Abstract
We evaluated phylogenetic clustering of bacterial and archaeal communities from redox-dynamic subtropical forest soils that were defined by 16S rRNA and rRNA gene sequences. We observed significant clustering for the RNA-based communities but not the DNA-based communities, as well as increasing clustering over time of the highly active taxa detected by only rRNA.
TEXT
Most microbial community analyses based on rRNA genes treat taxa as though they are independent. This implicitly assumes equal phylogenetic distances between taxa, although various distances of relatedness are well known. The distribution of relatedness among taxa is the community's phylogenetic structure, and it can be measured using phylogenetic tree branch lengths and taxon abundances (15, 27). Microbial communities, much like plants, arthropods, or vertebrates, tend to be more phylogenetically structured than would be expected by chance (3, 11, 12, 24). Phylogenetic structure can reveal contemporary ecological interactions (24). Negative interactions (like competition) result in overdispersion—assemblages made up of distantly related organisms—which is observed in plant communities (3, 24); positive ecological interactions (like environmental filtering) result in clustering. For example, phylogenetic clustering of bacteria is observed for rRNA gene copy numbers, suggesting phylogenetic coherence according to the growth rate trait, one ecological strategy (20, 22).
While the number of ribosomal operons within a genome is correlated with the growth rate potential under conditions of abundant resources (22), rRNA gene-based microbial community analysis can include DNA from dead or dormant populations (6), structural DNA from biofilms (21), or contaminating DNA (18, 23). Environmental rRNA can yield information on a community that is or has recently been active (10, 14, 25). rRNA gene abundances are not perfectly correlated with cell number, and rRNA abundances are even less so, but rRNA has the advantage of providing a wider dynamic range of abundance than rRNA genes, from tens of copies to 105 copies per cell depending upon the metabolic state (1, 7, 13, 19). Combining RNA- and DNA-based microbial community analyses may reveal a portion of the microbial community that is either active or primed to become active within the seed bank of organisms.
To evaluate differences in phylogenetic structure among DNA- and RNA-based communities, we analyzed data from an experiment designed to assess microbial adaptation to redox fluctuation in tropical forest soils (5). Soil cores sampled from Luquillo forest in Puerto Rico were placed in microcosms, and three biological replicates (4 cores each) were subjected to static anoxic, static oxic, or fluctuating redox conditions, alternating between 4 days of N2 and 4 days of air over 32 days (see Fig. S1 in the supplemental material). Redox treatments resulted in changes in nitrous oxide and methane production but continuously high carbon dioxide production, indicative of continuous activity (5). For high-density 16S rRNA microarray analysis, PhyloChip G2 was used to identify communities based on rRNA and rRNA genes. Here we use these data to compare the phylogenetic clusterings of the RNA- and DNA-based communities. The net relatedness index (NRI) and nearest taxon index (NTI) assess phylogenetic structure, incorporating phylogenetic distance (26) and taxon relative abundance as measured by PhyloChip hybridization scores (2). The NRI is based on mean pairwise distances among taxa and is a measure of the overall tree-wide structure, while the NTI is based on mean distance to the nearest neighbor and is a measure of local clustering. NRI or NTI values significantly greater than zero suggest greater community clustering compared to that of a null (random) model.
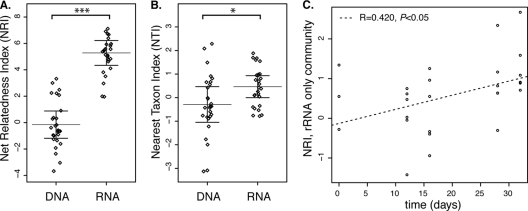
Communities defined by rRNA were significantly more clustered than rRNA gene-defined communities when measured by NRI (P < 0.001) (Fig. 1A) or NTI (P < 0.05) (Fig. 1B). Differences in clustering between RNA- and DNA-based communities were likely not due to differences in detection (16). The average richness values for the RNA- and DNA-measured communities were 1,230 and 1,494 taxa, respectively, and not significantly different. There was substantial overlap between the two communities, with about 80% of Bacteria and 60 to 90% of Archaea detected by RNA as well as DNA (see Table S1 in the supplemental material).
Fig 1.
The Phylocom package comstruct was used to calculate the NRIs and NTIs (26). The mean phylogenetic distance (MPD) and mean nearest phylogenetic taxon distance (MNTD) for each sample are compared to the MPD and MNTD values for null communities. The null model consists of species drawn randomly from the sample pool without replacement. Tree-wide clustering metric NRI (as measured by MPD) (A) and branch tip clustering metric NTI (as measured by MNTD) (B) of the rRNA- and rRNA gene-based microbial communities (RNA and DNA, respectively). Graphs display the data (jittered for clarity), with bars denoting the mean and one standard error of three replicates (n = 27). (C) Plot of net relatedness indexes (NRI) of the community detectable by rRNA but not by rRNA genes over time, with linear models displayed. The original data are also available in tabular format (see Table S3 in the supplemental material).
Significant clustering of the RNA-based community (NRI = 5.28 ± 0.98 [mean ± standard error]) but not the DNA-based community (NRI = −0.152 ± 1.04) suggests that the more active subset of the total community exhibits phylogenetic coherence within the tree defined by all taxa detected. Such phylogenetic coherence suggests that the functional characteristics responsible for activity may share some common evolutionary origins. Phylogenetic clustering was absent from DNA-based communities, suggesting a preservation of phylogenetic diversity among the seed bank populations.
Taxa detected by rRNA only, likely relatively rare taxa with high activity, showed significantly increasing NRIs over time (Fig. 1C; see also Table S2 in the supplemental material). Taxa detected in RNA communities but absent in DNA communities likely had DNA concentrations below our detection limit but measurable rRNA levels. This “RNA-only” population was 5.0 to 12.5% of the total detected bacteria and between 0.8 and 6.0% of archaea (see Table S1 in the supplemental material). The trend toward increasing clustering over time suggests a convergence in community phylogenetic structure among the highly active taxa due to selective pressure of the lab incubation itself, conditions which depart substantially from field conditions. That this trend was only observed with the RNA-only community may suggest that this high-activity, relatively rare set of bacteria has distinct strategies for stress response.
Phylogenetic structure is a net result of forces that shape communities. Significant clustering may be evidence of positive interactions, such as phenotypic attraction and spatial isolation; overdispersion can result from negative interactions, such as competition, predation, minimal niche overlap, or the presence of many niches (4, 27). We observed no phylogenetic community structure in DNA-based communities, suggesting that both positive and negative interactions have shaped the indigenous soil microbial community. Evidence for clustering in the RNA-based communities suggests phylogenetic coherence among those taxa actively producing ribosomes in response to dynamic environmental conditions. This may be a result of growth, a change in ribosome abundance within cells resulting in altered protein production, or both. This first comparison of phylogenetic clustering in both RNA- and DNA-based microbial communities demonstrates that this type of community analysis can enhance our understanding of the forces that shape complex microbial community structures in nature.
ACKNOWLEDGMENTS
We acknowledge Whendee Silver and Ellen Simms for helpful comments.
This work was funded in part by NSF grant DEB-0089783, a Seaborg Fellowship to K.M.D., and the DOE-LBNL contract DE-AC02-05CH11231.
Footnotes
Published ahead of print 27 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Binder BJ, Liu YC. 1998. Growth rate regulation of rRNA content of a marine Synechococcus (Cyanobacterium) strain. Appl. Environ. Microbiol. 64: 3346–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brodie EL, et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bryant JA, et al. 2008. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U. S. A. 105: 11505–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12: 693–715 [DOI] [PubMed] [Google Scholar]
- 5. DeAngelis KM, Silver WL, Thompson AW, Firestone MK. 2010. Microbial communities acclimate to recurring changes in soil redox potential status. Environ. Microbiol. 12: 3137–3149 [DOI] [PubMed] [Google Scholar]
- 6. De Nobili M, Contin M, Mondini C, Brookes P. 2001. Soil microbial biomass is triggered into activity by trace amounts of substrate. Soil Biol. Biochem. 33: 1163–1170 [Google Scholar]
- 7. Dortch Q, Roberts TL, Clayton J, Jr, Ahmed S. 1983. RNA/DNA ratios and DNA concentrations as indicators of growth rate and biomass in planktonic marine organisms. Mar. Ecol. Prog. Ser. 13: 61–71 [Google Scholar]
- 8. Reference deleted.
- 9. Reference deleted.
- 10. Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66: 5488–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horner-Devine MC, Bohannan BJM. 2006. Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: 100–108 [DOI] [PubMed] [Google Scholar]
- 12. Horner-Devine MC, et al. 2007. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353 [DOI] [PubMed] [Google Scholar]
- 13. Kerkhof L, Kemp P. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30: 253–260 [DOI] [PubMed] [Google Scholar]
- 14. Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9: 119–130 [DOI] [PubMed] [Google Scholar]
- 15. Reference deleted.
- 16. Manini E, Danovaro R. 2006. Synoptic determination of living/dead and active/dormant bacterial fractions in marine sediments. FEMS Microbiol. Ecol. 55: 416–423 [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Polz MF, Cavanaugh CM. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64: 3724–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poulsen L, Ballard G, Stahl D. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59: 1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rastogi R, Wu M, DasGupta I, Fox G. 2009. Visualization of ribosomal RNA operon copy number distribution. BMC Microbiol. 9: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steinberger R, Holden P. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71: 5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stevenson BS, Schmidt TM. 2004. Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 70: 6670–6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62: 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vamosi S, Heard S, Vamosi J, Webb C. 2009. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18: 572–592 [DOI] [PubMed] [Google Scholar]
- 25. Vandenkoornhuyse P, et al. 2007. Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc. Natl. Acad. Sci. U. S. A. 104: 16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Webb CO, Ackerly DD, Kembel SW. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24: 2098–2100 [DOI] [PubMed] [Google Scholar]
- 27. Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33: 475–505 [Google Scholar]