Abstract
The determination of the success of in situ bioremediation strategies is complex. By using controlled laboratory conditions, the influence of individual variables, such as U(VI), Cr(VI), and electron donors and acceptors on community structure, dynamics, and the metal-reducing potential can be studied. Triplicate anaerobic, continuous-flow reactors were inoculated with Cr(VI)-contaminated groundwater from the Hanford, WA, 100-H area, amended with lactate, and incubated for 95 days to obtain stable, enriched communities. The reactors were kept anaerobic with N2 gas (9 ml/min) flushing the headspace and were fed a defined medium amended with 30 mM lactate and 0.05 mM sulfate with a 48-h generation time. The resultant diversity decreased from 63 genera within 12 phyla to 11 bacterial genera (from 3 phyla) and 2 archaeal genera (from 1 phylum). Final communities were dominated by Pelosinus spp. and to a lesser degree, Acetobacterium spp., with low levels of other organisms, including methanogens. Four new strains of Pelosinus were isolated, with 3 strains being capable of Cr(VI) reduction while one also reduced U(VI). Under limited sulfate, it appeared that the sulfate reducers, including Desulfovibrio spp., were outcompeted. These results suggest that during times of electron acceptor limitation in situ, organisms such as Pelosinus spp. may outcompete the more-well-studied organisms while maintaining overall metal reduction rates and extents. Finally, lab-scale simulations can test new strategies on a smaller scale while facilitating community member isolation, so that a deeper understanding of community metabolism can be revealed.
INTRODUCTION
Microbial community structure and function are controlled by many physicochemical factors, including pH, temperature, electron donors and acceptors, and hydrology (13, 19, 22). Alterations of these conditions may shift community composition and select for members that can adapt to, and outcompete, other organisms under the different parameters (32, 50). Anthropogenic contamination adds an additional influence (24). Although communities at historically contaminated sites have exhibited lower biomass and diversity (22), specific electron donors can increase the microbial biomass and activity (3, 6, 59), but the specific interplay of these events is not well understood due to difficulties with in situ assessment and the lack of replicated stimulations (10).
Uranium [U(VI)] and chromium [Cr(VI)] are common metal contaminants, and they pose a lower human health risk when reduced to U(IV) (54) and Cr(III) (12). A wide diversity of metal-reducing bacteria have been isolated in pure and mixed cultures (4, 59), with reduction via direct enzymatic processes or indirectly through metabolic by-products, such as Fe(II) or sulfide (4, 28, 33). In metal-contaminated sites with an adequate carbon and electron source, subsurface communities exhibit a preference in processes where nitrate reduction tends to predominate (43), followed by Fe(III) and sulfate reduction (7).
Selection of specific compounds for stimulation of groundwater microbial communities can selectively affect community structure (3). Various electron donors have been used to stimulate specific biochemical activities at contaminated sites, including donors ranging from ethanol, methanol, glycerol, and lactate to more complex substrates, such as glycerol polylactate and humic acids (1, 5, 8, 12, 38, 50). Stimulation of in situ anaerobic microbial communities with lactate increases metal reduction rates (3, 4, 12) for the enrichment of Acidobacteria, Firmicutes, Deltaproteobacteria, and Betaproteobacteria (4). The Deltaproteobacteria are the most-well-recognized metal reducers, including sulfate-reducing bacteria (SRB) and Fe(III)-reducing bacteria (IRB) (17, 30, 31, 48), while Clostridium spp. within the Firmicutes also reduce U(VI) (15). Most laboratory studies use an ample amount of exogenous electron acceptor. However, in situ experiments rely on groundwater electron acceptor concentrations, which are occasionally depleted due to increased microbial activity or dilution from rainfall (12). The current work was performed to address which populations within a subsurface community contaminated with Cr(VI) would persist with nearly depleted sulfate concentrations in the absence of other electron acceptors so that we could examine their metal reduction potential. Lactate was chosen not only because it has been used in situ at Hanford site 100-H (12) but also because acetate would likely be generated to support a greater percentage of the initial community. Organisms such as Geobacter spp. and SRB might persist with low sulfate levels or via fermentation. Under low nitrate and sulfate conditions, fermenters such as the Firmicutes may be more likely to outcompete these organisms, but their capacity for metal reduction is unknown. If Firmicutes exhibit the capacity to reduce metals such as Cr(VI) and U(VI), then an underappreciated portion of the subsurface community capable of accomplishing these activities during times of depleted electron acceptors may have been revealed.
MATERIALS AND METHODS
Sampling and cultivation.
Groundwater samples were collected from well H-100 on the Department of Energy's Hanford site (Washington) (12). Samples (600 ml) were sealed under N2, placed on ice, and shipped to Oak Ridge National Laboratory. Upon arrival, 150 ml was removed as a reference and immediately frozen at −80°C. The remaining groundwater (450 ml) was inoculated into triplicate custom-built, anaerobic glass fermentation vessels (Allen Glass, Boulder, CO) as described previously (34), each receiving 150 ml, with working volumes of ∼650 ml (see Fig. S1 in the supplemental material). The reactors were supplied with medium from a single 19-liter carboy (10 liters of medium) via a peristaltic pump at a flow rate of 0.22 to 0.23 ml/min for a dilution rate/medium turnover of 0.487 day−1. The carboy was kept anaerobic via constant purging with filter-sterilized N2 gas.
Lactate-enriched CCM1 medium (53) was modified to not contain exogenous electron acceptors and was constantly stirred. Anaerobic conditions were maintained with N2 gas (7 to 9 ml min−1) flushing through the medium inlet drip tube, substantially decreasing biofilm development. Vessel temperature was maintained at 30°C ± 2°C by a recirculating water bath. Spent culture fluid and gas drained out of the vessel overflow vents into a closed collection vessel to maintain a constant volume. Exit gas passed through a Zn-acetate solution (1% [wt/vol]) to remove H2S before being vented into a chemical fume hood (see Fig. S1). Gas samples were taken with needles and syringes through vessel top ports sealed with butyl rubber stoppers. Liquid samples were taken biweekly throughout the 95-day experiment via syringe and a stainless steel cannula inserted through one of the stoppers.
Cell counts.
Microscopic cell counts were performed using the LIVE/DEAD bacteria viability kit (BacLight; Invitrogen, Eugene, OR) (20) and a Petroff Hausser counting chamber on a Zeiss Axioskop 2 plus microscope (Carl Zeiss Light Microscopy, Germany). For each temporal sample, 16 fields of view were counted, and the averages and standard deviations were calculated. Samples from the first 2 weeks of the experiment were concentrated via centrifugation and resuspended due to low cell counts (<1 × 107 cells/ml).
Metabolite analysis.
Filtered supernatants were acidified with 200 mM H2SO4 (5 mM, final concentration) before injection into a Hitachi Lachrom Elite high-performance liquid chromatography system (Hitachi High Technologies). Metabolites were separated on an Aminex HPX-87H column (Bio-Rad Laboratories) under isocratic temperature (40°C) and flow (0.5 ml/min) conditions, then passed through a refractive index (RI) detector (Hitachi L-2490). Metabolite identification used retention time comparisons to known standards, and quantification was calculated against linear standard curves. All standards were prepared in fresh culture medium to account for the interference of salts in the RI detector.
Fermenter gases were collected via sterilized Hamilton gas-tight syringes and injected into an Agilent 6850 gas chromatograph (Agilent Technologies) equipped with a thermal conductivity detector (TCD) for CO2 quantification. Analytes were separated on an HP-PLOT U column (30 m by 0.32 mm by 0.10 μm film; J&W Scientific, Agilent Technologies). Two HP-PLOT U columns were joined together for a total length of 60 m for optimized separation. Samples were injected into a 185°C split-splitless injector with a split ratio of 3:1 and an isocratic oven (70°C) with He carrier flow (7.0 ml min−1). The detector had a 10-ml/min helium makeup flow at 185°C, with the detector filament set for positive polarity.
Samples to detect CH4 concentrations were injected into an Agilent 6890 gas chromatograph equipped with a flame ionization detector (FID). Samples were separated on a DB-FFAP column (30 m by 0.32 mm by 0.5 μm film; J&W Scientific, Agilent Technologies) after passing through a 230°C split-splitless injector with the split ratio set to 3:1 and isocratic oven (50°C) and helium carrier flow (1.5 ml min−1). The FID had a hydrogen flow of 40 ml min−1, air flow of 450 ml min−1, and helium makeup flow of 45 ml min−1. The detector temperature was set at 230°C. Peak identifications were performed by comparisons with known standards, and compound quantification was calculated against individual linear standard curves.
DNA extraction and pyrosequencing of the bacterial and archaeal 16S rRNA genes.
For pyrosequencing analysis, 13-ml samples were collected every 2 weeks from reactor outflows, centrifuged, and stored at −80°C until analysis. Selected samples were analyzed at the conclusion of the experiment. Total community genomic DNA (cgDNA) was extracted using the PowerSoil DNA isolation kit (MoBio Labs, Inc., Carlsbad, CA). Pyrosequencing was conducted using the barcode tagging method described at the Ribosomal Database Project (RDP) Pyrosequencing Pipeline website (http://pyro.cme.msu.edu/index.jsp), and primers were designed for the hypervariable V4 region (∼200 to 210 bp) of the 16S rRNA gene for GS 454 FLX pyrosequencing (Roche Inc.) as described previously (52) using 50-μl PCR mixtures with high-fidelity AccuPrime Pfx DNA polymerase (Invitrogen, Carlsbad, CA). The PCR amplicons were purified using the AMPure solid-phase paramagnetic bead technology (Agencourt Bioscience Corporation, Beverly, MA). The PCR amplicon purity, concentration, and size were estimated using DNA 1000 reagents and an Agilent 2100 (Waldbronn, Germany) bioanalyzer. The reaction mixtures were paired according to DNA quantity and quality prior to performing emulsion reactions for sequencing on a 454 Life Sciences genome sequencer FLX (Roche Diagnostics, Indianapolis, IN) using the unidirection amplicon library sequencing protocol with the emPCR kit II (Roche). Primary processing of the raw 454 FLX data (∼100 Mb for bacteria) was conducted through the RDP Pyrosequencing Pipeline (9). Sequences were sorted by tag sequence, and the 16S rRNA gene primers with low-quality sequences were removed. A total of 68,481 high-quality (99% cutoff) sequences of 200 to 250 bp were obtained for 19 samples.
Archaeal sequences were analyzed similarly, except that amplification included ∼300 bp of the 16S rRNA Archaea gene with forward primer nucleotides containing modified U519F primer (46) fused to variable key tags for multiplexing (9) and to the 454 FLX sequencing primer A (5′-GCCTCCCTCGCGCCATCAGxxxxxxCAGYMGCCRCGGKAAHACC, where the x region represents the various key tags and the 16S rRNA primer is shown in bold). The reverse primer was a fusion of the 454 FLX sequencing primer B and a modified Arch806R primer (47) (5′-GCCTTGCCAGCCCGCTCAGGGACTACNSGGGTMTCTAAT, where the 16S rRNA region is shown in bold). Reaction products were sequenced on the 454 FLX apparatus. Raw data (∼36 Mb for archaea) was processed as above with 20,923 high-quality sequences of 290 to 300 bp obtained for 19 samples.
Quantitative PCR (qPCR) analyses.
SYBR green quantification of the 16S rRNA gene copy number was performed in a Bio-Rad CFX96 (Hercules, CA) thermal cycler on DNA extractions prepared as described above in duplicate. Both general archaeal and bacterial assays were performed in empirically optimized, 20-μl reaction mixtures. Archaeal assays used primers arc915f and arc1059r (Eurofins MWG Operon, Huntsville, AL) at 350 nM each and iQ Supermix at 1× with 2 μl of cgDNA. Amplification used 45 cycles and then a fluorescence reading. Following amplification, products were denatured (95°C for 10 s), and a melt curve was determined (60 to 95°C). Standard curves used Methanococcus maripaludis S2 gDNA diluted from 107 to 102 16S rRNA gene copies per reaction mixture (40).
Bacterial assays used primers Eub338 and Eub558 (Integrated DNA Technologies; Coralville, IA) at 500 nM each, iQ Supermix at 1×, and 2 μl of cgDNA. Amplification again used 45 cycles and a fluorescence reading. Following amplification, products were denatured (95°C for 10 s), and a melt curve was determined (50 to 95°C). Standard curves were constructed using Escherichia coli gDNA diluted from 108 to 103 16S rRNA gene copies per reaction mixture (14).
Phylogenetic analyses.
Bacterial and archaeal 16S rRNA sequences were assigned to a set of hierarchical taxa by using Naïve Bayesian rRNA classifier version 2.0 with a confidence threshold of 80% (http://rdp.cme.msu.edu/classifier/classifier.jsp) (57). Sequences from this study were subsequently aligned using the fast, secondary structure-aware Infernal aligner (36) and clustered by the complete linkage clustering method available with the RDP's Pyrosequencing Pipeline.
Further exploration into the shifts in the archaeal community present in the reactors was performed by clustering the sequences from each genus at a 97% confidence level. Clusters containing 10 or fewer sequences were eliminated due to possible sequence error or artifacts. Heat maps were constructed using relative abundance for each cluster from the total number of sequences, using Genesis version 1.7.6 software (Graz University of Technology, Graz, Austria).
Statistical analyses.
Constrained ordination techniques were utilized to identify patterns of sequence variation between reactors, sampling date, sequence relative abundance, and metabolite concentrations. Bacterial and archaeal sequences were combined in order to observe the overall increasing similarity during the reduced diversity of reactor communities. Although the pyrosequencing and amplification reaction products of bacteria and archaea were amplified and analyzed separately, ratios of the gene copy numbers obtained from qPCR analyses of the gDNA were used to determine the percentage that each domain contributed to the overall sample DNA concentration. Accordingly, the data could be combined based upon relative abundance to perform constrained ordination statistics. Sequence abundances for each genus were converted into weight percentages by dividing by the total abundance per sample; weight percentage values were natural log transformed (ln + 1). Relative abundances of bacterial and archaeal data were combined as the bacteria:archaea ratio as determined by qPCR. Detrended correspondence analysis (DCA), an indirect gradient analysis based on segment length, was performed to determine the modality of the sequence data. The analyses resulted in short (<2.0) segment lengths, indicating linear data sets. Therefore, redundancy analysis (RDA) was performed (CANOCO 4.5; Microcomputer Power). The RDA identified variation patterns among genera present in each reactor and correlated those patterns to predictor variables. Sequence data were used as response variables, and predictor variables were the measured metabolite data and cell counts. Forward selection of the predictor variables followed by Monte Carlo permutation tests were used to prevent artificial inflation of variation due to autocorrelation in the constrained ordination model (27).
Isolates.
At the conclusion of the experiment, isolates were obtained by either fluorescence-activated cell sorting (FACS) (18) or by serial dilution to extinction. Isolates obtained through FACS were cultured using the CCM1 medium. A fresh fermentation vessel sample (7.5 ml) was vigorously shaken and then diluted 1:100 in anaerobic, ice-cold PBS solution. Cells were sorted via forward/side scatter into the 48-well plate, with one cell per well using an InFlux flow cytometer (Cytopeia, Seattle, WA), and the plates were returned to the anaerobic glove bag. After 5 days of incubation, individual wells were screened for growth via adding bromothymol blue (final concentration, 150 μg/ml) to indicate a drop in pH, followed by microscopy. Wells displaying growth were marked as putative isolates and transferred to Balch tubes containing CCM1 medium and incubated at 30°C until visible growth occurred. FACS isolates were also grown with exogenous sulfate to determine if growth could be spurred by sulfate.
Attempts to obtain isolates were also carried out using serial dilutions and plating for sulfate- and Fe(III)-reducing bacteria as well as methanogens. The same medium was used with the following modifications: (i) Fe(III)-reducing bacteria were enriched with 30 mM acetate (rather than lactate) and 10 mM fumarate, (ii) SRB medium contained 10 mM sodium sulfate, (iii) tubes for methanogens used 30 mM acetate with 14 μM choline chloride. A second set of methanogen tubes were pressurized (3 lb/in2) with 80%–20% H2:CO2 gas. The headspace of all other tubes contained 80%–20% N2:CO2 gas at 3 lb/in2. The Balch tubes were sealed using butyl stoppers and aluminum crimp seals and made anaerobic by three cycles of vacuum to −20 lb/in2 and pressurizing with the appropriate gas to 3 lb/in2. All tubes were then autoclaved at 121°C for 20 min and allowed to cool before receiving postautoclave amendments as described above.
Serial dilutions (10-fold) were made from 10−1 to 10−8 in triplicate and incubated (30°C). After growth occurred, samples were taken from the 10−5 to 10−8 tubes and inoculated into fresh medium (giving dilutions of 10−6 to 10−9). Samples (100 μl) were plated with the same medium plus agar. Isolated colonies were collected and inoculated into Balch tubes and incubated (30°C). One more series of dilutions was made into fresh medium tubes to ensure the isolation of single organisms. Microscopy was used to verify that only one morphotype was present for each isolate.
Sequencing of isolates.
Genomic DNA was extracted from 100 μl of isolate culture by using a PowerSoil DNA isolation kit (MoBio Labs, Inc., Carlsbad, CA) or a liquid N2 grinding process (23). For bacterial isolates, 16S rRNA genes were amplified using universal bacterial primers 8F and 1492R (18). Methanogen isolate 16S rRNA genes were amplified using universal archaeal primers Ar21F and Ar958R. A separate amplification using bacterial primers was performed on the putative methanogen cultures to ensure that the cultures were devoid of bacteria.
16S rRNA amplification was performed using 10 ng gDNA template, 0.25 μM primers, 250 μM deoxynucleoside triphosphates, and 1 unit of Pfu DNA polymerase in 1× Pfu DNA polymerase reaction buffer [20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 10 mM (NH4)2SO4, 10 mM KCl, 0.1 mg ml−1 BSA, and 0.1% Triton X-100]. Universal primer 1100R was used for single-pass sequencing reactions of each bacterial isolate, and Ar958R was used to generate a single-pass sequencing product from the methanogen isolates. DNA sequences were determined using BigDye Terminator chemistry (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations and resolved using a 3730 DNA analyzer at a 5:1 dilution. Sequences were compared to those of known organisms using the Basic Local Alignment Search Tool (2) through the NCBI database.
Metal reduction assays.
Cultures of each isolate and Pelosinus fermentans strain R7 (DSM 17108) (43) were grown in lactate-enriched CCM1 medium in duplicate 1-liter pyrex bottles sealed with a rubber stopper under N2 headspace at 30°C. All manipulations were conducted in an anaerobic glove bag unless otherwise noted. Cultures were grown to log phase and centrifuged (8,000 rpm, 8 min at 4°C), washed 3 times with 30 mM lactate–30 mM NaHCO3 buffer (pH 6.8), and finally resuspended to 7.5 ml (11). Each metal reduction assay (detailed below) was performed in duplicate serum vials (sterile, degassed vials containing 30 mM lactate–30 mM NaHCO3 buffer) and contained 4 ml of the washed cells. Separate “no-cell” and heat-killed controls (Shewanella oneidensis MR1 and P. fermentans R7) were employed, while S. oneidensis MR-1 was used as the positive control (21, 37). Samples were taken at 0, 180, and 480 min.
Assays for soluble (FeIII) and solid iron (FeOOH) reduction contained 10 mM FeCl3 · 6H2O and 20 mM FeOOH, respectively, and used the ferrozine method (29). Chromate reduction assays used 60 μM potassium chromate and 60 μM potassium dichromate, individually, with the diphenlycarbazide method (60). U(VI) reduction assays used 0.5 mM uranyl acetate, and 0.25 ml of sample was added to 2.25 ml of 0.1 M HNO3, mixed with Uraplex (Chemchek Instruments Inc.), removed from the anaerobic chamber, and analyzed on a Kinetic phosphorescence analyzer (KPA; Chemcheck Instruments, Inc.) (49).
Nucleotide sequence accession number.
16S rRNA sequences from this study were deposited in the GenBank Short Read Archive database under accession number SRP003881.2.
RESULTS
Cell counts and qPCR quantification.
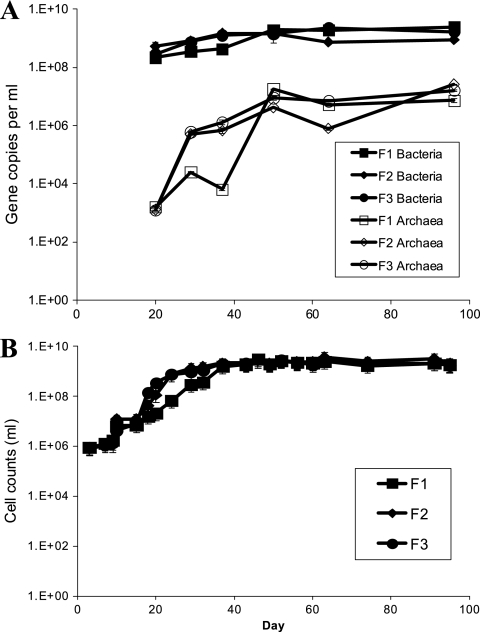
Direct cell counts from the initial groundwater sample showed 8.0 × 105 cells/ml, and this increased by day 7 in all three reactors to 1.08 × 106 to 1.28 × 106 cells/ml (Fig. 1A). By day 37, all three reactors reached cell densities greater than 1 × 109 cells/ml, and these densities were maintained throughout the experiment. Similar trends and values were determined with duplicate qPCR analysis and yielded a ratio of archaeal versus bacterial gene copies (for Archaea, slope of −3.461, reaction efficiency of 94.5%, R2 of 0.999; for Bacteria, slope of −3.419, reaction efficiency of 96.1%, R2 of 0.998) (Fig. 1B). Overall, the gene copy numbers per reactor were highly similar except for archaea, which decreased in reactor 1 for days 20 to 40 but then became similar to the other reactors by day 50. This similarity was sustained throughout the remainder of the experiment, indicating a steady state had been achieved for the reactors, although not necessarily for individual populations.
Fig 1.
qPCR quantification data (A) and cell counts (B) for microbial consortia in triplicate anaerobic continuous flow reactors inoculated with groundwater from Hanford well H-100. Average values with standard error bars are presented.
Metabolite analysis.
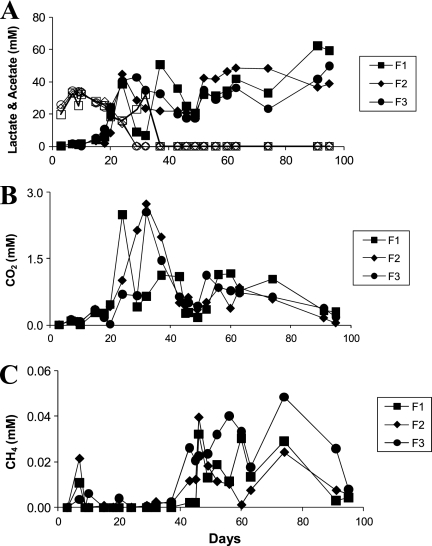
Lactate concentrations in each reactor decreased from the initial 30 mM present on day 3 to as low as 19.7 mM in reactor 1 and 25.0 mM in reactors 2 and 3 (Fig. 2A), but then the concentration returned to 30 mM by day 7. This was followed by a steady decrease, until lactate was below detection by day 32 in reactors 2 and 3 and by day 39 in reactor 1 (Fig. 2A), suggesting electron donor and carbon limitation. Acetate concentrations steadily increased to 38.4 to 44.5 mM by day 24 and to stoichiometric amounts during the rest of the cultivation. Other organic acid concentration, specifically of formate and pyruvate, were intermittently detected but always below 0.1 mM (data not shown).
Fig 2.
Concentrations of actetate (closed symbols) and lactate (open symbols) (A), CO2 (B), and CH4 (C) from the triplicate anaerobic continuous flow reactors inoculated with groundwater from Hanford well H-100.
Carbon dioxide was not detected until day 7, when concentrations ranged from 0.1 to 0.13 mM and fluctuated throughout the experiment (Fig. 2B). For example, at day 24, a divergence was observed with reactor 1 (2.49 mM), while reactors 2 and 3 showed concentrations from 0.7 to 1.0 mM, perhaps suggesting an increase in the fermenter population in reactor 1. However, by day 32, reactors 2 and 3 attained similar peak concentrations of 2.73 and 2.55 mM, respectively, while the concentration in reactor 1 had decreased to 0.64 mM. Methane was also initially detected on day 7 (0.01 to 0.02 mM) and appeared intermittently from days 9 through 37 (Fig. 2C). By day 43, methane ranged from 0.012 to 0.05 mM. Hydrogen in headspace was below detection limits throughout the experiment (<0.5 μM).
Microbial community composition of initial groundwater sample.
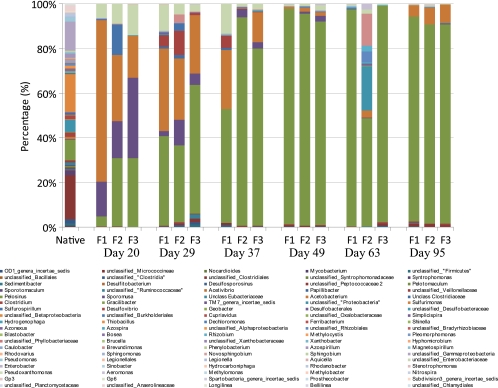
Pyrosequencing analysis of the initial groundwater sample yielded 2,351 bacterial sequences and 83 archaeal sequences identified through the RDP classifier at the 80% confidence threshold. Of the bacteria, 309 (13.1%) sequences were unclassified bacteria at the domain level. The remainder of the bacterial community was classified within 11 phyla with 661 (28.1%) sequences classified within 55 genera, while the remaining 1,381 (58.7%) sequences were grouped as unclassified classes, orders, or families within distinct phyla. The most abundant sequences grouped as “unclassified Clostridiales” (19.1%) (Fig. 3). While these sequences were classified as members of the phylum Clostridia and order Clostridiales at a confidence level of greater than 99%, the sequences were not comparable to other known sequences in the RDP database at a more specific level of classification. The next most abundant groups were the unclassified Betaproteobacteria (16.3%) and the unclassified Gammaproteobacteria (12.7%). With respect to more specific classifications, the most abundant genus was Pelosinus (8.9%), followed by genus Syntrophomonas (2.2%) and Pseudomonas (1.8%) (Fig. 3).
Fig 3.
The original and temporal changes in the bacterial community composition according to pyrosequencing analysis from selected dates from triplicate continuous flow bioreactors of lactate-enriched Hanford well H-100 groundwater sample where Pelosinus (green) and Acetobacterium (orange) became dominant.
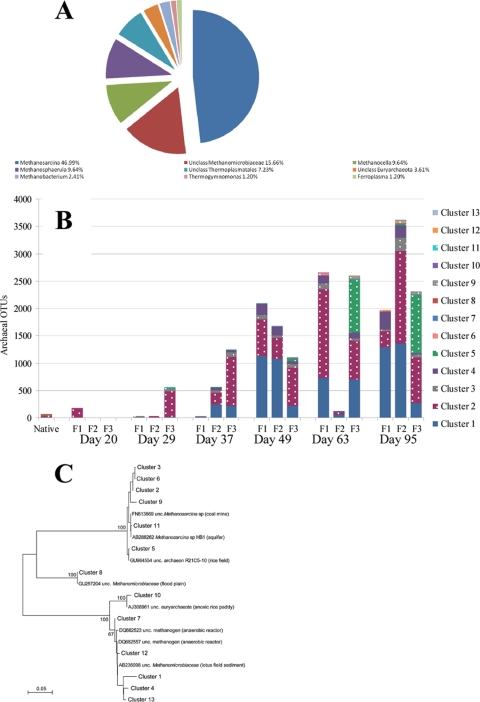
All 83 archaeal sequences in the initial groundwater samples were classified within the phylum Euryarchaeota. Members of the genus Methanosarcina comprised the majority of the sequences (47.0%), followed by unclassified Methanomicrobiaceae (15.7%). While these identifications were performed at a confidence level of >99%, determination of the classification at a more specific level was not possible. The genera Methanocella and Methanosphaerula each comprised 9.64% of the archaeal sequences, while Thermoplasmatales contributed 7.23%. The remaining groups, unclassified Euryarchaeota, Methanobacterium, Thermogymnomonas, and Ferroplasma, each possessed <5% of the sequences (Fig. 4A).
Fig 4.
(A and B) Original archaeal groundwater community (A) and temporal changes in archaeal OTU abundance (B) from selected dates from triplicate continuous flow bioreactors of the lactate-enriched Hanford well H-100 groundwater sample. Methanosarcina OTUs are dotted, and Methanobacteria OTUs are solid. (C) Distance tree of sequence representatives from the individual archaeal clusters (97% level).
Enriched microbial community composition.
Throughout the course of the cultivation, the diversity of the microbial community was reduced and a few distinct genera emerged as the dominant populations within the enriched community. By day 20, the bacterial community was dominated by Acetobacterium (18.6 to 72.3%), Sporumosa (15.6 to 36%), Pelosinus (4.4 to 30.8%), and unclassified Enterobacteriaceae (6.2 to 13.6%). By day 49 and throughout the remainder of the experiment, the community was dominated by Pelosinus spp. (48.4 to 97.2%) and to a lesser extent Acetobacterium spp. (1.0 to 8.3%) (Fig. 3). Although the triplicate reactors received the same medium and gas feed, there were temporal variations in the community compositions. Most notably was the community shift observed in reactor 2 on day 63 that was associated with increases in Rhizobium, Brevundimonas, Aeromonas, unclassified Rhizobiales, and unclassified Enterobacteriaceae which, in turn, decreased by day 95 and yielded a community very similar to reactors 1 and 3. However, by day 95, all 3 bacterial communities were highly similar in the percentages of the community that each genera represented, as shown by Pelosinus spp. (89.0 to 91.7%), Acetobacterium spp. (5.0 to 8.3%), unclassified Clostridiales spp. (1.1 to 1.3%), and unclassified Veillonellaceae spp. (0.3 to 0.4%).
Only two archaeal genera, the acetoclastic Methanosarcina and the hydrogenophagic Methanobacterium (within the Methanomicrobiaceae), maintained substantial populations but had notable temporal fluctuations between the reactors Early on, only a few archaeal sequences were detected, but by day 29 Methanosarcina was the dominant genera (93.7 to 99.0%). By day 37, Methanobacterium (21.3 to 99%) abundances increased, while Methanosarcina decreased (1 to 78.7%) (Fig. 4B). For the remainder of the experiment, the archaeal proportions remained relatively stable in reactors 2 and 3, while reactor 1 showed greater variation, resulting in the highest concentration of Methanobacterium (83.8%).
Due to these continued fluctuations, the archaeal compositions were further investigated by cluster analysis of all the archaeal OTU sequences and their temporal relative abundances. The sequences grouped into 13 clusters and three clades at the 97% confidence level, and the temporal abundance and sequence distribution from each cluster are displayed in Fig. 4B and C. The majority of sequences in the initial sample were found in clusters C2, C5, and C8 (22 to 35%) and to a lesser degree C1, C3, and C9 (1.6 to 3.2%). Clusters C4, C6, C7, and C10 to C13 were initially below detection limits based on the 97% cutoff value, but abundance variations were observed over time. For example, C4 was originally below detection but increased over time in reactors 1 and 2 (Fig. 4B). Similarly, while originally detectable, cluster C5 showed an even more dramatic increase late in the experiment in reactor 3. The other clusters (C6, C7, and C10 to 13) were rare or below detection limits throughout the course of the experiment.
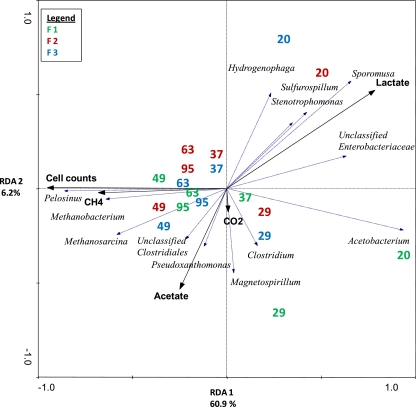
RDA was utilized to examine patterns of relative sequence abundance variation to measured descriptor variables (i.e., metabolites, cell counts, and gas concentrations) (Fig. 5). RDA axes 1 and 2 described 67.1% of the variation in microbial composition from each reactor (F = 19.98; P = 0.002). Samples taken from the reactors on day 20 grouped together in the triplot according to higher abundances of Hydrogenophaga, Sporomusa, Sulfurospirillum, Stenotrophomonas, Acetobacterium, and unclassified Enterobacteriaceae and were correlated with high concentrations of lactate (r = 0.7385). As the experiment progressed and cell counts (r = 0.8949) and methane (r = 0.6403), and acetate (r = 0.4644) concentrations increased, these correlated to greater relative abundances of Pelosinus, Methanobacterium, Methanosarcina, unclassified Clostridiales, and Pseudoxanthomonas. It was also notable that over the course of the experiment, the variation in the community composition within the triplicate reactors decreased, as shown by the decreasing distance from the origin of the sampling day values for each reactor (e.g., the closer grouping of the day 37 and later samples versus the day 20 and 29 samples) along the y axis.
Fig 5.
Triplot of RDA of the relative abundances of microbial genera determined by pyrosequencing analysis on selected dates from the triplicate continuous flow reactor experiment of the lactate-enriched Hanford well H-100 groundwater samples. Dashed arrows (blue) indicate genera associated with the variation in microbial community composition. Solid (black) arrows indicate metabolite data significantly associated with the variation.
Isolates.
A total of 16 isolates were obtained from FACS analysis and identified as being 99 to 100% identical to the 16S rRNA gene of Pelosinus fermentans strain R7 (DSM 17108) through the NCBI database (41). Although these isolates possessed 16S rRNA gene sequences similar to the type strain, they displayed different metabolic characteristics. All strains tested reduced soluble Fe(III), including Pelosinus fermentans strain R7 (Table 1). No strain reduced solid iron (FeOOH). FACS strain A11 also reduced U(VI), monochromate and dichromate. FACS strain B3 and Pelosinus fermentans strain R7 reduced monochromate and dichromate, while FACS strain B4 only reduced monochromate. All strains were tested for the ability to reduce sulfate, but none was capable of sulfidogenesis (data not shown). The enrichment cultures via serial dilutions for sulfate- and Fe(III)-reducing bacteria were cocultures after repreated streaking and serial dilutions. Further efforts to attain pure cultures are ongoing. Methanogens were isolated that appeared closely related (greater than 98% similarity) to Methanosarcina barkerii and uncultured members of Methanosarcina spp. (data not shown). Although unlikely to be new species, they may be among the first methanogenic isolates from these Hanford groundwaters.
Table 1.
Metal reduction assay results for Pelosinus fermentans type strain and Hanford isolates
| Isolate | Reduction of metal? |
||||
|---|---|---|---|---|---|
| Iron(III) | Solid iron (FeOOH) | Monochromate | Dichromate | Uranium | |
| Pelosinus fermentans strain A11 | Yes | No | Yes | Yes | Yes |
| Pelosinus fermentans strain A12 | Yes | No | No | No | No |
| Pelosinus fermentans strain B3 | Yes | No | Yes | Yes | No |
| Pelosinus fermentans strain B4 | Yes | No | Yes | No | No |
| Pelosinus fermentans strain R7a | Yes | No | Yes | Yes | No |
Type strain.
DISCUSSION
After in situ lactate amendments in the presence of abundant acceptors, U(VI) and Cr(VI) reduction typically coincides with increased sulfate- and Fe(III)-reducing populations (12, 54, 58). According to phylochip and other supporting data (12), lactate-utilizing SRB are followed by acetate-utilizing SRB. However, the selective pressure of lactate in the presence of low sulfate (50 μM) resulted in a shift in the in situ microbial community structure, where the extensively studied Desulfovibrio spp. and Geobacter spp. (16, 55) decreased from their initial 1.51% and 0.33%, respectively, to an average of 0.13% and 0.14%, respectively. It was instead the less-studied Pelosinus spp. that became dominant, followed by Acetobacterium spp. Within the archaea, both Methanobacterium and Methanosarcina genera appeared to outcompete others. This suggested that although there was ample lactate and acetate, sulfate concentrations were not sufficient to allow Desulfovibrionales to thrive. H2 was routinely near detection limits, and methanogenic populations were present as potential terminal electron acceptors throughout the 95 days. However, coupling of SRB with methanogens, although well documented (45, 53), did not appear to predominate in these reactors. Rather, SRB and Geobacter spp. were unable to compete and were displaced by a community dominated by Pelosinus spp. and Acetobacter spp.
Previous sediment-based experiments utilizing lactate amended, U(VI)-contaminated microcosms produced different results, in which Pelosinus spp. and Geothrix spp. became predominant (4). Long-term lactate enrichments using flowthrough contaminated sediment columns and microcosms resulted in increased Geobacter spp. and Desulfovibrio spp. (3, 42), while another study using lactate-enriched sediments from the same site found Pelosinus fermentans to be dominant with various Deltaproteobacteria showing increased abundance (20). The latter study also found that in acetate-based Fe(III)-reducing enrichments of saturated sediments, Desulfovibrio and Desulfomicrobium spp. dominated and not Geobacter spp., while Pelosinus spp. were predominant in nutrient-poor saturated, unsaturated, and acidic sediments. These observations suggest a competitive advantage for Pelosinus spp. via metabolic flexibility with respect to nutrient-poor, unsaturated, or lower-pH conditions (19). In each of these cases, ample electron acceptor(s) was available. In direct competition experiments between Acetobacterium, Desulfovibrio, and Veillonella with l-lactate, Desulfovibrio outcompeted Actetobacterium and Veillonella under lactate-limited conditions, while sufficient sulfate and Fe(III) were present (26). Neither electron acceptor was present in appreciable concentrations in the present study, but Desulfovibrio spp. have been successful during lactate limitation. Both Geobacter spp. and Desulfovibrio spp. were outcompeted here, suggesting that adequate electron acceptors may be important for the dominance of Geobacter spp. and Desulfovibrio spp., perhaps being more critical than electron donors and carbon sources. These direct competition results suggest that Desulfovibrio outcompeted Acetobacterium under lactate-limited conditions, and Pelosinus outcompeted Desulfovibrio here. Accordingly, Pelosinus outcompeting Acetobacterium under electron acceptor limitation, as occurred here, should not be surprising.
One reason that Pelosinus spp. may have overwhelmed the community by becoming 90.0 ± 1.5% of the final consortium is that it comprised ∼9% of the initial groundwater community; ∼6 times greater than Desulfovibrio spp. and ∼30 times greater than Geobacter spp. However, this also suggests that Pelosinus spp. displayed a fitness under the conditions tested and perhaps also in situ. Further, Acetobacterium spp. were a mere 0.09% of the community initially but became the second most abundant population at 6.9 ± 1.7%. It is worthy to note that Acetobacterium increased to 72% of the community by day 29, when lactate became limiting, and then this genus diminished, perhaps being outcompeted for lactate by Pelosinus.
The closest known organism to the Pelosinus strains isolated in this study is Pelosinus fermentans, which is to date one of only three cultured strains within this genus and is capable of fermenting lactate and coupling its oxidation to Fe(III) reduction (41). The second species, P. defluvii, cannot utilize lactate but can reduce Fe(III) although not sulfate (35), while P. fermentans strain UFO1, isolated from Oak Ridge sediments, can consume lactate and reduce Fe(III) as well as U(VI) (39). While Fe(III) and U(VI) reduction have been observed in Pelosinus isolates, no isolates have been shown to reduce Cr. Although the isolates obtained in this study were a >99% match to the P. fermentans type strain according to the 16S rRNA sequences, they demonstrated broad metal-reducing characteristics. Strain A12 can only reduce Fe(III), as opposed to strains A11, B3, and the type strain, which can reduce both mono- and dichromate, while strain B4 can only reduce monochromate. Not only is strain A11 capable of the above activities, but also it can reduce U(VI) similar to strain UFO1. Hence, not only did the genus Pelosinus become dominant under electron-accepting conditions over the more extensively studied metal reducers during electron acceptor limitation, but also the characterization of metal-reducing capabilities for four different strains now allows for an appreciation of the diversity of metal reduction within this genus. Given this information, further characterization of the functional and genomic capabilities of these isolates will be pursued.
With respect to the archaeal populations, several operational taxonomic units (OTUs) belonging to two methanogenic genera maintained populations throughout the experiment, the acetoclastic Methanosarcina spp. and the hydrogenotrophic Methanobacterium spp., suggesting multiple routes of carbon mineralization from fermentation by-products in situ. However, there was no obvious correlation between temporal species abundances and volatile fatty acid or gas concentrations, similar to findings from studies of anaerobic sludge and food waste reactors (25, 56). However, further investigation into archaeal community revealed that several OTUs, while not initially detectable in the original or early temporal samples, were able to maintain low-level populations despite numerous reactor turnovers and became robust in later time periods. Such organisms may be part of the “rare biosphere” that is not readily culturable or detected but can grow and become dominant if the proper microenvironment conditions are met (44), such as low gas and liquid flow rates.
In summary, comprehensive investigations such as these allow for the study of consequential succession in microbial communities from contaminated environments and allow determination of the relative importance of particular community populations via alteration of selected, imposed perturbations. The designed system for continuous steady-state enrichment over many generations followed by FACS or other isolation methods can facilitate these studies on a more specific level than is possible in situ. It also presents certain advantages for the cultivation of organisms that have been traditionally difficult to isolate but whose presence is routinely indicated by pyrosequencing. In the present case, the limitation of available electron acceptor(s) on a community that has been shown to transiently increase and decrease in sulfate- and Fe(III)-reducing organisms with concomitant Cr(VI) reduction in situ (12) was explored. While the abundance levels of Desulfovibrio spp. and Geobacter spp. were expected to be lower than in situ levels, it was unknown whether their numbers could increase via fermentation or if fermenting organisms would instead become dominant. With respect to metal reduction, it is encouraging that although the population matrix was substantially different during electron acceptor limitation, the resultant dominating species were capable of the complete reduction of in situ Cr(VI) levels as well as U(VI). This suggests that whether the contaminated areas are electron acceptor rich or depleted, the native microbial community may be capable of reducing and immobilizing oxidized metals and radionuclides, whether they result from plume movement into the area or from reoxidation of previously reduced Cr and U pools.
Supplementary Material
ACKNOWLEDGMENTS
This work, conducted by ENIGMA, was supported in part by the Office of Science, Office of Biological and Environmental Research, U.S. Department of Energy, under contract number DE-AC02-05CH11231 to Lawrence Berkeley National Laboratory. Oak Ridge National Laboratory is managed by University of Tennessee UT-Battelle LLC for the Department of Energy under contract number DE-AC05-00OR22725.
We thank Zamin Yang, Sue Carroll, and Marilyn Kerley for their technical support of this work.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print 20 January 2012
REFERENCES
- 1. Akob DM, et al. 2008. Functional diversity and electron donor dependence of microbial populations capable of U(VI) reduction in radionuclide-contaminated subsurface sediments. Appl. Environ. Microbiol. 74: 3159–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Bundschuh R, Olsen R, Hwa T. 2001. The estimation of statistical parameters for local alignment score distributions. Nucleic Acids Res. 29: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brodie EL, et al. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72: 6288–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burkhardt E, et al. 2010. Impact of biostimulated redox process on metal dynamics in an iron-rich creek soil of a former uranium mining area. Environ. Sci. Technol. 44: 177–183 [DOI] [PubMed] [Google Scholar]
- 5. Cardenas E, et al. 2008. Microbial communities in contaminated sediments, associated with bioremediation of uranium to submicromolar levels. Appl. Environ. Microbiol. 74: 3718–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang YJ, et al. 2005. Microbial incorporation of C-13-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39: 9039–9048 [DOI] [PubMed] [Google Scholar]
- 7. Chapelle FH, Lovley DR. 1992. Competitive exclusion of sulfate reduction by iron(III)-reducing bacteria: a mechanism for producing discrete zones of high-iron ground water. Ground Water 30: 29–36 [Google Scholar]
- 8. Coates JD, Cole KA, Chakraborty R, O'Connor SM, Achenbach LA. 2002. Diversity and ubiquity of bacteria capable of utilizing humic substances as electron donors for anaerobic respiration. Appl. Environ. Microbiol. 68: 2445–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards L, Kusel K, Drake H, Kostka JE. 2007. Electron flow in acidic subsurface sediments co-contaminated with nitrate and uranium. Geochim. Cosmochim. Acta 71: 643–654 [Google Scholar]
- 11. Elias DA, Suflita JM, McInerney MJ, Krumholz LR. 2004. Periplasmic cytochrome c3 of Desulfovibrio vulgaris is directly involved in H2-mediated metal but not sulfate reduction. Appl. Environ. Microbiol. 70: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faybishenko B, et al. 2008. In situ long-term reductive bioimmobilization of Cr(VI) in groundwater using hydrogen release compound. Environ. Sci. Technol. 42: 8478–8485 [DOI] [PubMed] [Google Scholar]
- 13. Fields MW, et al. 2006. Phylogenetic and functional biomakers as indicators of bacterial community responses to mixed-waste contamination. Environ. Sci. Technol. 40: 2601–2607 [DOI] [PubMed] [Google Scholar]
- 14. Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71: 4117–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francis AJ, Dodge CJ, Lu F, Halada GP, Clayton CR. 1994. ZPS and XANES studies of uranium reduction by Clostridium sp. Environ. Sci. Technol. 28: 636–639 [DOI] [PubMed] [Google Scholar]
- 16. Gadd GM. 2010. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156: 609–643 [DOI] [PubMed] [Google Scholar]
- 17. Ganesh R, Robinson KG, Reed GD, Sayler GS. 1997. Reduction of hexavalent uranium from organic complexes by sulfate- and iron-reducing bacteria. Appl. Environ. Microbiol. 63: 4385–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamilton-Brehm SD, et al. 2010. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansel CM, Fendorf S, Jardine PM, Francis CA. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74: 1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen AA, et al. 2007. Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, northern Norway. Environ. Microbiol. 9: 2870–2884 [DOI] [PubMed] [Google Scholar]
- 21. Heidelberg JF, et al. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20: 1118–1123 [DOI] [PubMed] [Google Scholar]
- 22. Hemme CL, et al. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 4: 660–672 [DOI] [PubMed] [Google Scholar]
- 23. Hurt RA, et al. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67: 4495–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hwang CC, et al. 2009. Bacterial community succession during in situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J. 3: 47–64 [DOI] [PubMed] [Google Scholar]
- 25. Ike M, et al. 2010. Microbial population dynamics during startup of a full-scale anaerobic digester treating industrial food waste in Kyoto eco-energy project. Bioresour. Technol. 101: 3952–3957 [DOI] [PubMed] [Google Scholar]
- 26. Laanbroek HJ, Geerligs HJ, Peijnenburg A, Siesling J. 1983. Competition for l-lactate between Desulfovibrio, Veillonella, and Acetobacterium species isolated from anaerobic intertidal sediments. Microb. Ecol. 9: 341–354 [DOI] [PubMed] [Google Scholar]
- 27. Leps J, Smilauer P. 2003. Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, United Kingdom: [Google Scholar]
- 28. Lovley DR. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1: 35–44 [DOI] [PubMed] [Google Scholar]
- 29. Lovley DR, Phillips EJP. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53: 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lovley DR, Phillips EJP, Gorby YA, Landa ER. 1991. Microbial reduction of uranium. Nature 350: 413–417 [Google Scholar]
- 31. Lovley DR, Roden EE, Phillips EJP, Woodward JC. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113: 41–53 [Google Scholar]
- 32. Luo WS, et al. 2007. Influence of bicarbonate, sulfate, and electron donors on biological reduction of uranium and microbial community composition. Appl. Microbiol. Biotechnol. 77: 713–721 [DOI] [PubMed] [Google Scholar]
- 33. Madden AS, et al. 2009. Donor-dependent extent of uranium reduction for bioremediation of contaminated sediment microcosms. J. Environ. Qual. 38: 53–60 [DOI] [PubMed] [Google Scholar]
- 34. Miller LD, et al. 2010. Establishment and metabolic analysis of a model microbial community for understanding trophic and electron accepting interactions of subsurface anaerobic environments. BMC Microbiol. 10: 149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moe WM, et al. Pelosinus defluvii sp. nov., isolated from chlorinated solvent contaminated groundwater, emended description of the genus Pelosinus, and transfer of Sporotalea propionica to Pelosinus propionicus comb. nov. Int. J. Syst. Evol. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 36. Nawrocki EP, Eddy SR. 2007. Query-dependent banding (QDB) for faster RNA similarity searches. PLOS Comput. Biol. 3: 540–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nealson KH, Belz A, McKee B. 2002. Breathing metals as a way of life: geobiology in action. Antonie Van Leeuwenhoek 81: 215–222 [DOI] [PubMed] [Google Scholar]
- 38. Petrie L, North NN, Dollhopf SL, Balkwill DL, Kostka JE. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69: 7467–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ray AE, et al. 2010. Evidence for multiple modes of uranium immobilization by an anaerobic bacterium. Geochim. Cosmochim. Acta 75: 2684–2695 [Google Scholar]
- 40. Reysenbach AL, et al. 2006. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 442: 444–447 [DOI] [PubMed] [Google Scholar]
- 41. Shelobolina ES, et al. 2007. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov., sp. nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 57: 126–135 [DOI] [PubMed] [Google Scholar]
- 42. Sitte J, et al. 2010. Microbial links between sulfate reduction and metal retention in uranium- and heavy metal-contaminated soil. Appl. Environ. Microbiol. 76: 3143–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith RL. 2002. Determining the terminal electron-accepting reaction in the saturated subsurface, p 743–752 In Hurst CJ. (ed), Manual of environmental microbiology, 2nd ed. American Society for Microbiology, Washington, DC: [Google Scholar]
- 44. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103: 12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stolyar S, et al. 2007. Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 3: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62: 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takai K, Horikoshi K. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66: 5066–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tebo BM, Obraztsova AY. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162: 193–198 [Google Scholar]
- 49. Truex MJ, Peyton BM, Valentine NB, Gorby YA. 1997. Kinetics of U(VI) reduction by a dissimilatory Fe(III)-reducing bacterium under non-growth conditions. Biotechnol. Bioeng. 55: 490–496 [DOI] [PubMed] [Google Scholar]
- 50. Van Nostrand JD, et al. 2009. GeoChip-based analysis of functional microbial communities during the reoxidation of a bioreduced uranium-contaminated aquifer. Environ. Microbiol. 11: 2611–2626 [DOI] [PubMed] [Google Scholar]
- 51. Vishnivetskaya TA, et al. 2010. Microbial community changes in response to ethanol or methanol amendments for U(VI) reduction. Appl. Environ. Microbiol. 76: 5728–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vishnivetskaya TA, et al. 2011. Mercury and other heavy metals influence bacterial community structure in contaminated Tennessee streams. Appl. Environ. Microbiol. 77: 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walker CB, et al. 2009. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191: 5793–5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wall JD, Krumholz LR. 2006. Uranium reduction. Annu. Rev. Microbiol. 60: 149–166 [DOI] [PubMed] [Google Scholar]
- 55. Wan JM, et al. 2008. Effects of organic carbon supply rates on uranium mobility in a previously bioreduced contaminated sediment. Environ. Sci. Technol. 42: 7573–7579 [DOI] [PubMed] [Google Scholar]
- 56. Wang H, et al. 2010. Development of microbial populations in the anaerobic hydrolysis of grass silage for methane production. FEMS Microbiol. Ecol. 72: 496–506 [DOI] [PubMed] [Google Scholar]
- 57. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu WM, et al. 2007. In situ bioreduction of uranium(VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ. Sci. Technol. 41: 5716–5723 [DOI] [PubMed] [Google Scholar]
- 59. Xu M, et al. 2010. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J. 4: 1060–1070 [DOI] [PubMed] [Google Scholar]
- 60. Zhang CL, Liu S, Logan J, Mazumder R, Phelps TJ. 1996. Enhancement of Fe(III), Co(III), and Cr(VI) reduction at elevated temperatures and by a thermophilic bacterium. Appl. Biochem. Biotechnol. 57–58: 923–932 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.