Abstract
The insertion sites of the conjugative transposon Tn916 in the anaerobic pathogen Clostridium difficile were determined using Illumina Solexa high-throughput DNA sequencing of Tn916 insertion libraries in two different clinical isolates: 630ΔE, an erythromycin-sensitive derivative of 630 (ribotype 012), and the ribotype 027 isolate R20291, which was responsible for a severe outbreak of C. difficile disease. A consensus 15-bp Tn916 insertion sequence was identified which was similar in both strains, although an extended consensus sequence was observed in R20291. A search of the C. difficile 630 genome showed that the Tn916 insertion motif was present 100,987 times, with approximately 63,000 of these motifs located in genes and 35,000 in intergenic regions. To test the usefulness of Tn916 as a mutagen, a functional screen allowed the isolation of a mutant. This mutant contained Tn916 inserted into a gene involved in flagellar biosynthesis.
INTRODUCTION
Tn916 is a conjugative transposon (or integrative conjugative element [ICE]) which was originally isolated from Enterococcus faecalis DS16 (11). Since its original isolation, the element has been transferred into or been found in a large number of different bacteria (for recent reviews, see references 24 and 25). Tn916 is a prototype of a large family of conjugative transposons called the Tn916/Tn1545 family. Typically, these elements have a modular organization with regions encoding conjugation, recombination, regulation, and accessory functions (24), with the last often being antibiotic resistance genes (tetracycline resistance in the case of Tn916). At 18 kb in length, Tn916 is the smallest of this family of genetic elements.
The first step in conjugative transposition of Tn916 is excision of the element to form a circular intermediate molecule. It is thought that a single strand of this is transferred, via nicking at a proven oriT site, to a suitable recipient (15). In the transconjugant, a second strand is synthesized and the element can then enter the genome via site-specific recombination (a diagrammatic representation of the process can be seen in reference 29). In the current model, transcription of the conjugation genes is upregulated in the presence of tetracycline, initially via a transcriptional attenuation mechanism (32), and this signal is subsequently amplified at the promoter Porf7 (7).
As well as being an important vector in the spread of antibiotic resistance, Tn916 has been used previously as a tool for mutagenesis in the pathogenic clostridia (1, 18) and for cloning genes into the bacterial chromosome (12, 22, 24). We have previously shown that the element has a highly preferred target in the genome of the nontoxigenic Clostridium difficile strain CD37 (34) but that the element enters the genome at multiple sites (13, 14, 26) in the toxigenic strain R20291, a ribotype 027 strain responsible for a severe outbreak of C. difficile-associated disease (23), and 630, a ribotype 012 strain and the first C. difficile strain to be completely sequenced (30). In order to determine the target site requirements of Tn916 and its usefulness as an insertional mutagen in C. difficile, we undertook Illumina Solexa high-throughput DNA sequencing of insertions of Tn916 in the two toxigenic C. difficile strains: R20291 and an erythromycin-sensitive derivative of 630, 630Δerm (14). Furthermore, to test the usefulness of Tn916 as a mutagen, a mutant R20291 strain with an insertion within a gene involved in flagellar biosynthesis was investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and transposons used in this study.
All bacterial strains, plasmids, and transposons are shown in Table 1.
Table 1.
Bacterial strains and transposons used in this study
| Bacterial strain or transposon | Comments | Reference or source |
|---|---|---|
| Clostridium difficile 630 | Sequenced strain of ribotype 012 | 35 |
| Clostridium difficile 630Δerm | Erythromycin-sensitive derivative of the genome strain | 14 |
| Clostridium difficile R20291 | Ribotype 027, responsible for the Stoke Mandeville outbreak in the United Kingdom | 31 |
| Clostridium difficile FM168 | Recipient strain CD37 containing Tn5397 and Tn916ΔE | 34 |
| Bacillus subtilis CU2189 | Recipient strain | 9 |
| Bacillus subtilis BS34A | B. subtilis CU2189::Tn916, containing a single copy of Tn916 | 26 |
| Bacillus subtilis BS59A | B. subtilis CU2189 containing a single copy of Tn916ΔE from FM168 | This study |
| Tn916 | Tetracycline resistance-encoding conjugative transposon | 11 |
| Tn916ΔE | Erythromycin resistance-encoding derivative of Tn916 | 27 |
Filter mating.
Tn916 was transferred from Bacillus subtilis BS34A to C. difficile R20291 and Tn916ΔE was transferred from B. subtilis BS59A to C. difficile 630Δerm on nitrocellulose membrane filters as described previously (14). Transconjugants were selected on brain heart infusion (BHI) agar containing 5% defibrinated horse blood (E&O Laboratories), Clostridium difficile selective supplement (Oxoid), and tetracycline (10 mg/liter) or erythromycin (50 mg/liter).
Testing the stability of Tn916 insertions.
Single colonies of C. difficile R20291 containing Tn916 and C. difficile 630Δerm containing Tn916ΔE were used to inoculate BHI broth, incubated for 48 h, and then subcultured (100 μl into 10 ml) into fresh broth. This was repeated 15 times in the absence of antibiotic selection. After the final subculture, the broth was diluted and plated onto antibiotic-free BHI agar. Single colonies were tested for their ability to grow on agar containing the appropriate antibiotic. This period of time (approximately 1 month) was chosen because it is likely to extend beyond the length of any in vivo experiments. Genomic DNA was isolated from the strains both before and after subculturing using the ArchivePure DNA yeast and Gram-positive bacteria kit (5Prime) according to the manufacturer's instructions. Southern blots were carried out using HyBond membrane filters, probed with an internal fragment of the Tn916 integrase gene (14), and detected with an ECL kit (Amersham, Little Chalfont, United Kingdom) according to the manufacturer's instructions.
Nucleotide sequencing.
Genomic DNA (gDNA) was prepared from a pool of 96 transconjugants of C. difficile 630Δerm::Tn916ΔE and from 100 transconjugants of C. difficile R20291::Tn916. Ten micrograms of each sample was processed separately for sequencing library preparation as previously described (16) except that the PCR step was completed before the gel size selection step. The PCR primers used were 5′-AATGATACGGCGACCACCGAGATCTACACATAAGTCCAGTTTTTATGCGGATAAC (forward) and 5′-CAAGCAGAAGACGGCATACGAGATCGGTACACTCTTTCCCTACACGACGCTCTTCCGATCT (reverse).
The amplified DNA fragment libraries were sequenced on a single-end Illumina flow cell with an Illumina GAII sequencer for 54 cycles of sequencing using a custom sequencing primer (5′-TCTACACATAAGTCCAGTTTTTATGCGGATAACTAGAT) and 2× hybridization buffer. This primer was designed such that the first 14 bp of each read was the Tn916 transposon sequence (5′-TTTTATGCTATTTT).
Sequence read analysis.
Custom Perl scripts were written to identify sequence reads with a 100% identical match to the 14-bp Tn916 transposon tag. All qualifying reads had this tag removed along with a further 10 bp to take into account any possible coupling sequence. Maq-0.6.8 (17) using the easyrun.pl option with all of the default parameters was used to map the remaining 30 bp to the relevant C. difficile genome sequence, either 630 (accession no. AM180355) (30) or R20291 (accession no. NC013316).
Mapped reads were filtered for mapping quality, and those with a score of >20 (CD630) or >10 (R20291) were taken forward. Each read mapped to a particular nucleotide position in the genome, named a Tn916 insertion site. All insertion sites were inspected for the frequency of reads, and sites with <20 reads were excluded from the CD630 list as potentially spurious. The read depth of the R20291 library was much lower; hence, sites with <10 reads were excluded as potentially spurious. Due to the potential presence of a coupling sequence, these insertion sites are up to 10 bp away from the true insertion site.
Insertion site motif.
Using the relevant reference C. difficile genome sequences, 20 base pairs upstream and downstream of each mapped Tn916 insertion site in the CD630Δerm and R20291 pools were retrieved. The CD630Δerm and R20291 40-bp sequences were submitted as two separate batches to MEME (2), specifying a distribution of 1 motif per sequence. For CD630Δerm, the output from MEME was loaded into FIMO (find individual motif occurences) (also part of the MEME suite at http://meme.nbcr.net/) to identify all occurrences of the predicted motif using C. difficile CD630 as the reference genome sequence.
Electron microscopy.
For negative staining, bacteria were grown overnight in BHI broth and a 1-ml aliquot was harvested by centrifugation for 1.5 min. A total of 0.95 ml of supernatant was removed, and the bacteria were gently resuspended in the remaining supernatant. Cells were negatively stained by a protocol adapted from McNab et al. (21). Specifically, for each sample, a Formvar/carbon-coated, 400 mesh, gold transmission electron microscopy (TEM) grid (Agar Scientific, Stansted, Essex, United Kingdom) was placed film side up on clean dental wax. Five microliters of prepared bacterial suspension was carefully dropped onto the film and left for 2 to 5 min. The grid was removed from the bacterial suspension, excess liquid was absorbed with a damp filter paper, and the grid was floated film side down on a 50-μl drop of 1% (wt/vol) methylamine tungstate (Sigma Chemical Co., Poole, Dorset, United Kingdom) for 30 s. After removal from the staining solution, excess stain was removed with dry filter paper and the grid was allowed to dry for 10 min prior to viewing in a Philips CM12 TEM (FEI, Eindhoven, Netherlands) at 80 kV.
RESULTS
Characterization of Tn916 insertions into the C. difficile R20291 and 630 genomes.
Tn916 and Tn916ΔE were transferred into C. difficile R20291 and C. difficile 630Δerm, respectively. Southern blotting of 20 of each of the transconjugants showed that the elements had entered the genomes at multiple sites, with no two patterns the same (results not shown). There was a single insertion of the Tn916 element in 12 630Δerm and 8 R20291 transconjugants, with multiple (ranging from 2 to 4) insertions in the remainder of the isolates.
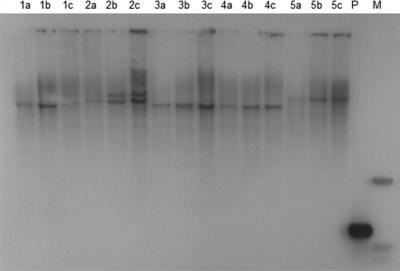
In order to test the stability of Tn916 within the C. difficile strains, 10 independent R20291::Tn916 and 10 independent 630Δerm::Tn916ΔE transconjugants were subcultured (see Materials and Methods) 15 times over a period of 4 weeks. All 20 isolates tested retained their resistance to tetracycline or erythromycin. A Southern blot analysis of genomic DNA isolated from these transconjugants both before and after subculturing showed identical hybridizing patterns, indicating that the Tn916 insertions were stable over the time period tested. The Southern blots for the 630Δerm::Tn916ΔE transconjugants are shown in Fig. 1 (the results for R20291 are not shown, but all transconjugants tested had stable insertions).
Fig 1.
Southern blots of insertions of Tn916ΔE into C. difficile 630Δerm. HindIII-digested C. difficile DNA probed with an internal fragment of the intTn gene. Numbers refer to DNA from isolate 1, isolate 2, etc. a, DNA from the original colony; b and c, DNA from the same isolate which has been subcultured 15 times (see Materials and Methods). Lane P contains the PCR product used as the probe (0.7 kb), and lane M contains the molecular weight marker. The hybridizing bands are 0.5 kb and 1 kb.
Mapping Tn916 insertion sites from C. difficile mutant pools.
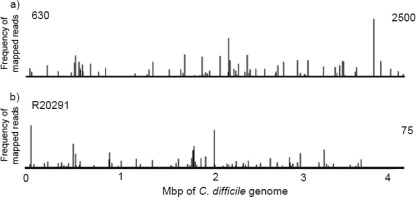
Individual mutant colonies from either C. difficile strain 630Δerm (96 mutants) or R20291 (100 mutants) were combined, and genomic DNA from each pool was isolated. To determine the Tn916 insertion sites within each pool, we sequenced the genome region adjacent to the transposon using the transposon-directed insertion site sequencing (TraDIS) method that is based on the Illumina system (16). For C. difficile 630, we generated 105,473 tagged sequence reads which mapped unambiguously to the 630 genome, demarking 112 unique insertion sites (19 coding and 93 noncoding). Insertions into protein-coding genes are shown in Table 2. For C. difficile R20291, we generated 2,944 reads which mapped to the R20291 genome in 102 unique insertion sites (27 coding and 75 noncoding). Insertions into protein-coding genes are shown in Table 3. As can be seen in Tables 2 and 3, the same gene is frequently targeted by the element at more than one site. Some insertion sites in the same gene are within one base of each other; this has been observed previously with Tn916 (28). Furthermore, homologous genes in the two strains have also been targeted (genes hit in both strains are shaded in Table 2). For each mutant pool, the number of insertion sites identified was greater than the number of mutants, confirming the presence of multiple Tn916 insertions in some mutants. The Tn916 insertions were distributed throughout the genomes (Fig. 2). Thus, Tn916 inserts into a variety of sites within the genomes of C. difficile 630 and R20291.
Table 2.
Insertions within ORFs in C. difficile 630a
| Position in genome | Read depth | Systematic IDb | Name | Start | End | Length | Strand | Product |
|---|---|---|---|---|---|---|---|---|
| 69341 | 679 | CD0044 | 69040 | 69513 | 473 | + | Putative membrane protein | |
| 370209 | 431 | CD0308 | 369853 | 370353 | 500 | + | Putative membrane protein | |
| 552402 | 1,762 | CD0463 | 552158 | 552796 | 638 | − | TetR family transcriptional regulator | |
| 1238338 | 447 | CD1045 | 1237997 | 1239049 | 1,052 | + | Putative membrane protein | |
| 1620690 | 914 | CD1400 | 1620152 | 1620745 | 593 | + | Conserved hypothetical protein | |
| 2311806 | 517 | CD2001 | 2311732 | 2312130 | 398 | − | Conserved hypothetical protein | |
| 2311818 | 2,120 | CD2001 | 2311732 | 2312130 | 398 | − | Conserved hypothetical protein | |
| 2511080 | 2,744 | CD2170 | 2510369 | 2511097 | 728 | + | Putative cNMP-binding regulatory protein | |
| 2704147 | 1,035 | CD2337 | 2703553 | 2704473 | 920 | − | Putative membrane protein | |
| 2856215 | 1,416 | CD2475 | 2856026 | 2857321 | 1,295 | − | Putative competence membrane protein | |
| 2856216 | 38 | CD2475 | 2856026 | 2857321 | 1,295 | − | Putative competence membrane protein | |
| 2922039 | 1,853 | CD2528 | 2921714 | 2922397 | 683 | + | Hypothetical protein | |
| 3196460 | 2,052 | CD2752 | 3195904 | 3196569 | 665 | + | Hypothetical protein | |
| 3376862 | 1,362 | CD2887 | 3375865 | 3377019 | 1,154 | + | Putative signaling protein | |
| 3507312 | 1,237 | CD3019 | 3506993 | 3507925 | 932 | − | Putative transporter | |
| 3557078 | 1,472 | CD3063 | 3556888 | 3558102 | 1,214 | − | Putative exported protein | |
| 3752367 | 1,224 | CD3205 | 3752057 | 3752650 | 593 | + | Putative nitroreductase | |
| 3947431 | 7,812 | CD3377 | mgtA | 3947397 | 3950060 | 2,663 | − | Magnesium-transporting ATPase, P-type 1 |
| 4028263 | 968 | CD3437 | cobS | 4028256 | 4029029 | 773 | − | Cobalamin synthase |
Genes that are also targets for Tn916 in R20291 are shaded.
ID, identification.
Table 3.
Insertions within ORFs in R20291
| Position | Read depth | Systematic IDa | CD630 orthologue | Start | End | Gene length | Strand | Product |
|---|---|---|---|---|---|---|---|---|
| 412998 | 25 | CDR20291_0344 | CD0339 | 412623 | 413360 | 737 | + | Two-component response regulator |
| 487710 | 21 | CDR20291_0404 | CD0463 | 487466 | 487999 | 533 | − | TetR family transcriptional regulator |
| 564605 | 27 | CDR20291_0467 | CD0542 | 564053 | 564631 | 578 | + | Putative chemotaxis-related protein-glutamate |
| 564606 | 31 | CDR20291_0467 | CD0542 | 564053 | 564631 | 578 | + | Putative chemotaxis-related protein-glutamate |
| 612718 | 14 | CDR20291_0506 | CD0579 | 612337 | 612915 | 578 | − | TetR family transcriptional regulator |
| 620016 | 29 | CDR20291_0512 | CD0588 | 619824 | 620111 | 287 | + | Hypothetical protein |
| 777867 | 13 | CDR20291_0628 | CD0702 | 777396 | 778079 | 683 | + | Putative membrane protein |
| 944547 | 36 | CDR20291_0773 | CD0843 | 943977 | 945212 | 1,235 | + | Putative glycosyl transferase |
| 1080916 | 31 | CDR20291_0886 | CD1030 | 1080844 | 1082100 | 1,256 | + | Putative glycosyl transferase |
| 1433939 | 32 | CDR20291_1206 | CD1363 | 1432968 | 1434032 | 1,064 | + | Putative phage protein |
| 1645610 | 13 | CDR20291_1389 | CD1540 | 1643765 | 1645645 | 1,880 | + | ABC transporter, permease protein |
| 1859883 | 12 | CDR20291_1570 | CD1673 | 1859074 | 1859898 | 824 | + | Putative membrane protein |
| 1919051 | 27 | CDR20291_1634 | CD1737 | 1918970 | 1920235 | 1,265 | + | Putative gluconate permease |
| 1934871 | 15 | CDR20291_1645 | CD1751 | 1932770 | 1935157 | 2,387 | + | Cell surface protein (putative cell surface-associated cysteine protease) |
| 2037368 | 11 | CDR20291_1737 | CD1842 | 2037161 | 2037907 | 746 | + | Putative membrane protein |
| 2037411 | 36 | CDR20291_1737 | CD1842 | 2037161 | 2037907 | 746 | + | Putative membrane protein |
| 2037412 | 11 | CDR20291_1737 | CD1842 | 2037161 | 2037907 | 746 | + | Putative membrane protein |
| 2143796 | 14 | CDR20291_1830 | CD1909 | 2143364 | 2143813 | 449 | + | Putative ethanolamine/propanediol utilization |
| 2339670 | 18 | CDR20291_2001 | CD2094 | 2339541 | 2341475 | 1,934 | − | Putative restriction enzyme |
| 2382608 | 29 | CDR20291_2033 | CD2126 | 2382398 | 2382655 | 257 | − | Putative membrane protein |
| 2382651 | 14 | CDR20291_2033 | CD2126 | 2382398 | 2382655 | 257 | − | Putative membrane protein |
| 2465352 | 21 | CDR20291_2101 | CD2195 | 2464855 | 2465370 | 515 | + | Ferritin |
| 2771369 | 15 | CDR20291_2368 | CD2475 | 2771181 | 2772483 | 1,302 | − | Putative competence membrane protein |
| 2771370 | 24 | CDR20291_2368 | CD2475 | 2771181 | 2772483 | 1,302 | − | Putative competence membrane protein |
| 2850359 | 15 | CDR20291_2431 | CD2544 | 2849896 | 2851011 | 1,115 | − | Putative membrane protein |
| 2990722 | 44 | CDR20291_2551 | CD2663 | 2989423 | 2991702 | 2,279 | − | Putative signaling protein |
| 3109990 | 60 | CDR20291_2642 | CD2752 | 3109477 | 3110142 | 665 | + | Hypothetical protein |
| 3110033 | 42 | CDR20291_2642 | CD2752 | 3109477 | 3110142 | 665 | + | Hypothetical protein |
| 3379053 | 20 | CDR20291_2855 | CD3019 | 3378774 | 3379706 | 932 | − | Putative transporter |
| 3379096 | 74 | CDR20291_2855 | CD3019 | 3378774 | 3379706 | 932 | − | Putative transporter |
| 3677207 | 14 | CDR20291_3079 | CD3220 | 3677190 | 3678047 | 857 | − | Putative methyltransferase |
| 3802754 | 35 | CDR20291_3188 | 3802387 | 3803703 | 1,316 | − | Sensor histidine kinase VirS |
ID, identification.
Fig 2.
Frequency and distribution of transposon-directed insertion site sequence reads across the entire genomes of C. difficile 630ΔErm (a) and R20291 (b). The y axis shows the number of mapped sequence reads scaled to 2,500 and 75 for 630 and R20291, respectively, within a window size of 3 bp.
Tn916 insertion motif discovery and analysis.
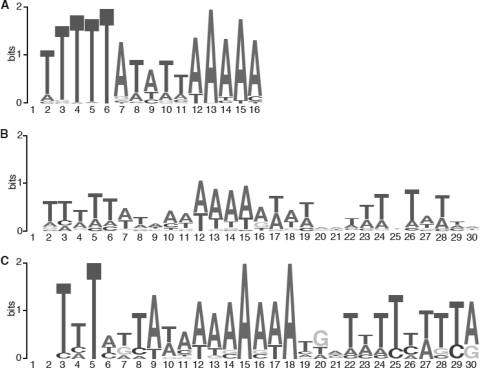
The Tn916 insertion sites predominantly mapped to noncoding regions (70 to 80%) in both R20291 and 630, even though only 18% of the genome is considered noncoding (30). These observations imply a bias for Tn916 insertion, perhaps due to a preference to insert into a DNA sequence motif. To address this possibility, the 40-bp regions around each insertion site in both the 630Δerm::Tn916ΔE and R20291 pools were submitted to MEME, an online tool for identifying similarities (motifs) in sequences. The 15-bp consensus motif 5′-TTTTTA[AT][AT][AT][AT]AAAAA (Fig. 3) was associated with all insertion sites, and there was an extension to the motif in strain R20291 (Fig. 3). Furthermore, when the same gene was targeted in both R20291 and 630, an extended consensus similar to the R202901 extended consensus was seen in both genomes (Fig. 3).
Fig 3.
Tn916 target site sequence logos generated by MEME (2) for nucleotide sequences around 112 Tn916 insertion sites in C. difficile 630ΔErm (A), around 102 Tn916 insertion sites in R20291 (B), and around 12 Tn916 insertion sites in the same genes in both genomes (C). The level of sequence conservation is indicated by the height of the letters (maximum of 2 bits at each position).
The output from the MEME search was used to find all occurrences of the 15-bp motif in the 630 reference genome sequence by using a second tool from the online MEME suite, FIMO (find individual motif occurrences). Using a P value cutoff of 0.01 (which finds all the insertion sites identified by TraDIS), there were 100,987 occurrences of the Tn916 insertion motif in the 630 genome sequence. Approximately 63,000 of these were within genes and ∼35,000 were intergenic (the remainder overlap the genic/intergenic borders). There appears to be a bias for Tn916 insertion sites within intergenic regions, since only ∼18,000 would be expected for randomly distributed motifs as there is less noncoding DNA than coding DNA (30). Thus, Tn916 preferentially inserts into a consensus sequence that is enriched within the intergenic regions of C. difficile 630.
Identification of a C. difficile R20291 mutant in flagellar biosynthesis.
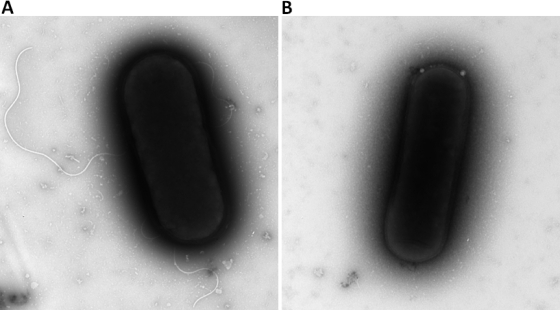
In order to test the ability of Tn916 to generate C. difficile mutants, we carried out a functional screen looking for sporulation-deficient insertional mutants and noticed a colony that had reduced growth following the heat-treatment assay. This R20291 isolate was further studied and found to contain an insertion within a putative flagellar biosynthesis gene (CDR20291_0231). In this mutant, Tn916 is present at the nucleotide (nt) position 289890 on the minus strand of the R20291 genome. BLAST searches with the predicted amino acid sequence revealed homology to proteins within the FlgN superfamily (Pfam 05130) (20) which are described as export chaperones involved in flagellar synthesis. In Salmonella enterica serovar Typhimurium, FlgN has been shown to act as a substrate-specific export chaperone which aids the incorporation of the hook-associated proteins into the growing flagellum (4). We therefore hypothesized that the Tn916 mutant would be unable to assemble flagella, and as predicted, electron microscopy showed that no flagella were present on this mutant strain (Fig. 4B). In contrast, the parental R20291 strain (Fig. 4A) and another mutant from the Tn916 library with the transposon inserted within a gene encoding a putative cobalamin biosynthesis kinase did produce flagella (data not shown).
Fig 4.
Electron micrograph of a negatively stained wild-type R20291 (A) and R20291 flagellar mutant (B). Flagella are present in the wild-type strain but absent from the flgN mutant (panel B). Magnification for wild-type R20291, ×15,000. Magnification for R20291 flagellar mutant, ×11,500.
DISCUSSION
Our previous work, based on Southern blot analysis, has shown that in C. difficile strains R20291 and 630, Tn916 inserts into the genome with no obvious site preference (13, 14). However, in this study, the availability of high-throughput genomic sequencing allowed a large number of genomic insertions in the two strains to be sequenced. The consensus insertion motif 5′-TTTTTA[AT][AT][AT][AT]AAAAA was identified in 630 and R20291. A similar sequence had previously been identified in other hosts, although this was based on much less sequencing data (28). This consensus motif appears 105 times in the 630 genome, with it appearing 60% of the time in coding sequences. However, the sequence analysis performed in this work showed that 60% of the insertions were in intergenic regions, indicating that factors other than DNA sequences are involved in target site selection. For example, in C. difficile CD37, there are many copies of the consensus target, yet one is highly preferred (34). Furthermore, in this work, we have shown that there is an extended consensus sequence in strain R20291 compared to that of 630, suggesting that there are differences between the two strains that influence target site selection by Tn916. This could be local variation in DNA topology, as it has been shown that Tn916 is likely to preferentially insert into targets that have a static bend (19).
A similar analysis of the insertion sites of Tn916 has also been performed in Butyrivibrio proteoclasticus B316 (10). As we have observed in C. difficile, a preference for insertion in intergenic sites was found in this genus. This is likely an important adaptation of this broad host range element to stop it from inactivating important host genes on arrival in a new host. A second, not mutually exclusive hypothesis is that these host genomes evolve to lose target sites which are present within open reading frames (ORFs) in order to protect their genes. The fact that C. difficile has such a large amount of intergenic DNA may be one of the reasons for the large number of integrative elements that are present within this host (5, 30). It appears that target site selection is an interaction of element and host properties.
Recently, a system for rapidly generating mutants in C. difficile, a derivative of the mariner transposon, has been developed (6). This element apparently enters the C. difficile genome at random and appears to be an easier method for generating random insertions than using Tn916. However, a detailed study of the insertion sites of mariner in C. difficile, such as that reported here for Tn916, has not yet been undertaken. Furthermore, in this work, we show that Tn916 preferentially targets intergenic regions which have been shown to have an important regulatory role in bacterial metabolism, an observation which could be useful if one wished to disrupt noncoding RNA sequences (3). In support of this notion, Chen et al. (8) have undertaken a systematic search of small noncoding RNA (sRNA) sequences in the clostridia, and the C. difficile sRNA sequences contained 2,134 Tn916 insertion motifs entirely within the sRNA sequences and a further 739 motifs that overlap into/out of an sRNA. In addition, Tn916 has been used to generate a metabolic mutant in a noncoding region of Clostridium proteoclasticum (33). A further advantage of Tn916 preferentially inserting into intergenic regions is that the element can be used to insert genes into the chromosome without disrupting coding sequences. We have previously shown that Tn916 can be used to insert heterologous DNA sequences into the C. difficile genome (22, 26). Tn916 enters the genomes of many different strains of C. difficile, indicating that it will be a generally useful tool for C. difficile research. The use of Tn916 as a mutagen for C. difficile was confirmed in this work, as a gene involved in flagellum production was inactivated by insertion of the element, resulting in a C. difficile R20291 strain which could not produce flagella.
ACKNOWLEDGMENTS
We thank the MRC (capacity building fellowship and grant number G0601176) and EU FP7 HYPERDIFF, contract 241446.
Footnotes
Published ahead of print 20 January 2012
REFERENCES
- 1. Awad MM, Rood JI. 1997. Isolation of alpha-toxin, theta-toxin and kappa-toxin mutants of Clostridium perfringens by Tn916 mutagenesis. Microb. Pathog. 22:275–284 [DOI] [PubMed] [Google Scholar]
- 2. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p 28–36 In Altman R, Brutlag D, Karp P, Lathrop R, Searls D. (ed), Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 3. Beisel CL, Storz G. 2010. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 34:866–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett JCQ, Thomas J, Fraser GM, Hughes C. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol. Microbiol. 39:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brouwer MSM, Warburton PJ, Roberts AP, Mullany P, Allan E. 2011. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One 6:e23014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cartman ST, Minton NP. 2010. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76:1103–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Celli J, Trieu-Cuot P. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103–117 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y, Indurthi DC, Jones SW, Papoutsakis ET. 2011. Small RNAs in the genus Clostridium. mBio 2(1):e00340–10 doi:10.1128/mBio.00340-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christie PJ, Korman RZ, Zahler SA, Adsit JC, Dunny GM. 1987. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J. Bacteriol. 169:2529–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cookson AL, et al. 2011. Transposition of Tn916 in the four replicons of the Butyrivibrio proteoclasticus B316(T) genome. FEMS Microbiol. Lett. 316:144–151 [DOI] [PubMed] [Google Scholar]
- 11. Franke AE, Clewell DB. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haraldsen JD, Sonenshein HL. 2003. Efficient sporulation in Clostridium difficile requires disruption of the σK gene. Mol. Microbiol. 48:811–821 [DOI] [PubMed] [Google Scholar]
- 13. Hussain HA, Roberts AP, Whalan R, Mullany P. 2010. Transposon mutagenesis in Clostridium difficile. Methods Mol. Biol. 646:203–211 [DOI] [PubMed] [Google Scholar]
- 14. Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54(Part 2):137–141 [DOI] [PubMed] [Google Scholar]
- 15. Jaworski DD, Clewell DB. 1995. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J. Bacteriol. 177:6644–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Langridge GC, et al. 2009. Simultaneous assay of every gene in Salmonella Typhi using one million transposon mutants. Genome Res. 19:2308–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin WJ, Johnson EA. 1991. Transposon Tn916 mutagenesis in Clostridium botulinum. Appl. Environ. Microbiol. 57:2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu F, Churchward G. 1995. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J. Bacteriol. 177:1938–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McNab R, et al. 1999. Cell wall-anchored CshA polypeptide (259 kilodaltons) inStreptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181:3087–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mullany P, Wilks M, Puckey L, Tabaqchali S. 1994. Gene cloning in Clostridium difficile using Tn916 as a shuttle conjugative transposon. Plasmid 31:320–323 [DOI] [PubMed] [Google Scholar]
- 23. O'Connor JR, Johnson S, Gerding DN. 2009. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136:1913–1924 [DOI] [PubMed] [Google Scholar]
- 24. Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251–258 [DOI] [PubMed] [Google Scholar]
- 25. Roberts AP, Mullany P. 2011. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 35:856–871 [DOI] [PubMed] [Google Scholar]
- 26. Roberts AP, et al. 2003. Development of an integrative vector for the expression of antisense RNA in Clostridium difficile. J. Microbiol. Methods 55:617–624 [DOI] [PubMed] [Google Scholar]
- 27. Rubens CE, Heggen LM. 1988. Tn916ΔE: a Tn916 transposon derivative expressing erythromycin resistance. Plasmid 20:137–142 [DOI] [PubMed] [Google Scholar]
- 28. Scott JR, Bringel F, Marra D, Van Alstine G, Rudy CK. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099–1108 [DOI] [PubMed] [Google Scholar]
- 29. Scott JR, Churchward GG. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367–397 [DOI] [PubMed] [Google Scholar]
- 30. Sebaihia M, et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 31. Stabler R, et al. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent strain. Genome Biol. 10:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su YA, He P, Clewell DB. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Villas-Bôas SG, et al. 2008. Phenotypic characterization of transposon-inserted mutants of Clostridium proteoclasticum B316T using extracellular metabolomics. J. Biotechnol. 20:55–63 [DOI] [PubMed] [Google Scholar]
- 34. Wang H, Roberts AP, Mullany P. 2000. DNA sequence of the insertional hot spot of Tn916 in the Clostridium difficile genome and discovery of a Tn916-like element in an environmental isolate integrated in the same hot spot. FEMS Microbiol. Lett. 192:15–20 [DOI] [PubMed] [Google Scholar]
- 35. Wüst J, Hardegger U. 1983. Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob. Agents Chemother. 23:784–786 [DOI] [PMC free article] [PubMed] [Google Scholar]