Abstract
Environmental conditions in the western Arctic Ocean range from constant light and nutrient depletion in summer to complete darkness and sea ice cover in winter. This seasonal environmental variation is likely to have an effect on the use of dissolved organic matter (DOM) by heterotrophic bacteria in surface water. However, this effect is not well studied and we know little about the activity of specific bacterial clades in the surface oceans. The use of DOM by three bacterial subgroups in both winter and summer was examined by microautoradiography combined with fluorescence in situ hybridization. We found selective use of substrates by these groups, although the abundances of Ant4D3 (Antarctic Gammaproteobacteria), Polaribacter (Bacteroidetes), and SAR11 (Alphaproteobacteria) were not different between summer and winter in the Beaufort and Chukchi Seas. The number of cells taking up glucose within all three bacterial groups decreased significantly from summer to winter, while the percentage of cells using leucine did not show a clear pattern between seasons. The uptake of the amino acid mix increased substantially from summer to winter by the Ant4D3 group, although such a large increase in uptake was not seen for the other two groups. Use of glucose by bacteria, but not use of leucine or the amino acid mix, related strongly to inorganic nutrients, chlorophyll a, and other environmental factors. Our results suggest a switch in use of dissolved organic substrates from summer to winter and that the three phylogenetic subgroups examined fill different niches in DOM use in the two seasons.
INTRODUCTION
Over the past few years, we have started to explore the relationship between bacterial community structure and activity in perennially cold marine waters. Heterotrophic bacterial activity is generally lower in polar than in temperate oceans even though primary production in polar oceans often reaches that of temperate oceans (29). In the Arctic Ocean and the Ross Sea, bacterial production is low year-round, with some seasonal variation (13, 26). In both the subarctic North Pacific Ocean and the more temperate North Atlantic Ocean, bacterial production is 10- to 50-fold higher than in polar regions and varies seasonally (14, 27). Although perennially low temperatures in polar ecosystems contribute to low heterotrophic bacterial activity, input of dissolved organic matter (DOM) is of greater importance (28, 29).
In contrast to bulk measurements of bacterial activity, measurements of single-cell activity that combine microautoradiography and fluorescence in situ hybridization (MAR-FISH) integrate both the number of cells actively using DOM within a community and their phylogenetic composition (9, 20, 40). In earlier single-cell studies of the temperate Delaware Bay marine ecosystem, heterotrophic bacteria in the Cytophaga-Flavobacteria, Alphaproteobacteria, and Gammaproteobacteria were shown to contribute markedly to the uptake of low-molecular-weight DOM, such as glucose, amino acids, and N-acetylglucosamine (8, 9, 16, 31). Many of the same broad taxa, such as Alphaproteobacteria and Gammaproteobacteria and the Cytophaga-Flavobacteria, are abundant in both polar and temperate oceans (1, 4, 11, 15, 16, 38). In two polar systems, the Arctic Ocean and coastal waters of the West Antarctic Peninsula (WAP), bacterioplankton preferred amino acids over other substrates by these same phylogenetic groups (5, 42).
However, our knowledge of the activity and distribution of bacterial clades, particularly in perennially cold marine waters, is still limited. Many studies have used MAR-FISH to measure single-cell activity of major bacterial groups, but few have examined subgroups within larger clades. In particular, subgroups such as Polaribacter and SAR11 have been examined in the Arctic Ocean only twice before (5, 33). Only one previous study has explored Ant4D3, a gammaproteobacterial clade that is present and active in the WAP (42) but which has not been examined in the Arctic Ocean. Glucose use is low by these three bacterial groups in the WAP (42), while uptake of leucine and the amino acid mixture is high. In contrast, glucose use by SAR11 is much higher in the Arctic and other high-latitude oceans (3, 5, 32), suggesting that DOM use by heterotrophs varies among polar regions.
In addition to the percentage of active cells, variation in the degree of activity within the active fraction can be determined by measuring silver grain area around each cell in microautoradiographic preparations. Silver grain area has been explored by only a few previous MAR-FISH studies. The silver grain area around an individual cell is proportional to how much radioactive compound is taken up by the cell (8, 40, 43). For example, the silver grain area of leucine-assimilating cells indicates that SAR11 accounts for a substantial portion of biomass production in the North Atlantic Ocean (31). Thus, examining silver grain clusters provides additional information about single-cell uptake of DOM.
Single-cell bacterial activity likely responds to physical properties such as light, inorganic nutrient availability, and surface temperature, which can change drastically from summer to winter seasons in polar waters. However, few studies have examined seasonal differences in bacterial production and use of DOM by heterotrophs in permanently cold waters (5, 6, 12). Heterotrophic bacterial production in both the WAP and Arctic Ocean is higher in summer than in either spring or fall, despite the low temperature (12, 26). High summer productivity corresponds with phytoplankton blooms and input of DOM (12, 39, 45). In a seasonal study, Alonso-Sáez et al. (5) found higher use of glucose by Alphaproteobacteria and Gammaproteobacteria in summer than in winter in Franklin Bay, Arctic Ocean, while use of amino acids and ATP was relatively high year-round. However, due to a limited number of studies, it is not yet clear whether one subgroup of bacteria contributes more than another to the seasonal differences in activity observed by Alonso-Sáez and colleagues (5).
In this study, we examined the abundance of specific bacterial groups and their uptake of three low-molecular-weight compounds, leucine, glucose, and a mix of amino acids, in two summer and two winter seasons in the coastal western Arctic Ocean. We measured abundance and activity of the Antarctic gammaproteobacterial clade Ant4D3. We expected that bacterial single-cell activity for all three compounds would be higher in summer than in winter. However, our results suggest that heterotrophic bacteria use DOM compounds differently between summer and winter.
MATERIALS AND METHODS
Sample collection.
Samples were collected offshore from Barrow, Alaska, in the Beaufort and Chukchi Seas. Sampling was conducted by small boat in summer and from a hole drilled in the sea ice in winter. Surface water (2-m depth) was pumped into 20-liter carboys and transported back to the shore-based lab in insulated containers. Details of sampling locations and biological oceanographic parameters have been previously described (7, 10) and are summarized in Table 1.
Table 1.
Biogeochemical properties of the Beaufort and Chukchi Seas in summer and wintera
| Date | Site | Temp (°C) | Leucine incorporation (pmol liter−1 h−1) | Chlorophyll a (μg liter−1) | Total prokaryotes (106 cells/ml) | Turnover rate constant (h−1) |
|
|---|---|---|---|---|---|---|---|
| Glucose | Amino acid | ||||||
| July 2007 | B1 | 2.5 | 38.1 ± 3.0 | 0.33 ± 0.03 | 2.09 | 0.017 | 0.013 |
| July 2007 | B2 | 3.9 | 32.0 ± 2.1 | 0.49 ± 0.00 | 1.74 | 0.018 | 0.011 |
| July 2007 | C1 | 4.6 | 33.0 ± 6.0 | 0.50 ± 0.02 | 1.58 | 0.013 | 0.006 |
| July 2007 | C2 | 5.5 | 26.3 ± 0.9 | 0.59 ± 0.02 | 1.92 | 0.014 | 0.011 |
| August 2008 | B3 | 1.4 | 24.0 ± 1.5 | 1.12 ± 0.02 | 1.07 | 0.011 | <0.001 |
| August 2008 | B4 | 1.2 | 21.4 ± 6.4 | 1.04 ± 0.07 | 1.27 | 0.011 | <0.001 |
| August 2008 | B5 | 3.2 | 25.5 ± 1.1 | 1.11 ± 0.08 | 1.18 | 0.064 | 0.001 |
| August 2008 | B6 | 4.2 | 19.2 ± 1.5 | 0.86 ± 0.03 | 1.33 | 0.078 | 0.001 |
| August 2008 | C6 | 2.1 | 28.1 ± 1.0 | 1.89 ± 0.02 | 0.93 | 0.012 | N/A |
| August 2008 | C7 | 6.2 | 10.9 ± 0.9 | 1.35 ± 0.03 | 0.93 | 0.008 | <0.001 |
| Summer avg | 3.5 | 28.0 ± 2.6 | 0.88 ± 0.03 | 1.4 ± 0.4 | 0.025 ± 0.02 | 0.005 ± 0.01 | |
| January 2008 | C3 | −1.8 | 3.0 ± 0.0 | 0.07 ± 0.00 | 0.42 | 0.001 | 0.003 |
| January 2008 | C4 | −1.8 | 2.1 ± 0.2 | 0.06 ± 0.00 | 0.47 | 0.033 | 0.009 |
| January 2008 | C5 | −1.8 | 2.2 ± 0.1 | 0.06 ± 0.01 | 0.75 | 0.006 | 0.008 |
| January 2009 | B7 | −1.7 | 2.5 ± 0.1 | 0.04 ± 0.00 | 0.52 | 0.001 | 0.002 |
| January 2009 | B8 | −1.7 | 2.4 ± 0.1 | 0.04 ± 0.00 | 0.57 | N/A | 0.004 |
| January 2009 | B9 | −1.7 | 2.2 ± 0.1 | 0.03 ± 0.01 | 0.47 | 0.001 | 0.002 |
| January 2009 | C8 | −1.7 | 12.9 ± 1.5 | 0.14 ± 0.02 | 0.22 | 0.001 | 0.003 |
| Winter avg | −1.7 | 3.9 ± 0.3 | 0.06 ± 0.01 | 0.49 ± 0.2 | 0.007 ± 0.01 | 0.005 ± 0.00 | |
B1 to B9 are nine samples collected in the Beaufort Sea. C1 to C8 are eight samples collected in the Chukchi Sea. Leucine incorporation, chlorophyll a, and average turnover rate constants are shown with ±standard deviations. N/A, not available.
Abundance of bacterial groups by catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH).
CARD-FISH was carried out as previously described (36). Briefly, seawater was fixed with 2% final concentration paraformaldehyde overnight at 4°C and filtered onto a 0.22-μm polycarbonate filter. Wedges were cut from each filter and coated with 0.01% (wt/vol) agarose (MetaPhor agarose, Lonza Group, Switzerland). Coated filters were dried, incubated for 60 min in lysozyme solution at 37°C, and incubated overnight with CARD-FISH probes. Horseradish peroxidase (HRP)-conjugated probes were used for larger groups: EUB338 (general bacteria), Alf968 (Alphaproteobacteria), CF319a (Cytophaga-Flavobacteria group), and Gam42a (Gammaproteobacteria), under conditions previously described (42). HRP probes for subgroups were SAR11 (Alphaproteobacteria) (34), Polaribacter (Cytophaga-Flavobacteria) (33), and Ant4D3 (Gammaproteobacteria) (42). A nonsense probe (NON338) was also used to evaluate nonspecific binding; this signal was <2% (33). After probe incubation, Cy3-tyramide amplification solution (Perkin Elmer) was added for 8 min. The filters were washed and prepared for imaging by mounting with a 4:1 mixture of Citifluor (Ted Pella) to Vectashield (Vector Labs), plus 4′,6-diamidino-2-phenylindole (DAPI; 0.5 ng liter−1).
Activity of subgroups measured by MAR-CARD-FISH.
Seawater was incubated with 3H-labeled compounds in separate bottles, i.e., leucine (20 nM final concentration of 100 to 150 Ci/mmol; Perkin Elmer), glucose (0.5 nM final concentration of 10 to 20 Ci/mmol; Perkin Elmer), and a mixture of 15 amino acids (total 0.5 nM final concentration of 55 Ci/mmol; American Radiolabeled Chemicals, Inc.), for 4 h in the dark. A killed incubation was run alongside each substrate as a negative control. Samples were fixed, filtered, and stored as previously described (42). After the CARD-FISH procedure, filters were taken to the dark room for microautoradiography as described previously (8). Filters were exposed to emulsion as follows: 1 day for leucine, 4 days for the amino acid mix, and 4 days for glucose. The number of DAPI-stained, Cy3 probe-positive, and silver grain-positive cells were quantified by epifluorescence microscopy (8). The silver grain background of the killed control was <3% for all samples.
Measuring turnover rate constants.
Seawater was incubated with 3H-labeled compounds using the same incubation procedure described for microautoradiography. After the incubation, cells were fixed with a 2% final concentration of paraformaldehyde overnight at 4°C and filtered onto two 0.22-μm polycarbonate filters. After extensive washing with deionized water, filters were dried and 7 ml of Ultima Gold (Perkin Elmer) scintillation cocktail was added. Radioactivity was measured using a liquid scintillation counter (LS 6500; Beckman Coulter). Turnover rate constants for glucose and amino acids were calculated by dividing the radioactivity taken up by the cells by the total radioactivity initially added.
Correlation analysis between environmental factors and the percentage of active cells.
We used pairwise correlation analyses to determine relationships between the environmental parameters measured at each sampling location and the percentage of active cells assessed by MAR-CARD-FISH. Values for total prokaryotes, chlorophyll a, inorganic nutrients, and bacterial production were log transformed before analysis. The percentage of active cells was arcsine transformed before analysis.
RESULTS
In two consecutive summers and winters, we measured bacterial single-cell activity and biogeochemical parameters of the surface water in both the Beaufort Sea and the Chukchi Sea (Table 1). Temperature of summer surface water ranged from 1.2 to 6.2°C. In winter, temperature under the sea ice was −1.8°C. In both seas, chlorophyll a content of surface waters was higher in summer than winter. Abundance of total prokaryotes decreased by 4-fold from summer to winter in the Beaufort Sea (Student's t test, P < 0.05; n = 9), although the difference was not significant in the Chukchi Sea. Bulk incorporation of leucine decreased from summer to winter by 10-fold (Student's t test, P < 0.05; n = 17). Average turnover rate constant for glucose in summer (0.025 ± 0.02 day−1) was significantly higher than in winter (0.007 ± 0.01 day−1) (Student's t test, P < 0.05; n = 16). In contrast, the turnover rate constant for amino acids in summer and winter did not change significantly (0.005 ± 0.003 d−1).
Abundance of bacterial subgroups.
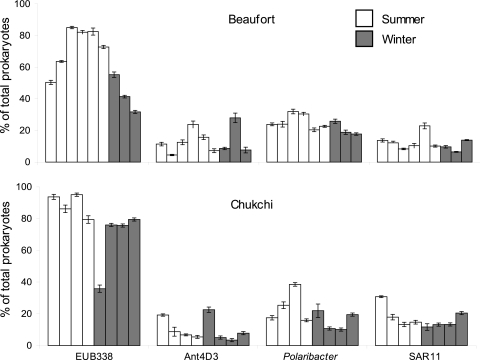
The average abundances of three large bacterial groups, Gammaproteobacteria, Cytophaga-Flavobacteria, and Alphaproteobacteria, were 33%, 20%, and 33% of the total prokaryotic community, respectively, and did not change substantially between summer and winter. We also examined the abundance of subgroups Ant4D3, Polaribacter, and SAR11, which are members of the larger bacterial groups Gammaproteobacteria, Cytophaga-Flavobacteria, and Alphaproteobacteria, respectively (Fig. 1). Polaribacter abundance averaged about 17%, which was not significantly greater than either SAR11 or Ant4D3 abundances. SAR11 abundance was about 15% of total prokaryotes for most samples. The Ant4D3 gammaproteobacterial subgroup was present in all locations, although its abundance ranged widely, from 3.4% to 28%. There was no obvious pattern in the relative abundance of the subgroups from summer to winter or between the two Arctic seas.
Fig 1.
Relative abundance of bacterial groups in the Beaufort Sea (top) and Chukchi Sea (bottom). Each bar is the abundance in one sample. There were nine samples from the Beaufort Sea (six summer and three winter samples) and eight samples from the Chukchi Sea (four summer and four winter samples). Error bars are ±standard errors (SE) of probe-positive cells from 10 fields of view. Probe-positive cells are expressed as a percentage of all cells.
Single-cell activity in summer and winter.
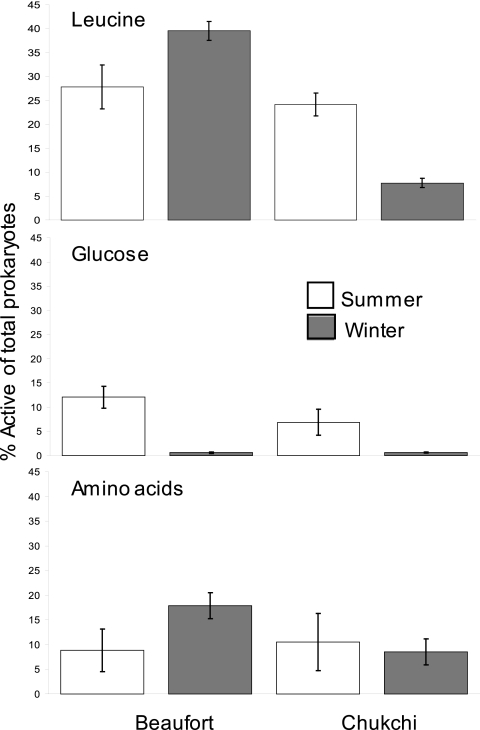
We used MAR-CARD-FISH to examine DOM uptake by various bacterial groups in summer and winter (Fig. 2). The percentage of active cells for leucine use significantly decreased by 2.5-fold from summer to winter in the Chukchi Sea (Student's t test, P < 0.05; n = 8), but this pattern was not observed for the Beaufort Sea. The percentage of cells using glucose also decreased markedly from over 6% (Chukchi) and 12% (Beaufort) in summer to less than 2% in winter. Overall, the fraction of cells actively using leucine (20 nM final concentration) was consistently higher (8 to 39%) than that for glucose (1 to 12%). In both seas, the percentage of cells active for leucine use was 2- to 4-fold higher than that for the amino acid mix (0.5 nM final concentration) in both seasons. Single-cell use of glucose was 10-fold lower than amino acid use in the winter (Student's t test, P < 0.05; n = 17), but the active fractions for the two compounds were comparable in summer.
Fig 2.
Percentage of total prokaryotes active in using leucine (top), glucose (middle), and amino acids (bottom). Bars are averages based on six summer and three winter samples from the Beaufort Sea and four summer and four winter samples from the Chukchi Sea. Error bars are 1 SE.
Single-cell activity of bacterial subgroups.
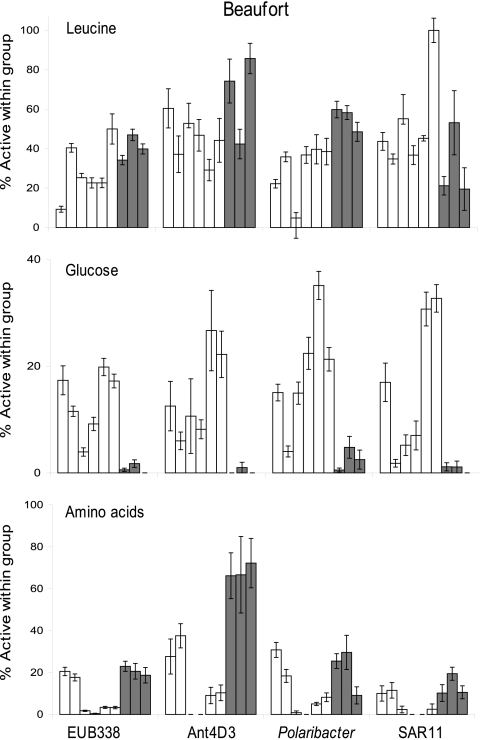
The three bacterial groups examined differed in their use of substrates in the Beaufort Sea (Fig. 3). Single-cell leucine use increased from summer to winter by the Polaribacter clade (30 to 56%) (Student's t test, P < 0.05; n = 9). In contrast, the fraction of leucine-active cells within the Ant4D3 and SAR11 groups did not change between seasons, remaining at about 56% and 40%, respectively. The fraction of cells taking up glucose decreased for the Ant4D3, Polaribacter, and SAR11 clades, from 14 to 18% in summer to less than 2% in winter. Surprisingly, the active fraction of Ant4D3 using the amino acid mix increased markedly from an average of 14% in summer to 68% in winter. The percentage of cells actively using amino acids within the SAR11 and Polaribacter clades were more similar to each other than to Ant4D3 in the two seasons, with overall averages of 7% and 14%, respectively.
Fig 3.
Percentage of cells within each bacterial group active in using leucine, glucose, and amino acids in the Beaufort Sea. Error bars are ±SE of probe-positive cells from 10 fields of view. Each bar represents one sampling site (six in summer and three in winter). No bar means that there were no active cells for that sample. Note the differences in scale for each graph.
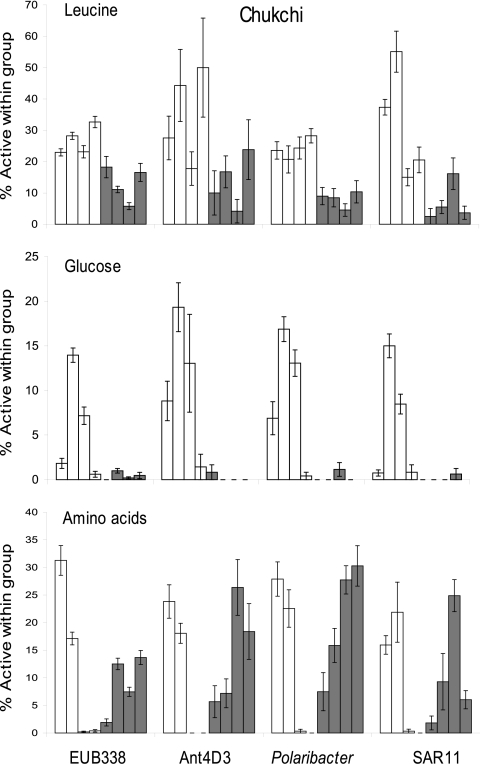
In the Chukchi Sea, single-cell activity of both leucine and glucose differed by season (Fig. 4), but single-cell activity of amino acids did not (Fig. 2). Within each of the bacterial groups examined, the fraction of cells using leucine decreased from 30% in summer to 9% in winter (Fig. 4). Similar to the Beaufort Sea, there was a significant decrease in glucose use from summer to winter in all three groups. The Ant4D3, Polaribacter, and SAR11 clades decreased from 11, 9, and 6%, respectively, to less than 1%. None of the three subgroups examined changed in the fraction actively using amino acids. Single-cell use of amino acids was less than 1% in two of the four summer samples (C6 and C7), while that in the other two summer samples was much higher (∼15%). Overall, the single-cell activity within the three subgroups varied with season and location in the Chukchi Sea.
Fig 4.
Percentage of cells within each bacterial group active in using leucine, glucose, and amino acids in the Chukchi Sea. Error bars are ±SE of probe-positive cells from 10 fields of view. Each bar represents one sampling site (four in summer and four in winter). No bar means that there were no active cells for that sample. Note the differences in scale for each graph.
Contribution of groups to activity and abundance of the bacterial community.
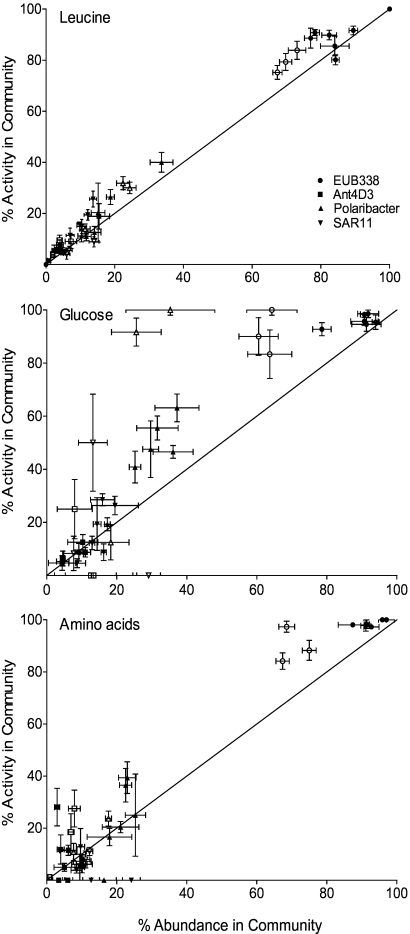
Previous studies have noted that the contribution of individual bacterial groups to total use of a compound may not necessarily reflect its abundance in the community (9, 16). We compared the relationship between the percentage of active cells belonging to a group and its relative abundance (Fig. 5). In general, the abundance of a particular group in the bacterial community matched its contribution to the uptake of all three compounds. Glucose use was more variable in both seasons than either leucine or amino acid mix use, and the summer samples were above the 1:1 line, indicating more single-cell glucose use by specific groups than abundance suggests. However, when all samples were examined together, there was no significant deviation from the 1:1 line for any of the groups (Student's t test, P > 0.05; n = 17).
Fig 5.
Relationship between abundance and activity of each phylogenetic subgroup for leucine, glucose, and amino acid uptake in the Beaufort Sea. The line represents the 1:1 line. Closed and open symbols are summer and winter samples, respectively, with ±SE.
Quantifying activity of individual cells by silver grain area.
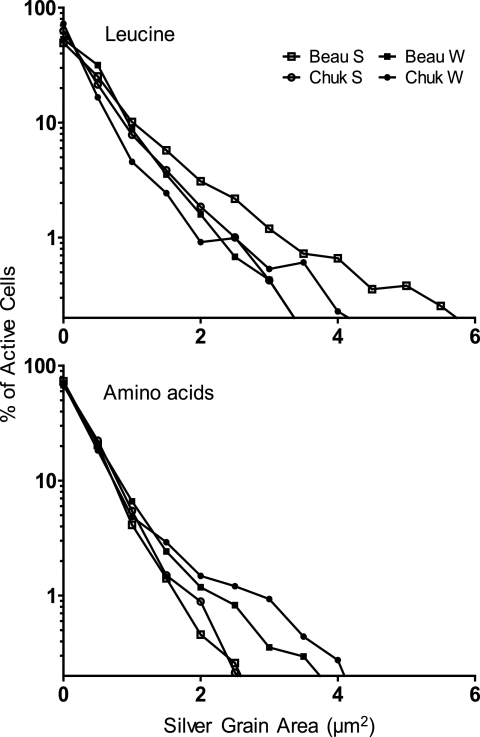
The area of the silver grain cluster around an active cell in microautoradiography is dependent on the amount of the radioactive compound taken up by the cell (40). To determine if all active cells within a community have similar leucine uptake levels in the two seasons, we examined the frequency distribution of silver grain area for each sampling season (Fig. 6). In all samples, only about 22% of leucine-active cells had silver grain clusters >2 μm2 in area, indicating that only a few cells were highly active, whereas the silver grain area around most cells was small (<1 μm2).
Fig 6.
Frequency distribution of silver grain area for all active cells using leucine in Beaufort (Beau) and Chukchi (Chuk) Seas. Silver grain area of active cells was sorted into 0.50-μm2 increments, and the relative number of cells per bin is plotted. Symbols represent the lower limit of a bin. For example, symbols at 0.5 represent cells with silver grain area from 0.50 to 0.99 μm2. Symbols at the 0 point represent cells with silver grain area from just above 0 to 0.49 μm2. S, summer; W, winter.
The silver grain area around the most active cells differed between summer and winter. For leucine in the Beaufort Sea, a greater number of cells had a silver grain cluster size between 4 and 6 μm2 in summer (11% of all active cells) than in winter (4%). The same trend was not observed in the Chukchi Sea (5% and 4% in summer and winter, respectively). For glucose single-cell activity in the Beaufort Sea, a larger fraction of active cells had a silver grain area within 4 to 6 μm2 in summer (7%) than in winter (0%), unlike in the Chukchi Sea (1% of active cells in summer and 9% of active cells in winter) (data not shown). Unexpectedly, we observed the opposite trend for the amino acid mix. In the Beaufort Sea, a greater number of cells had a silver grain cluster size between 4 and 6 μm2 in winter (4%) than in summer (2%). We observed this same trend in the Chukchi Sea when comparing winter (6%) to summer (2%).
Relationships between environmental parameters and single-cell bacterial activity.
We examined the relationships between environmental factors and the percentage of active cells for all three compounds (Table 2). We found no relationship between the percentage active for leucine and environmental factors, except a negative correlation with silicate. The fraction of cells using glucose was positively correlated with all environmental factors that increased in summer (total prokaryotes and chlorophyll a) and was negatively correlated with all factors that increased in winter, such as inorganic nutrients (Table 2). In contrast, single-cell use of amino acids was negatively correlated with abiotic factors that increased in summer and positively correlated with those that increased in winter. Of the three compounds, correlations between percent single-cell activity of glucose and environmental parameters were the highest.
Table 2.
Relationships between average percentage of active cells for each substrate and environmental measurementa
| Environmental parameter | Correlation with % of active cells in taking up indicated compound |
||
|---|---|---|---|
| Leucine | Glucose | Amino acid mix | |
| Temp | 0.21 | 0.62** | −0.08 |
| Salinity | −0.07 | −0.55* | −0.20 |
| Secchi disk depth | 0.29 | 0.71** | −0.02 |
| Total no. of prokaryotes | 0.32 | 0.77** | 0.08 |
| Chlorophyll a concn | 0.13 | 0.77** | −0.50* |
| PO43− concn | −0.41 | −0.64** | −0.31 |
| NH4+ concn | −0.23 | −0.81** | 0.05 |
| NO3+NO2 concn | −0.19 | −0.62** | 0.12 |
| Silicate concn | −0.50* | −0.76** | 0.02 |
| Bacterial production rate | 0.13 | 0.78** | −0.11 |
Amino acid mix is a mixture of 15 amino acids. Each number takes into account 17 samples. *, P < 0.05; **, P < 0.001.
DISCUSSION
The goal of this study was to compare the assimilation levels of organic carbon compounds in the western Arctic Ocean in the winter and summer and to investigate the abundance and activity of Polaribacter, SAR11, and Ant4D3 in these waters. We chose these three subgroups because only one previous study examined leucine use by Polaribacter and SAR11 in the Arctic Ocean basin (33) and glucose and amino acid use by SAR11 in Franklin Bay (5). Ant4D3 had been examined in the WAP (42) but not in the Arctic Ocean. Based on previous studies, the obvious hypothesis is that total uptake of all compounds would be higher in summer (4, 5, 12). While the percentage of cells using glucose was higher in the summer, unexpectedly the fraction of cells using leucine (20 nM final concentration) and the amino acid mix (0.5 nM final concentration) did not substantially differ between the two seasons. Our results suggest that bacterial single-cell uptake of leucine, glucose, and the amino acid mix is influenced by seasonal changes in the environment and that activity within the three bacterial groups, Ant4D3, Polaribacter, and SAR11, varied from summer to winter.
Ant4D3 was present and abundant in the western Arctic Ocean and WAP, consistent with previous studies suggesting that Ant4D3 is specialized to cold environments (19, 35, 42). The presence of Ant4D3 had not been shown before in the Arctic Ocean, except in a clone library of Laptev Sea samples (23). This Antarctic gammaproteobacterial group was much more abundant than we expected, even more abundant than in the WAP region where it was first discovered (19, 42). Preliminary results indicate low numbers (<4% of the prokaryotic community) of this subgroup in Delaware Bay surface waters. As Ant4D3 is abundant in both polar oceans, it may have a polar biogeography (42).
Both the SAR11 and Cytophaga-Flavobacteria groups are abundant in the Arctic Ocean, although the former was more abundant than the latter in the Franklin Bay and the Arctic basin (5, 33). Similar to our results, a recent study using 454 pyrosequencing of 16S rRNA genes found that SAR11 is present in both summer (12% of bacterial community) and winter (9% of bacterial community) in the Arctic Ocean (25). In general, SAR11 bacteria may compete better in oligotrophic environments such as the Sargasso Sea, where they are known to be abundant (30% of bacterial community) (34, 37), than in the Arctic Ocean, which is nutrient rich due to terrestrial input of detritus and organic matter (22). In contrast to SAR11, Cytophaga-Flavobacteria bacteria are abundant in cold aquatic systems (1, 44) and seem to thrive in the western Arctic Ocean. Accordingly, Polaribacter was abundant in our study, making up 17% of total prokaryotes, similar to that previously found in the Arctic Ocean shelf and basin, where it was 20% of total prokaryotes (33).
In general, single-cell uptake of leucine varied from summer to winter in our study. Surprisingly in the Beaufort Sea, the percentage of Polaribacter cells using leucine increased from summer to winter, although Ant4D3 and SAR11 single-cell leucine use did not change between seasons. Since leucine was added at a high concentration (20 nM) as in production assays, higher uptake by Polaribacter suggests that this group grew faster than either SAR11 or Ant4D3 in winter. Previous studies also found that the Bacteroidetes incorporate leucine and presumably grow faster than members of the Alphaproteobacteria in the subtropical North Atlantic Ocean (30). In particular, the Polaribacter group incorporates more leucine than SAR11 in both the Arctic Ocean basin and WAP polar waters (33, 42). In addition, the amount of leucine assimilated by individual cells varied (18, 40). According to the silver grain area analysis, a small fraction of the cells in summer incorporated twice as much leucine as the cells in winter. These data indicate that although the percentage of cells using leucine did not change drastically between seasons, the relative number of highly active cells decreased substantially in winter, suggesting an overall decrease in growth.
Single-cell use of glucose was 7- to 10-fold higher in summer than winter in both seas. These results are consistent with the bulk turnover rate constant for glucose, which was nearly 4-fold higher in summer than in winter Arctic waters. Similar to our results, glucose use is high in the Franklin Bay area of the Arctic Ocean in summer (5). In contrast to the Arctic Ocean, less than 5% of total prokaryotes use glucose in the summer months in WAP coastal waters (42). This difference in glucose uptake between seasons may be due to the differential input of nutrients from terrestrial runoff or from dying phytoplankton. In contrast to WAP waters, the Arctic Ocean system receives a significant input of terrestrial organic carbon and nutrients from rivers, the largest of which in our study area is the Mackenzie River (17, 21, 22). This input impacts the coastal locations, such as those we examined, by providing inorganic nutrients for phytoplankton growth. In turn, seasonal phytoplankton blooms release glucose and other DOM compounds which support the high uptake of glucose by heterotrophic bacteria (2, 41).
By late summer, polar surface waters are depleted in nutrients and labile DOM, as observed in other studies of the Arctic Ocean and the Ross Sea (12, 22). In this study, single-cell glucose uptake was correlated negatively to inorganic nutrients, which are low in summer and high in winter (10), the latter due to lower base flow of fresh water from rivers and reduced biological uptake (21). Because all three subgroups of bacteria used glucose more in the summer than winter, these results suggest a greater availability of glucose in summer. Uptake by SAR11 in the Arctic is more similar to the uptake of glucose by this group in the Delaware Bay than in the Sargasso Sea. In oligotrophic environments like the Sargasso, SAR11 can account for 50% of the glucose uptake (31), while in eutrophic environments such as the Delaware Bay, SAR11 accounts for 15 to 30% of glucose uptake (16). Both Bacteroidetes and Gammaproteobacteria are highly active for glucose in the western Arctic Ocean (5, 15).
Amino acids are a key source of nitrogen for cells and can support ∼50% of bacterial production in the oceans (24). Turnover rate constants for the amino acid mix were the same in summer and winter. However, we found summer to winter differences for the single-cell use of the amino acid mix in the two seas. According to silver grain area, more cells were active for the uptake of amino acids in summer but a few highly active cells incorporated 2-fold more of the amino acid mix in winter. Previous studies found that amino acid use varied seasonally and was always higher than the uptake of glucose or ATP (5, 15, 31). Alonso-Sáez and colleagues (5) observed relatively high single-cell amino acid uptake during March for Gammaproteobacteria but not for Bacteroidetes or Alphaproteobacteria in the Arctic Ocean. Similarly, we observed enhanced single-cell activity for amino acids in the Beaufort Sea in winter for Ant4D3 but not for Polaribacter or SAR11, suggesting that the three phylogenetic subgroups fill different niches in DOM use from summer to winter.
Use of organic compounds by bacteria in the western Arctic Ocean differed in summer and winter, when environmental parameters are drastically different. Using both bulk measurements and single-cell approaches, our study found clear summer to winter changes in the use of three DOM compounds by three subgroups of heterotrophic bacteria, including an Antarctic clade. Further work is needed to identify year-round DOM use by more bacterial subgroups at a higher temporal resolution in the rapidly changing coastal Arctic Ocean.
ACKNOWLEDGMENTS
This work was supported by NSF grants (OPP 0632233 and 0838830).
We thank Glenn Christman for his help in field sampling. Glenn Sheehan and Lewis Brower provided logistics support at the Barrow Arctic Science Consortium.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Abell GC, Bowman JP. 2005. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol. Ecol. 51:265–277 [DOI] [PubMed] [Google Scholar]
- 2. Alonso C, Pernthaler J. 2006. Concentration-dependent patterns of leucine incorporation by coastal picoplankton. Appl. Environ. Microbiol. 72:2141–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alonso C, Pernthaler J. 2005. Incorporation of glucose under anoxic conditions by bacterioplankton from coastal North Sea surface waters. Appl. Environ. Microbiol. 71:1709–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alonso-Sáez L, Gasol JM. 2007. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73:3528–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alonso-Sáez L, Sanchez O, Gasol JM, Balague V, Pedrós-Alió C. 2008. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ. Microbiol. 10:2444–2454 [DOI] [PubMed] [Google Scholar]
- 6. Bano N, Hollibaugh JT. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christman GD, Cottrell MT, Popp BN, Gier E, Kirchman DL. 2011. Abundance, diversity, and activity of ammonia-oxidizing prokaryotes in the coastal Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 77:2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cottrell MT, Kirchman DL. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168–178 [Google Scholar]
- 9. Cottrell MT, Kirchman DL. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cottrell MT, Kirchman DL. 2009. Photoheterotrophic microbes in the Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 75:4958–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delong EF, Franks DG, Alldredge AL. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924–934 [Google Scholar]
- 12. Ducklow H, et al. 2001. The seasonal development of the bacterioplankton bloom in the Ross Sea, Antarctica 1994–1997. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48:4199–4221 [Google Scholar]
- 13. Ducklow HW, et al. 2007. Marine pelagic ecosystems: the West Antarctic Peninsula. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 362:67–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ducklow HW, Kirchman DL, Quinby HL, Carlson CA, Dam HG. 1993. Stocks and dynamics of bacterioplankton carbon during the spring bloom in the eastern North Atlantic Ocean. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 40:245–263 [Google Scholar]
- 15. Elifantz H, Dittell AI, Cottrell MT, Kirchman DL. 2007. Dissolved organic matter assimilation by heterotrophic bacterial groups in the western Arctic Ocean. Aquatic Microb. Ecol. 50:39–49 [Google Scholar]
- 16. Elifantz H, Malmstrom RR, Cottrell MT, Kirchman DL. 2005. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware Estuary. Appl. Environ. Microbiol. 71:7799–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Emmerton CA, Lesack LFW, Vincent WF. 2008. Mackenzie River nutrient delivery to the Arctic Ocean and effects of the Mackenzie Delta during open water conditions. Global Biogeochem. Cycles doi:10.1029/2006GB002856 [Google Scholar]
- 18. Grossmann S. 1994. Bacterial activity in sea ice and open water of the Weddell Sea, Antarctica—a microautoradiographic study. Microb. Ecol. 28:1–18 [DOI] [PubMed] [Google Scholar]
- 19. Grzymski JJ, et al. 2006. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl. Environ. Microbiol. 72:1532–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herndl GJ, et al. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holmes RM, et al. 2011. Seasonal and annual fluxes of nutrients and organic matter from large rivers to the Arctic Ocean and surrounding seas. Estuar. Coast. doi:10.1007/s12237-011-9386-6 [Google Scholar]
- 22. Holmes RM, et al. 2008. Lability of DOC transported by Alaskan rivers to the Arctic Ocean. Geophys. Res. Lett. doi:doi:10.1029/2007GL03283 [Google Scholar]
- 23. Kellogg CTE, Deming JW. 2009. Comparison of free-living, suspended particle, and aggregate-associated bacterial and archaeal communities in the Laptev Sea. Aquatic Microb. Ecol. 57:1–18 [Google Scholar]
- 24. Kirchman DL. 2000. Uptake and regeneration of inorganic nutrients by marine heterotrophic bacteria, p 261–288 In Kirchman DL. (ed), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, NY [Google Scholar]
- 25. Kirchman DL, Cottrell MT, Lovejoy C. 2010. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12:1132–1143 [DOI] [PubMed] [Google Scholar]
- 26. Kirchman DL, et al. 2009. Standing stocks, production, and respiration of phytoplankton and heterotrophic bacteria in the western Arctic Ocean. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 56:1237–1248 [Google Scholar]
- 27. Kirchman DL, Keil RG, Simon M, Welschmeyer NA. 1993. Biomass and production of heterotrophic bacterioplankton in the oceanic subarctic Pacific. Deep Sea Res. Part 1 Oceanogr. Res. Pap. 40:967–988 [Google Scholar]
- 28. Kirchman DL, Malmstrom RR, Cottrell MT. 2005. Control of bacterial growth by temperature and organic matter in the Western Arctic. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 52:3386–3395 [Google Scholar]
- 29. Kirchman DL, Morán XAG, Ducklow H. 2009. Microbial growth in the polar oceans—role of temperature and potential impact of climate change. Nat. Rev. Microbiol. 7:451–459 [DOI] [PubMed] [Google Scholar]
- 30. Longnecker K, Lomas MW, Van Mooy BAS. 2010. Abundance and diversity of heterotrophic bacterial cells assimilating phosphate in the subtropical North Atlantic Ocean. Environ. Microbiol. 12:2773–2782 [DOI] [PubMed] [Google Scholar]
- 31. Malmstrom RR, Cottrell MT, Elifantz H, Kirchman DL. 2005. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl. Environ. Microbiol. 71:2979–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL. 2007. Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aquatic Microb. Ecol. 47:45–55 [Google Scholar]
- 34. Morris RM, et al. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810 [DOI] [PubMed] [Google Scholar]
- 35. Murray AE, Grzymski JJ. 2007. Diversity and genomics of Antarctic marine micro-organisms. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 362:2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 38. Schattenhofer M, et al. 2009. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ. Microbiol. 11:2078–2093 [DOI] [PubMed] [Google Scholar]
- 39. Sherr BF, Sherr EB. 2003. Community respiration/production and bacterial activity in the upper water column of the central Arctic Ocean. Deep Sea Res. Part 1 Oceanogr. Res. Pap. 50:529–542 [Google Scholar]
- 40. Sintes E, Herndl GJ. 2006. Quantifying substrate uptake by individual cells of marine bacterioplankton by catalyzed reporter deposition fluorescence in situ hybridization combined with microautoradiography. Appl. Environ. Microbiol. 72:7022–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skoog A, Whitehead K, Sperling F, Junge K. 2002. Microbial glucose uptake and growth along a horizontal nutrient gradient in the North Pacific. Limnol. Oceanogr. 47:1676–1683 [Google Scholar]
- 42. Straza TRA, Ducklow HW, Murray AE, Kirchman DL. 2010. Abundance and single-cell activity of bacterial groups in Antarctic coastal waters. Limnol. Oceanogr. 55:2526–2536 [Google Scholar]
- 43. Varela MM, van Aken HM, Sintes E, Reinthaler T, Herndl GJ. 2011. Contribution of Crenarchaeota and Bacteria to autotrophy in the North Atlantic interior. Environ. Microbiol. 13:1524–1533 [DOI] [PubMed] [Google Scholar]
- 44. Wells LE, Deming JW. 2003. Abundance of Bacteria, the Cytophaga-Flavobacterium cluster and Archaea in cold oligotrophic waters and nepheloid layers of the Northwest Passage, Canadian Archipelago. Aquatic Microb. Ecol. 31:19–31 [Google Scholar]
- 45. Yager PL, et al. 2001. Dynamic bacterial and viral response to an algal bloom at subzero temperatures. Limnol. Oceanogr. 46:790–801 [Google Scholar]