable hydrolase | −5.1 | 0.47 | −5.6 | 0.02 | −6.4 | 0.02 | −7.2 | 0.01 | ||
| PA2069 | X | Probable carbamoyl transferase | −6.3 | 0.21 | −13.3 | <0.01 | −16.0 | <0.01 | −17.5 | <0.01 | |
| PA2146 | Conserved hypothetical protein | −5.2 | 0.66 | −13.1 | <0.01 | −19.9 | <0.01 | −21.1 | <0.01 | ||
| PA2190 | Conserved hypothetical protein | −2.3 | 1 | −5.5 | <0.01 | −6.1 | <0.01 | −7.1 | <0.01 | ||
| PA2193 | hcnA | X | Hydrogen cyanide synthase HcnA | −1.3 | 1 | −2.6 | 0.04 | −6.0 | <0.01 | −9.1 | <0.01 |
| PA2194 | hcnB | X | Hydrogen cyanide synthase HcnB | −1.3 | 1 | −2.0 | 0.18 | −4.9 | 0.01 | −6.2 | <0.01 |
| PA2195 | hcnC | X | Hydrogen cyanide synthase HcnC | −1.1 | 1 | −1.7 | 0.42 | −3.7 | 0.05 | −6.4 | 0.02 |
| PA2300 | chiC | X | Chitinase | −8.7 | 0.38 | −28.1 | <0.01 | −48.2 | <0.01 | −57.3 | <0.01 |
| PA2366 | Uricase PuuD | −1.6 | 1 | −1.9 | 0.10 | −2.4 | 0.04 | −5.5 | <0.01 | ||
| PA2384 | Hypothetical protein | −2.3 | 0.81 | −3.1 | 0.03 | −3.7 | 0.02 | −6.7 | <0.01 | ||
| PA2433 | X | Hypothetical protein | −1.1 | 1 | −3.4 | 0.02 | −6.1 | <0.01 | −5.3 | <0.01 | |
| PA2566 | X | Conserved hypothetical protein | −2.9 | 0.30 | −4.7 | <0.01 | −6.6 | <0.01 | −8.8 | <0.01 | |
| PA2570 | lecA | X | LecA | −15.0 | 0.09 | −29.8 | <0.01 | −42.3 | <0.01 | −47.8 | <0.01 |
| PA2588 | X | Probable transcriptional regulator | −3.3 | 0.22 | −3.9 | <0.01 | −4.9 | <0.01 | −6.9 | <0.01 | |
| PA2939 | X | Probable aminopeptidase | −4.8 | 0.56 | −7.0 | 0.01 | −8.7 | <0.01 | −9.1 | <0.01 | |
| PA3326 | X | Probable Clp-family ATP-dependent protease | +1.0 | 1 | −1.9 | 0.05 | −5.0 | 0.01 | −8.8 | <0.01 | |
| PA3331 | X | Cytochrome P450 | +3.5 | 0.05 | +1.1 | 0.85 | −2.9 | 0.03 | −5.1 | <0.01 | |
| PA3361 | lecB | X | Fucose-binding lectin PA-IIL | −1.5 | 1 | −5.0 | 0.06 | −13.8 | <0.01 | −23.8 | <0.01 |
| PA3477 | rhlR | X | Transcriptional regulator RhlR | −1.8 | 0.23 | −2.3 | <0.01 | −3.2 | <0.01 | −5.3 | <0.01 |
| PA3478 | rhlB | X | Rhamnosyltransferase chain B | −2.1 | 0.75 | −6.4 | 0.02 | −18.0 | <0.01 | −42.1 | <0.01 |
| PA3479 | rhlA | X | Rhamnosyltransferase chain A | −1.8 | 0.65 | −5.8 | 0.02 | −22.2 | <0.01 | −59.0 | <0.01 |
| PA3520 | X | Hypothetical protein | −3.2 | 0.48 | −5.5 | <0.01 | −9.3 | <0.01 | −9.4 | <0.01 | |
| PA3692 | lptF | X | Lipotoxon F, LptF | +1.1 | 1 | −3.5 | 0.04 | −5.0 | <0.01 | −4.5 | <0.01 |
| PA3724 | lasB | X | Elastase LasB | −4.1 | 0.42 | −11.1 | <0.01 | −46.8 | <0.01 | −89.8 | <0.01 |
| PA3923 | Hypothetical protein | −3.4 | 0.88 | −5.0 | 0.04 | −7.2 | 0.02 | −9.0 | 0.01 | ||
| PA4129 | X | Hypothetical protein | −2.2 | 1 | −5.7 | 0.06 | −8.8 | <0.01 | −9.0 | <0.01 | |
| PA4130 | X | Probable sulfite or nitrite reductase | −2.1 | 1 | −5.1 | 0.05 | −7.0 | <0.01 | −6.8 | <0.01 | |
| PA4131 | X | Probable iron−sulfur protein | −2.7 | 0.24 | −4.9 | 0.01 | −6.7 | <0.01 | −6.9 | <0.01 | |
| PA4132 | X | Conserved hypothetical protein | −2.3 | 0.93 | −3.5 | 0.02 | −4.5 | <0.01 | −5.2 | <0.01 | |
| PA4133 | X | Cytochrome c oxidase subunit (cbb3 type) | −3.4 | 0.72 | −6.5 | <0.01 | −11.0 | <0.01 | −10.2 | <0.01 | |
| PA4134 | X | Hypothetical protein | −2.3 | 0.93 | −4.1 | 0.03 | −5.1 | 0.02 | −5.8 | <0.01 | |
| PA4141 | X | Hypothetical protein | −2.3 | 0.46 | −6.6 | 0.01 | −50.7 | 0.02 | −284.1 | <0.01 | |
| PA4142 | X | Probable secretion protein | −5.1 | 0.01 | −9.1 | <0.01 | −13.1 | <0.01 | −16.8 | <0.01 | |
| PA4175 | piv | X | Protease IV | −5.4 | 0.24 | −8.4 | <0.01 | −10.6 | <0.01 | −14.1 | <0.01 |
| PA4209 | phzM | X | Probable phenazine-specific methyltransferase | −1.5 | 1 | −3.1 | 0.12 | −5.8 | 0.02 | −7.2 | 0.02 |
| PA4211 | phzB1, phzB2 | X | Probable phenazine biosynthesis protein | −3.3 | 0.46 | −20.5 | 0.02 | −69.4 | <0.01 | −75.0 | <0.01 |
| PA4217 | phzS | X | Flavin-containing monooxygenase | −1.3 | 1 | −5.2 | 0.07 | −11.9 | <0.01 | −15.7 | <0.01 |
| PA4738 | X | Conserved hypothetical protein | +1.0 | 1 | −2.7 | 0.10 | −3.9 | <0.01 | −5.2 | <0.01 | |
| PA4739 | X | Conserved hypothetical protein | −1.0 | 1 | −3.2 | 0.04 | −4.7 | <0.01 | −6.9 | <0.01 | |
| PA5170 | arcD | Arginine/ornithine antiporter | −1.7 | 1 | −2.6 | 0.07 | −3.8 | 0.02 | −8.3 | <0.01 | |
| PA5171 | arcA | Arginine deiminase | −1.1 | 1 | −1.8 | 0.31 | −2.5 | 0.10 | −5.1 | 0.02 | |
| PA5172 | arcB | Ornithine carbamoyltransferase, catabolic | −1.3 | 1 | −2.1 | 0.27 | −2.5 | 0.14 | −5.0 | 0.03 | |
| PA5220 | X | Hypothetical protein | −2.5 | 1 | −4.3 | 0.03 | −5.8 | 0.02 | −8.3 | <0.01 | |
| PA5475 | Hypothetical protein | −1.5 | 1 | −2.0 | 0.24 | −2.9 | 0.11 | −5.2 | 0.03 | ||
Genes significantly (P < 0.05; Student's t test) downregulated (more than 5-fold) by iberin are indicated with gray shading. “X” entries in the QS column mark genes previously identified as QS regulated by Hentzer et al. (23). The results are based on three independent experiments. CoA, coenzyme A.
Of 64 genes that matched the specified criteria, 49 had previously been identified as QS genes (23). This means that roughly 76% of the most-downregulated genes are under the control of QS in P. aeruginosa; in general, the genes were downregulated in a concentration-dependent manner, which means that iberin is specific for QS-controlled genes. Among the particularly interesting genes downregulated by iberin were genes encoding different virulence factors, namely, rhlA (PA3479) and rhlB (PA3478), which are involved in production of rhamnolipids, piv (PA4175), which codes for protease IV, the cytotoxic galactophilic lectin lecA (PA2570), lasA (PA1871) and lasB (PA3724), which are involved in production of elastase, cbpD (PA0852) and chiC (PA2300), which encode a chitin binding protein precursor and chitinase, respectively, the phenazine-modifying genes phzM (PA4209), encoding a putative phenazine-specific methyltransferase, and phzS (PA4217), encoding a flavin-containing mooxygenase, which are involved in the biosynthesis of pyocyanin (40), the phzC-phzG operon (phzC-G; PA1901 to 1905), and phzAB (PA4210 to 4211), encoding phenazine biosynthesis proteins. All these genes contribute significantly to the virulence and pathogenicity of P. aeruginosa.
Only one of the central components of the QS network in P. aeruginosa, rhlR (PA3477), appears to have been significantly and potently (>5-fold) downregulated in the culture treated with 64 μg/ml iberin, whereas rhlI was only moderately (approximately 4-fold) downregulated, and lasR (<2-fold downregulated) and lasI (<2-fold upregulated) appear to have been nearly unaffected.
As shown by a comparison of the iberin QS-downregulated gene data with the analysis of expression profiles of the lasR and rhlR single and lasR-rhlR double mutants by Skindersoe et al. (56), expression of 70.5% of the genes required functional lasR and rhlR, 15.9% required a functional lasR, and 13.6% required a functional rhlR. In our survey, we used the definition of QS-regulated genes determined by Hentzer et al. (23).
Among the 51 genes that were significantly and highly upregulated in the presence of iberin (Table 6), expression of the three genes in the mexEF-oprN operon, encoding components of a resistance-nodulation-cell division (RND) efflux pump, was seen. mexE (PA2493), encoding a membrane fusion protein, mexF (PA2494), encoding an efflux transporter, and oprN (PA2495), encoding an outer membrane protein, were all highly upregulated (from approximately 21- to 100-fold) in the culture that had been treated with 64 μg/ml. According to Hentzer et al. (23), the mexEF genes are also upregulated in response to treatment with furanone C-30 (23). Furthermore, the sbp gene (PA0283), encoding a sulfate-binding protein precursor, was significantly upregulated (>5-fold) in the culture treated at 64 μg/ml. Several genes encoding components of ABC transporters and two genes encoding (probable) oxidoreductases were upregulated (>100-fold). Furthermore, five genes encoding hypothetical proteins were extensively upregulated (>100-fold).
Table 6.
Genes significantly upregulated (more than 5-fold) by iberina
| Probe set | Gene | Description of product | Upregulation by iberin at indicated concn (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8 |
16 |
32 |
64 |
|||||||
| Fold change | P value | Fold change | P value | Fold change | P value | Fold change | P value | |||
| PA0201 | Hypothetical protein | −1.1 | 1 | +1.4 | 0.46 | +3.3 | 0.05 | +9.0 | <0.01 | |
| PA0283 | sbp | Sulfate-binding protein precursor | +1.0 | 1 | +1.3 | 0.54 | +3.3 | 0.05 | +7.6 | 0.02 |
| PA0284 | Hypothetical protein | −1.1 | 1 | +1.8 | 0.53 | +5.6 | 0.11 | +11.7 | 0.04 | |
| PA0422 | Conserved hypothetical protein | +2.4 | 0.93 | +2.9 | 0.05 | +4.7 | 0.02 | +5.6 | 0.03 | |
| PA1210 | Conserved hypothetical protein | +4.4 | 0.10 | +5.7 | <0.01 | +9.5 | <0.01 | +14.8 | <0.01 | |
| PA1317 | cyoA | Cytochrome o ubiquinol oxidase subunit II | −1.3 | 1 | +1.3 | 0.46 | +3.8 | <0.01 | +6.4 | <0.01 |
| PA1319 | cyoC | Cytochrome o ubiquinol oxidase subunit III | +1.3 | 0.88 | +1.3 | 0.10 | +2.8 | <0.01 | +5.8 | <0.01 |
| PA1332 | Hypothetical protein | +1.3 | 0.89 | +2.0 | 0.02 | +3.6 | <0.01 | +9.8 | <0.01 | |
| PA1333 | Hypothetical protein | +3.3 | 0.49 | +4.8 | 0.03 | +9.1 | <0.01 | +16.4 | <0.01 | |
| PA1334 | Probable oxidoreductase | +21.8 | <0.01 | +53.6 | <0.01 | +99.9 | <0.01 | +142.4 | <0.01 | |
| PA1493 | cysP | Sulfate-binding protein of ABC transporter | −1.0 | 1 | +1.3 | 0.51 | +2.9 | 0.04 | +5.6 | 0.02 |
| PA1743 | Hypothetical protein | +1.2 | 1 | +2.2 | 0.05 | +5.6 | 0.06 | +16.1 | <0.01 | |
| PA1744 | Hypothetical protein | +2.9 | 0.66 | +8.9 | 0.03 | +27.8 | <0.01 | +80.4 | <0.01 | |
| PA1970 | Hypothetical protein | +8.2 | 0.50 | +16.9 | 0.01 | +39.6 | <0.01 | +100.5 | <0.01 | |
| PA2279 | arsC | ArsC protein | +1.2 | 1 | +1.6 | <0.01 | +2.4 | <0.01 | +5.6 | <0.01 |
| PA2359 | Probable transcriptional regulator | +1.0 | 1 | +1.3 | 0.26 | +2.2 | 0.03 | +5.7 | <0.01 | |
| PA2486 | Hypothetical protein | +6.2 | 0.66 | +12.8 | 0.04 | +35.7 | <0.01 | +95.4 | <0.01 | |
| PA2491 | Probable oxidoreductase | +4.6 | <0.01 | +7.6 | <0.01 | +16.2 | <0.01 | +30.1 | <0.01 | |
| PA2493 | mexE | Resistance-nodulation-cell division (RND) multidrug efflux membrane fusion protein MexE precursor | +5.0 | 0.41 | +18.8 | <0.01 | +56.8 | <0.01 | +101.5 | <0.01 |
| PA2494 | mexF | Resistance-nodulation-cell division (RND) multidrug efflux transporter MexF | +2.3 | 0.43 | +6.8 | <0.01 | +25.1 | <0.01 | +51.8 | <0.01 |
| PA2495 | oprN | Multidrug efflux outer membrane protein OprN precursor | +1.5 | 0.93 | +3.1 | <0.01 | +9.6 | <0.01 | +21.4 | <0.01 |
| PA2575 | Hypothetical protein | +6.6 | <0.01 | +10.1 | <0.01 | +15.5 | <0.01 | +21.1 | <0.01 | |
| PA2579 | kynA | l-Tryptophan:oxygen 2.3-oxidoreductase (decyclizing) KynA | +1.6 | 0.71 | +2.3 | 0.06 | +5.6 | <0.01 | +5.9 | <0.01 |
| PA2580 | Conserved hypothetical protein | +4.5 | 0.22 | +7.4 | <0.01 | +14.9 | <0.01 | +15.3 | <0.01 | |
| PA2759 | Hypothetical protein | +19.1 | 0.24 | +55.1 | <0.01 | +113.4 | <0.01 | +265.8 | <0.01 | |
| PA2811 | Probable permease of ABC-2 transporter | +1.3 | 1 | +2.3 | 0.01 | +5.6 | <0.01 | +11.6 | <0.01 | |
| PA2812 | Probable ATP-binding component of ABC transporter | +1.4 | 0.99 | +2.0 | <0.01 | +4.5 | <0.01 | +10.3 | <0.01 | |
| PA2813 | Probable glutathione S-transferase | +2.1 | 0.48 | +3.9 | <0.01 | +10.4 | <0.01 | +23.5 | <0.01 | |
| PA2845 | Hypothetical protein | +1.8 | 0.41 | +2.9 | <0.01 | +7.7 | <0.01 | +17.0 | <0.01 | |
| PA2932 | morB | morphinone reductase | −1.0 | 1 | +1.2 | 0.22 | +2.0 | 0.07 | +7.7 | <0.01 |
| PA3229 | Hypothetical protein | +24.9 | <0.01 | +63.7 | <0.01 | +132.1 | <0.01 | +213.0 | <0.01 | |
| PA3240 | Conserved hypothetical protein | +4.2 | 0.12 | +6.0 | <0.01 | +7.8 | <0.01 | +10.3 | <0.01 | |
| PA3443 | Probable permease of ABC transporter | −1.1 | 1 | +1.1 | 0.78 | +1.6 | 0.12 | +6.9 | 0.01 | |
| PA3444 | Conserved hypothetical protein | +1.1 | 1 | +1.5 | 0.14 | +2.7 | <0.01 | +11.2 | 0.01 | |
| PA3445 | Conserved hypothetical protein | +1.1 | 1 | +1.3 | 0.07 | +2.2 | 0.01 | +8.5 | 0.02 | |
| PA3446 | Conserved hypothetical protein | −1.2 | 1 | +1.2 | 0.43 | +2.4 | 0.07 | +12.1 | <0.01 | |
| PA3450 | Probable antioxidant protein | −1.1 | 1 | +1.7 | 0.25 | +4.9 | 0.03 | +12.3 | <0.01 | |
| PA3783 | Hypothetical protein | +4.2 | <0.01 | +4.4 | <0.01 | +7.0 | <0.01 | +9.3 | <0.01 | |
| PA3931 | Conserved hypothetical protein | −1.2 | 1 | +1.5 | 0.39 | +3.3 | 0.04 | +9.6 | <0.01 | |
| PA3938 | Probable periplasmic taurine-binding protein precursor | −1.2 | 1 | −1.2 | 0.64 | +1.7 | 0.13 | +5.0 | 0.02 | |
| PA4167 | Probable oxidoreductase | +34.2 | <0.01 | +71.4 | <0.01 | +128.8 | <0.01 | +142.6 | <0.01 | |
| PA4354 | Conserved hypothetical protein | +2.4 | 0.05 | +3.6 | <0.01 | +9.1 | <0.01 | +22.4 | <0.01 | |
| PA4356 | xenB | Xenobiotic reductase | +2.4 | 0.24 | +4.3 | <0.01 | +9.9 | <0.01 | +20.4 | <0.01 |
| PA4385 | groEL | GroEL protein | +3.2 | 0.48 | +3.4 | 0.03 | +4.7 | 0.01 | +5.1 | <0.01 |
| PA4623 | Hypothetical protein | +9.5 | 0.16 | +22.6 | <0.01 | +55.1 | <0.01 | +132.3 | <0.01 | |
| PA4761 | dnaK | DnaK protein | +2.8 | 0.66 | +2.8 | 0.05 | +4.1 | 0.02 | +5.7 | <0.01 |
| PA4881 | Hypothetical protein | +138.2 | 0.03 | +227.5 | <0.01 | +497.0 | <0.01 | +746.8 | <0.01 | |
| PA4918 | Hypothetical protein | +3.0 | 0.72 | +4.0 | 0.02 | +5.0 | <0.01 | +6.9 | <0.01 | |
| PA4919 | pncB1 | Nicotinate phosphoribosyltransferase | +3.0 | 0.47 | +3.2 | <0.01 | +4.0 | <0.01 | +7.2 | <0.01 |
| PA5082 | Probable binding protein component of ABC transporter | −1.2 | 1 | −1.0 | 0.91 | +1.6 | 0.38 | +7.4 | 0.02 | |
| PA5083 | Conserved hypothetical protein | −1.2 | 1 | −1.1 | 0.70 | +1.5 | 0.20 | +7.6 | 0.01 | |
Genes significantly (P < 0.05; Student's t test) upregulated more than 5-fold by iberin are indicated with gray shading. The results are based on three independent experiments.
Foreign-body infection model.
Mice were divided into two groups of 11 (iberin and placebo) 2 days before insertion of silicone implants precolonized with wild-type P. aeruginosa into the peritoneal cavity. The mice received treatments at 12-h intervals and were euthanized at day 3 postinsertion, at which time the implants were collected for determination of CFU per implant. No significant difference in clearances between the iberin-treated and placebo groups could be found (P < 0.94) (data not shown).
Structure/activity relationship.
To investigate the effect on gene expression exerted by molecular changes in the structure of iberin, four analogs (sulforaphane, alyssin, cheirolin, and iberverin) of iberin were tested for QSI activity in the lasB-gfp and rhlA-gfp screen and 50% inhibitory concentrations (IC50s) were calculated (Table 7). Cheirolin, sulforaphane, and iberverin showed considerable QSI activity, with IC50 values approximately 2 to 10 times higher than those seen with iberin, whereas alyssin showed very high IC50s. These data suggest that a sulfinyl or sulfonyl group is important for QSI activity and that the length of the carbon chain should consist of 3 carbons rather than 4 or 5. As shown by comparisons of iberin and cheirolin, a sulfinyl group seems to have a stronger QSI effect than a sulfonyl group. Iberverin and cheirolin exhibited a much stronger growth-inhibitory effect toward P. aeruginosa than the three other compounds. A structural difference is that iberverin and cheirolin contain a sulfur and sulfonyl group, respectively, whereas the three other compounds contain sulfinyl groups.
Table 7.
Chemical structures and IC50s of iberin and iberin analogs calculated for QSI monitor lasB-gfp and rhlA-gfp strainsa
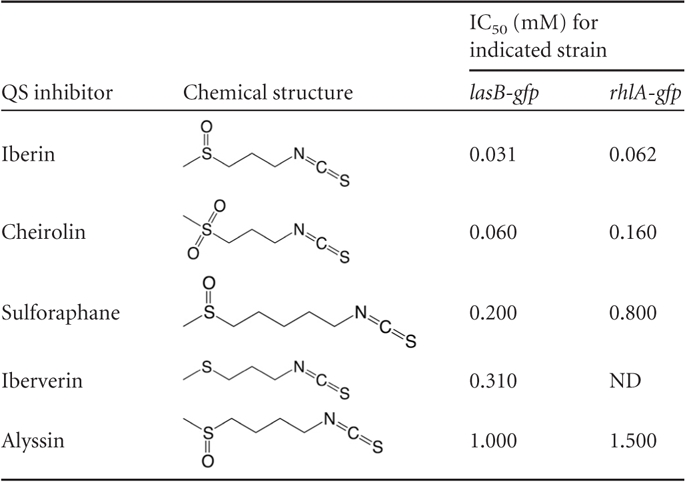
Chemical structures and IC50s of iberin and iberin analogs calculated for QSI monitor lasB-gfp and rhlA-gfp strains are shown. ND, not detectable because of growth inhibition.
DISCUSSION
The increased knowledge of QS systems gained in the last decades has paved the way for development of drugs that fight bacterial infections in a manner completely different from that of today's selection of antibiotics. Instead of attacking the bacterium's basal life processes, it has been proposed that inhibition of its pathogenicity would produce a more subtle method of infectious control that would reduce the harsh selection pressure seen with conventional antibiotics. Disrupting the QS signaling pathway is one possible way of attaining this goal, since production and secretion of important virulence factors in many bacteria, including P. aeruginosa, are not dependent on growth per se but are instead dependent on cell-cell signaling mediated through functional QS systems. This approach has been referred to as the antipathogenic drug principle by us (21).
In the present study, 69 common food products and plants were screened for QSI activity by the use of the QSIS1 screen. Thirteen of the tested crude extracts showed positive activity, and 3 of them showed positive activity in the P. aeruginosa QSI screens. This is likely to reflect that some QSIs may show specificity for the LuxIR QS system (operating the QSIS1 screen) compared with the RhlIR and LasIR QS systems operating in P. aeruginosa. The samples might also vary with respect to membrane permeability in the two strains. On the other hand, it is a possibility that by using the QSIS1 as an initial screen, extracts with activity exclusively toward P. aeruginosa were excluded. However, our experience from screening a multitude of extracts over the years has given us the impression that the QSIS1 screen is far more promiscuous compared with the P. aeruginosa QSI screens and therefore suitable as a general first screen.
The purification work on horseradish led to the isolation and identification of iberin as the QSI compound. Iberin inhibited the Lux QS system in the QSIS1 screen and was able to completely inhibit the expression of lasB without affecting growth in the P. aeruginosa screen at a concentration of 100 μM (16 μg/ml). Roughly 50% of gfp expression of the rhlA-gfp monitor was inhibited at the same concentration, without growth being affected. The rhlA gene is a part of the rhlAB operon encoding components for the biosynthesis of rhamnolipid (13). HPLC measurements of total rhamnolipid concentrations in batch cultures of P. aeruginosa treated with different concentrations of iberin confirmed that iberin attenuates the QS-controlled production of rhamnolipids in wild-type P. aeruginosa in a concentration-dependent manner, with a completely blockage at a concentration of 200 μM (32 μg/ml) iberin. This means that a treatment with iberin in a relative low concentration should have the potential to eliminate one of countermeasures employed by P. aeruginosa against the host immune system. The effect on gene expression in the P. aeruginosa screen was confirmed by RT-PCR measurements. Concentration-dependent downregulations of the rhlA and lasB genes were seen, with approximately 100-fold (32 μg/ml) and 230-fold (64 μg/ml) differences for the rhlA gene and approximately 220-fold (32 μg/ml) and 580-fold (64 μg/ml) for the lasB gene.
For a more comprehensive investigation of the effect of iberin on P. aeruginosa gene expression, we employed a microarray analysis with the same 4 concentrations of iberin as used for RT-PCR and rhamnolipid measurements. Sixty-four genes (approximately 1.1% of the total 5,570 P. aeruginosa genes) were significantly downregulated, and 49 (77%) of these genes are QS controlled, according to our previous work performed under identical experimental conditions (23). In comparison, the antimicrobial compounds patulin and penicillic acid produced by Penicillium species targeted 157 genes (49% QS-controlled genes) and 300 genes (34% QS-controlled genes), respectively (54). The synthetic furanone C-30 targeted 163 genes (80% QS-regulated genes) (23). Of the 49 QS-controlled genes downregulated by iberin, several encode important virulence factors, including lasA, lasB, rhlAB, chiC, lecA, piv, phzC-G, and phzAB. Of the genes encoding the central components of the QS system, only rhlR was significantly downregulated by iberin treatments. Patulin, penicillic acid, and furanone C-30 did not cause in any noticeable effect on lasRI or rhlRI gene expression (23, 54).
Competition experiments with reporter fusions in the E. coli background were employed to identify the mode of action of iberin. The experiments showed that iberin strongly repressed rhlA-lacZ expression but not lasB-gfp expression, which supports a model by which iberin affects the QS circuits by initially targeting and blocking binding of C4-HSL to the RhlR regulator protein. The study also showed that iberin repressed luxR-PluxI-gfp expression, which further supports the model of iberin being a competitive inhibitor of the binding sites of short- and medium-chain length AHL molecules. It is a general conception that molecules capable of competing with the native AHL molecules for binding to the regulator proteins need to have a structural similarity to the AHL in question. Iberin does not exhibit strong structural similarity to AHL molecules; to be able to classify iberin as a “classic” competitor for the AHL binding site, more investigations need to be carried out. Such experiments could entail iberin being tested with strains carrying specific mutations in the LuxR AHL binding site as previously done with furanones (33).
In order to assess structure/activity relationships, IC50s of iberin and four of its analogs were calculated. There are two functional iberin groups: isothiocyanate and sulfinyl. Taking into consideration that structural analogs of iberin that contain the sulfinyl (or sulfonyl) functional group and the shortest carbon chain (3 or 4 carbons) display the highest QSI activity suggests that the QSI activity of iberin can predominantly be attributed to the methylsulfinyl group and the length of the carbon chain. A possible contribution of the isothiocyanate group cannot be determined from these investigations; it might contribute indirectly to activity by other means. In the study of Amara et al. (3), the authors demonstrated that isothiocyanate-substituted analogs of 3-oxo-AHL bind covalently to the cys79 residue in the OdDHL binding pocket of LasR. The authors recorded a 3-fold reduction in expression of a lasI-luxCDABE reporter (present in a wild-type background) and a modest 1.5-fold reduction in pyocyanin production. Iberin, which exhibits QS-dependent regulation of synthesis very similar to that seen with pyocyanin, causes a significant reduction of rhamnolipid gene expression and synthesis. In addition, Amara et al. (3) reported no effect on the transcriptome of P. aeruginosa. The obvious lack of a thorough analysis of QS-regulated gene expression, taken together with the data provided by Amara et al. (3), supports a model in which the isothiocyanate group of iberin may contribute only insignificantly to the observed bioactivity of iberin and its derivatives against bacterial QS. The reported modest effect on two QS-controlled genes in combination with a sulfur-containing compound, ajoene, one of the major constituents of QSI compounds in garlic (T. H. Jakobsen, M. van Gennip, R. K. Phipps, M. S. Shanmugham, L. D. Christensen, M. Alhede, M. E. Skindersoe, T. B. Rasmussen, K. Friedrich, F. Uthe, P. Ø. Jensen, C. Moser, K. F. Nielsen, L. Eberl, T. O. Larsen, D. Tanner, N. Høiby, T. Bjarnsholt, and M. Givskov, submitted for publication), supports the view that QSI bioactivity may primarily relate to the sulfoxide group present in both molecules. In addition, Koch et al. (33) showed that, by replacement of a carbonyl group with a sulfur atom in 3-oxo-C6-HSL and C6-HSL, the molecules changed their activity from being agonists to antagonists of the Lux QS system, which further supports the notion of the importance of sulfur in relation to inhibitors of QS.
Iberin was tested in an in vivo foreign-body infection mouse model (10), and the results showed no significantly difference in bacterial clearance between treated and nontreated mice (data not shown). Previously published data from a study performed with furanone C-30 showed significant differences in clearance of P. aeruginosa from the implants in the treated mice (10). Also, ajoene showed a strong antimicrobial effect and promoted bacterial clearance in a pulmonary infectious mouse model (10a). It is therefore puzzling that iberin, with its strong, reductive effect on rhamnolipid production in vitro, is not capable of exerting a significant attenuation of bacterial populations in the foreign-body infection model. Rhamnolipids are considered to constitute a shield against attacking PMN leukocytes (2, 6, 30). Recent data published by us from work performed with similar infectious models (61) showed that an rhlA mutant of P. aeruginosa is attenuated. However, data provided by Amara et al. (3) suggest that the apparent deficiency of iberin as a functional QS inhibitor in vivo may in fact be attributable to the reactive isothiocyanate group, which may react with host-encoded proteins and therefore eliminate its bioactivity in vivo. One such eliminating agent could be BSA. The QSI activity of iberin was not lowered when it was tested for bioactivity in the presence of BSA. However, we recorded a major upregulation of mexEF in the culture treated with 64 μg/ml iberin, suggesting that the bacteria responded by inducing this important efflux pump and thereby exporting iberin from the cell.
According to SciFinder scholar searches, iberin has not been isolated from horseradish before, but it has been found in a variety of cruciferous vegetables, such as cabbage and broccoli (26, 51). Considering that the active compound identified from horseradish is an isothiocyanate, it is surprising that there are not more food products from the family of Brassicaceae (crucifers), such as radish, broccoli, and cauliflower, that showed QSI activity. They are commonly known to produce a variety of isothiocyanates, iberin being one of the most common ones (64). Furthermore, sulforaphane is found in broccoli, cauliflower, and Brussels sprouts and iberverin in cabbages (64). Cauliflower, garden cress, and radish did originally show activity on the QSIS1 but lost it when tested in the P. aeruginosa screen. This was presumably due to volatilization during the various drying or solvent removal processes. A diet rich in cruciferous vegetables has been found to be correlated with a decreased risk of development of common forms of cancer, including lung, colon, and breast cancer (4, 20, 63). Iberin has more specifically been found to inhibit growth and induce apoptosis of human neuroblastoma and glioblastoma cells as well as inducing phase II detoxification enzymes in rats (28, 29, 42). This makes iberin an obvious functional food ingredient, as it is a common constituent in the diet of people around the world and elicits activity not only as a QSI but also as an anticancer agent. It remains, however, unknown whether iberin is present in biologically relevant amounts in horseradish or whether horseradish contains iberin-stabilizing components that help to provide a natural prophylaxis against bacterial infections. Fresh garlic extract, for example, shows a much more pronounced effect on the transcriptome of P. aeruginosa (7) than synthesized ajoene (Jakobsen et al., submitted for publication). Garlic extracts target expression of a multitude of QS-regulated genes (50 genes in total compared with 11 genes targeted by synthesized ajoene). In fact, we have found that synthesized ajoene, upon subsequent purification, lost close to 100% of its activity in in vivo infectious models, as determined by LC-DAD-MS (our unpublished data). An epidemiological study on the correlation of intake of food products known to contain QSIs (such as horseradish, garlic, cabbage, etc.) and the incidence/severity of bacterial infections in immunocompromised patients (such as CF patients) would perhaps shed some light on the relevance of functional food in relation to a natural QSI prophylaxis.
ACKNOWLEDGMENTS
This work was supported by a grant from the Danish Strategic Research Council and by a grant from the Novo Nordisk Foundation to M.G.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1.Alanis AJ. 2005. Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36: 697–705 [DOI] [PubMed] [Google Scholar]
- 2.Alhede M, et al. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155: 3500–3508 [DOI] [PubMed] [Google Scholar]
- 3.Amara N, et al. 2009. Covalent inhibition of bacterial quorum sensing. J. Am. Chem. Soc. 131: 10610–10619 [DOI] [PubMed] [Google Scholar]
- 4.Ambrosone CB, et al. 2004. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. J. Nutr. 134: 1134–1138 [DOI] [PubMed] [Google Scholar]
- 5.Andersen JB, et al. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67: 575–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjarnsholt T, et al. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151: 373–383 [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt T, et al. 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151: 3873–3880 [DOI] [PubMed] [Google Scholar]
- 8.Bjarnsholt T, et al. 2010. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. Nat. Protoc. 5: 282–293 [DOI] [PubMed] [Google Scholar]
- 9.Choo JH, Rukayadi Y, Hwang JK. 2006. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 42: 637–641 [DOI] [PubMed] [Google Scholar]
- 10.Christensen LD, et al. 2007. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153: 2312–2320 [DOI] [PubMed] [Google Scholar]
- 10a.Christensen LD, et al. 2012. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. [Epub ahead of print.] doi:10.1093/jac/dks002 [DOI] [PubMed] [Google Scholar]
- 11.Clark DJ, Maaloe O. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23: 99–112 [Google Scholar]
- 12.Cox CD. 1993. Iron and the virulence of Pseudomonas aeruginosa, p 41–58 In Fick RB., Jr (ed), Pseudomonas aeruginosa the opportunist: pathogenesis and disease. CRC Press, Boca Raton, FL [Google Scholar]
- 13.Déziel E, Lépine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149: 2005–2013 [DOI] [PubMed] [Google Scholar]
- 14.Diggle SP, et al. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50: 29–43 [DOI] [PubMed] [Google Scholar]
- 15.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15: 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drenkard E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5: 1213–1219 [DOI] [PubMed] [Google Scholar]
- 17.Fazli M, et al. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47: 4084–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frederiksen B, Koch C, Høiby N. 1997. Antibiotic treatment of initial colonization with Pseudomonas aeruginosa postpones chronic infection and prevents deterioration of pulmonary function in cystic fibrosis. Pediatr. Pulmonol. 23: 330–335 [DOI] [PubMed] [Google Scholar]
- 19.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heber D, Bowerman S. 2001. Applying science to changing dietary patterns. J. Nutr. 131: 3078S–3081S [DOI] [PubMed] [Google Scholar]
- 21.Hentzer M, Eberl L, Nielsen J, Givskov M. 2003. Quorum sensing: a novel target for the treatment of biofilm infections. BioDrugs 17: 241–250 [DOI] [PubMed] [Google Scholar]
- 22.Hentzer M, et al. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148: 87–102 [DOI] [PubMed] [Google Scholar]
- 23.Hentzer M, et al. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22: 3803–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Høiby N. 1974. Epidemiological investigations of the respiratory tract bacteriology in patients with cystic fibrosis. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 82: 541–550 [PubMed] [Google Scholar]
- 25.Holloway BW, Morgan AF. 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40: 79–105 [DOI] [PubMed] [Google Scholar]
- 26.Howard LA, Jeffery EH, Wallig MA, Klein BP. 1997. Retention of phytochemicals in fresh and processed broccoli. J. Food Sci. 62: 1098–1104 [Google Scholar]
- 27.Hu FB, et al. 2002. Fish and omega-3 fatty acid intake and risk of coronary heart disease in women. JAMA 287: 1815–1821 [DOI] [PubMed] [Google Scholar]
- 28.Jadhav U, Ezhilarasan R, Vaughn SF, Berhow MA, Mohanam S. 2007. Dietary isothiocyanate iberin inhibits growth and induces apoptosis in human glioblastoma cells. J. Pharmacol. Sci. 103: 247–251 [DOI] [PubMed] [Google Scholar]
- 29.Jadhav U, Ezhilarasan R, Vaughn SF, Berhow MA, Mohanam S. 2007. Iberin induces cell cycle arrest and apoptosis in human neuroblastoma cells. Int. J. Mol. Med. 19: 353–361 [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen PO, et al. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153: 1329–1338 [DOI] [PubMed] [Google Scholar]
- 31.Kirketerp-Møller K, et al. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46: 2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjelleberg S, et al. 1997. Do marine natural products interfere with prokaryotic AHL regulatory systems? Aquat. Microb. Ecol. 13: 85–93 [Google Scholar]
- 33.Koch B, et al. 2005. The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology 151: 3589–3602 [DOI] [PubMed] [Google Scholar]
- 34.Koch C, Hoiby N. 1993. Pathogenesis of cystic fibrosis. Lancet 341: 1065–1069 [DOI] [PubMed] [Google Scholar]
- 35.Kore AM, Spencer GF, Wallig MA. 1993. Purification of the omega-(methylsulfinyl)alkyl glucosinolate hydrolysis products: 1-isothiocyanato-3-(methylsulfinyl)propane, 1-isothiocyanato-4-(methylsulfinyl)butane, 4-(methylsulfinyl)butanenitrile and 5-(methylsulfinyl)pentanenitrile from broccoli and Lesquerella fendleri. J. Agric. Food Chem. 41: 89–95 [Google Scholar]
- 36.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21: 1137–1146 [DOI] [PubMed] [Google Scholar]
- 37.Latifi A, et al. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17: 333–343 [DOI] [PubMed] [Google Scholar]
- 38.Lee YK, Salminen S. 1995. The coming of age of probiotics. Trends Food Sci. Technol. 6: 241–245 [Google Scholar]
- 39.Lépine F, Deziel E, Milot S, Rahme LG. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim. Biophys. Acta 1622: 36–41 [DOI] [PubMed] [Google Scholar]
- 40.Mavrodi DV, et al. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183: 6454–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKnight SL, Iglewski BH, Pesci EC. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182: 2702–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munday R, Munday CM. 2004. Induction of phase II detoxification enzymes in rats by plant-derived isothlocyanates: comparison of allyl isothiocyanate with sulforaphane and related compounds. J. Agric. Food Chem. 52: 1867–1871 [DOI] [PubMed] [Google Scholar]
- 43.Nielsen KF, Dalsgaard PW, Smedsgaard J, Larsen TO. 2005. Andrastins A-D, Penicillium roqueforti metabolites consistently produced in blue-mold-ripened cheese. J. Agric. Food Chem. 53: 2908–2913 [DOI] [PubMed] [Google Scholar]
- 44.Ochsner UA, Fiechter A, Reiser J. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269: 19787–19795 [PubMed] [Google Scholar]
- 45.Pantosti A, Moro ML. 2005. Antibiotic use: the crystal ball for predicting antibiotic resistance. Clin. Infect. Dis. 40: 1298–1300 [DOI] [PubMed] [Google Scholar]
- 46.Pearson JP, Passador L, Iglewski BH, Greenberg EP. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92: 1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179: 5756–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pesci EC, Iglewski BH. 1997. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 5: 132–135 [DOI] [PubMed] [Google Scholar]
- 49.Pesci EC, et al. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96: 11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesci EC, Pearson JP, Seed PC, Iglewski BH. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179: 3127–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prochazka Z, Severa Z. 1960. On the bound form of ascorbic acid 15. A new method of isolation of ascorbigen and iberin from savoy cabbage. Collect. Czechoslov. Chem. Commun. 25: 1100–1103 [Google Scholar]
- 52.Rahim R, et al. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40: 708–718 [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen TB, et al. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187: 1799–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmussen TB, et al. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151: 1325–1340 [DOI] [PubMed] [Google Scholar]
- 55.Rimm EB, Klatsky A, Grobbee D, Stampfer MJ. 1996. Review of moderate alcohol consumption and reduced risk of coronary heart disease: is the effect due to beer, wine, or spirits? BMJ 312: 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skindersoe ME, et al. 2008. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52: 3648–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swift S, Throup JP, Williams P, Salmond GPC, Stewart G. 1996. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21: 214–219 [PubMed] [Google Scholar]
- 58.Talbot GH, et al. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42: 657–668 [DOI] [PubMed] [Google Scholar]
- 59.Van Delden C, Iglewski BH. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4: 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanderkooi OG, Low DE, Green K, Powis JE, McGeer A. 2005. Predicting antimicrobial resistance in invasive pneumococcal infections. Clin. Infect. Dis. 40: 1288–1297 [DOI] [PubMed] [Google Scholar]
- 61.Van Gennip M, et al. 2009. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 117: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vattem DA, Mihalik K, Crixell SH, McLean RJC. 2007. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia 78: 302–310 [DOI] [PubMed] [Google Scholar]
- 63.Verhoeven DTH, Goldbohm RA, van Poppel G, Verhagen H, van den Brandt PA. 1996. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol. Biomarkers Prev. 5: 733–748 [PubMed] [Google Scholar]
- 64.WHO 2004. Cruciferous vegetables isothiocyanates and indoles. Handbook on cancer prevention, vol. 9 IARC, Lyon, France [Google Scholar]
- 65.Winson MK, et al. 1995. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92: 9427–9431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L, et al. 2009. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 53: 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]