Abstract
FK506 is an important 23-member polyketide macrolide with immunosuppressant activity. Its entire biosynthetic gene cluster was previously cloned from Streptomyces sp. strain KCTC 11604BP, and sequence analysis identified three putative regulatory genes, tcs2, tcs7, and fkbN, which encode proteins with high similarity to the AsnC family transcriptional regulators, LysR-type transcriptional regulators, and LAL family transcriptional regulators, respectively. Overexpression and in-frame deletion of tcs2 did not affect the production of FK506 or co-occurring FK520 compared to results for the wild-type strain, suggesting that tcs2 is not involved in their biosynthesis. fkbN overexpression improved the levels of FK506 and FK520 production by approximately 2.0-fold, and a deletion of fkbN caused the complete loss of FK506 and FK520 production. Although the overexpression of tcs7 decreased the levels of FK506 and FK520 production slightly, a deletion of tcs7 caused 1.9-fold and 1.5-fold increases in FK506 and FK520 production, respectively. Finally, fkbN overexpression in the tcs7 deletion strain resulted in a 4.0-fold (21 mg liter−1) increase in FK506 production compared to that by the wild-type strain. This suggests that fkbN encodes a positive regulatory protein essential for FK506/FK520 biosynthesis and that the gene product of tcs7 negatively regulates their biosynthesis, demonstrating the potential of exploiting this information for strain improvement. Semiquantitative reverse transcription-PCR (RT-PCR) analyses of the transcription levels of the FK506 biosynthetic genes in the wild-type and mutant strains proved that most of the FK506 biosynthetic genes are regulated by fkbN in a positive manner and negatively by tcs7.
INTRODUCTION
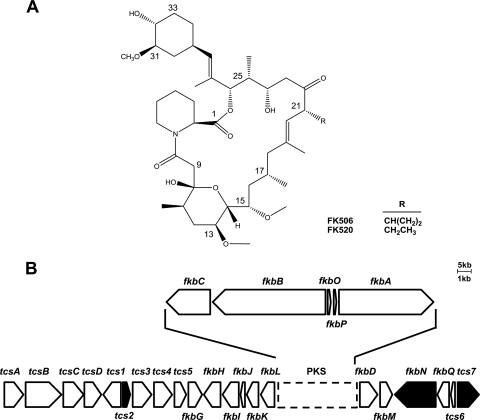
FK506 (Tacrolimus) is a clinically important drug used to prevent the rejection of transplanted organs and treat autoimmune diseases (28). FK506 also shows promising therapeutic potential as a neuroprotective (35) and neuroregenerative agent (7). FK506 and its structurally related macrolide polyketide FK520, in which the allyl side chain at carbon 21 of FK506 is replaced by an ethyl side chain, are 23-member polyketide macrolides produced by various Streptomyces species (16, 24) (Fig. 1A). Since the first isolation of FK506 from Streptomyces tsukubaensis (16), its biosynthetic gene cluster has been partially sequenced in Streptomyces sp. strain ATCC 55098 (MA6858) (26), Streptomyces sp. strain ATCC 53770 (MA6548) (25), and S. tsukubaensis NRRL 18488 (8). Recently, the sequences of the entire FK506 biosynthetic gene clusters from Streptomyces sp. ATCC 55098, Streptomyces sp. strain KCTC 11604BP, and Streptomyces kanamyceticus were reported (24). The complete biosynthetic gene cluster of FK520 was also isolated from Streptomyces hygroscopicus var. ascomyceticus ATCC 14891 (39). Because the only difference between FK506 and FK520 is the C-21 side chain, they are synthesized in an analogous manner using a hybrid polyketide synthase (PKS)/nonribosomal peptide synthetase (NRPS) system except that an allymalonyl-coenzyme A (allymalonyl-CoA) extender unit is loaded onto module 4 of FK506 PKS, whereas an ethylmalonyl-CoA is loaded onto the corresponding module of FK520 PKS (24). Therefore, 14 fkb genes, which contribute to the common structural elements of FK506 and FK520, and 1 putative regulatory gene (fkbN) are well maintained and organized identically in the FK506 and FK520 clusters. Four genes required for the synthesis of allymalonyl-CoA (tcsA, tcsB, tcsC, and tcsD) are present only in FK506 clusters, whereas genes for ethylmalonyl-CoA synthesis in the FK520 cluster (fkbE, fkbS, and fkbU) were not found in any of the FK506 clusters (24). Interestingly, one of the putative regulatory genes, fkbN, was observed in all FK506- and FK520-producing strains, but tcs2 and tcs7 were present only in the Streptomyces sp. KCTC 11604BP strain (Fig. 1B) (24). The tcs2, tcs7, and fkbN genes encoded an AsnC family transcriptional regulator (18), LysR-type transcriptional regulator (10), and LAL (large ATP-binding regulators of LuxR) family regulator (6, 30), respectively. However, the roles of these putative regulatory genes, including fkbN, found in both the FK506 and FK520 biosynthetic gene clusters have not been reported.
Fig 1.
Structures of FK506 and FK520 (A) and genetic organization of the FK506 biosynthetic gene cluster from Streptomyces sp. KCTC 11604BP (B). The putative regulatory genes (tcs2, fkbN, and tcs7) are black. tcsA, tcsB, tcsC, and tcsD, allylmalonyl-CoA biosynthetic genes; fkbG, fkbH, fkbI, fkbJ, and fkbK, methoxymalonyl-ACP biosynthetic genes; fkbA, fkbB, and fkbC, polyketide synthase (PKS) genes; fkbL and fkbP, pipecolate biosynthesis and PKS condensation-related genes; fkbO, dihydroxycyclohexanecarboxylic acid (DHCHC) biosynthetic gene; fkbQ, type II thioestease gene; fkbD and fkbM: post-PKS genes.
Generally, two types of regulators, pathway specific and global, control the production of several secondary metabolites produced by a Streptomyces strain. The pathway-specific regulatory genes are located on the biosynthetic gene clusters of secondary metabolites; they affect only the production of their own secondary metabolites directly. The global regulatory genes exert pleiotropic control over multiple aspects of secondary metabolism, such as antibiotic production and morphological differentiation. They are located outside the biosynthetic gene cluster and have an indirect influence on secondary metabolite production (2, 22, 37). Many of the pathway-specific regulators belong to the SARP (Streptomyces antibiotic regulatory protein) family regulators (2), as exemplified by ActII-ORF4, for actinorhodin biosynthesis, from Streptomyces coelicolor A3 (2) (1), and DnrI, for doxorubicin biosynthesis, from Streptomyces peucetius (33). On the other hand, other regulatory proteins belonging to other families, such as the LAL family, also function as pathway-specific regulators. In particular, genes encoding LAL family members, such as FkbN, have recently been found in several polyketide biosynthetic gene clusters (9, 12, 17, 19, 27, 29, 38, 39).
This study examined the functions of tcs2, tcs7, and fkbN in the regulation of FK506 biosynthesis in Streptomyces sp. KCTC 11604BP through the overexpression, in-frame deletion, complementation of tcs7 and fkbN deletion mutants, and transcriptional analysis of FK506 biosynthetic genes by semiquantitative reverse transcription-PCR (RT-PCR) in the wild-type and mutant strains. These results provide valuable information to help understand the pathway-specific regulatory mechanisms of fkbN and tcs7 in Streptomyces sp. KCTC 11604BP and demonstrate the potential of manipulating regulatory genes to increase the level of FK506 production in industrial production strains.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
All strains and plasmids used in this study are described in Table 1. Standard media and culture conditions were used (15, 31). Ampicillin (100 μg ml−1), apramycin (50 μg ml−1), and nalidixic acid (25 μg ml−1) were added selectively to the growth media as required. Spores of all strains were generated on ISP4 medium (34), and seed culture for production culture was prepared in liquid R2YE medium (15) with or without apramycin, as described previously (23). For FK506 and FK520 production, all strains were inoculated into baffled 250-ml flasks containing 50 ml R2YE medium with or without apramycin at pH 7.2 and with 500 μl of a seed culture suspension and grown on an orbital shaker (180 rpm) for 6 days at 28°C.
Table 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or fosmid | Characteristicsa | Source or reference(s) |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | Plasmids construction and general subcloning, F−recAlacZΔM15 | New England Biolabs |
| ET12567/pUZ8002 | Nonmethylating ET12567 containing nontransmissible RP4 derivative plasmid pUZ8002; Cmlr Tetr Kanr | 21 |
| Streptomyces | ||
| KCTC 11604BP | Wild-type FK506-producing strain | 24 |
| WT/pSET152(PermE*) | Wild-type stain with integrative plasmid pSET152(PermE*) | This study |
| WT/pTCS2 | Wild-type with integrative plasmid pTCS2; Aprar | This study |
| WT/pTCS7 | Wild-type with integrative plasmid pTCS7; Aprar | This study |
| WT/pFKBN | Wild-type with integrative plasmid pFKBN; Aprar | This study |
| Δtcs2 | Mutant of KCTC 11604BP with an in-frame deletion internal to tcs2 | 24 |
| Δtcs7 | Mutant of KCTC 11604BP with an in-frame deletion internal to tcs7 | This study |
| ΔfkbN | Mutant of KCTC 11604BP with an in-frame deletion internal to fkbN | This study |
| Δtcs7/pFKBN | Δtcs7 with integrative plasmid pFKBN; Aprar | This study |
| ΔfkbN/pTCS2 | ΔfkbN with integrative plasmid pTCS2; Aprar | This study |
| ΔfkbN/pTCS7 | ΔfkbN with integrative plasmid pTCS7; Aprar | This study |
| ΔfkbN/pFKBN | ΔfkbN with integrative plasmid pFKBN; Aprar | This study |
| Plasmids | ||
| pCCFOS1(fosmid) | Vector for genomic library construction; F−oriV Cmlr | Epicentre Biotechnol. |
| pGEM-T Easy | E. coli vector for cloning PCR products; Ampr | Promega |
| pSET152(PermE*) | Integrative plasmid containing PermE*, oriT, attP, ΦC31int, and aac(3)IV | 3, 31 |
| pKC1139 | Temperature-sensitive E. coli-Streptomyces shuttle vector containing oriT and aac(3)IV for gene disruption | 3 |
| pTCS2 | pSET152(PermE*)-based integrative plasmid containing single copy of tcs2; Aprar | This study |
| pTCS7 | pSET152(PermE*)-based integrative plasmid containing single copy of tcs7, Aprar | This study |
| pFKBN | pSET152(PermE*)-based integrative plasmid containing single copy of fkbN; Aprar | This study |
| pΔTCS2 | pKC1139 based deletion plasmid with in-frame deletion of 171 bp internal to tsc2 | This study |
| pΔTCS7 | pKC1139-based deletion plasmid with in-frame deletion of 888 bp internal to tsc7 | This study |
| pΔFKBN | pKC1139-based deletion plasmid with in-frame deletion of 2,772 bp internal to fkbN | This study |
| Fosmids | ||
| fos1004F01 | Fosmid clone; contains bases 1-40 and 366 of FK506 biosynthetic gene cluster | 24 |
| fos1006D05 | Fosmid clone; contains bases 58, 172–197, and 743 of FK506 biosynthetic gene cluster | 24 |
F−, does not carry the F plasmid; recA1, for reduced occurrence of unwanted recombination in cloned DNA; lacZΔM15, partial deletion of the lacZ gene, which allows α complementation of the β-galactosidase gene; Cmlr, chloramphenicol resistance; Tetr, tetracycline resistance; Kanr, kanamycin resistance; Aprar, apramycin resistance; Ampr, ampicillin resistance; oriV, origin of replication; oriT, origin of transfer; attP, plasmid ΦC31 attachment site; ΦC31 int, integrase gene; aac(3)IV, apramycin resistance gene; PermE*, mutated constitutive promoter.
Gene overexpression, disruption, and complementation.
DNA extraction and manipulation, as well as the transformation of Escherichia coli and Streptomyces, were performed using standard protocols (15, 31). The integrative E. coli-Streptomyces vector pSET152 (3) derivative containing the ermE* promoter (PermE*) (32) was used for gene expression in Streptomyces sp. KCTC 11604BP. For overexpression of the three putative regulatory genes (tcs2, tcs7, and fkbN) in Streptomyces sp. KCTC 11604BP, the overexpression plasmids were constructed by PCR amplification of the fragments of the three genes from fosmid DNA derived from Streptomyces sp. KCTC 11604BP (fos1004F01 and fos1006D05; GenBank accession no. HM116536). Primer pairs Tcs2OF/Tcs2OR, Tcs7OF/Tcs7OR, and FkbNOF/FkbNOR were designed to PCR amplify the DNA fragments containing tcs2, tcs7, and fkbN, respectively (see Table S1 in the supplemental material), and all primers for PCR amplification of tcs2, tcs7, and fkbN were designed to contain their natural ribosomal binding sites (RBS). A total of 3 PCR fragments (489 bp for tcs2, 1,452 bp for tcs7, and 2,793 bp for fkbN) were cloned separately using the pGEM-T Easy vector (Promega) and sequenced. After digestion with the appropriate restriction enzymes, the fragments were cloned into the BglII-XbaI site of the pSET152 derivative containing the ermE* promoter, yielding pTCS2, pTCS7, and pFKBN (Table 1). These plasmids were then transferred by conjugation from E. coli ET12567/pUZ8002 (21) to Streptomyces sp. KCTC 11604BP, as described elsewhere (23). Site-specific integration of the plasmid into the attB chromosomal site allowed selection of the apramycin-resistant exconjugants.
E. coli-Streptomyces shuttle vector pKC1139 (3) was used for in-frame gene deletion. The tcs2 deletion mutant (Δtcs2) was constructed previously (24). To delete tcs7 and fkbN, the deletion plasmids were constructed by PCR amplification of the left and right flanking fragments from fosmid DNA derived from Streptomyces sp. KCTC 11604BP (fos1004F01 and fos1006D05). The primer pairs Tcs7LF/Tcs7LR and FkbNLF/FkbNLR were designed to amplify the left flanking fragments of the target genes (see Table S1 in the supplemental material), whereas Tcs7RF/Tcs7RR and FkbNRF/FkbNRR were designed for the right flanking fragments (see Table S1 in the supplemental material). A total of four PCR fragments were cloned separately in the pGEM-T Easy vector and then sequenced. After digestion with the appropriate restriction enzymes, the fragments were cloned into pKC1139 that had been digested with HindIII-EcoRI or XbaI-EcoRV to construct the two different in-frame deletion plasmids, namely, pΔTCS7 and pΔFKBN (Table 1). These plasmids were then transferred to Streptomyces sp. KCTC 11604BP, as described above. The desired double-crossover mutants, namely, Δtcs7 and ΔfkbN, were selected as described previously (24), verified by PCR, and confirmed selectively by Southern blot analysis.
For complementation, the pTCS2, pTCS7, and pFKBN plasmids were introduced into the ΔfkbN strain by intergenic conjugation. The pTCS7 and pFKBN plasmids were also introduced into the Δtcs7 strain. Site-specific integration of the plasmids into the attB chromosomal site allowed the selection of apramycin-resistant exconjugants.
Analysis of cell growth and FK506 production.
To measure cell growth, samples (50 ml) of the fermentation broth were collected at 24-h intervals, starting at 24 h after inoculation. The mycelia were collected and weighed, as described previously (23). Wild-type, Δtcs2, Δtcs7, and ΔfkbN strains were grown in 50 ml of liquid R2YE medium on an orbital shaker (180 rpm) for 6 days at 28°C. WT/pSET152(PermE*), WT/pTCS2, WT/pTCS7, WT/pFKBN, ΔfkbN/pTCS2, ΔfkbN/pTCS7, ΔfkbN/pFKBN, and Δtcs7/pFKBN were grown in 50 ml of liquid R2YE medium containing 50 μg ml−1 apramycin, as described above. The levels of FK506 and FK520 production were determined by high-performance liquid chromatography (HPLC), as described previously (23). Authentic FK506 (Sigma-Aldrich) and FK520 (Santa Cruz Biotechnology) standards were used to construct a calibration curve of FK506 and FK520, respectively, by HPLC analysis. The levels of FK506 and FK520 production reported are the averages of two series of duplicate separate cultivations and extractions.
Isolation of mRNA and gene expression analysis by RT-PCR.
Total RNA was isolated from the wild-type and mutant strains of Streptomyces sp. KCTC 11604BP, which had been grown in liquid R2YE medium with or without apramycin, as described previously (14). The RNA preparations were treated with DNase I (Qiagen) to eliminate possible chromosomal DNA contamination. RT-PCR was performed with a Qiagen OneStep RT-PCR kit using 100 ng of the total RNA as the template. RT-PCR conditions were as follows: cDNA synthesis, 50°C for 30 min followed by 95°C for 15 min; amplification, 35 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 40 s. The cycle number for each gene was optimized in order to obtain enough visibility of the RT-PCR band and ensure that amplification was in the linear range and the results were semiquantitative. The primers were designed to generate PCR products of approximately 500 bp using the primers pairs listed in Table S1 in the supplemental material. With each pair of primers, the negative controls were carried out with Taq DNA polymerase (Stratagene) in the absence of reverse transcriptase to confirm that the amplified products were not derived from chromosomal DNA. The RT-PCR experiments were done in duplicate using RNA samples from two independent cultures.
RESULTS AND DISCUSSION
Sequence analysis of the putative regulatory genes present in the FK506 biosynthetic gene cluster.
The deduced products of tcs2 (155 amino acid residues) and tcs7 (474 amino acid residues) showed high sequence similarity to members of the AsnC family of transcriptional regulators (18) and LysR-type transcriptional regulators (10), respectively. The deduced product of fkbN (923 amino acid residues) showed high sequence similarity to members of the LAL family of transcription regulators, whose genes are often found in the polyketide biosynthetic gene clusters of Streptomyces (9, 12, 17, 19, 27, 29, 38, 39). Members of this family, which are typified by the regulator of the maltose regulon in E. coli, MalT (6, 30), are characterized by two distinct domains, the N-terminal ATP-binding domain and C-terminal LuxR-type HTH DNA binding domain (12, 38). An analysis of the FkbN deduced amino acid sequence revealed the presence of these two functional domains (data not shown). In addition, BLAST analysis of FkbN revealed its high identity/similarity to the known Streptomyces LAL-type regulatory proteins: 72%/83% with FkbN of the FK520 cluster (39), 59%/72% with RapH (19), 34%/47% with PikD (38), 37%/50% with GdmRII (9), 34%/43% with TmcN (12), and 32%/44% with AveR (17). One noteworthy feature of the fkbN gene is that, like some known regulatory genes, it contains one TTA codon, which is the rarest codon in the high-GC-content streptomycete genome. This suggests the possibility that FkbN may play a regulatory role in FK506 biosynthesis since the involvement of a TTA codon in regulating cell differentiation and secondary metabolism has been proposed for other actinomycetes (4, 20).
Overexpression of tcs2, tcs7, and fkbN.
To examine the function of the putative regulatory genes tcs2, tcs7, and fkbN, the genes were introduced into and overexpressed in the wild-type Streptomyces sp. KCTC 11604BP strain. To avoid the unexpected effect caused by the existence of multiple copies of the plasmid itself, the constructs pTCS2, pTCS7 and pFKBN were prepared based on the integrative expression plasmid pSET152 for overexpression (Table 1). As a control, pSET152 (PermE*) was introduced into the wild-type strain. There were no differences in FK506 production and growth or morphology between the exconjugants with pSET152 and the wild-type strain when grown in R2YE medium (data not shown).
As listed in Table 2, measurements of FK506 production of the WT/pFKBN strain showed that the FK506 titer increased significantly (11.1 mg liter−1), which is approximately 2.1 times higher than that in the wild-type strain after 5 days of cultivation. Moreover, the level of production of FK520, which is a by-product of FK506 biosynthesis generated by the misincorporation of ethylmalonyl-CoA instead of allylmalonyl-CoA at module 4 of FK506 PKS (24), was also increased approximately 2.0-fold in WT/pFKBN compared to the wild-type strain. In contrast, the titers of FK506 and FK520 in WT/pTCS7 were slightly lower than those observed in the wild-type strain. tcs2 overexpression did not have any observable effects on the levels of FK506 or FK520 production. This is consistent with a previous study, which reported that the in-frame deletion of tcs2 did not affect the biosynthesis of FK506 or co-occurring FK520 (24). This indicates that FkbN plays a significant role as a positive regulator and Tcs2 plays no observable role in FK506 and FK520 biosynthesis in Streptomyces sp. KCTC 11604BP. However, the role of Tcs7 could not be clearly determined by its overexpression. Furthermore, a LAL family regulator, FkbN, in the FK506 biosynthetic cluster of Streptomyces sp. KCTC 11604BP, probably caused the increased expression of FK506 biosynthetic genes, as reported for other LAL family regulators (13, 14).
Table 2.
FK506 and FK520 titers from Streptomyces sp. KCTC 11604BP and mutant strains
| Macrolide | Antibiotic titer (mg liter−1)a ± SD for strain: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | WT/pTCS2 | WT/pTCS7 | WT/pFKBN | Δtcs2 | Δtcs7 | ΔfkbN | ΔfkbN/pTCS2 | ΔfkbN/pTCS7 | ΔfkbN/pFKBN | |
| FK506 | 5.22 ± 0.65 | 5.18 ± 0.27 | 4.51 ± 0.98 | 11.1 ± 0.48 | 5.12 ± 0.51 | 9.89 ± 0.98 | NDb | ND | ND | 5.24 ± 0.71 |
| FK520 | 0.65 ± 0.06 | 0.61 ± 0.03 | 0.59 ± 0.08 | 1.39 ± 0.04 | 0.61 ± 0.08 | 0.97 ± 0.82 | ND | ND | ND | 0.79 ± 0.09 |
Standard error values are the results of two series of duplicate data acquisition.
ND, not detected.
Inactivation of tcs7 and fkbN by in-frame deletion and complementation of the deletion mutants.
To confirm the regulatory roles of tcs7 and fkbN in FK506 biosynthesis, they were inactivated by an in-frame deletion to avoid any polar effect. All in-frame deletion mutants generated using pKC1139 were confirmed by Southern hybridization (see Fig. S1, S2, and S3 in the supplemental material). The mutant strains Δtcs2, Δtcs7, and ΔfkbN showed no change in their morphological phenotypes, compared to the wild-type strain, when grown on solid ISP4 and R2YE plates (data not shown). Interestingly, although the Δtcs2 strain showed no observable changes in FK506 or FK520 production as reported previously (24), the Δtcs7 strain gave rise to an approximately 1.9-fold increase in the level of FK506 production compared to the wild-type strain (Table 2). In addition, the Δtcs7 strain showed a similar increase in FK520 production. These results provide strong evidence that tcs7 is a negative regulator of FK506 or co-occurring FK520 biosynthesis. In particular, the ΔfkbN strain could not produce FK506 and FK520 (Table 2), suggesting that fkbN plays a key role as a positive regulator of the FK506 biosynthetic pathway.
To confirm that the inactivation of fkbN was directly responsible for eliminating the production FK506 and FK520, plasmid-based complementation experiments in the ΔfkbN strain were carried out by reintroducing fkbN as well as tcs2 and tcs7, plus their putative RBS regions using pSET152(PermE*). Complementation plasmids pTCS2, pTCS7, and pFKBN were transferred from E. coli ET12567/pUZ8002 to the ΔfkbN strain. HPLC analysis confirmed that FK506 and FK520 productivity was restored to levels comparable to those observed in the wild-type strain in only the ΔfkbN strain containing pFKBN (Table 2). This demonstrates that the absence of fkbN is the sole reason for the loss of FK506 or co-occurring FK520 productivity in the ΔfkbN strain and that FkbN is a positive regulator. As expected, the overexpression of tcs2 and tcs7 did not alter the phenotype of the ΔfkbN strain. Similarly, self-complementation of Δtcs7 by pTCS7 lowered the production of FK506 and FK520 to the levels in the wild-type strain (data not shown), showing that the deletion of tcs7 is responsible for the enhanced production of FK506 and FK520.
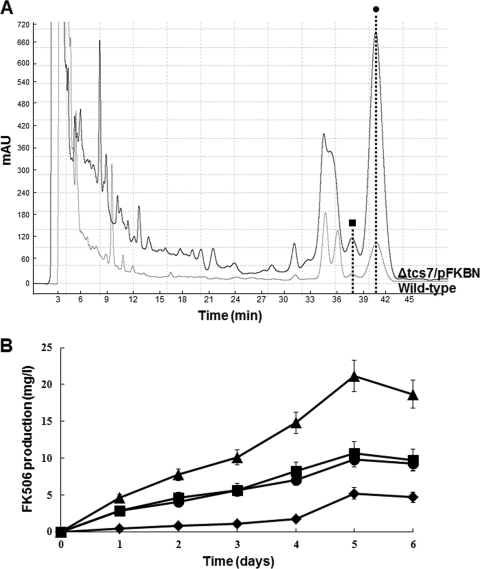
To further exploit the positive role of FkbN and the negative role Tcs7 in increasing FK506 biosynthesis, the fkbN gene was overexpressed in the Δtcs7 strain (Δtcs7/pFKBN). HPLC analysis of the culture broth extract of the Δtcs7/pFKBN strain revealed a considerable increase in the FK506 titer, approximately 4.0-fold (21 mg liter−1) higher than the level present in the wild-type strain (Fig. 2A and B). The FK506 production time courses of the wild-type, Δtcs7, WT/pFKBN, and Δtcs7/pFKBN strains in the R2YE medium revealed the maximal levels of FK506 at approximately 5 days (Fig. 2B). Moreover, there were no observed differences in cell mass or growth rate between the strains (data not shown), suggesting that fkbN and tcs7 are not associated with the primary metabolism or cell differentiation. Overall, gene deletion and the self- or cross-complementation results confirmed that fkbN is a positive regulatory gene and tcs7 is a negative regulatory gene in FK506 biosynthesis.
Fig 2.
(A) HPLC analyses from ethyl acetate-extracted broths from the Δtcs7/pFKBN strain (black line) and wild-type strain (gray line). The dotted lines indicate the identity of FK506 (circle) and FK520 (square). (B) Time course for FK506 production in R2YE medium. Triangles, FK506 concentration from the Δtcs7/pFKBN strain; circles, FK506 concentration from the Δtcs7 strain; squares, FK506 concentration from the WT/pFKBN strain; rhombuses, FK506 concentration from the wild-type strain.
Transcription analysis of the FK506 biosynthetic gene cluster in the wild-type, ΔfkbN, Δtcs7, and Δtcs7/pFKBN strains.
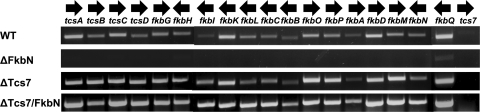
Transcription analysis of the FK506 biosynthetic gene cluster was carried out by semiquantitative RT-PCR in the wild-type, ΔfkbN, Δtcs7, and Δtcs7/pFKBN strains, except for tcs1 to tcs5, fkbJ, and tcs6. The nucleotide sequences of fkbJ and tcs6 are too short to perform RT-PCR, and it was shown previously that the tcs1 to tcs5 genes are not involved in the biosynthesis of FK506 (31). Total RNA was extracted from the wild-type and mutant strains after 72 h of cultivation (when the level of FK506 production began to increase notably in the wild-type strain) and was used as the template for gene expression analysis by RT-PCR. The primers for RT-PCR were specific to the sequences within the FK506 biosynthetic genes and were designed to generate cDNAs of approximately 500 bp (see Table S1 in the supplemental material). Figure 3 presents the results of these experiments. In the case of the wild-type strain, the transcripts of all genes, except for tcs7, in the FK506 cluster were observed at 72 h, suggesting that the silencing of tcs7 might precede the onset of FK506 biosynthesis. This observation is consistent with the time course of FK506 production (Fig. 2B). While FK506 biosynthesis increased slowly during the first 4 days of cultivation and then relatively rapidly between days 4 and 5 in the wild-type strain, FK506 production in the Δtcs7 and Δtcs7/pFKBN strains increased steadily over the 5 days. A similar phenomenon was reported in the case of tylosin biosynthesis. In Streptomyces fradiae, the negative regulatory gene tylQ was silenced after the onset of tylosin production but was expressed in the early phases of fermentation (36). In the ΔfkbN strain, all the genes investigated were switched off, except for fkbQ (type II thioesterase gene), for which a very faint band was observed. This shows that most of the genes of the FK506 gene cluster are not transcribed when fkbN is absent. The tcs7 transcript was not detected in the ΔfkbN strain, as in the wild-type strain, suggesting that the presence or absence of the positive regulatory gene fkbN did not observably affect the expression of the negative regulatory gene tcs7. There were observable increases in the expression levels of most genes of the FK506 cluster in the Δtcs7 strain except for fkbL, fkbC, fkbB, fkbA, and the positive regulatory gene fkbN. A further increase in the transcriptional level of the FK506 biosynthetic genes along with fkbN occurred in the Δtcs7/pFKBN strain, which is consistent with the significant enhancement of FK506 production in this mutant strain. Overall, the results of transcription analysis prove that fkbN encodes a LAL family pathway-specific positive transcriptional regulator for FK506 biosynthesis in Streptomyces sp. KCTC 11604BP and Tcs7 negatively controls the transcription of FK506 biosynthetic genes by a currently unknown mechanism.
Fig 3.
Transcriptional analysis of the FK506 biosynthetic genes in wild-type (WT) Streptomyces sp. KCTC 11604BP and in ΔfkbN, Δtcs7, and Δtcs7/pFKBN strains. The total RNAs were extracted from mycelia harvested after 72 h of cultivation.
Many LAL family pathway-specific positive regulators, including FkbN, play key roles in the production of secondary metabolites but have different modes of regulation. For example, TcmN of the tautomycetin biosynthetic gene cluster in Streptomyces sp. strain CK4412 (12) and PikD of the pikromycin cluster in S. venezuelae (38) are the only pathway-specific activators in each biosynthetic gene cluster. The LAL family regulator LipReg4 of the lipomycin cluster is under the control of a two-component signal transduction system (LipReg2 and Lipreg1). The LipReg1-LipReg2 pair possesses a hierarchically higher position and can regulate lipReg4 transcription (11). In the case of avermectin biosynthesis, the negative effects by the TetR family regulator AveI may be mediated indirectly by the LAL-type positive regulator AveR in Streptomyces avermitilis (5). The results presented herein may suggest a new mode of regulation involving a LAL family regulator and a LysR-type negative regulator, even though more-detailed regulatory mechanisms need to be examined by additional experiments, such as quantitative real-time RT-PCR or electrophoretic mobility shift assay (EMSA) analysis.
This study provides a valuable initial understanding of the regulatory factors governing the biosynthesis of FK506. In addition, the significantly enhanced production of FK506 (up to 4.0-fold) in the genetically engineered strain Δtcs7/pFKBN, in which the positive regulatory gene was overexpressed in the background of negative regulatory gene deletion, demonstrates the potential of this approach for strain development. Although these results were obtained using the wild-type strain, the application of this strategy to higher-producing industrial strains might allow the development of more improved strains.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Research Laboratory (NRL) program (R0A-2008-000-20030-0) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST), Republic of Korea, the Advanced Biomass R&D Center (ABC), funded by MEST (ABC-2010-0029800), Seoul R&BD Program (ST110024), Korea Research Foundation Grant (KRF-2006-005-J04001), and a grant from the Next-Generation BioGreen 21 Program (PJ0080932011), Rural Development Administration, Republic of Korea.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arias P, Fernández-Moreno MA, Malpartida F. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 181: 6958–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8: 208–215 [DOI] [PubMed] [Google Scholar]
- 3.Bierman M, et al. 1992. Plasmids cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116: 43–49 [DOI] [PubMed] [Google Scholar]
- 4.Chater KF, Chandra G. 2008. The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in streptomyces. J. Microbiol. 46: 1–11 [DOI] [PubMed] [Google Scholar]
- 5.Chen L, et al. 2008. Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl. Microbiol. Biotechnol. 80: 277–286 [DOI] [PubMed] [Google Scholar]
- 6.De Schrijver A, De Mot R. 1999. A subfamily of MalT-related ATP-dependent regulator in the LuxR family. Microbiology 145: 1287–1288 [DOI] [PubMed] [Google Scholar]
- 7.Gold BG. 2000. Neuroimmunophilin ligands: evaluation of their therapeutic potential for the treatment of neurological disorders. Expert Opin. Invest. Drugs 9: 2331–2342 [DOI] [PubMed] [Google Scholar]
- 8.Goranovic D, et al. 2010. Origin of the allyl group in FK506 biosynthesis. J. Biol. Chem. 285: 14292–14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He W, Lei J, Liu Y, Wang Y. 2008. The LuxR family members GdmRI and GdmRII are positive regulators of geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Arch. Microbiol. 189: 501–510 [DOI] [PubMed] [Google Scholar]
- 10.Henikoff S, Haughn GW, Calvo JM, Wallace JC. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. U. S. A. 85: 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horbal L, et al. 2010. Characterization and analysis of the regulatory network involved in control of lipomycin biosynthesis in Streptomyces aureofaciens Tü117. Appl. Microbiol. Biotechnol. 85: 1069–1079 [DOI] [PubMed] [Google Scholar]
- 12.Hur YA, Choi S-S, Sherman DH, Kim ES. 2008. Identification of TcmN as a pathway-specific positive regulator of tautomycetin biosynthesis in Streptomyces sp. CK4412. Microbiology 154: 2912–2919 [DOI] [PubMed] [Google Scholar]
- 13.Jeon H-G, Seo J, Lee MJ, Han K, Kim ES. 2011. Analysis and functional expression of NPP pathway-specific regulatory genes in Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 38: 573–579 [DOI] [PubMed] [Google Scholar]
- 14.Jung WS, et al. 2008. Enhanced heterologous production of desosaminyl macrolides and their hydroxylated derivatives by overexpression of the pikD regulatory gene in Streptomyces venezuelae. Appl. Environ. Microbiol. 74: 1972–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 16.Kino T, et al. 1987. FK-506, a novel immunosuppressant isolated from a Streptomyces. I. Fermentation, isolation, and physico-chemical and biological characteristics. J. Antibiot. (Tokyo) 40: 1249–1255 [DOI] [PubMed] [Google Scholar]
- 17.Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T. 2009. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 82: 1089–1096 [DOI] [PubMed] [Google Scholar]
- 18.Kölling R, Lother H. 1985. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J. Bacteriol. 164: 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuscer E, et al. 2007. Role of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 189: 4756–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leskiw BK, Lawlor EJ, Fernandez-Abalos LM, Chater KF. 1991. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative Streptomyces mutants. Proc. Natl. Acad. Sci. U. S. A. 88: 2461–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacNeil DJ, et al. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111: 61–68 [DOI] [PubMed] [Google Scholar]
- 22.Martin JF, Liras P. 2010. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 13: 263–273 [DOI] [PubMed] [Google Scholar]
- 23.Mo S, Ban YH, Park JW, Yoo YJ, Yoon YJ. 2009. Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. J. Ind. Microbiol. Biotechnol. 36: 1473–1482 [DOI] [PubMed] [Google Scholar]
- 24.Mo S, et al. 2011. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J. Am. Chem. Soc. 133: 976–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motamedi H, Cai SJ, Shafiee A, Elliston KO. 1997. Structural organization of a multifunctional polyketide synthase involved in the biosynthesis of the macrolide immunosuppressant FK506. Eur. J. Biochem. 244: 74–80 [DOI] [PubMed] [Google Scholar]
- 26.Motamedi H, et al. 1996. Characterization of methyltransferase and hydroxylase genes involved in the biosynthesis of the immunosuppressants FK506 and FK520. J. Bacterial. 178: 5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliynyk M, et al. 2003. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol. Microbiol. 49: 1179–1190 [DOI] [PubMed] [Google Scholar]
- 28.Parsons WH, Sigal NH, Wyvratt MJ. 1993. FK506 a novel immunosuppressant. Ann. N. Y. Acad. Sci. 685: 22–36 [DOI] [PubMed] [Google Scholar]
- 29.Rascher A, Hu Z, Buchanan GO, Reid R, Hutchinson CR. 2005. Insights into the biosynthesis of the benzoquinone ansamycins geldanamycin and herbimycin, obtained by gene sequencing and disruption. Appl. Environ. Microbiol. 71: 4862–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richet E, Raibaud O. 1989. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 8: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. 2001. Molecular cloning. a laboratory manual, 3rd ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Schmitt-John T, Engels JE. 1992. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 36: 493–498 [DOI] [PubMed] [Google Scholar]
- 33.Sheldon PJ, Busarow SB, Hutchinson CR. 2002. Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol. Microbiol. 44: 449–460 [DOI] [PubMed] [Google Scholar]
- 34.Shirling EB, Gottlieb D. 1966. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16: 313–340 [Google Scholar]
- 35.Sierra-Paredes G, Sierra-Marcuno G. 2008. Ascomycin and FK506: pharmacology and therapeutic potential as anticonvulsants and neuroprotectabts. CNS Neurosci. Ther. 14: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stratigopoulos G, Cundliffe E. 2002. Inactivation of a transcriptional repressor during empirical improvement of the tylosin producer, Streptomyces fradiae. J. Ind. Microbiol. Biotechnol. 28: 219–224 [DOI] [PubMed] [Google Scholar]
- 37.Takano E. 2006. Gamma-butyrolactones: Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9: 287–294 [DOI] [PubMed] [Google Scholar]
- 38.Wilson DJ, Xue Y, Reynolds KA, Sherman DH. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183: 3468–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu K, Chung L, Revill WP, Katz L, Reeves CD. 2000. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251: 81–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.