Abstract
Virion-associated peptidoglycan hydrolases have potential as antimicrobial agents due to their ability to lyse Gram-positive bacteria on contact. In this work, our aim was to improve the lytic activity of HydH5, a virion-associated peptidoglycan hydrolase from the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. Full-length HydH5 and two truncated derivatives containing only the CHAP (cysteine, histidine-dependent amidohydrolase/peptidase) domain exhibited high lytic activity against live S. aureus cells. In addition, three different fusion proteins were created between lysostaphin and HydH5, each of which showed higher staphylolytic activity than the parental enzyme or its deletion construct. Both parental and fusion proteins lysed S. aureus cells in zymograms and plate lysis and turbidity reduction assays. In plate lysis assays, HydH5 and its derivative fusions lysed bovine and human S. aureus strains, the methicillin-resistant S. aureus (MRSA) strain N315, and human Staphylococcus epidermidis strains. Several nonstaphylococcal bacteria were not affected. HydH5 and its derivative fusion proteins displayed antimicrobial synergy with the endolysin LysH5 in vitro, suggesting that the two enzymes have distinct cut sites and, thus, may be more efficient in combination for the elimination of staphylococcal infections.
INTRODUCTION
Staphylococcus aureus is a notorious pathogen that causes numerous pathologies, including food poisoning, toxic shock syndrome, endocarditis, and skin and wound infections (27). The emergence of multidrug-resistant strains, especially methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA), is raising serious concerns due to their high frequency in both nosocomial and community-acquired settings (21). Antibiotic resistance in combination with other important virulence determinants, such as surface-located binding proteins to facilitate adhesion to host tissue, as well as many mechanisms to evade attack by human host defenses, makes S. aureus a threatening pathogen (36), and novel therapeutic agents are sorely needed.
Bacteriophages are a feasible alternative to antibiotics for the treatment of bacterial infections (31). In addition, phage peptidoglycan (PG) hydrolases have demonstrated valuable antimicrobial activities against Gram-positive pathogens (11, 16). Most PG hydrolases are modular proteins composed of a C-terminal cell wall-binding (CWB) domain and N-terminal catalytic domain(s) (10). Specifically, there are three major PG hydrolase activities, namely, (i) glycosidase, (ii) amidase, and (iii) endopeptidase activities.
Recent results with phage therapy in animal models have driven much interest in phages and phage-encoded proteins to treat infections (9, 35). These studies clearly show the efficacy of phages and lysins in killing human pathogenic bacteria in animal models (11, 31). Specifically, several assays have been performed against S. aureus bacteremia. The intraperitoneal administration of phages phiMR11 and phiMR25 rescued mice inoculated with a lethal dose of S. aureus (18, 30). Moreover, in a rabbit model of wound infection caused by S. aureus, phages prevented abscess formation (44). Phage lytic proteins also showed successful results. The intraperitoneal administration of the endolysin MV-L from phage phiMR11 protected mice against septic death from MRSA (38). In other animal models, bacteremia in unprotected mice reached colony counts of ∼107 CFU/ml within 3.5 h after challenge, whereas the administration of the lytic enzyme LysGH15 1 h after MRSA injection was sufficient to protect mice, with the mean colony count being less than 104 CFU/ml (14). Furthermore, the activity of phage lytic proteins may be increased by using them in combination with other antimicrobials. Both in vitro (2, 5, 13) and in vivo synergy (5) between phage endolysin constructs and antibiotics or bacteriocins against S. aureus have been reported.
In addition to endolysins, other phage-encoded proteins (virion-associated peptidoglycan hydrolases) have potential as antimicrobials. Some bacteriophage virions harbor peptidoglycan (PG) hydrolases that facilitate the entry of phage DNA across the bacterial cell envelope during infection (32). They are also responsible for “lysis from without,” a phenomenon caused by some phages when adsorbed onto the host cell at very high numbers (6). This type of PG hydrolase lytic activity has been described from a variety of different phage particles infecting S. aureus (32, 39, 43), Lactococcus lactis (20), Escherichia coli (19, 33), and Salmonella (42).
Recently, a PG hydrolase, HydH5, encoded by phage vB_SauS-phiIPLA88 (phiIPLA88) has been identified and characterized (40). HydH5 has an N-terminal CHAP (cysteine, histidine-dependent amidohydrolase/peptidase) lytic domain and a C-terminal LYZ2 (lysozyme subfamily 2) lytic domain, but it does not have a recognized cell wall binding domain. The full-length 634-amino-acid HydH5 and a truncated version harboring just one lytic domain (and a 6×His tag) have been overproduced in E. coli. The purified proteins are able to lyse viable S. aureus cells. HydH5 is highly thermostable since it showed antimicrobial activity after heat treatment (100°C for 5 min) (40).
The endolysin LysH5 encoded by phiIPLA88 has three putative domains, an N-terminal CHAP domain, an amidase-2 domain, and a C-terminal SH3b CWB domain. LysH5 is able to inhibit the growth of S. aureus in milk (34) and showed a synergistic antimicrobial effect with the bacteriocin nisin (13).
Several truncated derivatives and fusion proteins were created in order to identify the essential domains and enhance the antimicrobial activity of HydH5 against S. aureus. Possible synergistic effects between HydH5 and the endolysin LysH5 were tested to explore alternative enzybiotic-based strategies to fight S. aureus infections.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Vector constructs were performed in E. coli DH5α cells (Invitrogen, Carlsbad, CA) and induced in E. coli BL21(DE3) (EMD Biosciences, San Diego, CA). The bacterial strains used to determine the lytic spectrum of our constructs are listed in Table 1. S. aureus Sa9 was used as the indicator strain for lytic activity (34). The S. aureus bovine isolates were from in-house stocks (IPLA-CSIC), the S. aureus clinical isolates were from J. E. Suárez (University of Oviedo, Spain), the S. epidermidis strains were from S. Delgado (IPLA-CSIC), and MRSA strain N315 was from Gabi Bierbaum (University of Bonn, Germany). All Staphylococcus strains were grown in tryptic soy broth (TSB; Difco, Franklin Lakes, NJ) at 37°C with shaking or in TSB plates containing 2% (wt/vol) bacteriological agar (TSA). Bacillus, Streptococcus, Listeria, and Enterococcus strains were grown in 2× yeast tryptone (YT) at 37°C with shaking. The Clostridium strain was grown in brain heart infusion (BHI; Scharlau, Barcelona, Spain) at 30°C under static conditions and anaerobiosis (Anaerocult A; Merck, Darmstadt, Germany). The Lactococcus strain was grown in M17 medium (Scharlau, Barcelona, Spain) at 30°C under static conditions. Leuconostoc and Lactobacillus strains were grown in MRS (Scharlau, Barcelona, Spain) at 30°C under static conditions.
Table 1.
Lytic spectrum of the peptidoglycan hydrolase HydH5, its derivative fusions, and lysostaphin
| Strain | Proteina |
||||
|---|---|---|---|---|---|
| HydH5 | HydH5Lyso | HydH5SH3b | CHAPSH3b | Lysostaphin | |
| S. aureus strains | |||||
| Bovine | |||||
| Sa9 | + | ++ | +++ | +++ | +++ |
| Sa6 | − | +++ | +++ | +++ | +++ |
| Sa11 | + | +++ | +++ | +++ | +++ |
| AFG1 | + | ++ | +++ | +++ | +++ |
| AC9 | + | + | +++ | +++ | +++ |
| Human | |||||
| MRSA strain N315 | − | ++ | +++ | +++ | +++ |
| 96 | − | + | ++ | +++ | +++ |
| 143 | − | + | + | ++ | +++ |
| 445 | − | ++ | +++ | +++ | +++ |
| c4 | + | ++ | +++ | +++ | +++ |
| S. epidermidis strains | |||||
| B | − | ++ | + | +++ | ++ |
| C213 | − | ++ | +++ | +++ | ++ |
| Z2LDC17 | + | + | ++ | +++ | − |
| DH3LIK | − | ++ | + | +++ | ++ |
| Bacillus cereus ATCC 9139 | − | − | − | − | ND |
| Streptococcus pneumoniae R6 | − | − | − | − | ND |
| Clostridium tyrobutyricum CECT 4012 | − | − | − | − | ND |
| Lactococcus lactis IPLA 947 | − | − | − | − | ND |
| Leuconostoc lactis IPLA 567 | − | − | − | − | ND |
| Lactobacillus citreum IPLA 979 | − | − | − | − | ND |
| Listeria monocytogenes ScottA | − | − | − | − | ND |
| Enterococcus faecalis CNRZ 1352 | − | − | − | − | ND |
Bacterial strains were exposed to 2 μM each protein. +++, strong lytic halo; ++, medium; +, weak; −, no halo; ND, not determined.
Plasmid constructs and DNA manipulation.
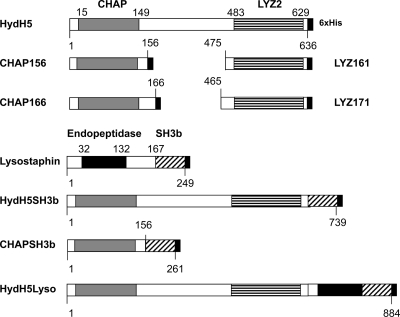
Bacteriophage vB_SauS-phiIPLA88 gene orf58 (open reading frame 58) encoding HydH5 (GenBank accession number ACJ64586) was codon optimized (orf58-opt) based on the E. coli codon usage (23) and commercially synthesized (Genescript, Piscataway, NJ). Then, orf58-opt was synthesized with a 5′ NdeI and a 3′ XhoI restriction enzyme site and the resultant fragment was subcloned between the NdeI and XhoI sites in the multicloning site of the inducible expression vector pET21a (EMD Biosciences, San Diego, CA), which introduces a C-terminal 6×His tag. All constructions in this work were created in this vector and therefore have two additional amino acid residues introduced at the C terminus, corresponding to the XhoI site (Leu-Glu), followed by the 6×His tag (Fig. 1). Truncated versions of HydH5 were constructed via PCR amplification by introducing cloning sites with PCR primers (Table 2).
Fig 1.
Schematic representation of HydH5 deletion and fusion constructs. The numbers indicate the initial and final amino acids in the domains as determined by using the Pfam domain database. Gray box, CHAP domain; horizontal stripes, LYZ2 domain; diagonal stripes, SH3b domain; large black box, endopeptidase domain; small black box, 6×His tag.
Table 2.
Primers used in this study
| Protein | Primer | Sequencea | Restriction site | Length (amino acids) |
|---|---|---|---|---|
| CHAP156 domain | CHAP156R | 5′-CTGACTGCTCGAGTTTGTCCGGGTG-3′ | XhoI | 156 |
| pET21aBglII-F | 5′-CGTAGAGGATCGAGATCTCGATC-3′ | |||
| CHAP166 domain | CHAP166R | 5′-ATCGACTGCTCGAGTTTCGGCACCGG-3′ | XhoI | 166 |
| pET21aBglII-F | 5′-CGTAGAGGATCGAGATCTCGATC-3′ | |||
| LYZ161 domain | LYZ161F | 5′-ACTGACTGCATATGGTTAGCGTCTCC-3′ | NdeI | 475 |
| pET21aStyI-R | 5′-CGTTTAGAGGCCCCAAGGGGTTATG-3′ | |||
| LYZ171 domain | LYZ171F | 5′-ACTGACTGCATATGCAAATGCTGAAC-3′ | NdeI | 465 |
| pET21aStyI-R | 5′-CGTTTAGAGGCCCCAAGGGGTTATG-3′ | |||
| Lysostaphin | Full Lyso SalI-F | 5′-ATCATCGTCGACGCTGCAACACATGAACATTCAGCAC-3′ | SalI | |
| pET21aStyI-R | 5′-CGTTTAGAGGCCCCAAGGGGTTATG-3′ | |||
| SH3b domain | Lyso SalI144-F | 5′-GGAAAAGCAGTCGACACAGTAACTCC-3′ | SalI | |
| pET21aStyI-R | 5′-CGTTTAGAGGCCCCAAGGGGTTATG-3′ |
The location of the restriction enzyme site (underlined) relative to the nearest amino acid residue in the protein is indicated.
Protein purification.
Protein purification was performed as previously described (8) with the following modifications: exponentially growing cultures induced by IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 1 mM) were incubated at 10°C for 20 h. Then, 500-ml pellets were sonicated for 5 min using an automatic pulsing sonicator (Bronson Sonifier; Bronson Sonic Power Co., Danbury, CT). Protein purification was carried out by Ni-nitrilotriacetic acid nickel column chromatography (Qiagen, Valencia, CA). The wash and elution profiles were empirically determined to be 20 ml of 10 mM imidazole, 40 ml of 20 mM imidazole and elution with 1 ml of 250 mM imidazole in phosphate-buffered saline (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) with 30% glycerol to prevent precipitation of the purified protein. Then, all samples were converted to HydH5 activity buffer {50 mM HEPES, 0.5 M NDSB (nondetergent sulfobetaine)-201, 0.25 mM CaCl2, 0.25 mM MnCl2, 0.25 mM MgCl2, 1 mM TCEP [Tris(2-carboxyethyl)phosphine], 24 mM NaCl, 1 mM KCl, pH 7.5} (39) containing 30% glycerol using preequilibrated Zeba desalting columns (Thermo Fisher Scientific, Rockford, IL).
SDS-PAGE and zymogram analysis.
The purity of HydH5 constructs was evaluated in 15% (vol/wt) SDS–PAGE in Tris-glycine buffer at 150 V for 1.5 h using Criterion precast gels (Bio-Rad, Inc., Hercules, CA). Zymogram assays were performed as previously described (40). SDS gels were stained via conventional Coomassie staining, and zymograms were soaked for 30 min in distilled water to remove SDS and then incubated at room temperature in water for 15 min to detect areas of clearing in the turbid gel where a lytic protein is localized.
Quantification of lytic activity.
Turbidity reduction assay was performed against live S. aureus Sa9 cells prepared as previously described (3, 8). Specific activities of the proteins were expressed as the ΔOD600 min−1 μM−1 (OD600, optical density at 600 nm). Plate lysis was performed as previously described (8). Serial 1:2 dilutions (in a range of 4 μM to 0.06 μM) of each construct in HydH5 activity buffer were obtained. Then, drops (10 μl) of these dilutions were spotted onto freshly plated, air-dried lawns of S. aureus Sa9, MRSA N315, or the strains in Table 1 that were grown to an OD600 of 0.5.
Synergy.
Checkerboard minimum lytic concentration (MLC) tests were performed between LysH5 (34) and HydH5 and between LysH5- (GenBank accession number EU573240) and HydH5-derived constructs using live S. aureus Sa9 cell suspensions in HydH5 activity buffer at a final OD600 of ∼0.8. MLC is defined as the lowest concentration of enzyme required to drop the turbidity of S. aureus strain Sa9 from an OD600 of 0.9 to 0.1 after 15 min of incubation at 37°C (13). The ranges of enzyme concentrations were as follows: LysH5, 2.5 μg/ml to 0.04 μg/ml; HydH5, 56.7 μg/ml to 0.06 μg/ml; HydH5Lyso, 25.1 μg/ml to 0.02 μg/ml; HydH5SH3b, 13.5 μg/ml to 0.01 μg/ml; and CHAPSH3b, 17.34 μg/ml to 0.03 μg/ml. The fractional inhibitory concentration (FIC) was calculated as the minimum lytic concentration (MLC) of the antimicrobial in combination divided by the MLC of the antimicrobial acting alone. Strong synergy exists if the sum of the two FICs (∑FIC = FICA + FICB) is <0.5 (15). All experiments were performed in duplicate.
RESULTS
Fusion to lysostaphin sequences increases the HydH5 and CHAP lytic activity.
To determine whether both CHAP and LYZ2 as single domains showed increased lytic activity toward S. aureus Sa9 over the activity previously obtained (40), four deletion constructs were generated. A schematic representation of all constructs is presented in Fig. 1. In order to ensure proper folding, each construct contained a single lytic domain with an additional 7 or 17 amino acids flanking the catalytic domain (CHAP156, CHAP166, LYZ161, and LYZ171) in order to help maintain the normal protein environment of the domain and thus enhance correct folding. In addition, due to the lack of a known CWB domain in HydH5, we proceeded to determine whether the addition of a CWB domain might increase the lytic activities of HydH5 and CHAP. Three different fusion proteins were created with lysostaphin and HydH5 (Fig. 1). Initially, the lysostaphin binding domain SH3b was fused to the full-length HydH5 protein, resulting in a protein with two catalytic domains and one CWB domain (HydH5SH3b). A second construct was obtained by fusion of the CHAP156 domain to the lysostaphin SH3b domain (CHAPSH3b). Finally, the full-length lysostaphin and HydH5 were fused in order to obtain a protein with three catalytic domains and a CWB (HydH5Lyso). All the fusion proteins could be detected and purified with the exception of LYZ161 and LYZ171, whose products could not be detected after nickel column purification of the E. coli cultures.
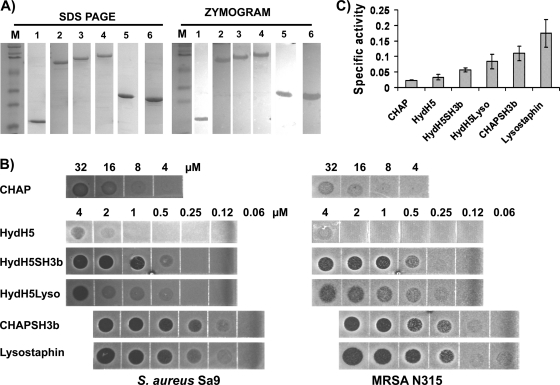
Due to reports that PG hydrolase lytic activity is not always quantitatively comparable between multiple assays (22), the lytic activities of the other proteins (HydH5, CHAP156, CHAP166, HydH5SH3b, CHAPSH3b, and HydH5Lyso) were determined in three antimicrobial assays, zymogram and plate lysis and turbidity reduction assays. As shown in Fig. 2A, all nickel column-purified proteins were >95% pure and created a single zone of clearing in the zymogram, consistent with the predicted molecular masses and the positions of the purified proteins in the SDS-PAGE. The activities of both truncated constructs (CHAP156 and CHAP166) turned out to be the same regardless of the length of the lytic domain protein fragment. Therefore, only the results obtained with CHAP156 are presented.
Fig 2.
Antimicrobial activities of HydH5 and deletion and fusion constructs. (A) SDS-PAGE and zymogram analysis of 5-μg nickel affinity-purified proteins. Lane M, standard molecular mass marker in kDa (prestained SDS-PAGE standards, broad range; Bio-Rad Laboratories); lane 1, CHAP (19.6 kDa); lane 2, HydH5 (73.6 kDa); lane 3, HydH5SH3b (85.1 kDa); lane 4, HydH5Lyso (100.8 kDa); lane 5, CHAPSH3b (30.4 kDa); lane 6, lysostaphin (28.1 kDa). (B) Plate lysis assay of HydH5 and derived proteins using mid-log-phase growing cells of S. aureus Sa9 and MRSA strain N315. (C) Turbidity reduction assay results using 1 μM each protein. Specific activity is expressed as ΔOD600 min−1 μM−1. Error bars show the means ± standard deviations of three independent assays.
In the plate lysis assay (Fig. 2B), the full-length HydH5 showed clearing of S. aureus at ≥2 μM, while the CHAP domain required higher concentrations (≥16 μM). All the fusion proteins showed significantly greater activities than the parental protein HydH5, even against the MRSA strain N315. HydH5 has clear staphylolytic activity at 4 μM, while the fusion proteins showed lytic activity at much lower concentrations (1 μM). Fusion of SH3b to the CHAP domain also increased CHAP activity against both S. aureus Sa9 and S. aureus N315, by 64-fold (Fig. 2B).
The proteins were also tested by turbidity reduction assay (Fig. 2C). This assay was performed using 1 μM purified protein. The full-length recombinant HydH5, synthesized from the E. coli codon-optimized version of orf58-opt, showed lytic activity in this assay that was in contrast to the previous observations of HydH5 from the standard orf58 (40). The recombinant HydH5 was able to lyse live S. aureus Sa9 cells and had a specific activity of 0.033 ± 0.009 ΔOD600 min−1 μM−1 (mean ± standard deviation) (Fig. 2C). Similar results were obtained for the CHAP domain, with an ∼1.4-fold reduction in specific activity (0.023 ± 0.001 ΔOD600 min−1 μM−1) observed compared to the activity of full-length HydH5. The fusion proteins HydH5Lyso and HydH5SH3b showed 2.5- and 1.7-fold higher specific activities than the parental protein HydH5, respectively. HydH5Lyso had 1.5-fold higher activity than HydH5SH3b. CHAPSH3b showed an activity 4.8-fold greater than that of the CHAP domain. The specific activity of CHAPSH3b was calculated to be 0.109 ± 0.023 ΔOD600 min−1 μM−1, the highest lytic activity obtained from the HydH5 fusions. Therefore, C-terminal lysostaphin fusions conferred an enhanced staphylolytic activity to HydH5 and its catalytic domain CHAP.
The lytic activity of HydH5 and its derivative fusions is specific for staphylococci.
In order to determine the specificity of the HydH5 and its derivative fusions, different bacterial cell suspensions were exposed to 2 μM protein solutions for the plate lysis assay. We found that 10-μl amounts of the HydH5-derived fusions HydH5SH3b and CHAPSH3b at a concentration of 2 μM were able to lyse all S. aureus and S. epidermidis strains (14 tested; Table 1). In contrast, 10 μl of HydH5Lyso at 2 μM was able to lyse all the S. aureus strains and only 2 of 4 S. epidermidis strains. S. epidermidis lysis could be achieved at 4 μM (data not shown). A variety of bacteria representing several genera (Bacillus, Streptococcus, Clostridium, Lactococcus, Leuconostoc, Lactobacillus, Listeria, and Enterococcus) were not affected by any of the staphylolytic proteins. It was observed that the degree of lytic activity on S. epidermidis strains was generally lower than that on S. aureus strains. Moreover, the spectrum of species lysed was not consistent for all proteins despite a similar origin of the domains. HydH5 and HydH5Lyso were less active in this assay, with CHAPSH3b showing higher lytic activities against S. epidermidis strains (Table 1). In addition to the lytic activity differences between species, host strain origin may also play a role in determining the spectrum of susceptible species. There is a trend for the HydH5-derived enzymes to show greater lysis against bovine-associated staphylococci than clinical isolates. This is consistent with the fact that phiIPLA88 was isolated from a dairy environment. The HydH5 parental enzyme is active on 4 of the 5 bovine isolates and is only active on 1 of the 5 human S. aureus isolates. The trend is diminished but still apparent with the fusion enzymes (HydH5Lyso and HydH5SH3b) (Table 1).
HydH5 and its fusions act synergistically with LysH5 against S. aureus.
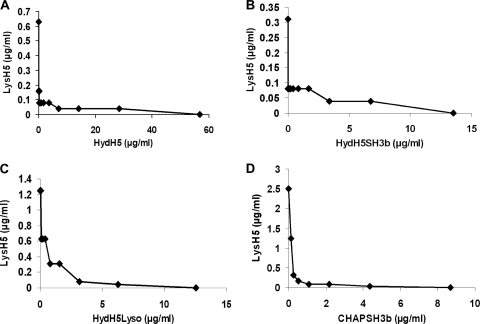
To determine the interaction of HydH5 and its derivative fusions with the endolysin LysH5, a standard checkerboard dilution assay was performed using live S. aureus Sa9 cells in HydH5 activity buffer. Initially, we determined the MLCs of the proteins; these were 56.7 μg/ml for HydH5, 12.5 μg/ml for HydH5Lyso, 13.5 μg/ml for HydH5SH3b, 8.7 μg/ml for CHAPSH3b, and 2.5 μg/ml for LysH5. When HydH5 or one of its derived fusions was combined with LysH5, a synergistic effect was observed in all combinations. Representative graphs of the synergistic interaction between LysH5 and HydH5 and between LysH5 and each HydH5-lysostaphin construct are shown in Fig. 3. The average ∑FIC values for the four protein combinations were calculated from the results of the checkerboard test (Fig. 3). All these values are indicative of strong synergistic interaction, and mixtures of these enzymes are more inhibitory for S. aureus growth.
Fig 3.
Synergistic effect in checkerboard assay between endolysin LysH5 and HydH5 fusion constructs. Minimum lytic concentrations (MLC) of each protein in the presence of subinhibitory concentrations of endolysin LysH5 are indicated. (A) HydH5, 56.7 μg/ml to 0.06 μg/ml, ∑FIC 0.065 ± 0.001. (B) HydH5SH3b, 13.5 μg/ml to 0.01 μg/ml, ∑FIC 0.130 ± 0.000. (C) HydH5Lyso, 25.1 μg/ml to 0.02 μg/ml, ∑FIC 0.049 ± 0.023. (D) CHAPSH3b, 17.34 μg/ml to 0.03 μg/ml, ∑FIC 0.063 ± 0.022.
DISCUSSION
With the goals of increasing the antimicrobial activity of HydH5 and determining the spectrum of species susceptible to lysis with the lytic domains of this enzyme, we performed deletion analysis and created multiple lytic domain fusion proteins with the most active HydH5 lytic domains. Through deletion analysis, we had identified in HydH5 the protein domains that are essential for its activity and showed that both the CHAP and LYZ2 domains possess lytic activity against S. aureus cells (40). Several reports have shown that endolysins can be truncated and some individual catalytic domains retain lytic activity, with some of them showing higher specific activity than the full-length protein (2, 4, 7, 17). However, as was previously shown, some endolysin catalytic domains are almost inactive, such as the glycosidase domains from the lambdaSa2 prophage endolysin (8) or the B30 endolysin (7) and the amidase domain from LysK (2). Our results indicated that the HydH5 CHAP domain construct has approximately 1.4-fold lower specific activity than the full-length HydH5, suggesting that the LYZ2 domain might also contribute to the full-length protein's activity.
We and others have shown previously that virion-associated PG hydrolases lacking a defined CWB domain can also bind to the cell surface (39, 40). Known CWB domains have been shown to be necessary for accurate cell wall recognition and subsequent lytic activity for some PG hydrolases, such as lysostaphin (1), ALE-1 (28), Listeria monocytogenes endolysins Ply118 and Ply500 (25), and Streptococcus pneumoniae CPL-1 (37). However, there are numerous reports of C-terminally deleted lysin constructs with deleted CWB domains where the N-terminal lytic domain maintains its activity in the absence of its CWB domain (2, 4, 7, 17, 41). Thus, a defined CWB domain is not a necessity for full lytic activity. This binding does not rule out the possibility that the inter-lytic domain region might harbor a heretofore-unidentified CWB domain, as suggested previously for the B30 endolysin (7), or that catalytic domains of both endolysins and virion-associated enzymes can recognize and bind to their substrate through amino acid residues in their active sites.
Although not always a necessity for baseline activity, some endolysins require a cell binding domain (CBD) to achieve high levels of activity (8). Data obtained with HydH5 and the derived fusions are consistent with these results. In the present work, we observed that the fusion of a CBD increased the activity in both the single CHAP domain and the full-length protein. This suggests that proteins in solution may need a CBD to optimally recognize their substrate. Despite these similarities, it should not be ignored that virion-associated enzymes are not predicted to be free in solution (as opposed to endolysins) but, rather, are expected to be bound into the matrix of the virion particle. In nature, CBDs on virion-associated enzymes might not be as essential as for endolysins because the phage particle might help deliver the virion-associated enzymes directly to their substrate. There are, of course, other explanations for CHAPSH3b having a higher level of activity than HydH5, such as a higher conformational stability of this construct. It is also possible to speculate that there might be some putative regulation exerted by one catalytic domain over the other (as described previously for the SH3b CBD [26]) or that a preferred substrate for one domain might be generated by the catalytic action of the other. Despite the potential similarities in function and structure of phage lytic proteins, it should not be ignored that the lytic activity of virion-associated PG hydrolases must be tightly regulated to avoid deleterious damage to the cell wall during the infection process.
Our hypothesis to justify the addition of a CWB domain considers the fact that a phage structural protein, HydH5, is probably brought into close proximity to the cell wall and the PG substrate by the intact virion. However, when considering HydH5 as a soluble single-protein antimicrobial, the scenario is quite different and the addition of a CWB domain might improve its lytic activity. In the HydH5SH3b construct, the lysostaphin SH3b domain was fused to full-length HydH5, which increased the lytic activity 1.7-fold, suggesting that the CWB domain helped the lytic domain degrade the PG substrate. A similar result was obtained when the native Cpl-7 cell wall binding domains of the streptococcal LambdaSa2 endolysin were replaced by the staphylococcal SH3b domain from lysostaphin or LysK, resulting in a 5-fold increase in staphylolytic activity of the LambdaSa2 CHAP endopeptidase domain (3). In the HydH5 CHAPSH3b construct, the lysostaphin SH3b domain was fused to the CHAP domain, resulting in a 4.8-fold increase over the lytic activity of the CHAP-only construct (CHAP) and a 3.3-fold increase compared to the lytic activity of full-length HydH5. These results indicate the importance of the SH3b CWB domain in recognizing the bacterial cell wall, taking into account that only one catalytic domain (CHAP) fused to the SH3b domain was sufficient to obtain the highest specific activity of the various constructs.
Moreover, addition of the full-length lysostaphin to HydH5 (HydH5Lyso) also achieved activity levels greater than those of the parental protein (HydH5). This result further indicates the modular nature of these protein domains and supports our hypothesis that the addition of new catalytic domains to HydH5 can improve its lytic activity. The three different catalytic domains are believed to each attack a different PG bond. The presence of multiple unique domains theoretically decreases the likelihood of the development of bacterial resistance (10). The increased activity also suggests that each domain is modular, as expected (12), and although the activity of the final construct does not achieve the sum of the parental lytic activities, each domain probably achieves a near-native conformation.
HydH5 and its derivative fusions were assessed for their ability to lyse a range of staphylococcal and nonstaphylococcal strains. No previous data have been reported regarding the lytic spectrum of S. aureus virion-associated PG hydrolases. All proteins were active against staphylococcal strains, but no other genus was lysed at detectable levels. Our results indicate a similar lytic spectrum for both HydH5 and its derivative fusions. These results are in agreement with descriptions to date of S. aureus endolysins that demonstrate a lytic spectrum limited to staphylococci. However, a different level of lytic activity was observed for each species/strain. In general, S. aureus strains were more sensitive to lysis than S. epidermidis strains. Within the S. aureus strains, we observed a trend wherein bovine strains were more sensitive than clinical strains. This could be due to the shared origin of these strains and the phage from which HydH5 originated, since both share a source from a dairy environment. A similar result was previously observed for endolysin LysH5 (34). Similarly, lysostaphin is known to have reduced activity against S. epidermidis compared to its activity against S. aureus (45). Although we do not yet have direct biochemical proof that all three lytic domains are functional in the HydH5Lyso construct, the reduced activity against S. epidermidis compared to the activity against S. aureus and the increased activity against human isolates while maintaining high activity against bovine strains are consistent with the combined lytic range of both HydH5 and lysostaphin and suggest that both protein fragments are functional in this fusion construct.
Synergistic interactions between antimicrobial compounds have the potential to be exploited in order to increase effectiveness against the target bacteria, thereby reducing the required dose of each antimicrobial compound while simultaneously decreasing the likelihood of the development of antimicrobial resistance. The combination of the two lytic bacteriophage enzymes HydH5 and LysH5 appears to have synergistic activity on S. aureus Sa9. HydH5-derived fusions also showed this positive interaction. The basis of synergy between PG hydrolases might be explained by the hydrolytic activity of one enzyme loosening the PG structure, thus facilitating better access of the second enzyme to its target or, potentially, by creating a better substrate for the second, “weaker” domain. It is encouraging that the strongest in vitro synergy was observed between LysH5 and HydH5Lyso, suggesting that this combination might be protective in vivo. However, in the absence of biochemical data to indicate which domains are active in the fusion and the parental LysH5, we are at a loss to fully identify those domains that are responsible for the observed antimicrobial synergy. However, these results are in agreement with previous studies indicating synergy between Pal and Cpl-1 lysins against S. pneumoniae (24) and the synergy between endolysin LysK and the bacteriocin lysostaphin against S. aureus. A 33% reduction in LysK concentration was obtained in the presence of lysostaphin (2). Another bacteriocin, nisin, enhanced the antimicrobial activity of LysH5 8-fold on S. aureus cell suspensions (13). Comparable results were obtained by combination with antibiotics (5, 29).
In summary, we report the development of novel chimeric PG hydrolases with improved lytic activities against S. aureus and S. epidermidis, including the MRSA strain N315. The effectiveness of HydH5 and its derivative fusions when used in combination with LysH5, another dairy-derived antistaphylococcal protein, were remarkable. We expect that these constructs will provide new weapons to combat multidrug-resistant S. aureus infections in both dairy and clinical environments.
ACKNOWLEDGMENTS
This research study was supported by nonfunded cooperative agreement no. 58-1265-0-092FN (Agricultural Research Service and Cooperator Instituto de Productos Lácteos de Asturias, CSIC, Spain) and by grants AGL2009-13144-C02-01 (Ministry of Science and Innovation, Spain), IB08-052 (Science, Technology and Innovation Programme, Principado de Asturias, Spain), and PIE200970I090 (CSIC, Spain). L.R.-R. is a fellow of the Science, Technology and Innovation Programme (Principado de Asturias, Spain). This work was supported in part by NIH grant 1RO1AI075077-01A1, NRI grant 2007-35204-18395, and U.S. State Department funds, all awarded to D.M.D.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print 20 January 2012
REFERENCES
- 1.Baba T, Schneewind O. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789–4797 [PMC free article] [PubMed] [Google Scholar]
- 2.Becker SC, Foster-Frey J, Donovan DM. 2008. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 287:185–191 [DOI] [PubMed] [Google Scholar]
- 3.Becker SC, et al. 2009. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol. Lett. 294:52–60 [DOI] [PubMed] [Google Scholar]
- 4.Cheng Q, Fischetti VA. 2007. Mutagenesis of a bacteriophage lytic enzyme PlyGBS significantly increases its antibacterial activity against group B streptococci. Appl. Microbiol. Biotechnol. 74:1284–1291 [DOI] [PubMed] [Google Scholar]
- 5.Daniel A, et al. 2010. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:1603–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbrück M. 1940. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 23:643–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan DM, et al. 2006. The cell lysis activity of the Streptococcus agalactiae bacteriophage B30 endolysin relies on the cysteine, histidine-dependent amidohydrolase/peptidase domain. Appl. Environ. Microbiol. 72:5108–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donovan DM, Foster-Frey J. 2008. LambdaSa2 prophage endolysin requires Cpl-7-binding domains and amidase-5 domain for antimicrobial lysis of streptococci. FEMS Microbiol. Lett. 287:22–33 [DOI] [PubMed] [Google Scholar]
- 9.Fenton M, Ross P, McAuliffe O, O'Mahony J, Coffey A. 2010. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs 1:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti VA. 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13:491–496 [DOI] [PubMed] [Google Scholar]
- 11.Fischetti VA. 2010. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García P, García JL, García E, Sánchez-Puelles JM, López R. 1990. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene 86:81–88 [DOI] [PubMed] [Google Scholar]
- 13.García P, Martínez B, Rodríguez L, Rodríguez A. 2010. Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. Int. J. Food Microbiol. 141:151–155 [DOI] [PubMed] [Google Scholar]
- 14.Gu J, et al. 2011. LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 49:111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall MJ, Middleton RF, Westmacott D. 1983. The fractional inhibitory concentration (FIC) index as a measure of synergy. J. Antimicrob. Chemother. 11:427–433 [DOI] [PubMed] [Google Scholar]
- 16.Hermoso JA, García JL, García P. 2007. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 10:461–472 [DOI] [PubMed] [Google Scholar]
- 17.Horgan M, et al. 2009. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 75:872–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshiba H, et al. 2010. Isolation and characterization of a novel Staphylococcus aureus bacteriophage, phiMR25, and its therapeutic potential. Arch. Virol. 155:545–552 [DOI] [PubMed] [Google Scholar]
- 19.Kanamaru S, et al. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553–557 [DOI] [PubMed] [Google Scholar]
- 20.Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köck R, et al. 2010. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 15:19688. [DOI] [PubMed] [Google Scholar]
- 22.Kusuma CM, Kokai-Kun JF. 2005. Comparison of four methods for determining lysostaphin susceptibility of various strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3256–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laird MW, et al. 2005. Optimization of BLyS production and purification from Escherichia coli. Protein Expr. Purif. 39:237–246 [DOI] [PubMed] [Google Scholar]
- 24.Loeffler JM, Fischetti VA. 2003. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob. Agents Chemother. 47:375–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335–349 [DOI] [PubMed] [Google Scholar]
- 26.Low LY, Yang C, Perego M, Osterman A, Liddington RC. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 280:35433–35439 [DOI] [PubMed] [Google Scholar]
- 27.Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 28.Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, Sakon J. 2006. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 281:549–558 [DOI] [PubMed] [Google Scholar]
- 29.Manoharadas S, Witte A, Bläsi U. 2009. Antimicrobial activity of a chimeric enzybiotic towards Staphylococcus aureus. J. Biotechnol. 139:118–123 [DOI] [PubMed] [Google Scholar]
- 30.Matsuzaki S, et al. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J. Infect. Dis. 187:613–624 [DOI] [PubMed] [Google Scholar]
- 31.Matsuzaki S, et al. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 11:211–219 [DOI] [PubMed] [Google Scholar]
- 32.Moak M, Molineux IJ. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169–1183 [DOI] [PubMed] [Google Scholar]
- 33.Molineux IJ. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1–8 [DOI] [PubMed] [Google Scholar]
- 34.Obeso JM, Martínez B, Rodríguez A, García P. 2008. Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 128:212–218 [DOI] [PubMed] [Google Scholar]
- 35.O'Flaherty S, Ross RP, Coffey A. 2009. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33:801–819 [DOI] [PubMed] [Google Scholar]
- 36.Otto M. 2010. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64:143–162 [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Dorado I, et al. 2007. Elucidation of the molecular recognition of bacterial cell wall by modular pneumococcal phage endolysin CPL-1. J. Biol. Chem. 282:24990–24999 [DOI] [PubMed] [Google Scholar]
- 38.Rashel M, et al. 2007. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J. Infect. Dis. 196:1237–1247 [DOI] [PubMed] [Google Scholar]
- 39.Rashel M, et al. 2008. Tail-associated structural protein gp61 of Staphylococcus aureus phage ΦMR11 has bifunctional lytic activity. FEMS Microbiol. Lett. 284:9–16 [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez L, et al. 2011. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol. 11:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sass P, Bierbaum G. 2007. Lytic activity of recombinant bacteriophage phi11 and phi12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl. Environ. Microbiol. 73:347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinbacher S, et al. 1997. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267:865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takac M, Blasi U. 2005. Phage P68 virion-associated protein 17 displays activity against clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 49:2934–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wills QF, Kerrigan C, Soothill JS. 2005. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob. Agents Chemother. 49:1220–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zygmunt WA, Browder HP, Tavormina PA. 1968. Susceptibility of coagulase-negative staphylococci to lysostaphin and other antibiotics. Appl. Microbiol. 16:1168–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]