Abstract
Iridescence is a property of structural color that is occasionally encountered in higher eukaryotes but that has been poorly documented in the prokaryotic kingdom. In the present work, we describe a marine bacterium, identified as Cellulophaga lytica, isolated from the surface of an anemone, that exhibits bright green iridescent colonies under direct epi-illumination. This phenomenon has not previously been investigated in detail. In this study, color changes of C. lytica colonies were observed at various angles of direct illumination or observation. Its iridescent green appearance was dominant on various growth media. Red and violet colors were also discerned on colony edges. Remarkable C. lytica bacterial iridescence was revealed and characterized using high-resolution optical spectrometry. In addition to this, by culturing other bacterial strains to which various forms of faintly iridescent traits have previously been attributed, we identify four principal appearance characteristics of structural color in prokaryotes. A new general classification of bacterial iridescence is therefore proposed in this study. Furthermore, a specific separate class is described for iridescent C. lytica strains because they exhibit what is so far a unique intense glitter-like iridescence in reflection. C. lytica is the first prokaryote discovered to produce the same sort of intense iridescence under direct illumination as that associated with higher eukaryotes, like some insects and birds. Due to the nature of bacterial biology, cultivation, and ubiquity, this discovery may be of significant interest for both ecological and nanoscience endeavors.
INTRODUCTION
The use of light and color is fundamentally important in biological systems. While the majority of animal and plant colored appearances are generated through pigmentary processes (20), some employ micron- and submicron-sized physical structures as a means to generate color appearance effects (71). These structures can interact with incident light to create preferential scattering of specific spectral colors which can generate the most intense and often functionally targeted optical effects. If the structures are spatially arranged with very periodic geometry, then the color appearance of the animal or plant takes on an often strongly angle-dependent character and the colored appearance is defined as iridescent (18, 35, 36, 78, 41, 71). Animals exhibiting iridescence or, more generally, structural color have been the subject of keen interest to both the biology and physics fields. Structural color has been particularly well studied in Insecta (22, 47, 65, 70, 72, 73), in fishes (34, 40), and in Aves (17, 25, 58, 67). One virus system has also been linked to structural coloration (79). In the marine environment, iridescence has been reported in crabs (49), seashells (6, 38), squid (68), ctenophores (76), macroalgae (21, 51), and diatoms (23, 45).
Iridescence in prokaryotes has been poorly documented since its first observation, made in 1904 by Preisz (57). Until now, the phenomenon has been observed only on colonies or concentrated cell suspensions (53, 54, 57), and detailed illustrations are limited. Various general terms such as “shine,” “sheen,” “glistening,” “metallic effect,” “bright colors,” “luster,” “glow,” “glisten,” or “rainbow-like” were employed to describe the visual effects observed (15, 19, 28). Confusion with fluorescence has also been made (8, 13, 29). The lack of such precision has created difficulty in the accurate description of the visual characteristics of bacterial iridescence.
The two most described iridescent bacteria are Pseudomonas aeruginosa (9, 14, 15, 16, 28, 31, 77, 80) and Haemophilus influenzae (8, 19, 42, 44, 55). Two distinct observation methods were used, direct epi-illumination and transillumination, respectively, for P. aeruginosa and H. influenzae. The type of iridescence observed in certain strains of P. aeruginosa was described as a metallic iridescence and has been linked to cell lysis (16, 28, 80) and/or to the production of quinoline derivatives (9, 10, 77). A silvery appearance was also recently mentioned in Aneurinibacillus migulanus type III (2). In H. influenzae, colonies of capsulated cells display all spectral colors from red to blue under oblique transmitted light (transillumination). This type of iridescence was reported in several bacteria, including Listeria marthii (24), Pasteurella multocida (7, 29), coli-typhoid group bacteria (53), Listeria monocytogenes (39), and Alcanivorax balearicus (63). Several attempts were made to explain the transmitted iridescence, notably, by using spectral observations (19, 26, 44, 53, 55, 56). The phenomenon has been ascribed to either orderly arranged cells (19, 52, 53) or randomly arranged cells (27, 56). In these older works, both diffraction grating (19, 44, 52, 53, 56) and film effect (27) theories were proposed, but none could be confirmed.
Taken all together, the literature data suggest that bacterial iridescence is at best a loosely defined phenomenon that lacks rigorous description and understanding. Moreover, an intense structural color similar to some insect and vertebrate iridescence has not yet been documented in the bacterial kingdom. In this study, a marine bacterium forming intense structurally colored colonies that are spectrally brilliant in reflection has been isolated and described. Furthermore, in order to clarify the state of the art, we have compared the optical effects in a broad range of bacteria using both epi-illumination and transillumination. Special attention was given to the strains previously described as “iridescent.”
MATERIALS AND METHODS
Sample collection and bacterial isolation.
Collection of samples was performed on Chassiron lighthouse rocky shore at Oléron Island, west Atlantic coast of France (46°02′48″N, 1°24′37″W) in December 2009. Various marine organisms (macroalgae, sponges, anemones, crustaceans, mollusks, starfishes, and fishes) were collected with plastic gloves, transported in sterile plastic bags (to avoid terrestrial contamination), and processed immediately for microbiological studies. Tissues from the marine organisms were washed thoroughly with sterile artificial seawater (ASW; Instant Ocean) in order to remove loosely attached epibionts. Two-centimeter-square tissue specimens were then imprinted on marine agar (MA) purchased from Dutscher (Laboratorios Conda, S.A. Pronadisa) (64). Plates were examined visually after aerobic incubation for 24 h at 20°C or 30°C.
Taxonomic identification.
Genetic sequencing identified the isolated bacterial strain. Primers used for rRNA 16S gene sequencing were F1 (5′-AGAGTTTGATCCTGGCTCAG-3′), R1 (5′-GTATTACCGCGGCTGCTGGCAC-3′), F2 (5′-CTCCTACGGGAGGCAG-3′), and R2 (5′-GACACGAGCTGACGACA-3′) (75). Primers used for the 23S rRNA and the internal transcribed spacer 2 (ITS2) area were 23SF (5′-AACCCGTTGACGTTGAAAAG-3′), 23SR (5′-CTTGCTTTTCTCGGAGGATG-3′), ITSF (5′-TAGAGGTCGGCAGTTCGAGT-3′), and ITSR (5′-ATCTTCAATATGCCGGGTTG-3′). The sequences were compared with the sequences available in the NCBI database and LeBibi database (http://umr5558-sud-str1.univ-lyon1.fr/lebibi/lebibi.cgi) by using the BLAST service to determine their phylogenetic identity.
Culture of C. lytica.
The isolated marine bacterium was cultivated at 20°C or 25°C on three solid media. MA medium was employed preferentially for analysis of iridescence. Cytophaga agar (CYT) and low-nutrient (LN) media were made with ASW (30 g · liter−1; Instant Ocean). CYT medium contained 1 g of tryptone, 0.5 g of yeast extract, 0.5 g of CaCl2 · H2O, 0.5 g of MgSO4 · H2O, and 15 g of agar in 1 liter of ASW. In this medium, casein was replaced by tryptone because C. lytica does not degrade casein (33). LN medium contained only 15 g of agar in 1 liter of ASW (32).
Bacterial strains and culture media used for iridescence comparison.
Bacterial strains with their respective culture conditions are detailed in Table S1 in the supplemental material.
(i) Bacterial strains.
A total of 74 strains were compared. Bacteria described as iridescent in previous literature were Haemophilus influenzae (8, 19, 42, 44, 55), Pseudomonas aeruginosa (9, 14, 30, 77, 80), Alcanivorax balearicus (63), Aneurinibacillus migulanus (2), Listeria marthii (24), Listeria monocytogenes (39), Bordetella trematum (69), Salmonella typhi (43), Mannheimia haemolytica (Pasteurella mastitidis) (31), and Pasteurella multocida (7, 12, 29). Since iridescence was mentioned in bacterial groups such as coli-typhoid (46, 53), cocci (52), or bacilli (19, 56), strains of the following species were included: Staphylococcus spp., Bacillus spp., Pseudomonas stutzeri, Salmonella spp., Yersinia spp., Proteus vulgaris, Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli. Control bacteria, described as not being iridescent, were Micrococcus luteus, Lactococcus lactis, Stenotrophomonas maltophilia, Streptococcus pyogenes, Enterobacter cloacae, and two marine strains, Vibrio anguillarum and Vibrio lentus. Among the tested bacteria, several clinical strains were selected since bacterial iridescence has been associated with pathogenicity (30, 44, 55). Finally, four Cellulophaga lytica strains were compared, including the DSM 7489 strain corresponding to the LIM-21T strain, for which the complete genomic sequence has recently been published (50).
(ii) Culture media.
Appropriate media for iridescence observations were selected from literature data or were defined by experimental assays. Ready-to-use media (Dutscher) were nutrient agar (NA) (53), brain heart agar (BHA), and tryptic soy agar (TSA) for Aneurinibacillus migulanus (2), Luria-Bertani (LB) for Pseudomonas culture (9), and MA for marine strains such as Vibrio spp. and C. lytica. Prepared media were tryptose agar (TrypA) with 20 g of tryptose, 1 g of glucose, 5 g of NaCl, and 15 g of agar per liter for Listeria spp. (39) and Tween-peptone agar (TPA) with 10 g of Tween 20, 10 g of peptone, 5 g of NaCl, 0.1 g of CaCl2 · H2O, and 15 g of agar per liter for Alcanivorax balearicus (63). For Haemophilus influenzae cultures, Levinthal's XV medium (Lev XV) was prepared by mixing 10 g of peptone A, 10 g of meat extract, 5 g of NaCl, and 20 g of agar per liter, with a supplement of 15 mg of X (hemin) and V (NAD+) factors added after autoclaving (19, 55).
Macroscopic examination of bacterial iridescence. (i) Epi- and transillumination methods.
Iridescence of bacterial colonies was observed with the aid of a streaking procedure. One colony from a 24-h-old plate was subcultured in duplicate plates by drawing thin 5-cm linear streaks. After 24 h incubation, cultures were photographed in a dark room using two experimental arrangements of oblique epi-illumination and transillumination (see Fig. S1 in the supplemental material). The camera was a Canon Powershot A650 IS image stabilizer AiAF on the Av program. The lens was a macro, large size (12.1 megapixels) used in superfin mode. Illumination was with an E14 220- to 240-V, 11-W bulb (532 lumen at 2,700 K). For oblique epi-illumination, the plate was placed on a black backing. The optical axis of the camera formed an angle of 45° with the center of the plate. The light was fixed obliquely with an angle α of 67.5° from the plate. For transillumination measurements, samples were photographed from an angle of 45° above the petri dish with the light source directly behind it (i.e., normal incidence illumination in transmission).
(ii) Examination of C. lytica color changes.
In order to observe the color changes as a function of the illumination angle, the epi-illumination setup was employed. Pictures were taken alternatively at five different angles of incident light. Angle α values were 22.5°, 67.5°, 90°, 112.5°, or 135°. For these experiments, C. lytica was grown at 20°C (instead of 25°C) to observe all colorations more effectively.
Microscopic examination of C. lytica colony colors.
Detailed observations of colored colonies were performed under epi-illumination by using a numeric Keyence microscope (VHX-1000E). A VHX-1100 camera with a VH-Z20R/Z20W objective lens with adjustable magnifications of ×20, ×30, ×50, ×100 ×200, and ×400, the last one with a specific tool doubling the magnification, was used. To avoid specular reflections, the VH-S30 supporting mount of the camera was oriented at a 60° angle from the plate. The DEPTH UP/3D tool corresponding to the depth-from-defocus (DFD) process was employed at high magnification to focus on all optical fields and to improve image quality. In order to observe transitory colorations, the Keyence device was equipped with a VH-K20 lens ring. The support of the camera was oriented at a 90° angle. By moving the ring from right to left, three positions of illumination were used, namely, high, intermediate, and low light incidence angles.
Physical measurement of C. lytica (microspectrophotometry).
Illumination was directed onto the sample through an Ocean Optics UV-visible-near infrared optical fiber that was connected to an Ocean Optics HPX-2000 light source that spans approximately 300 nm to 850 nm. The reflected light was collected using a similar optical fiber that was itself connected to an Ocean Optics USB4000-UV-visible spectrometer (see Fig. S2 in the supplemental material). The angles of illumination and of detection could be separately set and controlled to a resolution of 0.5°. For a series of chosen fixed illumination angles, the collection fiber was stepped in 2° angle steps in an arc over the sample, and reflection spectra were recorded at each angular position. In this way, the dependence of reflected color with angle and, hence, the extent of each sample's iridescence could be measured and assessed (72, 74).
RESULTS
Isolation and identification of a marine bacterium with a glitter-like color appearance.
While searching for new cultivable epibiotic bacteria in the marine environment, we isolated a Gram-negative bacterium from the surface of a red anemone (Actinia equine) (Fig. 1A). Colonies exhibited bright iridescent reflected color when grown on MA plates and viewed under epi-illumination (Fig. 1B). The iridescence was not visible when colonies were resuspended or cells were grown in liquid media (data not shown). Iridescent green was the dominant color, but red and blue-violet were also observed at the colonies' peripheral edges. The MA-grown colonies' color appearance comprised submillimeter-sized centers of color of varying brightness distributed across the iridescent region. This gave the colonies' color reflection and intensity a glitter-like character.
Fig 1.
Observations of the marine environment-isolated Cellulophaga lytica. (A) The first isolation plate shows colored C. lytica colonies together with agarolytic and white bacterial colonies; (B) the second shows a pure culture of C. lytica observed under direct epi-illumination allowing examination of the intense structural color. In both cases, C. lytica was grown aerobically at 25°C on MA.
The marine strain was taxonomically identified by performing both 16S rRNA and 16S to 23S (16S-23S) ITS sequence analyses. The strain was phylogenetically affiliated with the Cytophaga-Flavobacterium-Bacteroides (CFB) group and the Flavobacteriaceae family and was identified as Cellulophaga lytica (CP002534, DSM 7489) (33, 37) with 16S rRNA, 23S rRNA, and ITS sequence similarities of 100%, 100%, and 99%, respectively. A thorough analysis of literature data showed that a “metallic tinge” of the colonies was previously mentioned for the affiliated strain C. lytica ATCC 23178T (DSM 7489 = CIP 103822 = LIM-21T) (33). The relative strain LIM-21T was recently genome sequenced, but no shiny effect was detailed in the description of its morphological appearance (50).
Coloration of C. lytica colonies on different culture media.
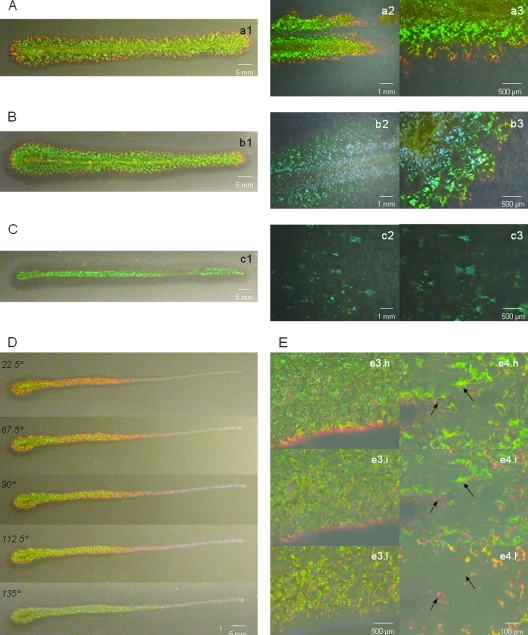
Agarolytic, mucous colonies with gliding motility and the bright glitter-like color centers effect were common characteristics for all media (Fig. 2a1, b1, and c1). On MA, colonies' color appearances comprised yellow pigmentation and principally a green structural color. At the peripheral growth zones, yellow, red, and then violet were observed (Fig. 2a1). This color gradation was confirmed using the Keyence microscope (Fig. 2a2 and a3; see Movie S1 in the supplemental material). Colonies grown on CYT were larger and less pigmented (Fig. 2b1). Blue was observed in the inner zone of the colony, and green, yellow, red, and violet were also visible (Fig. 2b2 and b3). Less growth occurred on the LN medium (Fig. 2c1). Colonies were translucent in this medium, and only green iridescence was discernible (Fig. 2c2 and c3; see Movie S2 in the supplemental material).
Fig 2.
Macroscopic and microscopic observation of C. lytica colonies' coloration. Colonies were pictured after 24 h growth on MA (A, D, and E), CYT (B), and LN (C). The inoculation was a thin 5-cm linear streak. Gliding motility can be identified as the spreading zone from the colony center. Bacterial agarolysis corresponds to the dark halo visible on colony edges. Pictures were taken under epi-illumination with a light angle of 67.5° at the macroscopic level (a1 to c1) or using Keyence microscope examination with 60° light incidence using a ×30 objective lens (a2 to c2) and a ×100 objective lens (a3 to c3). Evaluation of color changes at the macroscopic level was performed at diverse illumination angles (D). At the microscopic level (E), examination was performed at high (h), intermediate (i), and low (l) angles of incidence light with a ×100 (e3) and a ×400 (e4) objective lens. Arrows indicate positions of the color center appearing and disappearing as glitters.
Angle dependence of C. lytica colonies' coloration.
On MA, by changing the illumination angle from 22.5° to 135°, red and violet zones of the colony became green, while the central bright green region became blue or noniridescent (Fig. 2D). This angle dependence of reflected color, defined as iridescence, was the first direct evidence for a structural mechanism as the origin of the color. Color changes were also examined microscopically using the Keyence microscope (Fig. 2E; see Movie S3 in the supplemental material). Pictures demonstrated that bright green iridescence was predominant when illumination was close to grazing incidence (Fig. 2e3.h and e4.h). Violet-to-red color changes were observed at colony edges (Fig. 2e3.h and e3.l). Different iridescent color centers appeared and disappeared when the illumination position was modified from high to low incidence (black arrows in Fig. 2e4.h and e4.l). The image associated with an intermediate angle of illumination exhibited color centers which overlapped (Fig. 2e3.i and e4.i).
Physical evidence of C. lytica iridescence.
The data presented in Fig. 3 show optical reflection bands that unequivocally represent the iridescence of C. lytica bacterial colonies by the change in their color with angle. For instance, under illumination at an angle of −70°, the principal reflected color is green over a 70° angle range (−60° to +10°). However, within this angle range, the peak reflection wavelength changes continuously from approximately 550 nm to approximately 500 nm. This band of reflected color extends still further toward higher positive angles, the peak wavelength of which decreases to approximately 410 nm at a scattered angle of +70°. Three other reflected bands of color are shown on this map of reflectance data: each of these shows peak wavelengths that are also angle dependent: two at near-UV wavelengths and one in the near infrared.
Fig 3.
Color map showing the angle-dependent spectral reflectance of Cellulophaga lytica and confirming its iridescent appearance. The C. lytica sample was illuminated at a fixed light angle of −70°. Scattered wavelengths from 300 nm to 850 nm were recorded at different detection angles from −80° to 85° with 2° angle step resolution (the illumination plane and the detection plane were offset from each other by 3° to enable unobstructed detection over the full angle range). The color scale indicates the relative intensity of reflectance. The following emitted colors are given by the indicated wavelength value: UV, <400 nm; violet, 400 to 435 nm; blue, 435 to 490 nm; cyan, 490 to 520 nm; green, 520 to 560 nm; yellow, 560 to 590 nm; orange, 590 to 620 nm; red, 620 to 700 nm; and infrared, >700 nm.
Overall comparison of bacterial iridescence.
For a better understanding of bacterial structural color effects, we examined the iridescence of a broad range of bacterial strains. The extended classification of these bacterial optical effects is presented in Table S1 in the supplemental material, with a selection of images presented in Fig. 4. We propose a model of four separate bacterial iridescence categories: rainbow-diffuse (D) and rainbow-edge (R) appearances under transillumination and metallic (M) and glitter-like (G) appearances under epi-illumination.
Fig 4.
Examples of bacterial colonies belonging to different structural color categories. Observations were processed on epi- and transillumination. Iridescence categories are rainbow-diffuse (D), consisting of diffuse colors of the light spectrum; rainbow-edge (E), consisting of shining light spectrum color only on edges; metallic (M), consisting of silvery luster; and glitter-like (G), consisting of iridescent green in the middle and red and violet on the colony edges. Culture conditions are informed in Table S1 in the supplemental material.
The rainbow-diffuse category comprises bacterial colonies that exhibited all spectral colors ranging from red to blue only under the condition of transillumination (Fig. 4). Various color intensities were observed within this category. A large number of bacterial strains also displayed this visual effect (see Table S1 in the supplemental material). The rainbow-edge iridescence was visible only on colonies' edges. This phenomenon does not appear to have previously been described in literature. A few strains, namely, Bacillus cereus (Fig. 4), Stenotrophomonas maltophilia, Klebsiella pneumoniae, and Aneurinibacillus migulanus, displayed rainbow-edge iridescence, with these four expressing a common characteristic of thick and opaque colonies.
The metallic category comprised colonies exhibiting a silvery appearance under epi-illumination. As described in the literature (9), the ΔlasR mutant of P. aeruginosa exhibited a faintly silver appearance (Fig. 4). The metallic appearance of Aneurinibacillus migulanus type III previously described (2) could not be reproduced (see Table S1 in the supplemental material).
C. lytica strains were not iridescent under transillumination (Fig. 4). Their glitter-like iridescence is characterized principally by an intense green iridescent reflection. This novel iridescence is significantly higher in intensity than that of the bacterial structural coloration of all other three categories (Fig. 4).The iridescence of C. lytica was also found in another strain, DSM 2040, but not in the two strains CIP 103822 and DSM 2039. Moreover, the genome-sequenced strain DSM 7489 displayed only very-low-intensity iridescent color.
DISCUSSION
A marine bacterium exhibiting a bright iridescently colored colony appearance has been isolated in this study. Although other forms of bacterial iridescence have been described in selected literature, the phenomenon has never been comprehensively investigated or discussed.
The comparison of diverse bacteria by two illumination protocols, transillumination and epi-illumination, enabled the classification of four categories of bacterial iridescence. The rainbow-diffuse iridescence was common in particular in smooth colonies and was present in mucous, capsulated, and pathogenic bacteria. This iridescence, which has never been explained, has previously been used as an easily observed criterion to discriminate between capsulated and noncapsulated strains of H. influenzae (26, 54, 55). The rainbow-edge iridescence was less common and might occur only at specific thicknesses of the colonies.
Only a few P. aeruginosa strains exhibited the metallic appearance. Surprisingly, two P. aeruginosa strains (ATCC 27853 and a clinical mucous strain) had both rainbow-diffuse (under transillumination) and metallic (under epi-illumination) iridescence. Metallic reflections in P. aeruginosa 14 ΔlasR Δpqsh have been linked to the accumulation of the 4-hydroxy-2-heptylquinoline molecule (9, 10, 77). However, no explanation as to how the accumulated molecule creates a metallic-looking reflection has yet been presented. Since metallic appearance is not associated with a change of color with angle, the term “metallic iridescence” should not be used.
A novel iridescence category for the appearance of isolated C. lytica was discovered and termed “glitter-like” iridescence. The practical measurement of a broad range of spectrophotometric reflection data on C. lytica colonies has enabled us to prove this structural color and to construct the map of wavelength-dispersive reflection bands. These represent a clear iridescence effect, namely, a change of reflected color with angle.
Interestingly, certain C. lytica strains appeared noniridescent. The sequenced strain LIM-21T (ATCC 23178T = DSM 7489) (50) exhibited low-intensity iridescence. Described to be identical in the bacterial collection banks, the strains DSM 7489 and CIP 103822 were found to have different colony morphologies; this has possibly led to their different iridescent characteristics. The C. lytica organism isolated in this study has the most intense glitter-like appearance.
The iridescence of C. lytica was mentioned only superficially in two studies. Colonies of C. lytica ATCC 23178T with “metallic tinge” were evoked (33). The term “iridescent” was used only once in an algicidal bioactivity study of C. lytica ASM 21 (66). “Greenish metallic iridescence” was mentioned in the Cellulophaga genus in Bergey's Manual of Systematic Bacteriology (5). It is noteworthy that Cellulophaga (Cytophaga) lytica was first related to the group Bacteroides and the order Cytophagales before its reclassification within the Cytophaga-Flavobacterium-Bacteroides (CFB) group and the order Flavobacteriales (33). The unique illustration of C. lytica colony in the book The Prokaryotes does not show iridescence but shows only common yellow-pigmented colonies. Another picture of a Cytophaga species showed a very weak red color appearance described as iridescence (62). Since that date, “reddish-greenish iridescence” has been employed as a descriptor for strains belonging to the order Cytophagales in the second edition of The Prokaryotes or in Bergey's manual but without additional explanations (37, 59, 60). Moreover, no mention or illustration of iridescence was found in the most recent editions (3, 4, 61). Glitter-like iridescence within the genus Cellulophaga and in the family Flavobacteriaceae is under investigation.
Structures responsible for the coherent scattering that creates the C. lytica iridescence are under investigation by electron microscopy; however, specialized preparation protocols are needed and under development for observation of the micron- and submicron-scale biofilm structures in their original state. However, since iridescence involves periodicity, then intercellular communication mechanisms may be involved in the multicellular organization (1, 11). Although these mechanisms are still unknown, it is possible that iridescence implies associated biological roles for spatial organization that offer advantage for the ensemble population. In addition, the iridescence of C. lytica colonies was observed under epi-illumination. This manner of illumination is more natural and ubiquitous than transillumination and may also indicate potential ecobiological roles for the phenomenon.
In many higher organisms, structural colors have been strongly linked to biological functions associated with conspecific and interspecific communication purposes. However, these same structures can also serve noncommunication functions such as those related to thermoregulation, UV protection, light filtering, water repellency, mechanical friction reduction, or desiccation prevention (18). In lower organisms such as diatoms, the strong light manipulation associated with the periodic nanostructure on diatom frustule walls might influence the collection of more light into the photoreceptors for more optimized photosynthetic efficiencies (23, 45, 48). In contrast to these examples, the functional roles of iridescence in bacteria have never been explored. Also unanswered is whether bacterial iridescence occurs in natural habitats. C. lytica's iridescence might provide a selective advantage under the relatively extreme conditions (high salinity, temperature variation, desiccation, and light exposure) of its habitats.
Supplementary Material
ACKNOWLEDGMENTS
Betty Kientz was a Ph.D. student with a grant from the Ministère de la Recherche et de l'Enseignement Supérieur. Peter Vukusic acknowledges the support of AFOSR grant FA9550-10-1-0020.
We are indebted to Deborah Hogan (Dartmouth Medical School), Olivier Gaillot (CHU Lille, Lille, France), and Jocelyne Caillon (CHU Nantes, Nantes, France) for providing bacterial strains. H. Agogue's expert technical assistance with preliminary 16S rRNA gene sequence analysis is gratefully acknowledged.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bassler BL. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109: 421–424 [DOI] [PubMed] [Google Scholar]
- 2. Berditsch M, Afonin S, Ulrich AS. 2007. The ability of Aneurinibacillus migulanus (Bacillus brevis) to produce the antibiotic gramicidin S is correlated with phenotype variation. Appl. Environ. Microbiol. 73: 6620–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bernardet J-F, Nakagawa Y. 2006. An introduction to the family Flavobacteriaceae, p 455–480 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes, 3rd ed, vol 7 Springer, New York, NY: [Google Scholar]
- 4. Bowman JP. 2006. The marine clade of the family Flavobacteriaceae: the genera Aequorivita, Arenibacter, Cellulophaga, Croceibacter, Formosa, Gelidibacter, Gillisia, Maribacter, Mesonia, Muricauda, Polaribacter, Psychroflexus, Psychroserpens, Robiginitalea, Salegentibacter, p 677–694 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes, 3rd ed, vol 7 Springer, New York, NY: [Google Scholar]
- 5. Bowman JP. 2011. Genus IX. Cellulophaga Johansen, Nielsen and Sjøholm 1999, 1238VP, p 176–180 In Krieg NR, et al. (ed), Bergey's manual of systematic bacteriology, the Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, 2nd ed, vol 4 Springer, New York, NY: [Google Scholar]
- 6. Brink DJ, van der Berg NG, Botha AJ. 2002. Iridescent colors on seashells: an optical and structural investigation of Helcion pruinosus. Appl. Optics 41: 717–722 [DOI] [PubMed] [Google Scholar]
- 7. Brogden KA. 1980. Physiological and serological characteristics of 48 Pasteurella multocida cultures from rabbits. J. Clin. Microbiol. 11: 646–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandler CA, Fothergill LD, Dingle JH. 1939. The pattern of dissociation in Haemophilus influenzae. J. Bacteriol. 37: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cugini C, Morales DK, Hogan DA. 2010. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156: 3096–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Argenio DA, et al. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64: 512–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies DG, et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298 [DOI] [PubMed] [Google Scholar]
- 12. DeAngelis PL, Jing W, Drake RR, Achyuthan AM. 1998. Identification and molecular cloning of a unique hyaluronan synthase from Pasteurella multocida. J. Biol. Chem. 273: 8454–8458 [DOI] [PubMed] [Google Scholar]
- 13. De Kruif PH. 1921. Dissociation of microbic species. I. Coexistence of individuals of different degrees of virulence in cultures of the Bacillus of rabbit septicemia. J. Exp. Med. 33: 773–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dickinson L. 1948. The bacteriophages of Pseudomonas pyocyanea. 1. The effect of various substances upon their development. J. Gen. Microbiol. 2: 154–161 [Google Scholar]
- 15. Dickinson L, Codd S. 1952. The bacteriophages of Pseudomonas pyocyanea. 2. Bacteriophage reproduction in an iridescent strain. J. Gen. Microbiol. 6: 1–13 [DOI] [PubMed] [Google Scholar]
- 16. Don PA, van den Ende M. 1950. A preliminary study of the bacteriophages of Pseudomonas aeruginosa. J. Hyg. (Lond.) (48): 196–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doucet SM, Shawkey MD, Hill GE, Montgomerie R. 2006. Iridescent plumage in satin bowerbirds: structure, mechanisms and nanostructural predictors of individual variation in colour. J. Exp. Biol. 209: 380–390 [DOI] [PubMed] [Google Scholar]
- 18. Doucet SM, Meadows MG. 2009. Iridescence: a functional perspective. J. R. Soc. Interface 6(Suppl 2): S115–S132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Engboek HC. 1950. The phenomenon of iridescence in bacterial cultures, with particular reference to Pfeiffer's bacillus. Acta Pathol. Microbiol. 27: 388–393 [Google Scholar]
- 20. Fox DL. 1976. Animal biochromes and structural colours: physical, chemical, distributional & physiological features of coloured bodies in the animal world, 2nd ed University of California Press, Berkeley, CA: [Google Scholar]
- 21. Gerwick WH, Lang NJ. 1977. Structural, chemical and ecological studies on iridescence in Iridaea (Rhodophyta). J. Phycol. 13: 121–127 [Google Scholar]
- 22. Ghiradella H. 1991. Light and color on the wing: structural colors in butterflies and moths. Appl. Optics 30: 3492–3500 [DOI] [PubMed] [Google Scholar]
- 23. Gordon R, Losic D, Tiffany MA, Nagy SS, Sterrenburg FAS. 2009. The glass menagerie: diatoms for novel applications in nanotechnology. Trends Biotechnol. 27: 116–127 [DOI] [PubMed] [Google Scholar]
- 24. Graves LM, et al. 2010. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60: 1280–1288 [DOI] [PubMed] [Google Scholar]
- 25. Greenewalt CH, Brandt W, Friel DD. 1960. The iridescent colors of hummingbird feathers. Proc. Am. Philos. Soc. 104: 249–253 [Google Scholar]
- 26. Guillot M. 1941. Relation entre certaines propriétés optiques des bactéries et leur forme géométrique vraie. Diffraction de la lumière par les cultures et symétrie de la bactérie. C. R. Hebd. Seances Acad. Sci. 1100–1103 [Google Scholar]
- 27. Guillot M. 1942. Critères physiques d'homogénéité d'une culture de microbes vivants. C. R. Soc. Biol. 720–721 [Google Scholar]
- 28. Hadley P. 1924. Transmissible lysis of Bacillus pyocyaneus. J. Infect. Dis. 34: 260–304 [Google Scholar]
- 29. Heddleston KL, Wessman G. 1975. Characteristics of Pasteurella multocida of human origin. J. Clin. Microbiol. 1: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffman LR, et al. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8: 66–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jasmin AM. 1945. An improved staining method for demonstrating bacterial capsules, with particular reference to Pasteurella. J. Bacteriol. 50: 361–363 [PubMed] [Google Scholar]
- 32. Jensen PR, Kauffman CA, Fenical W. 1996. High recovery of culturable bacteria from the surfaces of marine algae. Mar. Biol. 126: 1–7 [Google Scholar]
- 33. Johansen JE, Nielsen P, Sjøholm C. 1999. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of [Cytophaga] lytica to Cellulophaga lytica gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 49: 1231–1240 [DOI] [PubMed] [Google Scholar]
- 34. Land M. 1972. The physics and biology of animal reflectors. Prog. Biophys. Mol. Biol. 24: 75–106 [DOI] [PubMed] [Google Scholar]
- 35. Lee DW. 1991. Ultrastructural basis and function of iridescent blue colour of fruits in Elaeocarpus. Nature 349: 260–262 [Google Scholar]
- 36. Lee DW, Taylor GT, Irvine AK. 2000. Structural fruit coloration in Delarbea michieana (Araliaceae). Int. J. Plant Sci. 161: 297–300 [DOI] [PubMed] [Google Scholar]
- 37. Lewin RA, Lounsbery DM. 1969. Isolation, cultivation and characterization of flexibacteria. J. Gen. Microbiol. 58: 145–170 [DOI] [PubMed] [Google Scholar]
- 38. Liu Y, Shigley J, Hurwit K. 1999. Iridescent color of a shell of the mollusk Pinctada margaritifera caused by diffraction. Opt. Express 4: 177–182 [DOI] [PubMed] [Google Scholar]
- 39. Luppi A, Rocourt J, Bucci G, Maini P. 1986. Isolement de Listeria de l'eau du Pô. Boll. Ist. Sieroter. Milan 65: 108–111 [PubMed] [Google Scholar]
- 40. Lythgoe JN, Shand J. 1989. The structural basis for iridescent colour changes in dermal and corneal iridophores in fish. J. Exp. Biol. 141: 313–325 [Google Scholar]
- 41. Meadows MG, et al. 2009. Iridescence: views from many angles. J. R. Soc. Interface 6(Suppl 2): S107–S113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mulder J. 1939. Haemophilus influenzae of the respiratory type as a cause of purulent meningitis. J. Pathol. Bacteriol. 48: 175–185 [Google Scholar]
- 43. Nicolle P, Jude A, Le Minor L. 1950. Relation entre l'intensité de l'irisation présentée par certaines colonies de Salmonella et leur constitution antigénique. Ann. Inst. Pasteur (Paris) 78: 572–582 [PubMed] [Google Scholar]
- 44. Nogrady G, Guérault A. 1964. Conditions favorisant l'iridescence bacterienne et l'observation du phénomène. Rev. Can. Biol. 23: 367–373 [PubMed] [Google Scholar]
- 45. Noyes J, Sumper M, Vukusic P. 2008. Light manipulation in a marine diatom. J. Mater. Res. 23: 3229–3232 [Google Scholar]
- 46. Ogg JE, Zelle MR. 1957. Isolation and characterization of a large cell possibly polyploid strain of Escherichia coli. J. Bacteriol. 74: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parker AR. 1998. The diversity and implications of animal structural colours. J. Exp. Biol. 201: 2343–2347 [DOI] [PubMed] [Google Scholar]
- 48. Parker AR, Townley HE. 2007. Biomimetics of photonic nanostructures. Nat. Nanotechnol. 2: 347–353 [DOI] [PubMed] [Google Scholar]
- 49. Parker AR, Mckenzie DR, Ahyong ST. 1998. A unique form of light reflector and the evolution of signalling in Ovalipes (Crustacea: Decapoda: Portunidae). Proc. R. Soc. Lond. B Biol. 265: 861–867 [Google Scholar]
- 50. Pati A, et al. 2011. Complete genome sequence of Cellulophaga lytica type strain (LIM-21T). Stand. Genomic Sci. 4: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pellegrini L, Pellegrini M. 1982. Iridescent bodies of Cystoseira stricta Sauvageau (Phaeophyta, Fucales): their fine structure, development and nature in vegetative cells. Phycologia 21: 34–46 [Google Scholar]
- 52. Pijper A. 1918. Diffraction-phenomena in cultures of microöganisms. Med. J. S. Afr. 14: 211–218 [Google Scholar]
- 53. Pijper A. 1923. Diffraction in biological structures. I. The structure of colonies of the coli-typhoid group. S. Afr. Med. Rec. 17: 243–248 [Google Scholar]
- 54. Pijper A. 1952. Dimensional differentiation, filtration and separation of bacteria. J. Pathol. Bacteriol. 64: 529–538 [DOI] [PubMed] [Google Scholar]
- 55. Pittman M. 1931. Variation and type specificity in the bacterial species Haemophilus influenzae. J. Exp. Med. 53: 471–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ponder E. 1934. Diffraction patterns produced by bacteria. J. Exp. Biol. 11: 54–57 [Google Scholar]
- 57. Preisz H. 1904. Studien über Morphologie und Biologie des Milzbrandbacillus (mit besonderer Berücksichtigung der Sporenbildung auch bei anderen Bacillen). Zentralbl. Bakteriol. Parasitenkd. Orig. 35: 280–293 [Google Scholar]
- 58. Prum RO, Torres R. 2003. Structural colouration of avian skin: convergent evolution of coherently scattering dermal collagen arrays. J. Exp. Biol. 206: 2409–2429 [DOI] [PubMed] [Google Scholar]
- 59. Reichenbach H. 1989. Genus I. Cytophaga Winogradsky 1929, 577, AL emend, p 2015–2050 In Staley JT, Bryant MP, Pfennig N, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 3 Williams & Wilkins, Baltimore, MD: [Google Scholar]
- 60. Reichenbach H. 1992. The order Cytophagales, p 3631–3675 In Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H. (ed), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed Springer-Verlag, New York, NY: [Google Scholar]
- 61. Reichenbach H. 2006. The order Cytophagales, p 549–590 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 7 Springer, New York, NY: [Google Scholar]
- 62. Reichenbach H, Dworkin M. 1981. The order Cytophagales (with addenda on the genera Herpetosiphon, Saprospira, and Flexithrix), p 356–379 In Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG. (ed), The prokaryotes: a handbook on habitats, isolation, and identification of bacteria, 1st ed, vol 1 Springer-Verlag, Berlin, Germany: [Google Scholar]
- 63. Rivas R, et al. 2007. Alcanivorax balearicus sp. nov., isolated from Lake Martel. Int. J. Syst. Evol. Microbiol. 57: 1331–1335 [DOI] [PubMed] [Google Scholar]
- 64. Schulz B, Guske S, Dammann U, Boyle C. 1998. Endophyte-host interactions. II. Defining symbiosis of the endophyte-host interaction. Symbiosis 25: 213–227 [Google Scholar]
- 65. Seago AE, Brady P, Vigneron J-P, Schultz TD. 2009. Gold bugs and beyond: a review of iridescence and structural colour mechanisms in beetles (Coleoptera). J. R. Soc. Interface 6(Suppl 2): S165–S184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Skerratt JH, Bowman JP, Hallegraeff G, James S, Nichols PD. 2002. Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar. Ecol. Prog. Ser. 244: 1–15 [Google Scholar]
- 67. Stavenga DG, Leertouwer HL, Marshall NJ, Osorio D. 2011. Dramatic colour changes in a bird of paradise caused by uniquely structured breast feather barbules. Proc. R. Soc. Lond. B Biol. 278: 2098–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sutherland RL, Mäthger LM, Hanlon RT, Urbas AM, Stone MO. 2008. Cephalopod coloration model. I. Squid chromatophores and iridophores. J. Opt. Soc. Am. A 25: 588–599 [DOI] [PubMed] [Google Scholar]
- 69. Vandamme P, et al. 1996. Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int. J. Syst. Bacteriol. 46: 849–858 [DOI] [PubMed] [Google Scholar]
- 70. Vukusic P, Wootton RJ, Sambles JR. 2004. Remarkable iridescence in the hindwings of the damselfly Neurobasis chinensis chinensis (Linnaeus) (Zygoptera: Calopterygidae). Proc. R. Soc. Lond. B Biol. 271: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vukusic P, Sambles JR. 2003. Photonic structures in biology. Nature 424: 852–855 [DOI] [PubMed] [Google Scholar]
- 72. Vukusic P, Sambles JR, Lawrence CR, Wootton RJ. 1999. Quantified interference and diffraction in single Morpho butterfly scales. Proc. R. Soc. Lond. B Biol. 266: 1403–1411 [Google Scholar]
- 73. Vukusic P, Sambles JR, Lawrence CR, Wootton RJ. 2001. Structural colour: now you see it-now you don't. Nature 410: 36 [DOI] [PubMed] [Google Scholar]
- 74. Vukusic P, Stavenga DG. 2009. Physical methods for investigating structural colours in biological systems. J. R. Soc. Interface 6(Suppl 2): S133–S148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173: 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Welch V, Vigneron J-P, Lousse V, Parker A. 2006. Optical properties of the iridescent organ of the comb-jellyfish Beroë cucumis (Ctenophora). Phys. Rev. E 73041916: 1–7 [DOI] [PubMed] [Google Scholar]
- 77. Wensinck F, van Dalen A, Wedema M. 1967. Iridescent material and the effect of iron on its production by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 33: 73–86 [DOI] [PubMed] [Google Scholar]
- 78. Whitney MW, et al. 2009. Floral iridescence, produced by diffractive optics, acts as a cue for animal pollinators. Science 323: 130–133 [DOI] [PubMed] [Google Scholar]
- 79. Williams RC, Smith KM. 1958. The polyhedral form of the Tipula iridescent virus. Biochim. Biophys. Acta 28: 464–469 [DOI] [PubMed] [Google Scholar]
- 80. Zierdt CH. 1971. Autolytic nature of iridescent lysis in Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 37: 319–337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.