Abstract
Fusarium graminearum (Gibberella zeae) is an important pathogen of wheat, maize, barley, and rice in South Korea, and harvested grain often is contaminated with trichothecenes such as deoxynivalenol and nivalenol. In this study, we examined 568 isolates of F. graminearum collected from maize at eight locations in South Korea. We used amplified fragment length polymorphisms (AFLPs) to identify four lineages (2, 3, 6, and 7); lineage 7 was the most common (75%), followed by lineage 6 (12%), lineage 3 (12%), and lineage 2 (1%). The genetic identity among populations was high (>0.98), and the effective migration rate between locations was higher than that between lineages. Female fertility varied by lineage: all lineage 7 isolates were fertile, while 70%, 26%, and 14% of the isolates in lineages 6, 3, and 2, respectively, were fertile. All lineage 3 and lineage 7 isolates produced deoxynivalenol, whereas most lineage 2 and 6 isolates produced nivalenol. Genotypic diversity in lineage 3 and lineage 6 populations is similar to that found in previously described Korean rice populations, but genotypic diversity in lineage 7 is much lower, even though similar levels of gene flow occur between lineage 7 populations. We conclude that lineage 7 was relatively recently introduced into South Korea, perhaps accompanying imported maize seeds.
INTRODUCTION
Fusarium graminearum Schwabe (teleomorph: Gibberella zeae) is an important pathogen of several cereal crops. It causes seedling blight, brown foot rot, and head blight of wheat, barley, and rice and stalk and ear rots of maize (19). Head blight and ear rot reduce the yield of grain, and the harvested grain often is contaminated with mycotoxins such as trichothecenes and zearalenone (6). Cereals contaminated with trichothecenes are associated with feed refusal, vomiting, diarrhea, dermatitis, and hemorrhage in farm animals (6). Trichothecenes also contribute to the virulence of F. graminearum for host plants (5, 6, 28).
In South Korea, maize is third in importance to rice and barley and is grown primarily in Gangwon Province, which is located in the middle-eastern part of the country. Although incidents of mycotoxicoses due to consumption of moldy maize have not been reported, we have found that maize samples from this region are heavily contaminated with Fusarium mycotoxins such as trichothecenes, zearalenone, and fumonisins (12, 33). Fusarium graminearum strains producing either deoxynivalenol (DON) or nivalenol (NIV) are present in the region, but strains producing DON are the most common (12). An earlier report described Korean populations of F. graminearum with high levels of vegetative self-incompatibility (i.e., isolates that are not vegetatively compatible with themselves) and relatively little genetic variation (23). This pattern is quite different from that found in the United States (11, 31, 33, 34) and China (9). Unfortunately, the previous Korean study was limited in scope and lacked both the large number of isolates and the more exhaustive phylogenetic analyses that are commonly found in more recent studies of this topic. Therefore, a more in-depth study was needed to understand the population structure of F. graminearum in South Korea.
The population structure of F. graminearum has been studied in different geographic regions using multilocus molecular markers, including restriction fragment length polymorphisms (9, 13), amplified fragment length polymorphisms (AFLPs) (31, 44, 45), and PCR-based random amplified polymorphic DNAs (3, 4). In most cases, populations of F. graminearum have high levels of genotypic diversity, but populations of F. graminearum from rice in South Korea have relatively low levels of genotypic diversity (14). As strains in populations from rice are usually members of lineage 6 and those from maize populations members of lineage 7 (14, 18), the patterns observed in the rice populations may not carry over to the maize populations.
Our objectives in this study were (i) to determine which lineages were present in maize fields in Gangwon Province, (ii) to determine if lineage was correlated with fertility or toxin production, and (iii) to determine if genetic variability was distributed as expected in a sexually reproducing population. A more detailed understanding of the population structure could provide insight into the evolutionary behavior of this organism and guide resistance breeding strategies and plant quarantine regulations.
MATERIALS AND METHODS
Fungal isolates.
Diseased maize ears were collected in November 1999 from 40 farmers' fields in eight maize-producing regions in Gangwon Province, South Korea. Seeds with symptoms of Fusarium ear rot were removed from the ears with forceps. One hundred seeds from each field were soaked in 2% sodium hypochlorite for 2 min, rinsed in sterile water, transferred to potato dextrose agar (PDA; Difco Laboratories, Detroit, MI), and incubated at 25°C for 4 to 7 days. Fusarium isolates, which were identified by their carmine-red pigmentation, were transferred to homemade PDA (20% potato extract, 2% dextrose, agar 1.5%) and carnation leaf agar (CLA) (19) and incubated under fluorescent lamps (cool white, 5,000 lx) at 25°C. We recovered 809 isolates, purified them by subculturing single macroconidia, and stored them as spore suspensions in 15% glycerol at −80°C. Isolates were maintained for short periods of time on slants of PDA as needed. A total of 568 isolates were morphologically identified as F. graminearum based on the characters described by Leslie and Summerell (19). These isolates were used in AFLP fingerprinting. An additional set of 24 standard isolates previously identified as belonging to F. graminearum lineages 1 to 7, Fusarium pseudograminearum, F. sporotrichioides, F. culmorum, and F. crookwellense (24) served as controls.
Nomenclature.
O'Donnell et al. (24) divided F. graminearum into seven phylogenetic lineages using DNA sequences of six single-copy genes, and these seven lineages and four additional lineages were elevated to species status (34), with at least three additional lineages or species described since (7, 25, 43). Members of all lineages are cross-fertile with lineage 7 tester strains and in some cases with strains of other lineages (1, 2, 15, 21), which suggests that all of the lineages belong to a single biological species. In this report we use the lineage numbers rather than proposed names for the phylogenetic species to emphasize the close relationships among the groups. The lineages we identified in this study were given the following names by O'Donnell et al. (26): lineage 2, F. meridionale; lineage 3, F. boothii; lineage 6, F. asiaticum; and lineage 7, F. graminearum sensu stricto.
DNA isolation.
Small pieces of F. graminearum cultures were transferred from PDA to complete medium (CM) broth (19) and incubated on an orbital shaker (150 rpm) for 72 h at 25°C. Mycelia were harvested by filtration through nongauze milk filters, and DNA was extracted with a cetyltrimethylammonium bromide (CTAB) procedure (19). The concentration of each DNA sample was adjusted to 20 μg/ml for the AFLP analyses.
AFLP.
AFLPs were generated by the protocol of Vos et al. (40), as modified by Leslie and Summerell (19), using the primer pairs Eco+AA/Mse+AT, Eco+CC/Mse+CG, and Eco+TG/Mse+TT. The EcoRI primers in the final specific amplification reactions were end labeled with [γ-33P]ATP (Amersham Biosciences Korea Ltd., Seoul, South Korea). AFLP fragments were separated in 6% denaturing polyacrylamide gels (Long Ranger; FMC Scientific, Rockland, ME) in 1× TBE buffer [100 mM Tris base, 100 mM boric acid, and 2 mM EDTA (pH 8.0)]. Dried gels were exposed to X-ray film (Kodak Biomax MS Film, Rochester, NY) for 2 to 5 days at room temperature to identify DNA bands. We manually scored polymorphic AFLP bands ranging from 200 to 1,000 bp in length. We estimated the lengths of the AFLP fragments by comparisons with a low-mass ladder (Life Technologies Inc., Bethesda, MD) DNA standard that also was end labeled with [γ-33P]ATP.
Amplification, cloning, and sequencing of Tri101 and MAT1-1.
We arbitrarily selected 101 isolates from the 568 F. graminearum isolates and sequenced two genes, Tri101 and MAT1-1, following PCR amplification (24). The amplified PCR products were purified with a DNA purification kit (Promega, Madison, WI) and then sequenced directly at the National Instrumentation Center for Environmental Management (NICEM, Seoul National University, Seoul, South Korea) using an ABI377 DNA sequencer (Applied Biosystems Inc., Foster City, CA).
Analyses of AFLP and DNA sequence data.
AFLP profiles were scored manually for the presence or absence of bands and compared with designated tester strains of known lineage (24). We assumed that bands of the same molecular size in different individuals were identical. Each band was treated as a single independent locus with two alleles, and unresolved bands or missing data were scored as ambiguous. We estimated allele frequencies at polymorphic loci, the Nm values (effective migration rates), and the genetic identity among populations by using the shareware program POPGENE, version 1.32 (42; free program available at: http://www.ualberta.ca/∼fyeh). AFLP haplotypes (putative clones) within populations were identified by analyzing the binary data with the unweighted pair grouping by mathematical average (UPGMA) subroutine of PAUP 4.10b (37). Bootstrap analyses (1,000 iterations) were conducted on the resulting UPGMA tree to assess the support for any resulting subgroups. We also estimated genotype diversity (Ĝ) for each population as described by Milgroom (22) and normalized the index for each population by dividing each estimated Ĝ value by the number of genotypes identified from that population.
Fertility tests.
Fusarium graminearum is a homothallic fungus, and strains originating from a single conidium can complete the entire life cycle. These strains are termed self-fertile. Strains that are not self-fertile may cross with strains that are female fertile, but this form of “male” fertility was not measured in this study.
Each isolate was inoculated on carrot agar (19) and incubated at 25°C under fluorescent lamps with a 12-h photoperiod. Plates were arranged right side up in a single layer on the incubator shelves. Seven days after inoculation, 1 ml of a sterile 2.5% Tween 60 (Sigma-Aldrich Corp., St. Louis, MO) solution was added to each plate, and aerial mycelia were knocked down with a sterile bent glass rod to induce sexual reproduction. Plates were returned to the incubator for an additional 2 weeks. Perithecia were observed by eye. The presence of asci and ascospores within the perithecia was confirmed in water mounts of squashed perithecia that were observed with a light microscope. The fertility test was repeated three times with three plates of each culture per replicate.
PCR assay of the DON/NIV genotype.
PCR primers for determining DON and NIV genotypes—GzTri7/p1, GzTri7/p2, GzTri13/p1, and GzTri13/p2—were derived from the Tri7 and Tri13 genes in the trichothecene biosynthetic gene cluster and used in PCR amplifications of these genes as previously described (16, 17). Amplified fragments were separated by electrophoresis on 1.2% agarose gels. F. graminearum lineage 7 strain H-11 (DON producer from maize) and lineage 6 strain 88-1 (NIV producer from barley) were used as standards for the two lineages (16).
Toxin analysis.
Erlenmeyer flasks (500 ml), each containing 100 g of rice and 60 ml of distilled water, were autoclaved for 1 h, allowed to cool to room temperature for 24 h, and then autoclaved again. The autoclaved rice was inoculated with mycelial plugs from a rapidly growing fungal culture. Rice cultures were harvested after 3 weeks of incubation at 25°C, and each ground culture (20 g) was extracted with 160 ml of acetonitrile-water (3:1, vol/vol) as previously described (32). The extract was filtered through Whatman no. 1 filter paper, and 80 ml of the filtrate was defatted with 80 ml of n-hexane and concentrated to dryness. The residue was dissolved in 2 ml of methanol, of which 10 μl was spotted on thin-layer chromatography plates coated with silica gel 60 (Merck, Darmstadt, Germany), and the plates were developed with chloroform-methanol (9:1, vol/vol). DON and NIV were visualized by spraying the plates with p-anisaldehyde–sulfuric acid and heating them at 110°C. For detection of either DON or NIV, standard compounds were purchased from Sigma-Aldrich (St. Louis, MO).
RESULTS
AFLP analysis.
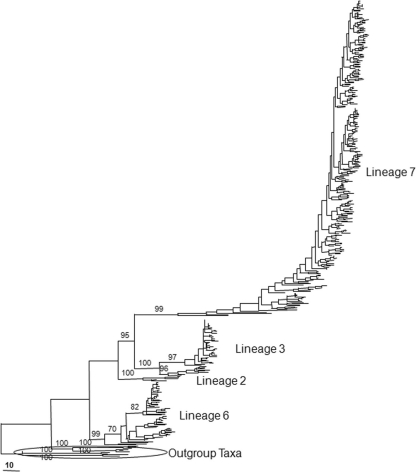
Three primer pairs resulted in 248 AFLP bands from the 568 Korean isolates, of which 169 (68%) were polymorphic. A UPGMA tree was constructed that contained the 568 field isolates and 24 standard isolates previously described for lineages 1 to 7. Other species, including F. culmorum, F. crookwellense, F. pseudograminearum, and F. sporotrichioides, were significant outliers relative to the F. graminearum strains (Fig. 1). The isolates clustered into four distinct groups corresponding to lineages 2, 3, 6, and 7. The similarity of Korean lineage 7 isolates to the standard tester isolates for this lineage ranged from 91 to 96% for the lineage 6 isolates, 87 to 95% for the lineage 3 isolates, 85 to 92%, and 87 to 89% for the lineage 2 isolates. Lineage 7 was the most common group (75%), followed by lineages 6 and 3 at 12%, and finally lineage 2 at 1%. Lineage 7 isolates were recovered from all eight sites, lineage 6 isolates from seven of eight sites, lineage 3 isolates from three sites, and lineage 2 isolates from two sites (Table 1).
Fig 1.
UPGMA network of AFLP fingerprint similarity among F. graminearum isolates after censoring clonal haplotypes. Bootstrap values (1,000 iterations) for clusters of strains that received >70% support are indicated above those branches.
Table 1.
Composition of eight F. graminearum populations from South Korea by lineage and location
| Location | No. of: |
|||||
|---|---|---|---|---|---|---|
| Isolates | Haplotypes | Strains |
||||
| Lineage7 | Lineage6 | Lineage3 | Lineage2 | |||
| Jecheon | 12 | 6 | 12 | |||
| Danyang | 52 | 33 | 42 | 10 | ||
| Hoengseong | 52 | 36 | 38 | 14 | ||
| Jeongseon | 197 | 106 | 146 | 13 | 32 | 6 |
| Gangneung | 12 | 9 | 7 | 5 | ||
| Pyeongchang | 119 | 74 | 83 | 8 | 27 | 1 |
| Wonju | 47 | 30 | 32 | 15 | ||
| Yeongwol | 77 | 56 | 65 | 5 | 7 | |
| Total | 568 | 347 | 425 | 70 | 66 | 7 |
Population structure and genetic diversity.
The 568 isolates from the eight populations were assigned to one of 347 unique haplotypes (Table 1), 270 of which were represented by a single strain and 77 of which were detected more than once. Of the 77 multiply represented haplotypes, 74 were found at only one location and 3 were found in two locations. The largest of the multimember haplotypes contained 19 isolates. The AFLP patterns of the isolates from six populations (those from Jecheon and Gangneung were excluded due to small sample sizes) were used to evaluate population differentiation. Nei's unbiased measures of genetic identity among the six populations were high, ranging from 0.985 to 0.998 (Table 2), indicating little genetic differentiation. The effective migration rate (Nm) ranged from 9 to 50 among the six populations and 7 to 18 among the six populations when only lineage 7 isolates were considered (Table 2).
Table 2.
Nei's unbiased measures of genetic identity and effective migration rate (Nm) for populations of F. graminearum from maize growing at six locations in South Korea
| Location | Genetic identity or Nma |
|||||
|---|---|---|---|---|---|---|
| Danyang | Hoengseong | Jeongseon | Pyeongchang | Wonju | Yeongwol | |
| Danyang | 0.995 (0.996) | 0.992 (0.996) | 0.989 (0.996) | 0.993 (0.995) | 0.994 (0.995) | |
| Hoengseong | 20 (10) | 0.989 (0.996) | 0.986 (0.995) | 0.997 (0.994) | 0.989 (0.995) | |
| Jeongseon | 13 (9) | 11 (12) | 0.998 (0.997) | 0.986 (0.995) | 0.996 (0.996) | |
| Pyeongchang | 10 (9) | 9 (8) | 50 (14) | 0.985 (0.996) | 0.995 (0.998) | |
| Wonju | 15 (8) | 30 (7) | 9 (8) | 8 (9) | 0.987 (0.994) | |
| Yeongwol | 15 (8) | 10 (10) | 24 (12) | 22 (18) | 8 (7) | |
Genetic identity (above the diagonal) and Nm (below the diagonal) values are based on 169 polymorphic AFLP loci. Values are for all lineages from each location; values in parentheses are for lineage 7 isolates only.
We also pooled isolates from the various lineages and treated them as separate populations to determine the genetic identity between them and the genetic exchange occurring among them. The genetic identity among lineages ranged from 0.803 to 0.837 and the Nm value was 0.3 (Table 3), indicating that gene flow was more limited among lineages than it was between locations.
Table 3.
Nei's unbiased measures of genetic identity and effective migration rate (Nm) for three F. graminearum lineages from maize in South Korea
| Lineage | Genetic identity or Nma |
||
|---|---|---|---|
| Lineage 7 | Lineage 6 | Lineage 3 | |
| Lineage 7 | 0.818 | 0.837 | |
| Lineage 6 | 0.3 | 0.803 | |
| Lineage 3 | 0.3 | 0.3 | |
Genetic identity (above the diagonal) and Nm (below the diagonal) values are based on 169 polymorphic AFLP loci.
Genotypic diversity also varied by location. The lowest genotypic diversity (20%) was in the Danyang population and the highest (97%) was in the Jeongseon population (Table 4). Genotypic diversity within lineages varied by lineage. The lowest genotypic diversity was in lineage 7 (4%), whereas the genotypic diversities of lineages 3 and 6 were 34% and 37%, respectively (Table 4).
Table 4.
Estimates of genotypic diversity and linkage disequilibrium in Korean populations of F. graminearum
| Population | No. ofa: |
Ĝa,b |
Ĝ/na,c |
% of locus pairs in linkage disequilibrium ata,d: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | Haplotypes | Polymorphic loci |
Private alleles |
All | >5% | All | >5% |
P < 0.05 |
P < 0.01 |
|||||
| All | >5%e | All | >5% | All | >5% | All | >5% | |||||||
| Locationf | ||||||||||||||
| All | 544 (406) | 332 (227) | 169 (125) | 90 (35) | —g | — | — | — | — | — | — | — | — | — |
| DY | 52 (42) | 33 (26) | 86 (41) | 70 (30) | 0 (1) | 0 (0) | 7 (5) | 4 (5) | 0.20 (0.19) | 0.15 (0.20) | 63 (23) | 68 (24) | 51 (17) | 57 (16) |
| HS | 52 (38) | 36 (26) | 105 (84) | 72 (32) | 0 (5) | 0 (0) | 4 (2) | 4 (2) | 0.11 (0.09) | 0.11 (0.07) | 48 (28) | 64 (18) | 39 (22) | 57 (11) |
| JS | 197 (146) | 106 (69) | 143 (65) | 89 (34) | 8 (6) | 0 (0) | 3 (2) | 3 (2) | 0.03 (0.02) | 0.03 (0.02) | 48 (23) | 69 (30) | 40 (18) | 59 (21) |
| PC | 119 (83) | 74 (47) | 138 (66) | 90 (35) | 3 (4) | 0 (0) | 2 (3) | 3 (2) | 0.04 (0.05) | 0.05 (0.04) | 42 (23) | 61 (30) | 36 (17) | 54 (19) |
| WJ | 47 (32) | 30 (18) | 96 (32) | 73 (24) | 2 (0) | 0 (0) | 2 (4) | 3 (1) | 0.11 (0.14) | 0.12 (0.09) | 54 (25) | 67 (35) | 34 (18) | 57 (25) |
| YW | 77 (65) | 56 (44) | 129 (79) | 90 (35) | 3 (3) | 0 (0) | 3 (4) | 4 (3) | 0.06 (0.06) | 0.07 (0.06) | 37 (19) | 46 (23) | 31 (15) | 40 (14) |
| Lineageh | ||||||||||||||
| All | 561 | 340 | 158 | 98 | — | — | — | — | — | — | — | — | — | — |
| 3 | 66 | 46 | 68 | 51 | 8 | 6 | 16 | 3 | 0.34 | 0.09 | 24 | 33 | 15 | 21 |
| 6 | 70 | 54 | 96 | 68 | 9 | 1 | 20 | 14 | 0.37 | 0.29 | 15 | 19 | 11 | 14 |
| 7 | 425 | 240 | 125 | 84 | 16 | 4 | 10 | 9 | 0.04 | 0.04 | 21 | 25 | 18 | 10 |
Numbers in parentheses are for lineage 7 isolates in each group only.
Calculated as described by Milgroom (22) from comparisons of AFLP allelic data at polymorphic loci. Ĝ = 1/Σpi2, where pi is the observed frequency of the ith multilocus genotype in a population.
Calculated by dividing Ĝ by the number of AFLP haplotypes observed in each population.
Linkage disequilibrium was detected by POPGENE version 1.32 (44).
Calculated from the loci for which the frequency of both alleles was >5%.
Two populations, Jecheon and Gangneung, were excluded in these analyses due to small sample sizes. DY, Danyang; HS, Hoengseong; JS, Jeongseon; PC, Pyeongchang; WJ, Wonju; YW, Yeongwol.
—, not calculated.
Lineage 2 was excluded from these analyses due to the small number of isolates.
Sequencing of Tri101 and MAT1-1 genes.
Tri101 sequences of 1108 nucleotides and MAT1-1 sequences of 1571 nucleotides were aligned using CLUSTAL W (39) and were analyzed phylogenetically by using the neighbor-joining method. The dendrogram clustered the isolates into four groups which were concordant with the AFLP data. As a result of analysis of Tri101 sequence data, a branch containing five lineage 2 isolates received 100% bootstrap support, a branch containing 27 lineage 3 isolates received 100% support, a branch containing 37 lineage 6 isolates received 99% support, and a branch containing 32 lineage 7 isolates received 82% support. This topology was concordant with that resulting from an analysis of the MAT1-1 sequences (data not shown).
Fertility.
All of the strains in lineage 7 produced perithecia homothallically on carrot agar. A smaller proportion of the other lineages were self-fertile: lineage 6, 71%; lineage 3, 26%; and lineage 2, 14%.
Trichothecene production.
Based on PCR assays with the Tri7- and Tri13-specific primer pairs, all 425 isolates of lineage 7 had the DON-DON genotype at Tri7 and Tri13 (Table 5). Most lineage 6 strains and all lineage 2 strains had the NIV-NIV genotype, and all lineage 3 strains had the NIV-DON genotype.
Table 5.
Mycotoxin production by strains of F. graminearum collected from maize in South Korea
We also assayed the strains chemically to test the accuracy of the predictions made by the PCR assays. Approximately 13% of the strains tested produced neither DON nor NIV. Strains with the DON-DON and NIV-NIV genotypes always produced the predicted trichothecene, if they produced any trichothecenes. The single lineage 6 strain with the DON-NIV genotype produced NIV. All 66 of the lineage 3 strains had the NIV-DON genotype. The majority (41 strains) of these strains produced DON, while the rest produced no detectable trichothecenes (Table 5).
DISCUSSION
Fusarium graminearum is a complex species with a global distribution that contains a number of phylogenetic lineages. The biological significance of the distinctions between these phylogenetic lineages remains to be elucidated, although there are some general differences in trichothecene production and perhaps in host preference. The Korean populations of F. graminearum are of interest because they contain representatives of multiple phylogenetic lineages and because they occur on hosts that are part of different cropping systems. Rice populations of F. graminearum are dominated by strains belonging to lineage 6 that produce nivalenol in South Korea (14, 18). Maize populations, primarily from North America or Europe, are dominated by strains belonging to lineage 7 and produce deoxynivalenol (24, 38). However, recently lineage 6 population was reported in southern Louisiana of the United States (10).
The strains of F. graminearum we evaluated could generally be associated with one of four existing lineages based on AFLP banding patterns and on DNA sequences of portions of the Tri101 and MAT1-1 loci. Most (75%) of the strains belonged to lineage 7, followed by a similar number (12%) of strains belonging to each of lineages 3 and 6, and a few strains (1%) belonging to lineage 2. This pattern of lineage frequency was unexpected, since lineage 6 usually dominates in Asia (14, 29, 35). Genetic exchange can occur between the lineages but at only ∼10% of the rate of exchange that occurs between populations composed solely of strains that belong to lineage 7. Thus, genetic homogenization of the lineages through the production of interlineage hybrids is possible in the field in South Korea, if the hybrids are sufficiently fit to survive.
Interlineage hybrids.
Strains resulting from interlineage hybrids are likely to be both rare and difficult to detect. Estimating the frequencies of hybrids between lineage 7 and one of other lineages (7/x) from the data in the present study (Table 1) (lineage 7, 74.8%; lineage 6, 12.3%; lineage 3, 11.6%; and lineage 2, 1.2%), then 37.7% (2 × 0.748 × 0.252) of the potential crosses should result from interlineage crosses between a lineage 7 strain and one of the other lineages (and 56.0% from lineage 7 × lineage 7 crosses and 6.2% from the other lineages crossing with one of the other lineages or themselves). Although lineage 6 is more fertile than lineage 7 on rice, lineage 7 isolates usually are the most fertile on maize. If lineage 7 parents are presumed to be the female parents of all of the crosses that occur, then there would be no interlineage crosses between lineages 2, 3, and 6, and only half of the predicted 7/x crosses would occur. In the population, the frequency of 7/x crosses would change from 37.7% to 18.8/(18.8 + 56), or 25.1% of the crosses. These numbers assume that all of the crosses that occur in these populations are outcrosses, which need not be true, since the fungus is homothallic and self-fertile. In F. verticillioides, the proportion of the population outcrossing annually is 1 to 3% of the total population (20), which suffices to maintain high levels of genotypic diversity. If outcrossing occurs at a similar frequency in G. zeae populations, then 0.025 to 0.075% of the crosses will result from 7/x crosses.
Identifying a hybrid isolate also is difficult. To be certain that an isolate is a hybrid, between 25% and 75% of its genome should be the same as that of one of the lineages. Isolates with genomes that are more than 75% similar to one of the parental lineages will probably be scored as members of the lineage rather than as hybrids. With four chromosomes, the proportion of hybrids could be calculated by following approximate chromosome segregation. Initially, most progeny (7/8) of an interlineage cross would be identifiable as hybrids, but their total numbers would be at most 0.02 to 0.065% of the population. To be 95% certain of detecting at least one hybrid strain, between 4,500 (0.065%) and 15,000 (0.2%) isolates must be analyzed. If only lineage 7 strains are available as female parents, then after one generation of outcrossing in this population, the number of hybrids will be reduced by 25%, and by 10 generations the number of hybrids remaining will be <5% of the original number. If recombination occurs on the chromosomes, then the decay back to lineage 7 will be more rapid than calculated here, as these calculations are based on no recombination within the chromosomes. Thus, identifying hybrid strains in field populations of G. zeae, even if more than one lineage is present at reasonable frequencies, requires the analysis of a very large number of isolates, even if random mating between members of the different lineages occurs.
Korean maize populations.
The heterogeneous population structure of F. graminearum from maize may be indicative of the evolutionary history of the pathogen in South Korea. A previous study showed that populations of F. graminearum lineage 6 from rice in South Korea had low levels of genotypic diversity (30 to 58%) (14) compared with populations reported from China (9) and the United States (44, 45). The genotypic diversity values for the lineage 6 and lineage 3 populations in this study were similar to those of the previously analyzed rice populations from South Korea. However, the genotypic diversity of the lineage 7 population in this study was extremely low (4%), indicating that the structure of the lineage 7 populations is much more clonal than that of other lineages in South Korea. This result is not consistent with the hypothesis that lineage 7 populations are more genotypically diverse because they have higher fertility than other lineages and differs from reports for lineage 7 populations in Argentina (30) and the United States (44) which had much higher levels of genotypic diversity (>95%). One possible explanation for this unexpected result is that lineage 7 strains were recently introduced to South Korea with maize and rapidly adapted to maize fields, perhaps occupying a niche that might otherwise be filled by strains from lineages 3 or 6. The geographic distribution of lineages in South Korea supports this hypothesis. Maize is a major crop in Gangwon Province, which has allowed lineage 7 strains to dominate the local maize populations. In contrast, lineage 6 dominates in southern provinces of South Korea, because little maize is grown and rice is the major crop available for colonization.
Outcrossing and female fertility.
Genetic exchange occurs among lineage 7 populations as frequently as it does between other lineages, even though the lineage 7 populations have relatively high genetic identity and relatively low genotypic diversity. The Nm value among lineage 7 populations was relatively high compared with that of lineage 6 populations from rice in South Korea (14). This result suggests that outcrossing between different lineages in South Korea is limited under field conditions, perhaps by relative location, even though outcrossing occurs under laboratory conditions (1, 15).
Ascospores of F. graminearum are thought to be an important factor for the primary infection of wheat (27, 36), but the relative importance of these spores as inocula in rice and maize systems is not known. We used the production of perithecia and ascospores by cultures growing on carrot agar under laboratory conditions as a means of estimating the fertility of the strains being evaluated, even though lineage 6 strains are known to be more fertile when they are grown on rice than on carrots (14). Lineage 7 strains in the United States are nearly 100% self-fertile under these conditions, as were the Korean strains of lineage 7 tested for this study. Strains of the other three lineages evaluated in this study were significantly less self-fertile than were the strains of lineage 7. There are several possible explanations for these observations. First, the laboratory tests could underestimate fertility under field conditions, as was hypothesized previously (14). Alternatively, ascospores and the sexual stage might not be as important in the disease and life cycles of strains belonging to lineages 2, 3 and 6. If ascospores are the inoculum source, then there will be selection pressure for all of the strains to be sexually fertile as females, which in a homothallic fungus such as F. graminearum means that they are self-fertile. If there is significant asexual reproduction and/or dispersal via asexual conidia, then there will be selection for the production of asexual spores, i.e., conidia, and against female fertility (20). This scenario suggests that ascospores are not essential for the survival of the strains in these three lineages, perhaps because the strains belonging to these lineages use alternative methods to infect the host or to survive the off-season. If the lineage 7 strains are relatively new to South Korea, then selection against female fertility may not yet have progressed to the point of being detectable in the population.
Trichothecene production.
Trichothecene production need not be correlated with phylogenetic lineage (41). In the populations we sampled, all of the lineage 3 and lineage 7 isolates produced deoxynivalenol, and all lineage 2 isolates and all but one lineage 6 isolate produced nivalenol (Table 5). Strains with a DON-DON or NIV-NIV genotype produced the predicted toxin, if they produced any trichothecenes. All lineage 7 isolates had the DON-DON genotype in Tri7 and Tri13, all except two lineage 6 isolates had the NIV-NIV genotype, and all lineage 3 isolates had the NIV-DON genotype. If the Tri7 and Tri13 genotypes differ, then the chemotype predicted by the Tri13 genotype is to be expected, if any trichothecenes are produced. Strains with these mixed genotypes probably cannot synthesize the full range of DON or NIV derivatives that could be synthesized by strains with either the DON-DON or NIV-NIV genotype. In wheat and maize, trichothecene biosynthesis alters strain aggressiveness (28), with DON-producing strains being perceived as more virulent than NIV-producing strains (8). Trichothecene production is not thought to be important in aggressiveness toward rice. Thus, the generally higher level of toxicity of NIV toward microbes and other organisms is probably selected instead and could be the reason that lineage 6 strains from rice in South Korea are primarily NIV producers (14).
Most (>90%) of the strains we evaluated from maize produce DON, if they produce any detectable trichothecenes. These results are consistent with the hypothesis that strains pathogenic to maize usually are DON producers. There are at least two possible explanations for our results. One explanation is that selection for DON producers has occurred within the F. graminearum population on maize. Alternatively, the lineage 7 strains, which compose the bulk of the population and are exclusively DON producers, could be a relatively recent introduction from outside South Korea—perhaps from North America. Indeed, most maize seeds for maize production in Gangwon Province are imported from North America. In this scenario, the high level of DON producers could be attributed to fact that North American populations of F. graminearum are nearly exclusively DON producers and the idea that the lineage 7 isolates accompanied maize from North America to South Korea relatively recently. If the population went through a bottleneck in the process of moving across the Pacific Ocean, then the observed relatively low levels of genotypic diversity also can be explained.
In conclusion, our results present a view of populations of F. graminearum from maize that is different from that reported elsewhere. Some of the most important differences include the absence of some lineages and the presence of multiple lineages within the population, the presence of a significant number of strains that produce NIV and not DON, and the relatively low levels of genotypic diversity observed. The similarity of the strains belonging to lineages 3 and 6 to populations already known to occur in South Korea on rice suggests that these strains are of local Korean origin and have drifted to maize. The lineage 7 strains, however, could represent a relatively recent introduction into South Korea. The lack of the detection of this lineage from an extensive survey of strains from rice is consistent with this explanation. The possible hybridization of lineage 7 strains with those from lineages 3 and 6 also may occur here and could provide critical insights into the selection pressures on these genetically isolated lineages when they are found together in a common site. The nonzero value of Nm that results when the lineages are treated as separate populations suggests that such an exchange may have already begun. The speed with which such interbreeding occurs will depend on the relative importance of the sexual stage under the conditions that prevail in South Korea. The relatively high number of self-sterile strains in lineages 3 and 6 suggests that sexual reproduction in these lineages is not as important as it is in lineage 7. If these differences act to reduce the number of non-self perithecia, then they also will act to reduce the amount of interbreeding between the lineages and could slow the interbreeding process even further. Monitoring the F. graminearum populations in this region, whether they are from maize or other hosts, could be critical in assessing the ways in which this pathogen makes evolutionary and economically important responses to changes in host and environment.
ACKNOWLEDGMENTS
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2011-0000963); the Crop Functional Genomics Center (CG1140) of the 21st Century Frontier Research Program, funded by the Korean Ministry of Science and Technology; and the USDA Wheat and Barley Scab Initiative.
Footnotes
Published ahead of print 27 January 2012
This is article no. 12-174-J from the Kansas Agricultural Experiment Station, Manhattan, Kansas, USA.
REFERENCES
- 1. Bowden RL, Leslie JF. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182–188 [DOI] [PubMed] [Google Scholar]
- 2. Bowden RL, Leslie JF, Lee J, Lee Y-W. 2006. Cross fertility of lineages in Fusarium graminearum (Gibberella zeae), p 54–60 In Ban T, Lewis JM, Phipps EE. (ed), The global Fusarium initiative for international collaboration A strategic planning workshop held at CIMMYT, El Batan, Mexico [Google Scholar]
- 3. Carter JP, Rezanoor HN, Desjardins AE, Nicholson P. 2000. Variation in Fusarium graminearum isolates from Nepal associated with their host of origin. Plant Pathol. 49:452–460 [Google Scholar]
- 4. Carter JP, et al. 2002. Variation in pathogenicity associated with the genetic diversity of Fusarium graminearum. Eur. J. Plant Pathol. 108:573–583 [Google Scholar]
- 5. Cumagun CJR, Bowden RL, Jurgenson JE, Leslie JF, Miedaner T. 2004. Genetic mapping of pathogenicity and aggressiveness of Gibberella zeae (Fusarium graminearum) towards wheat. Phytopathology 94:520–526 [DOI] [PubMed] [Google Scholar]
- 6. Desjardins AE. 2006. Fusarium mycotoxins: chemistry, genetics, and biology. APS Press, St. Paul, MN [Google Scholar]
- 7. Desjardins AE, Proctor RH. 2011. Genetic diversity and trichothecene chemotypes of the Fusarium graminearum clade isolated from maize in Nepal and identification of a putative new lineage. Fungal Biol. 115:38–48 [DOI] [PubMed] [Google Scholar]
- 8. Desjardins AE, et al. 2008. Gibberella ear rot of maize (Zea mays) in Nepal: distribution of the mycotoxins nivalenol and deoxynivalenol in naturally and experimentally infected maize. J. Agric. Food Chem. 56:5428–5436 [DOI] [PubMed] [Google Scholar]
- 9. Gale LR, Chen L-F, Hernick CA, Takamura K, Kistler HC. 2002. Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology 92:1315–1322 [DOI] [PubMed] [Google Scholar]
- 10. Gale LR, et al. 2011. Nivalenol-type populations of Fusarium graminearum and F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology 101:124–134 [DOI] [PubMed] [Google Scholar]
- 11. Gale LR, Ward TJ, Balmas V, Kistler HC. 2007. Population subdivision of Fusarium graminearum sensu strict in the upper midwestern United States. Phytopathology 97:1434–1439 [DOI] [PubMed] [Google Scholar]
- 12. Kim J-C, Kang H-J, Lee D-H, Lee Y-W, Yoshizawa T. 1993. Natural occurrence of Fusarium mycotoxins (trichothecenes and zearalenone) in barley and maize in Korea. Appl. Environ. Microbiol. 59:3798–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laday M, et al. 2004. Mitochondrial DNA diversity and lineage determination of European isolates of Fusarium graminearum (Gibberella zeae). Eur. J. Plant Pathol. 110:545–550 [Google Scholar]
- 14. Lee J, Chang I-Y, Kim H, Yun S- H, Leslie JF, Lee Y-W. 2009. Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl. Environ. Microbiol. 75:3289–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J, Lee T, Lee Y-W, Yun S-H, Turgeon BG. 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50:145–152 [DOI] [PubMed] [Google Scholar]
- 16. Lee T, et al. 2001. Identification of deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae using PCR. Appl. Environ. Microbiol. 67:2966–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee T, Han Y-K, Kim K-H, Yun S-H, Lee Y-W. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 68:2148–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee Y-W, et al. 2004. Lineage composition and trichothecenes production of Gibberella zeae population in Korea, p 117–122 In Yoshizawa T. (ed), New horizons of mycotoxicology for assuring food safety. Japanese Association of Mycotoxicology, Kagawa, Japan [Google Scholar]
- 19. Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Blackwell Professional, Ames, IA [Google Scholar]
- 20. Leslie JF, Klein KK. 1996. Female fertility and mating-type effects on effective population size and evolution in filamentous fungi. Genetics 144:557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leslie JF, Bowden RL. 2008. Fusarium graminearum: when species concepts collide. Cereal Res. Commun. 36(Suppl. B):609–615 [Google Scholar]
- 22. Milgroom MG. 1996. Recombination and the multilocus structure of fungal populations. Annu. Rev. Phytopathol. 43:457–477 [DOI] [PubMed] [Google Scholar]
- 23. Moon J-H, Lee Y-H, Lee Y-W. 1999. Vegetative compatibility groups in Fusarium graminearum isolates from corn and barley in Korea. Plant Pathol. J. 15:53–56 [Google Scholar]
- 24. O'Donnell K, Kistler HC, Tacke BK, Casper HH. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. U. S. A. 97:7905–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Donnell K, et al. 2008. Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genet. Biol. 45:1514–1522 [DOI] [PubMed] [Google Scholar]
- 26. O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 41:600–623 [DOI] [PubMed] [Google Scholar]
- 27. Parry DW, Jenkinson P, McLeod L. 1995. Fusarium ear blight (scab) in small grains: a review. Plant Pathol. 44:207–238 [Google Scholar]
- 28. Proctor RH, Hohn TM, McCormick SP. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593–601 [DOI] [PubMed] [Google Scholar]
- 29. Qu B, et al. 2008. Comparison of genetic diversity and pathogenicity of Fusarium head blight pathogens from China and Europe by SSCP and seedling assays on wheat. Plant Pathol. 57:642–651 [Google Scholar]
- 30. Ramirez ML, et al. 2007. Population genetic structure of Gibberella zeae isolated from wheat in Argentina. Food Addit. Contam. 24:1115–1120 [DOI] [PubMed] [Google Scholar]
- 31. Schmale DG, III, et al. 2006. Genetic structure of atmospheric populations of Gibberella zeae. Phytopathology 96:1021–1026 [DOI] [PubMed] [Google Scholar]
- 32. Seo J-A, Kim J-C, Lee D-H, Lee Y-W. 1996. Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia 134:31–37 [DOI] [PubMed] [Google Scholar]
- 33. Sohn HB, Seo J-A, Lee Y-W. 1999. Co-occurrence of Fusarium mycotoxins in mouldy and healthy corn from Korea. Food Addit. Contam. 16:153–158 [DOI] [PubMed] [Google Scholar]
- 34. Starkey DE, et al. 2007. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 44:1191–1204 [DOI] [PubMed] [Google Scholar]
- 35. Suga H, et al. 2008. Molecular characterization of the Fusarium graminearum species complex in Japan. Phytopathology 98:159–166 [DOI] [PubMed] [Google Scholar]
- 36. Sutton JC. 1982. Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can. J. Plant Pathol. 4:195–209. [Google Scholar]
- 37. Swofford DL. 1999. PAUP. Phylogenetic analysis using parsimony, version 4.0. Sinauer Associates, Sunderland, MA [Google Scholar]
- 38. Szécsi Á, Bartók T, Varga M, Magyar M, Á Mesterházy. 2005. Determination of trichothecene chemotypes of Fusarium graminearum strains isolated in Hungary. J. Phytopathol. 153:445–448 [Google Scholar]
- 39. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vos P, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O'Donnell K. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. U. S. A. 99:9278–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeh FC, Yang RC, Boyle TBJ, Mao JX. 1997. POPGENE: Microsoft Windows®-based freeware for population genetic analysis, version 1.32. University of Alberta and Center for International Forestry Research, Edmonton, Canada [Google Scholar]
- 43. Yli-Mattila T, et al. 2009. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 101:841–852 [DOI] [PubMed] [Google Scholar]
- 44. Zeller KA, Bowden RL, Leslie JF. 2003. Diversity of epidemic populations of Gibberella zeae from small quadrats in Kansas and North Dakota. Phytopathology 93:874–880 [DOI] [PubMed] [Google Scholar]
- 45. Zeller KA, Bowden RL, Leslie JF. 2004. Population differentiation and recombination in wheat scab populations of Gibberella zeae from the United States. Mol. Ecol. 13:563–571 [DOI] [PubMed] [Google Scholar]