Abstract
Soils were incubated for 80 days in a continuously labeled 14CO2 atmosphere to measure the amount of labeled C incorporated into the microbial biomass. Microbial assimilation of 14C differed between soils and accounted for 0.12% to 0.59% of soil organic carbon (SOC). Assuming a terrestrial area of 1.4 × 108 km2, this represents a potential global sequestration of 0.6 to 4.9 Pg C year−1. Estimated global C sequestration rates suggest a “missing sink” for carbon of between 2 and 3 Pg C year−1. To determine whether 14CO2 incorporation was mediated by autotrophic microorganisms, the diversity and abundance of CO2-fixing bacteria and algae were investigated using clone library sequencing, terminal restriction fragment length polymorphism (T-RFLP), and quantitative PCR (qPCR) of the ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) gene (cbbL). Phylogenetic analysis showed that the dominant cbbL-containing bacteria were Azospirillum lipoferum, Rhodopseudomonas palustris, Bradyrhizobium japonicum, Ralstonia eutropha, and cbbL-containing chromophytic algae of the genera Xanthophyta and Bacillariophyta. Multivariate analyses of T-RFLP profiles revealed significant differences in cbbL-containing microbial communities between soils. Differences in cbbL gene diversity were shown to be correlated with differences in SOC content. Bacterial and algal cbbL gene abundances were between 106 and 108 and 103 to 105 copies g−1 soil, respectively. Bacterial cbbL abundance was shown to be positively correlated with RubisCO activity (r = 0.853; P < 0.05), and both cbbL abundance and RubisCO activity were significantly related to the synthesis rates of [14C]SOC (r = 0.967 and 0.946, respectively; P < 0.01). These data offer new insights into the importance of microbial autotrophy in terrestrial C cycling.
INTRODUCTION
Global warming in the last 100 years is believed to be closely associated with increases in the concentration of atmospheric CO2, due primarily to the increased use of fossil fuels since the industrial revolution (26). Terrestrial ecosystems have been recognized as major sinks for global CO2 emissions and are of significant interest because of their potential to mitigate atmospheric CO2 (17). It has been estimated that the soil C pool is about twice as large as the atmospheric pool (6). The Calvin-Benson-Bassham cycle is the major and most widely distributed pathway for CO2 fixation. In terrestrial ecosystems, the Calvin cycle is found in diverse organisms, from bacteria and algae to green plants (35). Ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) catalyzes the first, rate-limiting step in the Calvin cycle that enables 102 Pg of inorganic atmospheric CO2 to be converted into organic cellular constituents every year (34). RubisCO is central to primary productivity on land and in the oceans and has been well characterized in aquatic environments and in plant and pure cultured microorganisms (15). However, microbial autotrophs can be difficult to culture, and relatively few studies have reported on RubisCO activities in soils.
RubisCO is found in four forms (I, II, III, and IV) with each form having a different structure, catalytic activity, and O2 sensitivity (35). Form I RubisCOs are composed of four large subunit dimers containing catalytically active amino acid residues, as well as eight additional small subunits. Form II RubisCOs exist only in certain photosynthetic bacteria and in peridinin-containing dinoflagellates. In addition to forms I and II, there are two groups of structurally related proteins that have been designated form III and form IV RubisCOs that consist only of large subunits and lack several of the key active-site residues necessary for the carboxylation of ribulose 1,5-bisphosphate (RuBP) (10). Of the different forms, form I RubisCOs are thought to predominate in soils and are found in plants, algae, cyanobacteria, and autotrophic bacteria (38). The key active-site residue in the large subunit of form I RubisCO is encoded by the cbbL gene (16). Phylogenetic studies on the cbbL gene sequences have shown that cbbL genes encoding form I RubisCO can be further assigned to one of four clades, IA to ID (27, 35). These clades encompass the obligate autotrophic bacteria, including some marine cyanobacteria (Synechococcus and Prochlorococcus) (form IA RubisCO), the facultative autotrophic bacteria (form IC), green plants, green algae, and some cyanobacterial sequences (form IB), and the chromophytic algae (form ID 35). As such, RubisCO form I-encoding cbbL genes have been used as functional markers for molecular ecological studies of CO2 assimilative autotrophs in aquatic systems (27). However, the genetic diversity of CO2 fixation in soil microbial autotrophs has not been studied in any detail (32, 33).
Autotrophic microorganisms and macroalgae are known to contribute significantly to CO2 assimilation in aquatic systems such as the oceans and wetlands (31) but have not generally been thought to have a key role in CO2 fixation and sequestration in soils. This is despite the fact that microbial autotrophs have been reported in a number of soil studies. O'Donnell et al. (24), working with soil and compost isolates, have shown that certain actinomycetes, predominantly Streptomyces, can grow on carbon monoxide as their sole carbon source by first oxidizing CO to CO2 and then fixing CO2 using RubisCO and phosphoribulose kinase (2). Other studies have shown that microbial autotrophy is evident in many taxa, including aerobic, CO-oxidizing bacteria such as Bradyrhizobium japonicum, Sinorhizobium meliloti, and purple sulfide-oxidizing bacteria such as Chromatium vinosum (23, 32, 40).
Thus, while organisms with the genetic potential to fix CO2 are widespread in soils, their capacity to fix CO2 and their importance in terrestrial C cycling have yet to be fully investigated. In this paper, cbbL gene diversity and RubisCO activity were investigated in six different soils where 14CO2 incubation studies had shown significant incorporation of 14C into the soil microbial biomass. The significance of the work in terms of global carbon cycling and the potential for microbial autotrophy is discussed.
MATERIALS AND METHODS
Soil preparation.
Three paddy soils (P1, P2, P3) and three upland soils (U1, U2, and U3) were sampled from the subtropical region of China. The sampling sites of the soils had a mean annual temperature of approximately 16.8°C and an annual rainfall of about 1,400 mm. For each soil, a bulk sample was collected from the plough layer (depth, 0 to 20 cm), hand-sorted to remove visible plant residues, and then sieved by passage through a 5-mm mesh. Soil pH was determined at a soil-to-H2O ratio of 1:2.5 (wt/vol) using a pH meter (Delta 320, Mettler-Toledo Instruments Ltd., China), and clay content was determined using the pipette method (22). Soil organic C (SOC) content and total N were measured by dry combustion using an elemental analyzer (Vario MAX C/N, Elementar, Germany), and cation exchange capacity (CEC) was measured by titration (28). Site information and soil properties were as shown in Table 1. Prior to use, the paddy soils P1, P2, and P3 were flooded with distilled water while the other soils were adjusted to 45% of field water holding capacity. To accommodate changes in microbial activity following disturbance (3), all soils were left to equilibrate prior to analysis for 10 days after flooding or rewetting.
Table 1.
Characteristics of the paddy and upland soils used in this study
| Soila | Site/position | Soil type | pH | CEC (mol kg−1) | Clay content (%) | SOC (g kg−1) | Total N (g kg−1) |
|---|---|---|---|---|---|---|---|
| P1 | 113°11′E, 28°8′N | Fluvisol | 5.2 | 6.2 | 25.0 | 15.9 | 1.56 |
| P2 | 113°11′E, 28°8′N | Fluvisol | 4.7 | 7.2 | 11.3 | 14.8 | 1.64 |
| P3 | 111°31′E, 29°13′N | Ultisol | 5.1 | 12.2 | 11.5 | 17.9 | 1.74 |
| U1 | 113°3′E, 28°12′N | Fluvisol | 6.0 | 11.9 | 34.1 | 16.9 | 1.88 |
| U2 | 113°5′E, 28°12′N | Ultisol | 4.6 | 10.3 | 41.6 | 9.07 | 1.19 |
| U3 | 111°31′E, 29°13′N | Ultisol | 5.2 | 11.5 | 43.9 | 5.63 | 0.76 |
P1, P2, and P3 were paddy soils, and U1, U2, and U3 were upland soils.
Incubation with 14C-labeled CO2.
For each soil, two sets of microcosms with four replicates each were prepared by weighing 1.0 kg fresh soil (in an oven-dried form) into plastic containers (10 cm in diameter and 22 cm in height). One set was used as the control and was covered with a sheet of 0.7-cm-thick dark plastic foam which blocked light but allowed aeration. Both sets of microcosms were transferred to the growth chamber (area, 80 by 250 cm; height, 120 cm; 29 April 2009, China patent no. ZL2006100197402) and incubated for 80 days in a continuously 14CO2-labeled atmosphere generated from NaH14CO3 solution (1.0 M at a radioactivity of 1.68 × 104 Bq μg−1 C). The incubation system also provided artificial light (approximately 500 mmol photons m−2 s−1 parabolic aluminum reflector [PAR]) between 8:00 a.m. and 8:00 p.m., maintained temperatures of 22°C between 8:00 p.m. and 8:00 a.m. and 32°C between 8:00 a.m. and 8:00 p.m., and held relative humidity at 80 to 90% throughout the incubation period. At the end of the 80-day incubation, soils were removed from the microcosms, mixed thoroughly, and then divided into three separate portions. One portion was immersed immediately in liquid nitrogen, freeze-dried, and stored in 10-ml sterile centrifuge tubes at −70°C for future DNA extraction, cbbL amplification, and RubisCO activity analyses. Another was air-dried, ground to pass through a 100-mesh filter, and used for [14C]SOC determination (43). The remaining portion was used immediately for determination of 14C-labeled microbial biomass carbon ([14C]MBC) (41). The synthesis rates (RS) of [14C]SOC (g C m−2 day−1) were calculated using the formula
| (1) |
where D represents the internal diameter (m) of the container and T represents the incubation time (80 days), respectively.
Microbial DNA extraction.
Microbial DNA was extracted in triplicate from 0.5 g freeze-dried soil using the SDS-GITC-PEG method described by Chen et al. (5). The extracted pellet was dissolved in 50 μl sterilized H2O and the DNA concentration determined using a spectrophotometer (Nanodrop; PeqLab, Germany). Prior to real-time quantitative PCR (qPCR), the DNA solution was purified using a DNA purification kit (Tiangen, China) and diluted to a final concentration of approximately 5 ng μl−1 DNA using sterilized H2O to provide the soil DNA template.
PCR amplification and terminal restriction fragment length polymorphism (T-RFLP) determination of cbbL genes.
Bacterial cbbL genes (IA and IC) and chromophytic algal cbbL genes (ID) were separately amplified by PCR using the primers described by Nanba et al. (23) and Paul et al. (27), using forward primers labeled at the 5′ end with 6-carboxy-fluorescein (FAM) (Invitrogen, China).
Briefly, each individual PCR mixture (in triplicate) contained approximately 50 ng soil DNA, 100 pmol of the primers, 200 μM deoxynucleoside triphosphates (dNTPs) (Tiangen, China), 1 U Taq polymerase (Tiangen, China) in 1× reaction buffer provided with the enzyme (Tiangen, China), made up to 50 μl with sterilized H2O. Amplification was done in a Mastercycler PCR machine (model 5333; Eppendorf, Germany) using programs of 3 min at 95°C, 35 cycles of 1 min at 95°C, 1 min at 62°C, and 1.5 min at 72°C and ending with a 10-min extension at 72°C (bacterial cbbL gene) and 3 min at 95°C, 35 cycles of 1 min at 95°C, 1 min at 52°C, and 1.5 min at 72°C and finally with a 10-min extension at 72°C (chromophytic algal cbbL gene).
Labeled PCR products were purified using an agarose gel DNA purification kit (Tiangen, China). Portions (10 μl) of the purified solution (approximately 30 ng μl−1) were separately digested with enzyme HhaI (for bacterial cbbL gene fragments) and RsaI (for the chromophytic algal cbbL gene fragments; TaKaRa, Japan). Digestion products were then analyzed using an automated sequencer (model 373A; Applied Biosystems, Weiterstadt, Germany) by the Sunny Company, China.
Data comprising 48 T-RFLP profiles (24 soils incubated in light with two gene targets) were processed using the Gene Scan analysis software package (version 2.1; Applied Biosystems). The relative abundance (Ap) of each T-RF was calculated from Ap = ni/N × 100, where ni represents the peak height of one distinct T-RF and N is the sum of all peak heights in the profile. Minor peaks, where the relative abundance was <1%, were regarded as background noise (19) and not used in the statistical analysis. T-RFs greater than 10% of the total peak height were regarded as dominant fragments. Canonical correspondence analysis (CCA) of the T-RFLP profiles was done in CANOCO 4.5 for Windows (Microcomputer Power, Ithaca, NY) (39) to assess the relationship between microbial community structure and soil physiochemical parameters SOC, total N (TN), CEC, pH, and C:N ratio.
Cloning and sequencing of cbbL genes.
The same primer sets that were used for the T-RFLP analysis, without the fluorescent labels, were used to separately amplify the bacterial and chromophytic algal cbbL genes in the DNA templates of soils P1 and U1 (the representatives for paddy and upland soil, respectively). PCR products were purified and cloned into Escherichia coli DH5α using the pGEM-T easy vector system (Promega, Mannheim, Germany), followed by “blue-white screening.” Randomly selected clones were then screened for positive inserts by PCR using the SP6 and T7 primers (29) and sequenced by BGI (Wuhan, China). The sequences of cbbL clones were checked for close relatives and taxonomic assignment to known cbbL sequences using BLAST (1). Sequence identity of >97% was defined as an operational taxonomic unit (OTU). Bacterial and chromophytic algal cbbL clone sequences were each aligned using Clustal X 1.83, and two neighbor-joining trees were produced from the alignments using MEGA 4.0 (37). Bootstrap analysis of 1,000 replicates was used to estimate the stability of tree topologies.
Quantification of cbbL genes.
Bacterial and chromophytic algal cbbL genes in the DNA template (1 μl) of all of the soils were analyzed separately by real-time qPCR (21) using the primers described above. For each of the microbial types, triplicate reactions were prepared in 384-well PCR microplates (Axygen) containing 1× SYBR Premix ExTaq (Takara Bio Inc., Shiga, Japan), 0.15 μM each primer (Invitrogen, China), 1× ROX (provided with SYBR Premix ExTaq), and 1 μl soil DNA template (5 ng μl−1) and then amplified in an ABI Prism 9700 real-time PCR system (PerkinElmer, Applied Biosystems) using a thermal protocol for both bacterial and chromophytic algal cbbL genes: 30 s at 95°C followed by 40 cycles of 10 s at 95°C, 40 s at 60°C, and 30 s at 72°C. A standard curve ranging from 102 to 108 bacterial or chromophytic algal cbbL copies μl−1 was generated using 10-fold serial dilutions of plasmids linearized by SalI (TaKaRa, Japan) with 1010 copies μl−1 partial bacterial or chromophytic cbbL sequence from environmental samples. A set of the reaction mixtures for the standard curve (1 μl) was also prepared and carried out in parallel with that for the soil cbbL genes. The cbbL copies in the reaction mixtures of soils were automatically analyzed using the SDS 2.3 software provided with the real-time PCR system. The efficiency of real-time qPCRs was 95% for bacterial cbbL and 96% for algal cbbL gene (based on the slope of the standard curve).
RubisCO enzyme activity analysis.
Total soil protein was extracted after 80 days of soil incubation using the method described by Takai et al. (36). Freeze-dried soil samples (2 g, four replicates) were placed in 10-ml centrifuge tubes and suspended in 6 ml protein extract containing 100 mM Tris-HCl (pH 7.8) and 1 mM dithiothreitol (DTT) (Sigma). The soil suspension was thoroughly disrupted by ultrasonication (JY92-II Scientz, China) in an ice bath and centrifuged at 20,000 × g for 20 min at 4°C. The supernatant was amended with solid ammonium sulfate to reach 80% saturation, stirred for 30 min, and then centrifuged at 4°C (20,000 × g, 20 min). The resultant pellets were dissolved in 50 μl extract and used for determining RubisCO activity.
RubisCO activity in the protein extracts was measured in a reaction mixture (1.5 ml) containing the buffer system, substrates, cofactor, and coupling enzymes prepared as described by Takai et al. (36). After incubation at 30°C in a water bath for 10 min to recover enzyme activity, the absorbance of reaction mix (Eo) was determined at 340 nm using a spectrophotometer (UV-2450; Shimadzu, Japan). Following the addition of substrate (50 μl 25 mM ribulose bisphosphate), the absorbance (Et) was determined at a reaction time of 30 s. RubisCO activity (nmol CO2 g−1 soil min−1) was calculated using the formula
| (2) |
where V represents the volume of the reaction mixture (ml); ε, the absorbency (6.22 × 10−3 ml nmol−1 cm−1); d, the optical path length of the cuvette (cm); t, the reaction time (min); and w, the weight of soil (g).
Statistical analysis.
Data were processed using Excel 2000 for the means and the standard errors. Multiple comparisons of significant differences were made using one-way analysis of variance (ANOVA) followed by a Duncan test (P < 0.05). Correlation analyses were done using the Pearson correlation method with significance defined at the 0.05 level unless otherwise stated. All analyses were performed using SPSS 13.0 software for Windows XP.
Nucleotide sequence accession numbers.
Clones determined in the present study have been deposited in GenBank under accession numbers HQ174564 to HQ174665 for the bacterial cbbL gene and HQ184440 to HQ184456 for the chromophytic algal cbbL gene.
RESULTS
Amount of 14CO2 assimilation in different soils during incubation.
After 80 days of incubation, the amounts of 14CO2 incorporated into the soil organic carbon and microbial mass were determined. Radioactivity in soils incubated in the dark was barely detectable, while significant amounts of [14C]SOC and [14C]MBC were recovered from soils incubated in the light (Table 2). The amounts of [14C]SOC and [14C]MBC ranged from 8.44 (U3) to 64.61 (P3) mg kg−1 and from 1.55 (U3) to 10.36 (P2) mg kg−1, respectively. The soils differed significantly in terms of incorporation of SOC and MBC (Table 2). The amounts of [14C]SOC and [14C]MBC incorporated were highly correlated (r = 0.945; P < 0.05) and were generally higher in paddy than in upland soil (P < 0.05). Correlation analysis also showed that the synthesis rates of [14C]SOC were closely related to RubisCO activities and the abundance of cbbL genes in the soils (r = 0.946 and 0.967, respectively; P < 0.01).
Table 2.
Amounts of [14C]SOC and [14C]MBC and synthesis rates of [14C]SOC in six cropland soils
| Soila | Amt (mg kg−1)b |
Synthesis rates of [14C]SOC (g C m−2 day−1) | |
|---|---|---|---|
| [14C]SOC | [14C]MBC | ||
| P1 | 46.41 ± 3.56 B | 4.52 ± 0.27 C | 0.074 ± 0.003 |
| P2 | 64.61 ± 2.65 A | 10.36 ± 1.46 A | 0.103 ± 0.002 |
| P3 | 60.99 ± 7.33 A | 7.61 ± 0.42 B | 0.097 ± 0.006 |
| U1 | 21.94 ± 3.36 C | 3.74 ± 0.38 C | 0.035 ± 0.003 |
| U2 | 26.36 ± 2.08 C | 4.75 ± 0.57 C | 0.042 ± 0.002 |
| U3 | 8.44 ± 1.52 D | 1.55 ± 0.23 D | 0.013 ± 0.001 |
Soils are described in Table 1.
Means followed by the same letter are not significantly different (P > 0.05) between the different soils.
Phylogenetic analysis of cbbL gene clones from soils.
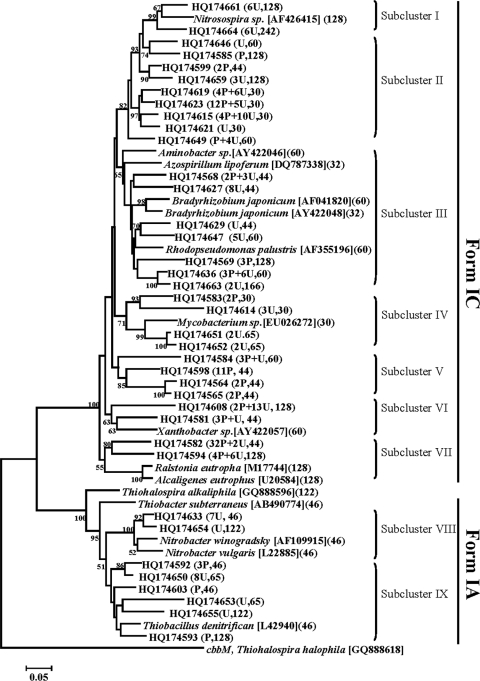
A bacterial cbbL gene phylogenetic tree was constructed from the 38 bacterial cbbL sequences (OTUs) of the test soils and 16 selected, published bacterial cbbL reference sequences from GenBank. The bacterial cbbL gene sequences were recovered in two clades, form IA and form IC (Fig. 1). Of the bacterial cbbL sequences, 88% were grouped in clade IC. This clade encompassed seven subclusters (I to VII), of which subclusters II, III and VII contained 70% of the form IC cbbL clones and accounted for 28%, 19% and 23% of the total form IC cbbL genes, respectively. Except for subcluster II, their distribution between soils was uneven. For example, in subcluster III, about 70% of the clones were from upland soil, whereas in subcluster VII the clones were mainly from paddy soil. The remaining four subclusters contained only 30% of the form IC cbbL genes, and their distribution between soils did not appear to follow any specific pattern. Clones in subclusters I, IV, and VI were mostly recovered from the upland soil, while subcluster V clones were predominantly from paddy soil. In contrast to the form IC cbbL sequences, the form IA cbbL sequences were less diverse and formed only two subclusters, VIII and IX, which were mostly from the upland soil. Additionally, six of the nine subclusters grouped with the cultivated cbbL-containing bacteria. In the form IC clade, the major subcluster III grouped with known sequences of Azospirillum lipoferum (DQ787338), Aminobacter sp. (AY422046), Bradyrhizobium japonicum (AY422048 and AF041820), and Rhodopseudomonas palustris (AF355196), and subcluster VII grouped with Ralstonia eutropha (M17744) and Alcaligenes eutrophus (EU20584). In the form IA clade, subcluster VIII included known cbbL sequences from Nitrobacter winogradskyi (AF109915) and Nitrobacter vulgaris (L22885) while subcluster IX included Thiobacillus denitrificans (L42940) gene sequences.
Fig 1.
Phylogenetic tree of bacterial cbbL sequences from soils with the cbbM gene from Thiohalospira halophila (accession no. GQ888618) used as an outgroup. Bootstrap values (>50%) are indicated at the branch points. The paddy soil (P) and upland soil (U) are shown in brackets, and the numbers before each soil abbreviation represent the respective number of clones. The numbers after soil abbreviation indicate the respective sizes of the T-RFs after in silico analysis with HhaI. The scale bar represents a 5% estimated sequence divergence.
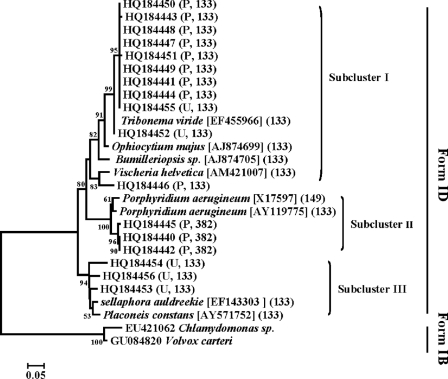
Amplified chromophytic algal cbbL fragments and related reference sequences formed three subclusters that demonstrated a relatively large diversity of chromophytic algae and included Xanthophyta, Bacillariophyta, and Rhodophyta (Fig. 2). Subclusters I and II accounted for 82% of the total and were dominated by clones from paddy soils. Subclusters I and II included known cbbL sequences from Tribonema viride (EF455966) and Porphyridium aerugineum (X17597 and AY119775), respectively. The remaining clade, subcluster III, contained only 18% of the total clones and included sequences primarily from upland soil. Comparison with known sequences showed similarities with Sellaphora auldreekie (EF143303) and Placoneis constans (AY571752). Overall, the phylogenetic analysis indicated that there were distinct differences in cbbL gene diversity between soils.
Fig 2.
Phylogenetic tree of algal cbbL sequences from soils with the Chlamydomonas sp. (EU421062) and Volvox carteri (GU084820) used as outgroups. Bootstrap values (>50%) are indicated at the branch points. The paddy soil (P) and upland soil (U) are shown in brackets, and the numbers before each soil abbreviation represent the respective number of clones. The numbers after soil abbreviation indicate the respective sizes of the T-RFs after in silico analysis with RsaI. The scale bar represents a 5% estimated sequence divergence.
T-RFLP analyses of the cbbL-containing bacterial and algal community compositions.
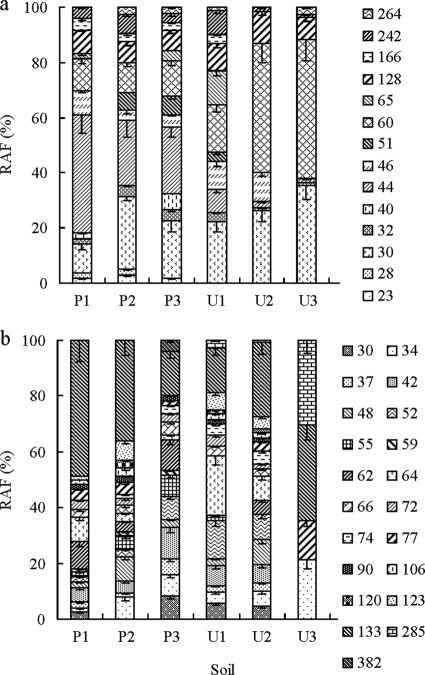
A total of 14 T-RFs were identified and used to compare bacterial cbbL communities (Fig. 3a). Soil type had an impact on both the presence and the relative abundance of different bacterial cbbL T-RFs. For example, the relative abundance of the T-RF of 30 bp in length (T-RF 30bp) was highest in soil U3 and lowest in soil P1 whereas T-RF 60bp was highest in U3 and lowest in P3 (Fig. 3a). T-RFs 46bp and 128bp were major components in soils U1 and U2, with relative abundances significantly higher than those in other soils except for P1 (P < 0.05). Some general trends in cbbL distribution were also evident between paddy and upland soils. The values for T-RF 30bp (22.1 to 35.5%) and T-RF 60bp (17.1 to 50.3%) in upland soils were generally higher than those of paddy soils (10.6 to 26.4% and 11.0 to 12.7%, respectively) and for T-RF 60bp the differences between soils were significant (P < 0.05). In contrast, T-RF 44bp was a major component of the cbbL gene diversity in all paddy soils but not in upland soils. Notwithstanding the limitations of extrapolating from TRFs to organism identity, the in silico analysis indicated that the dominant T-RFs 30bp, 60bp, 44bp, and 128bp were most closely related to facultatively autotrophic bacteria such as Rhodopseudomonas palustris, Bradyrhizobium japonicum, and Ralstonia eutropha. This is consistent with the results of the cloning analyses, where 88% of the cloned cbbL sequences clustered with RubisCO form 1C (facultative autotrophs) and with known sequences of Bradyrhizobium, Rhodopseudomonas, and Ralstonia. Such sequences were detected in all soils and comprised a major part of the cbbL-containing bacterial community. Obligate autotrophic bacterial cbbL gene sequences (form 1A), as represented by T-RF 46bp, from organisms such as Thiobacillus denitrificans, Nitrobacter winogradskyi, and Nitrobacter vulgaris, were a relatively small fraction of the autotrophic bacteria in these soils. Sequences representing these genera were also recovered in the clone library (Fig. 1), and the differences in relative abundance may indicate that bacterial CO2 fixation in soils is more prevalent in facultative than in obligate autotrophs.
Fig 3.
Average relative abundances of bacterial cbbL T-RFs (a) and algal T-RFs (b) from soils with light incubation. Different shadings on the graph have been used to indicate differences in fragment size. Bars indicate the standard errors of the means (n = 4).
Comparison of 24 chromophytic algal cbbL T-RFLP profiles (Fig. 3b) showed that nine T-RFs (34bp, 42bp, 48bp, 52bp, 62bp, 64bp, 77bp, 133bp, and 285bp) presented as major components of the cbbL gene diversity in these soils. The relative abundance of these T-RFs varied markedly with soil type. For example, the T-RFs 42bp and 62bp were major components in paddy soils while the T-RFs 34bp, 48bp, 77bp, and 285bp dominated in upland soils. The T-RF 133bp was detected in all soils, and in general the relative abundance of T-RF 133bp in paddy soils (mean = 33.5%; coefficient of variation [CV] = 49%) was higher than in upland soils (mean = 25.6%; CV = 36%). In silico comparison of fragment length showed that only two of the T-RFs from these soils showed fragment lengths that were similar to those of cbbL sequences from known chromophytic algae. The 133bp fragment was similar to that from Xanthophyta and Bacillariophyta, while the 382bp fragment could be putatively identified as originating from Rhodophyta. These taxa are also represented in the clone library (Fig. 2).
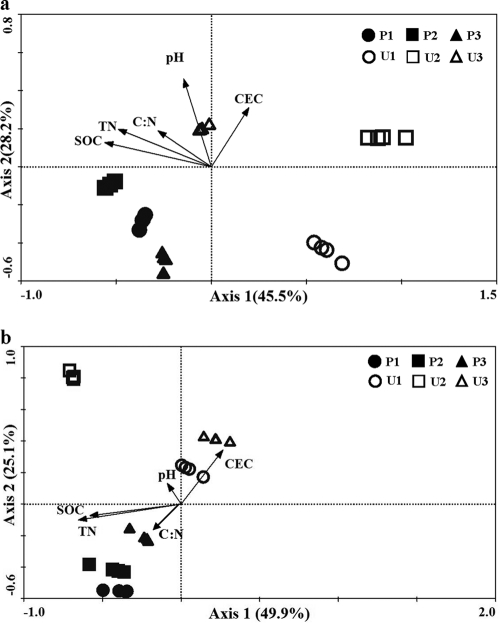
The comparative analysis (CCA) showed that the compositions of cbbL-containing bacterial and algal communities in upland soils were clearly differentiated from each other while the compositions of the paddy soil samples were relatively close. The variation among different cbbL-containing bacterial and algal community compositions was most highly correlated with SOC (as determined at P = 0.002 using a Monte Carlo permutation test within the CCA analysis) (Fig. 4).
Fig 4.
CCA analyses of the T-RFLP profiles for the bacterial (a) and chromophytic algal (b) cbbL gene diversity from the soils with light incubation. Arrows represent the environmental variables.
Abundances of bacterial and non-green-algal cbbL genes in soils.
From the soils incubated in light, the bacterial cbbL abundance ranged from 4.2 × 106 to 1.25 × 108 copies g−1 soil and the chromophytic algal cbbL abundance ranged from 8.2 × 103 to 1.8 × 106 copies g−1 soil (Table 3). In general, the copy number of the bacterial cbbL was higher (14 to 1,964 times) than that of the chromophytic algal cbbL. Soil type was shown to have a significant effect on the abundance of bacterial and chromophytic algal cbbL, with paddy soils having a greater number (4 to 30 times) of bacterial cbbL copies than upland soils. The highest bacterial cbbL abundance was observed in P2 and P3, and the lowest was in U3. Although the copy numbers for the chromophytic algal cbbL gene were generally lower, they also showed similar differences by soil type, with the highest chromophytic algal signals found in P3 and the lowest in U3. After 80 days of incubation, significantly lower cbbL abundances of bacteria and chromophytic algae were detected in soils incubated in the dark compared to those incubated in the light, suggesting that cbbL fixing potential declined when soils were incubated in the dark.
Table 3.
Abundance of bacterial and chromophytic algal cbbL genes in the different soils
| Soila | Abundanceb |
|||
|---|---|---|---|---|
| Bacteria (108 copies g−1 soil) |
Algae (106 copies g−1 soil) |
|||
| Light | Dark | Light | Dark | |
| P1 | 0.99 ± 0.02 B | 0.59 ± 0.02 | 0.62 ± 0.06 B | 0.37 ± 0.04 |
| P2 | 1.17 ± 0.03 A | 0.58 ± 0.03 | 1.39 ± 0.12 A | 0.22 ± 0.01 |
| P3 | 1.25 ± 0.02 A | 0.62 ± 0.02 | 1.40 ± 0.27 A | 0.56 ± 0.02 |
| U1 | 0.22 ± 0.03 C | 0.11 ± 0.02 | 1.08 ± 0.17 A | 0.69 ± 0.04 |
| U2 | 0.14 ± 0.01 D | 0.07 ± 0.01 | 0.01 ± 0.00 C | ND |
| U3 | 0.04 ± 0.01 E | 0.02 ± 0.00 | 0.01 ± 0.00 C | ND |
See Table 1 for soils.
Means followed by the same letter are not significantly different (P > 0.05) between different soil types. In all cases, the mean with light incubation was significantly (P < 0.05) larger than that with dark incubation for the same soil. ND, not detectable.
RubisCO enzyme activity in different soils.
After 80 days of incubation, RubisCO activity was barely detectable in the soils incubated in the dark while significant RubisCO activity was measured in soils incubated in light. This activity ranged from 0.77 to 6.73 nmol CO2 g−1 soil min−1 (Table 4). There were significant differences in RubisCO activity between soils. The highest RubisCO activity was measured in the paddy soil P2, with the lowest seen in the upland soil, U3. In general, measured RubisCO activities in paddy soils were higher than those in upland soils. Correlation analysis showed that the activity of RubisCO was closely related to the abundance of bacterial cbbL (r = 0.853; P < 0.05). However, no significant correlation was observed between RubisCO activity and chromophytic algal cbbL abundance (P > 0.05).
Table 4.
RubisCO activity in the soils incubated for 80 days
| Soila | RubisCO (nmol CO2 g−1 soil min−1)b |
|
|---|---|---|
| Light | Dark | |
| P1 | 3.65 ± 0.40 C | 0.58 ± 0.09 |
| P2 | 6.73 ± 0.48 A | 0.89 ± 0.17 |
| P3 | 5.55 ± 0.76 B | 1.66 ± 0.42 |
| U1 | 3.29 ± 0.64 C | 1.89 ± 0.27 |
| U2 | 3.09 ± 0.29 C | ND |
| U3 | 0.77 ± 0.28 D | ND |
See Table 1 for soils.
Means followed by the same letter are not significantly different (P > 0.05) between different soil types. The mean with light incubation was significantly (P < 0.05) larger than that with dark incubation for the same soil. ND, not detectable.
DISCUSSION
The work reported here shows that the diversity of bacterial and chromophytic algal cbbL genes encoding RubisCO in soils offers significant potential for the microbial assimilation of atmospheric CO2. Carney et al. (4) considered that C emission and absorption in terrestrial ecosystems in response to global climate change do not balance. Thus, the quantity of CO2 lost from soil is always greater than that resulting from CO2 uptake. If correct, this suggests that there is a “missing C sink” of about 2 to 3 Pg C year−1 at the global scale (14). Although the work reported here needs to be extended to include many more soils and soil types, the calculated rate of assimilation of organic C is approximately 0.013 to 0.103 g C m−2 day−1 (12 h light exposure). This would equate to a global rate for microbial synthesis of organic C of 4.9 to 37.5 g C m−2, or 0.68 to 4.9 Pg per annum, assuming a total terrestrial area of 1.4 × 108 km2. This means that microbial autotrophy could account for up to 4% of the total CO2 fixed by terrestrial ecosystems each year. Since there was significant 14CO2 assimilation only when soils were incubated in the light, with almost no uptake when incubated in the dark, it seems reasonable to assume that the microbial CO2 assimilation processes were predominantly phototrophic and, as such, were driven primarily by autotrophs (including photo- and chemoautotrophic microbes) rather than heterotrophs.
The measurement of RubisCO enzyme activity in these soils was shown to be closely related to the synthesis rate of SOC (r = 0.946; P < 0.01) and was positively correlated (r = 0.853; P < 0.05) with the abundance of the bacterial cbbL gene. There is no significant correlation with the abundance of the chromophytic algal genes supporting the hypothesis that facultatively autotrophic bacteria were the major microbial groups involved in CO2 assimilation. The diversity and abundance of these bacteria differed between soils and were correlated with soil use and SOC content, suggesting that soil management and cropping regime might be manipulated to enhance soil C sequestration by enhancing the growth of facultatively anaerobic bacteria. This hypothesis is supported by previous investigations that also showed a link between bacterial cbbL gene diversity and local edaphic factors such as plant cover, land use, and fertilizer management (23, 33, 40). However, the dominant factors regulating the diversity of CO2-fixing bacteria in soils and the potential for such systems to sequester carbon need further evaluation.
The work reported here has shown that the diversity and abundance of microbial autotrophs varies with soil type and land use. Of the physicochemical properties measured, the multivariate statistical analysis showed that the major influence on cbbL gene diversity and, by inference, on soil bacterial autotrophs is SOC. Soil organic matter is the most active fraction in soil and is a key determinant of soil fertility and productivity (9, 25, 42). SOC also stabilizes soil structure (7) and is an important source of carbon for microbial growth and of nutrients such as phosphorus, sulfur, calcium, magnesium, and trace elements (30). The finding that total gene copy number (qPCR) was lower in soils with low levels of SOC could be due to the limited availability of the C needed to support growth, since facultatively autotrophic bacteria, and not obligate autotrophs, were the more dominant (32, 33). The size of the cbbL-containing chromophytic algal community was related in a similar way to SOC levels. The algal cbbL sequences recovered in this study belonged to the genera Xanthophyta and Bacillariophyta. Although the algae were typically autotrophic organisms, some were mixotrophs and as such derive energy from both photosynthesis and the uptake of organic carbon either by osmotrophy, mixotrophy, or phagotrophy (18). In soils, the decomposition of soil organic matter can provide carbon dioxide for algal photosynthesis, with the transformation of soil organic matter providing rich organic substrates and key nutrients for mixotrophic algal growth. These findings are in general agreement with recent studies on nutrient dynamics and their impact on microbial community structure of bacteria and phytoplankton in different ecosystems (11, 32).
Since management can be used to moderate SOC levels in soils, the interaction between SOC levels and the CO2 assimilation activities of facultatively autotrophic bacteria warrants further investigation. For example, Ralstonia eutropha and Alcaligenes eutrophus were identified both in the clone libraries (Fig. 1, subcluster VII) and from the T-RFLP analyses as significant components of the microbial community in paddy soils. Ralstonia eutropha and Alcaligenes eutrophus are known to produce and sequester polyhydroxyalkanoates that can accumulate intracellularly to approximately 90% of the cell's dry weight (44). These polyhydroxyalkanoates are not readily turned over in flooded soils and are largely protected from mineralization in paddy soils by complexation with active iron oxides. This capacity for organic matter complexation and protection is considerably reduced in upland soils, where the amounts of active iron are generally lower. Taken together, the increase in RubisCO activity, the increase in facultative autotrophic bacteria, and the production of highly stable forms of soil carbon (polyhydroxyalkanoate) make paddy soils and other inundated systems potential land use options for maximizing the retention and sequestration of atmospheric CO2. However, additional work is needed to confirm the link between the changes in the relative abundance of cbbL genes and the capacity for paddy soils to fix CO2. These studies include the need for more detailed isotope tracer studies (13) and an investigation of alternate CO2 assimilation pathways such as the 3-hydroxypropionate cycle, the reductive citric acid cycle, and the reductive acetyl-CoA pathway (8, 12, 20).
A combination of RubisCO activity measurements, cbbL gene diversity analysis, and real-time qPCR has highlighted the potential for cropping systems (e.g., paddy and upland) soils to sequester CO2. These studies also suggest that the role of facultatively autotrophic bacteria and chromophytic algae in the biogeochemical cycling of soil C has probably been underestimated. T-RFLP and sequence analysis of clone libraries have shown that the major T-RF components among the bacterial autotrophs were indicative of taxa known to accumulate significant amounts of stable C in the form of polyhydroxyalkanoates and indicate that land management in arable and paddy systems that optimizes facultatively autotrophic activity could be a viable option for enhancing the sequestration and stabilization of carbon in soils.
ACKNOWLEDGMENTS
This study was jointly supported by the Knowledge Innovation Program of the Chinese Academy of Sciences (grant no. KZCX3-SW-437, ISACX-LYQY-QN-1103), the Strategic Priority Research Program-Climate Change: Carbon Budget and Related Issues of the Chinese Academy of Sciences (grant no. XDA05050505), the National Natural Science Foundation of China (40901124, 41090283), and the International S&T cooperation program of China (grant no. 2011DFA30770).
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1.Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell JM, Falconer C, Colby J, Williams E. 1987. CO metabolism by a thermophilic actinomycete, Streptomyces strain G26. J. Gen. Microbiol. 133:3445–3456 [Google Scholar]
- 3.Butterly CR, McNeill AM, Baldock JA, Marschner P. 2011. Rapid changes in carbon and phosphorus after rewetting of dry soil. Biol. Fertil. Soils 47:41–50 [Google Scholar]
- 4.Carney KM, Hungate BA, Drake BG, Megonigal JP. 2007. Altered soil microbial community at elevated CO2 leads to loss of soil carbon. Proc. Natl. Acad. Sci. U. S. A. 104:4990–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. 2010. Impact of long-term fertilization on the composition of denitrifier communities based on nitrite reductase analysis in a paddy soil. Microb. Ecol. 60:850–861 [DOI] [PubMed] [Google Scholar]
- 6.Falkowski P, et al. 2000. The global carbon cycle: a test of our knowledge of earth as a system. Science 290:291–296 [DOI] [PubMed] [Google Scholar]
- 7.Fonte SJ, Barrios E, Six J. 2010. Earthworms, soil fertility and aggregate-associated soil organic matter dynamics in the Quesungual agroforestry system. Geoderma 155:320–328. [Google Scholar]
- 8.Fuchs G. 1989. Alternative pathways of autotrophic CO2 fixation, p 365–382. In Schlegel HG, Bowien B. (ed), Autotrophic bacteria. Springer-Verlag KG, Berlin, Germany [Google Scholar]
- 9.Ge T, et al. 2011. Chemical properties, microbial biomass, and activity differ between soils of organic and conventional horticultural systems under greenhouse and open field management: a case study. J. Soils Sediment. 11:25–36 [Google Scholar]
- 10.Hanson TE, Tabita FR. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 98:4397–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heil CA, Revilla M, Glibert PM, Murasko S. 2007. Nutrient quality drives differential phytoplankton community composition on the southwest Florida shelf. Limnol. Oceanogr. 52:1067–1078 [Google Scholar]
- 12.Holo H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch. Microbiol. 151:252–256 [Google Scholar]
- 13.Jenkins SN, et al. 2010. Taxon-specific responses of soil bacteria to the addition of low level C inputs. Soil Biol. Biochem. 42:1624–1631 [Google Scholar]
- 14.Karim A, Veizer J, Barth J. 2008. Net ecosystem production in the Great Lakes basin and its implications for the North American missing carbon sink: a hydrologic and stable isotope approach. Global Planet. Change 61:15–27 [Google Scholar]
- 15.Kellogg EA, Juliano ND. 1997. The structure and function of RuBisCO and their implications for systematic studies. Am. J. Bot. 84:413–428 [PubMed] [Google Scholar]
- 16.Kusian B, Bowien B. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135–155 [DOI] [PubMed] [Google Scholar]
- 17.Lal R. 2008. Carbon sequestration. Philos. Trans. R. Soc. B. 363:815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litchman E, Klausmeier CA. 2008. Trait-based community ecology of phytoplankton. Annu. Rev. Ecol. Evol. Syst. 39:615–639 [Google Scholar]
- 19.Lukow T, Dunfield PF, Liesack W. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure with in an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241–247 [DOI] [PubMed] [Google Scholar]
- 20.Menendez C, et al. 1999. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouritzen P, et al. 2005. ProbeLibrary: a new method for faster design and execution of quantitative real-time PCR. Nat. Methods 2:313–316 [Google Scholar]
- 22.Müller T, Höper H. 2004. Soil organic matter turnover as a function of the soil clay content: consequences for model applications. Soil Biol. Biochem. 36:877–888 [Google Scholar]
- 23.Nanba K, King GM, Dunfield K. 2004. Analysis of facultative lithotrophic distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. Appl. Environ. Microbiol. 70:2245–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell AG, Falconer C, Goodfellow M, Ward AC, Williams E. 1993. Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie Van Leeuwenhoek 64:325–340 [DOI] [PubMed] [Google Scholar]
- 25.O'Donnell AG, Young IM, Rushton SP, Shirley MD, Crawford JW. 2007. Visualization, modelling and prediction in soil microbiology. Nat. Rev. Microbiol. 5:689–699 [DOI] [PubMed] [Google Scholar]
- 26.Pachauri RK, Reisinger A. 2007. Climate change 2007: synthesis report. Intergovernmental Panel on Climate Change, Cambridge, United Kingdom [Google Scholar]
- 27.Paul J, Alfreider A, Wawrik B. 2000. Micro- and macrodiversity in rbcL sequences in ambient phytoplankton populations from the southeastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 198:9–18 [Google Scholar]
- 28.Rhoades JD. 1982. Cation exchangeable capacity, p 149–165. In Pace AL, Miller RH, Keeney DR. (ed), Methods of soil analysis, part 2: chemical and microbiological properties, 2nd ed American Society of Agronomy, Inc., and Soil Science Society of America, Inc., Madison, WI. [Google Scholar]
- 29.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30.Saul-Tcherkas V, Steinberger Y. 2009. Substrate utilization patterns of desert soil microbial communities in response to xeric and mesic conditions. Soil Biol. Biochem. 41:1882–1893 [Google Scholar]
- 31.Savage DF, Afonso B, Chen AH, Silver PA. 2010. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327:1258–1261 [DOI] [PubMed] [Google Scholar]
- 32.Selesi D, Schmid M, Hartmann A. 2005. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl. Environ. Microbiol. 71:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selesi D, Pattis I, Schmid M, Kandeler E, Hartmann A. 2007. Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J. Microbiol. Methods 69:497–503 [DOI] [PubMed] [Google Scholar]
- 34.Siegenthaler U, Sarmiento JL. 1993. Atmospheric carbon dioxide and the ocean. Nature 365:119–125 [Google Scholar]
- 35.Tabita FR. 1999. Microbial ribulose-1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Rev. 60:1–28 [Google Scholar]
- 36.Takai K, et al. 2005. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl. Environ. Microbiol. 71:7310–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 38.Tcherkez GGB, Farquhar GD, Andrews TJ. 2006. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U. S. A. 103:7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ter Braak CJF. 1996. Unimodal models to relate species to environment. Agricultural Mathematics Group, Wageningen, The Netherlands [Google Scholar]
- 40.Tolli J, King GM. 2005. Diversity and structure of bacterial chemolithotrophic communities in pine forest and agroecosystem soils. Appl. Environ. Microbiol. 71:8411–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vance ED, Brookes PC, Jenkinson DS. 1987. Microbial biomass measurements in forest soils: determination of kc values and tests of hypotheses to explain the failure of the chloroform fumigation-incubation method in acid soils. Soil Biol. Biochem. 19:689–696 [Google Scholar]
- 42.Wu J. 2011. Carbon accumulation in paddy ecosystems in sub-tropical China: evidence from landscape studies. Eur. J. Soil Sci. 62:29–34 [Google Scholar]
- 43.Wu J, O'Donnell AG. 1997. Procedure for the simultaneous analysis of total and radioactive carbon in soil and plant materials. Soil Biol. Biochem. 29:199–202 [Google Scholar]
- 44.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969). comb. nov. Microbiol. Immunol. 39:897–904. [DOI] [PubMed] [Google Scholar]