Abstract
16S rRNA amplicon libraries from a haloarchaeal bloom in the hypersaline Dead Sea in 1992 were analyzed together with the 2007 residual population and simulated blooms in experimental mesocosms. Significant population shifts were observed during the bloom, and surprisingly a signature from the bloom was retained 15 years later.
TEXT
Due to the diversion of its source waters and the pumping of water into evaporation ponds, the salinity of the Dead Sea has been increasing for the past century (6). Unlike the Dead Sea of the mid-19th century, the modern Dead Sea, at a salinity of approximately 347 g/liter and dominated by divalent cations, is a truly inhospitable environment, with little cellular activity and cell counts below 5 × 105 cells/ml (13). Only twice in recent years (1980, 1992) have the surface waters of the Dead Sea been diluted sufficiently to permit the establishment of a bloom of the halophilic alga Dunaliella. The 1992 Dunaliella bloom began in April, peaked in May, and then declined rapidly (11). A coinciding halobacterial bloom peaked in June at approximately 3.5 × 107 cells/ml (11) and persisted at more than 107 cells/ml through September.
With the Dead Sea continuing to decline, plans exist for the construction of a Red Sea-Dead Sea canal that would introduce copious quantities of marine water to the hypersaline Dead Sea (7, 12). Consequently, researchers have been filling artificial pools with mixtures of Red Sea and Dead Sea waters (12). These pools inevitably form Dunaliella and halobacterial blooms and allow scientists to investigate the implications of the Red Sea-Dead Sea canal as well as the nature of blooms inoculated with modern Dead Sea water.
At no recent point in time has the surface of the Dead Sea been devoid of haloarchaeal life. Thus, it is difficult to ascertain how much of the haloarchaeal blooms originate from interbloom populations and how much is introduced via the surrounding environs. Similarly it is unclear to what extent the interbloom populations reflect the early and end stages of the blooms. Recent studies using metagenomic and amplicon-based techniques have begun to take in-depth looks at the compositions of both the bloom and residual populations (2, 16). Both studies concluded that the modern Dead Sea is inhabited primarily by uncultured representatives. Additionally, Bodaker et al. (2) utilized a collection of short (∼60-bp) amplicons from the V6 region of the 16S rRNA gene to conclude that a number of the taxa represented in the June 1992 sample were not present in the residual population.
Here, in an effort to understand the evolution of a halobacterial bloom, we have sequenced a suite of 16S rRNA amplicons from a variety of Dead Sea and simulated bloom environments. Our samples consist of material collected from the Dead Sea in June 1992, September 1992, and March 2007 and four Dead Sea mesocosm pools, enabling us for the first time to investigate in depth the origin and evolution of a Dead Sea haloarchaeal bloom. The June 1992 and March 2007 samples were collected and processed according to the protocols of Bodaker et al. (2), and the September 1992 sample was collected by following the protocols of Rhodes et al. (16). Both 1992 samples were stored at −80°C until processing. The Dead Sea mesocosm samples were collected in July 2007 from pools 1, 2, 9, and 10 at the Dead Sea Works research station, Sedom, Israel (see Table S1 in the supplemental material). For each pool, 250 ml of water was pumped through a 0.7-μm-pore-size glass fiber filter. The filters were immediately placed on ice, shipped on dry ice, and stored at −80°C until the fall of 2010, when DNA was extracted using the protocols of Macalady et al. (8). The unavoidable discrepancies in sample collection and processing may have imparted an unquantified bias in the results. However, given the significant differences in microbial populations between samples processed in the same manner and the similarities between the September 1992 bloom and the pool 9 sample, we believe that the impact is minimal.
For all samples, the V6 to V9 regions of the 16S rRNA gene were amplified using the primer 926F (5′-AAACTYAAAKGAATTGRCGG-3′), the reverse complement of 907R (9), and by adding a degeneracy at the 5′ end of 1392R (5′-ACGGGCGGTGTGTRC-3′) (5). These primers were independently developed for amplification from oil sands with promising results (14), and unpublished results from our lab have shown good and phylogenetically diverse amplification of both bacteria and archaea. The approximately 500-base sequence was amplified and purified according to the procedure of Rhodes et al. (16).
The samples were sequenced in multiplex on a quarter plate of a 454 FLX Titanium sequencer (454 Life Sciences). Quality control and analysis were performed using home-written scripts, the mothur version 1.20.0 platform (18), and UCHIME (4). Sequences were restricted to the first 305 bases spanning the V6 and V7 regions, and pairwise distances were calculated. The sequences were clustered using the average neighbor method. The clusterings were used to calculate both the inverse Simpson index and a subsampled inverse Simpson index (1,400 sequences). Both calculations were repeated 1,000 times (Table 1). The Simpson diversity indices revealed that at the strain (3% similarity) and species (5% similarity) levels, the March 2007 sample displayed the greatest diversity, followed by the June 1992 and the September 1992 samples. The artificial pools all displayed less diversity than the natural samples. However, at the genus level, the early stages of the 1992 halobacterial bloom contained greater diversity than the residual 2007 population. Thus, while the residual population was relatively diverse in strains, it was relatively poor with respect to broader phylogenetic categories. The overwhelming challenges presented by the modern Dead Sea appear to have restricted the residual microbial community to a select few branches. Nevertheless, within each branch there was considerable diversity.
Table 1.
Salinity, sample size, subsampled inverse Simpson index, and inverse Simpson index for amplicon data setsa
| Sample | Salinity (g/liter) | No. of sequences | Subsampled inverse Simpson index |
Inverse Simpson index |
||||
|---|---|---|---|---|---|---|---|---|
| 0.03 (strain) | 0.05 (species) | 0.1 (genus) | 0.03 (strain) | 0.05 (species) | 0.1 (genus) | |||
| June 1992 | 249 | 12,733 | 7.45 ± 0.56 | 7.31 ± 0.56 | 6.36 ± 0.38 | 7.46 ± 0.20 | 7.33 ± 0.19 | 6.37 ± 0.13 |
| September 1992 | 288 | 8,904 | 5.58 ± 0.30 | 5.43 ± 0.30 | 4.22 ± 0.18 | 5.59 ± 0.13 | 5.45 ± 0.13 | 4.22 ± 0.08 |
| March 2007 | 347 | 25,471 | 10.90 ± 0.70 | 9.20 ± 0.54 | 4.09 ± 0.22 | 10.98 ± 0.17 | 9.24 ± 0.13 | 4.10 ± 0.05 |
| Pool 1 | 287 | 3,188 | 2.97 ± 0.18 | 2.92 ± 0.18 | 2.67 ± 0.12 | 2.97 ± 0.15 | 2.92 ± 0.15 | 2.67 ± 0.11 |
| Pool 2 | 279 | 1,452 | 4.82 ± 0.08 | 4.78 ± 0.08 | 4.06 ± 0.04 | 4.83 ± 0.41 | 4.79 ± 0.40 | 4.07 ± 0.25 |
| Pool 9 | 225 | 4,655 | 3.90 ± 0.18 | 3.82 ± 0.18 | 3.42 ± 0.14 | 3.91 ± 0.12 | 3.82 ± 0.12 | 3.42 ± 0.09 |
| Pool 10 | 261 | 17,693 | 2.63 ± 0.20 | 2.61 ± 0.18 | 2.03 ± 0.12 | 2.63 ± 0.06 | 2.62 ± 0.06 | 2.02 ± 0.04 |
The values were calculated at the 3% , 5%, and 10% levels, which we approximate as the strain, species, and genus levels for the primarily hypervariable portions of the 16S rRNA gene that we sequenced. Ninety-five percent confidence intervals are provided for both the subsampled inverse Simpson index and the inverse Simpson index.
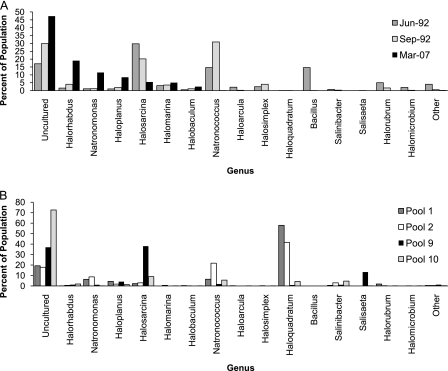
The operational taxonomic units (OTUs) were assigned to taxa at the 3% distance level using a combined Silva-RDP database (3, 15). The proportions of the major taxa for each sample were then tabulated (Fig. 1). All samples contained considerable portions of uncultured microorganisms. In particular, a single sequence displaying 99% homology to an uncultured haloarchaeon from an evaporitic crust in Guerrero Negro, Mexico (17), comprised over 25% of the data set and over 68% of pool 10.
Fig 1.
Bar charts depicting the percentages of the major genera comprising the microbial populations of the Dead Sea samples (A) and the mesocosm pool samples (B). Genera are organized according to their prevalence in the March 2007 interbloom sample.
Taxonomic assignment also revealed a number of population shifts between the samples. The identifiable portions of the haloarchaeal populations in both Dead Sea bloom samples were dominated by Halosarcina and the alkaliphilic Natronococcus (Fig. 1A). The March 2007 population, however, was dominated by Halorhabdus and the alkaliphilic Natronomonas. The dominance of alkaliphilic microbes confirms prior results but is unusual, as the pH of the bloom waters was near neutral (6.9 to 7.2) (2, 16). For the March 2007 Dead Sea sample, our taxonomic assignment largely confirms the results obtained by Bodaker et al. (2) from fosmid and environmental clones. However, with respect to the early stages of the 1992 bloom, our amplicon data sets revealed different and greater taxonomic diversity. It therefore appears that the residual Dead Sea population was not merely a reflection of the bloom population and that the introduction of Red Sea brine to the Dead Sea would cause a significant population shift.
The bacterial Dead Sea populations displayed a more pronounced shift. Only in the June 1992 sample did bacteria compose a significant proportion of the microbial community. Bacillus-related microbes comprised over 13% of the June 1992 sample, and the genera Burkholderia, Chryseobacterium, and Salinibacter were also identified. In total, while more than 18% of the June 1992 microbial community was bacterial in origin, the September 1992 and March 2007 samples each contained less than 0.5% bacteria. Finally, a limited number of amplicons from each sample were related to the newly identified halophilic lineage, the Nanohaloarchaea (10).
Due to their artificial nature and different initiation times and initial settings, the mesocosm pools could be expected to display differing microbial communities (Fig. 1B). While the more saline and younger pools, pools 1 and 2, were dominated primarily by Haloquadratum, the less saline and older pools, pools 9 and 10, were dominated primarily by uncultured haloarchaea. Pool 9, the oldest and least saline of the pools, also contained significant quantities of Halosarcina and the bacterium Salisaeta, which was originally isolated from a Sedom mesocosm pool (19). All other pools contained only trace amounts of bacteria. It therefore looks as if only pool 9 accurately reflected the early stages of the 1992 bloom and that a considerable dilution of the Dead Sea surface with Red Sea water would be required to promote the growth of bacterial halophiles and to return the Dead Sea to the peak bloom state of 1992.
Additionally, we had previously sequenced a metagenome from the March 2007 Dead Sea (16). The library contained 142 fragments of the 16S rRNA gene (1). As these sequences were sequenced directly from extracted DNA, they potentially offer the least biased representation of the Dead Sea microbiome. Of the 142 sequences, 133 originated in haloarchaea, 7 originated in bacteria, and 2 were undeterminable. The metagenomic 16S fragments proved difficult to accurately assign to genera. Nevertheless, to an extent the metagenomic sequences confirm the results of the March 2007 amplicon data set. Both the genera Halorhabdus (18 sequences) and Natronomonas (20 sequences) were well represented among the assignable sequences. Also present were Haloplanus, Halosarcina, and Halomarina.
Finally, we assessed the seeding potentials of each sample. The various natural and artificial Dead Sea environments are intimately linked with one another. The June 1992 Dead Sea evolved into the September 1992 sample over the course of 3 months. The September 1992 Dead Sea deteriorated into the March 2007 sample over the course of 15 years. Lastly, the mesocosm pools were constructed with nonbloom Dead Sea water akin to the March 2007 Dead Sea. At the same time, the various environments were exposed to microbial input from the surrounding environs. Thus, our samples represent some combination of inherited and introduced biota. We therefore determined the percent community composition of the “sink” community that could be assembled from propagules occurring in the “source” microbial community (Table 2).
Table 2.
Percentages of the microbial populations of the sink environments that can be constructed by OTUs occurring in the source environmentsa
| Sink sample | % of microbial population constructed from source sampleb |
||||||
|---|---|---|---|---|---|---|---|
| June 1992 | September 1992 | March 2007 | Pool 10 | Pool 9 | Pool 2 | Pool 1 | |
| June 1992 | 100.0 | 81.6 | 26.5 | 68.4 | 65.9 | 66.0 | 61.7 |
| September 1992 | 98.1 | 100.0 | 41.4 | 91.6 | 86.7 | 87.3 | 80.1 |
| March 2007 | 58.6 | 63.5 | 100.0 | 33.4 | 23.3 | 22.4 | 18.1 |
| Pool 10 | 82.5 | 86.9 | 73.7 | 100.0 | 84.7 | 94.4 | 92.1 |
| Pool 9 | 77.4 | 90.4 | 51.7 | 94.2 | 100.0 | 75.6 | 74.4 |
| Pool 2 | 46.3 | 70.4 | 33.7 | 93.0 | 44.2 | 100.0 | 89.2 |
| Pool 1 | 50.0 | 45.0 | 24.6 | 89.5 | 51.0 | 92.7 | 100.0 |
This analysis was performed at the 0% or “unique” level. Similar results were obtained for the 3% and 5% difference levels.
Percentage of microbial population in the sink environment constructed by OTUs occurring in the source environment (where the samples above each column represent the source samples and the samples to the left of each row represent the sink samples). For example, 63.5% of the March 2007 sample could be constructed using the September 1992 sample as a source sample.
The best source for the September 1992 sample was the June 1992 sample. Similarly the best source for the March 2007 sample was the September 1992 sample. Thus, despite the decrease in diversity from June to September, the September 1992 sample did a better job of seeding the residual population. This suggests that even after 15 years, the residual Dead Sea population is still partly a reflection of the later stages of the 1992 bloom. However, despite the degree of sequence diversity observed in the March 2007 Dead Sea, the March 2007 sample was a poor source for the mesocosm pools. The inocula for these blooms were therefore either microbes present at astonishingly low concentrations in the surface of the modern Dead Sea or were introduced from elsewhere, most likely via Aeolian deposition. Consequently, we conclude that while some of the bloom taxa are preserved in the surface waters of the residual Dead Sea, a significant proportion of bloom taxa are unable to survive the harsh interbloom conditions and must be reintroduced from elsewhere.
Nucleotide sequence accession number.
Sequence data for the samples are available through the NCBI SRA via accession number SRA 049809.
Supplementary Material
ACKNOWLEDGMENTS
We thank Oded Béjà, Idan Bodaker, Lynn Tomsho, and the late Moti Gonen for their assistance. We further thank the Geological Survey of Israel and the Dead Sea Works, Ltd., for maintaining the mesocosms.
This work was supported by the Agriculture and Food Research Initiative Competitive Grants Program grant no. 2010-65110-20488 from the USDA National Institute of Food and Agriculture and by the NASA Astrobiology Institute under NASA-Ames cooperative agreement NNA09DA76A (C.H.H.). The 454 facility at the Pennsylvania State University Center for Genome Analysis is partly funded using Tobacco Settlement Funds provided by the Pennsylvania Department of Health.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bodaker I, et al. 2010. Comparative community genomics in the Dead Sea: an increasingly extreme environment. ISME J. 4:399–407 [DOI] [PubMed] [Google Scholar]
- 3. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Field K, et al. 1988. Molecular phylogeny of the animal kingdom. Science 239:748–753 [DOI] [PubMed] [Google Scholar]
- 6. Gavrieli I, Beyth M, Yechieli Y. 1999. The Dead Sea—a terminal lake in the Dead Sea rift: a short overview, p 121–127 In Oren A. (ed), Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Raton, FL [Google Scholar]
- 7. Gavrieli I, Oren A. 2004. The Dead Sea as a dying lake, p 287–305 In Nihoul JCJ, Zavialov PO, Micklin PP. (ed), Dying and dead seas. Climatic versus anthropic causes. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 8. Macalady JL, et al. 2008. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2:590–601 [DOI] [PubMed] [Google Scholar]
- 9. Muyzer G, Hottentrager S, Teske A, Wawer C. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA. A new molecular approach to analyze the genetic diversity of mixed microbial communities, p 3.4.4.1–3.4.4.22 In Akkermans ADL, van Elsas JD, de Bruijn FJ. (ed), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 10. Narasingarao P, et al. 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 6:81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oren A. 1993. The Dead Sea—alive again. Experientia 49:518–522 [Google Scholar]
- 12. Oren A, et al. 2004. Biological effects of dilution of Dead Sea brine with seawater: implications for the planning of the Red Sea-Dead Sea “Peace Conduit.” J. Mar. Syst. 46:121–131 [Google Scholar]
- 13. Oren A, Ginzburg M, Ginzburg BZ, Hochstein LI, Volcani BE. 1990. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Int. J. Syst. Bacteriol. 40:209–210 [DOI] [PubMed] [Google Scholar]
- 14.Park HS, et al. Microbially influenced corrosion in oil sands water handling facilities, paper. Corrosion; NACE International, Houston, TX: 2011. 2011. p. 11226. [Google Scholar]
- 15. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhodes ME, Fitz-Gibbon ST, Oren A, House CH. 2010. Amino acid signatures of salinity on an environmental scale with a focus on the Dead Sea. Environ. Microbiol. 12:2613–2623 [DOI] [PubMed] [Google Scholar]
- 17. Sahl JW, Pace NR, Spear JR. 2008. Comparative molecular analysis of endoevaporitic microbial communities. Appl. Environ. Microbiol. 74:6444–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaisman N, Oren A. 2009. Salisaeta longa gen. nov., sp. nov., a red, halophilic member of the Bacteroidetes. Int. J. Syst. Evol. Microbiol. 59:2571–2574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.