Abstract
The ancient soilborne plant vascular pathogen Ralstonia solanacearum has evolved and adapted to cause severe damage in an unusually wide range of plants. In order to better describe and understand these adaptations, strains with very similar lifestyles and host specializations are grouped into ecotypes. We used comparative genomic hybridization (CGH) to investigate three particular ecotypes in the American phylotype II group: (i) brown rot strains from phylotypes IIB-1 and IIB-2, historically known as race 3 biovar 2 and clonal; (ii) new pathogenic variants from phylotype IIB-4NPB that lack pathogenicity for banana but can infect many other plant species; and (iii) Moko disease-causing strains from phylotypes IIB-3, IIB-4, and IIA-6, historically known as race 2, that cause wilt on banana, plantain, and Heliconia spp. We compared the genomes of 72 R. solanacearum strains, mainly from the three major ecotypes of phylotype II, using a newly developed pangenomic microarray to decipher their population structure and gain clues about the epidemiology of these ecotypes. Strain phylogeny and population structure were reconstructed. The results revealed a phylogeographic structure within brown rot strains, allowing us to distinguish European outbreak strains of Andean and African origins. The pangenomic CGH data also demonstrated that Moko ecotype IIB-4 is phylogenetically distinct from the emerging IIB-4NPB strains. These findings improved our understanding of the epidemiology of important ecotypes in phylotype II and will be useful for evolutionary analyses and the development of new DNA-based diagnostic tools.
INTRODUCTION
Ralstonia solanacearum (Smith) Yabuuchi et al. (45), a highly destructive and widespread bacterial plant pathogen, is surely one of the most successful vascular bacteria. This soilborne xylem inhabitant encompasses thousands of different strains distributed worldwide and causes bacterial wilt disease in more than 50 botanical families (21). As a highly genetically and phenotypically heterogeneous plant pathogen species, R. solanacearum is an excellent case for study in order to understand genomic evolution mechanisms in general and plant adaptation in particular. Classification of R. solanacearum has undergone many changes during the past 20 years. Historically, the biodiversity of strains was characterized by the race and biovar system, based on phenotypic traits (2–4). Nevertheless, this classification evolved with the multiple techniques employed to assay genomic differences among closely related bacterial species, strains, or lineages. The new tools developed during the genomics era, combined with phenotypic and geospatial data, allow the assessment of relationships between widely heterogeneous organisms as regards their physiology, ecology, and gene content (7, 35). The latest hierarchical classification scheme based on partial sequence analysis resulted in better understanding of the phylogeny within this species complex; classifications were unified into four distinct phylotypes, relating to the geographical origins of strains (13, 41). This phylotype classification largely correlates with the geographic origin and evolutionary past of strains (41), which are assigned to Asian (phylotype I), American (phylotype II), African (phylotype III), and Indonesian (phylotype IV) phylotypes. Phylotype IV hosts the two closely related species Ralstonia syzygii (the agent of Sumatra disease of clove) and the banana blood disease bacterium (BDB) (36, 40).
The breakthrough of DNA-based technologies brought new insights into the diversity and evolution of pathogens (22), and novel classifications emerged. R. solanacearum is no exception; many studies revisited its diversity at the genus level (8, 9, 40), the species complex level (14, 26, 41), or the ecological level (10, 37, 39). Hence, as a high-throughput alternative to the full sequencing of entire bacterial genomes, the pangenomic comparative genomic hybridization (CGH) microarray developed in this study was designed for fine investigation of the phylogenetic diversity of R. solanacearum.
A former study used a CGH microarray approach to estimate the distribution of genes among 18 R. solanacearum strains distributed in the phylogeny (19). That microarray was designed from the GMI1000 genome sequence (phylotype I) and encompassed about 5,000 oligonucleotides. Data from the whole genome confirmed the distribution of the phylogeny into four distinct phylotypes and brought a first estimation of the core genome content. However, the design of that CGH microarray was restricted to one phylotype I strain only, inducing a bias against estimating specific genes of the three other phylotypes. On the basis of that study, going one step further, we chose here to develop a pangenomic microarray from available sequenced genomes of six strains, distributed in the phylogeny of R. solanacearum.
As a first step, a genomic database was constituted around R. solanacearum, three genomes of which were recently fully sequenced and annotated (30), in addition to the three other genomes already available (18, 31). In addition to confirming the previous phylotype classification, this work highlighted the remarkable heterogeneity of this bacterial species between phylotypes and the probable need to further reshape its classification into at least three genomic species (30). Also, since the ancestral Ralstonia prototype is assumed to be a plant pathogen (15, 19), study of the extent of the diversity of this genus, along with the evolutionary pathways involved, is of major importance for a broad understanding of pathogen evolution.
We hypothesized that there was much more biodiversity to discover from genomic analysis of phylotype II strains of R. solanacearum, namely, cold-tolerant potato brown rot strains (IIB-1), previously recognized to be clonal after various neutral marker approaches (5, 27, 34), tropical Moko disease-causing strains (IIB-4), and emerging strains (IIB-4NPB). We thus focus on phylogenetically closely related phylotype II groups of strains with well-characterized and diverging ecological and phenotypical traits, in an attempt to reconstruct their epidemiological pathways along with the acquisition of their lifestyles.
MATERIALS AND METHODS
Bacterial strains.
A set of 72 R. solanacearum strains was selected to cover the known genetic diversity within the R. solanacearum species complex, especially in phylotype II (n = 60) (see Table S1 in the supplemental material). Pathotypes of strains were previously assessed (5) on genetic resources obtained from potato (Solanum tuberosum), tomato (Solanum lycopersicum), eggplant (Solanum melongena), and banana (Musa spp.). Ralstonia pickettii strain LMG5942T was included as an outgroup. Strains were obtained from different bacterial collections maintained at the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD; Saint Pierre, Réunion Island; Le Lamentin, Martinique, French West Indies), Laboratoire National de la Protection des Végétaux (LNPV; Angers, France), Institut National de la Recherche Agronomique (INRA; Rennes, France), University of Queensland (Brisbane, Australia), and Collection Française de Bactéries Phytopathogènes (CFBP; Angers, France).
Probe design and microarray manufacture.
Biological probes (n = 10,761) were designed from six sequenced and fully annotated genomes of R. solanacearum: GMI1000 (31), Molk2 and IPO1609 (18), CMR15, CFBP2957, and PSI07 (30). These six genomic sequences are publicly available through the online MaGe interface (www.genoscope.cns.fr/agc/microscope/about/collabprojects.php?P_id=67). Probe design was performed by Imaxio (Clermont-Ferrand, France) using the following strategy. Two 60-mer probes were designed from each coding sequence (CDS). Three groups of probes, called “specific,” “core,” or “variable” according to their target specificity, were constituted. The “specific” group was composed of probes specifically targeting a strain; these probes were encoded as “CFBP,” “CMR,” “GMI,” “IPO,” “MOLK,” or “PSI” depending on the target. The “core” group comprised probes targeting all orthologous genes in the six sequenced genomes. Finally, the “variable” group was composed of probes targeting genes in at least two, but not all, sequenced genomes. To avoid redundancy on orthologous genes, probe filtering was performed using the BLASTN algorithm (1), based on an 80% minimum match, a melting temperature (Tm) of 77°C ± 9°C, and a G+C content of 57% ± 20% (see Fig. S1 in the supplemental material). Hence, only one probe was designed per CDS or among orthologous CDSs. Microarrays were manufactured by Agilent Technologies (Santa Clara, CA) using in situ synthesis. The final set of probes, randomly implemented on the array surface, was composed of 3,317 “core,” 3,631 “variable,” and 3,963 “specific” probes. Hybridization quality and interslide reproducibility were assessed with 10 replicates of 300 biological probes: 275 “core” group and 25 “variable” group probes, along with the microarray manufacturer controls. Negative controls were also added, with 35 probes designed to target the close relative Cupriavidus taiwanensis and 7 random-sequence-based probes designed to be noncomplementary with any sequenced R. solanacearum genomes.
DNA labeling and hybridization.
Overnight liquid cultures were pelleted at 5,400 rpm and were washed with 500 ml of 1 M NaCl before genomic DNA was purified by using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Genomic DNA was labeled with using Cy3 or Cy5 dye, according to the method of Guidot et al. (19).
Labeled products were purified by using a CyScribe GFX purification kit (GE Healthcare, Bucks, United Kingdom) according to the manufacturer's recommendations. DNA was adjusted at a concentration of 70 ng · μl−1 in high-performance liquid chromatography (HPLC)-grade water using a NanoDrop 8000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), for an average dye labeling concentration of 12 pmol · liter−1. Hybridizations were done overnight (for approximately 16 h) in the G2545A microarray hybridization oven (Agilent Technologies) by following the 8×15K Custom CGH microarray protocol from Agilent Technologies (reference G4410-90010).
Hybridizations of the six sequenced genomes were repeated three times to assess interslide reproducibility, and strain CMR15 genomic DNA labeled with Cy5 was included in each hybridization as a reference for further reproducibility tests. Hybridizations of the other strains were not repeated to verify reproducibility (see Results).
Image scanning and data analysis.
Slides were scanned at a 5-μm resolution using the G2565CA scanner managed by Scan Control software, version 8.5 (Agilent Technologies). Data were extracted from the 16-bit tagged-image format file (TIFF) image with Feature Extraction software, version 10.5.1.1 (Agilent Technologies).
Data manipulation and statistical analysis were performed with the free statistical software R, version 2.11.1 (29). Spot intensities were calculated as the difference between the median foreground and background intensities, normalized according to the standard normal deviation method, along with a base 2 logarithm. The dispersion of the mass distribution was assessed using a kernel density method with a Gaussian kernel (32, 33). To distinguish between the positive and negative responses, a response threshold was estimated independently for each hybridization as the intensity value of the minimum density between the two peaks of the spot distribution. This response threshold allowed us to estimate two responses of probes based on their binary signals: a signal of 1 represented the presence of a gene, and a signal of zero represented its absence. All further analyses were performed on this binary data set.
Phylogeny and population structure reconstruction.
The phylogeny of R. solanacearum was reconstructed using MrBayes, version 3.2, with the binary evolution model implemented in and allowing for variation of substitution rates among sites. Two runs with four Markov chains were conducted simultaneously for 3,000,000 generations starting from random initial trees, sampled every 500 generations. Variations in the maximum-likelihood (ML) scores for these samples were examined graphically with Tracer, version 1.5 (A. Rambaut and A. J. Drummond, 2007; http://beast.bio.ed.ac.uk/Tracer). After discarding of the trees generated prior to the stabilization of ML scores (burn in, 10%), the consensus phylogeny and posterior probability of the nodes were determined. Trees were edited using FigTree, version 1.3.1 (A. Rambaut, 2007; http://tree.bio.ed.ac.uk/software/figtree/).
Population structure was estimated using STRUCTURE, version 2.3.3 (28). This iterative-model-based analysis aimed to assess the population structure with the assignment of individuals to “K” clusters, allowing for admixture. To infer the number of groups, a fully Bayesian process (28) was run with different values for the number of clusters (K). Analysis lengths were set to a burning period of 50,000 iterations followed by 100,000 iterations of simulation. A total of 20 independent simulations were performed, with K ranging from 1 to 10. STRUCTURE would attribute a probability, Pr(X|K) given the data (X), and the log Pr(X|K) was used to determine the more likely number of clusters by following the method described by Evanno et al. (11). STRUCTURE software also gives the assignment probabilities of each individual for each cluster, and we then inferred the most probable groups for each individual.
RESULTS
Microarray validation.
The specificity and sensitivity of the pangenomic CGH microarray were estimated through the gene content by comparing results from the genome sequences and microarray probing. First, all probes designed as negative controls were always given a negative response. The data showed that more than 98.1% of the genes found by the sequencing of the six reference strains were retrieved by the pangenomic microarray, but 1.9% of probe responses could be considered false-negative responses. The proportion of false-positive probes was estimated to be lower than 4.8% and corresponded to positive probes matching a genome different from that for which they were designed.
The reproducibility of microarray hybridizations was assessed using the replicated probes, including the 300 replicated probes on each array, which showed an interslide reproducibility of 97.6%, and the 85 replicated CMR15 (III-29) strains labeled with Cy5, which showed an interslide reproducibility of 93.4% and an intraslide reproducibility of 99.6% on the same array. Comparison of the 3 replicates of Cy3-labeled CMR15 with the 3 replicates of Cy5-labeled CMR15 showed a reproducibility of 96.7%.
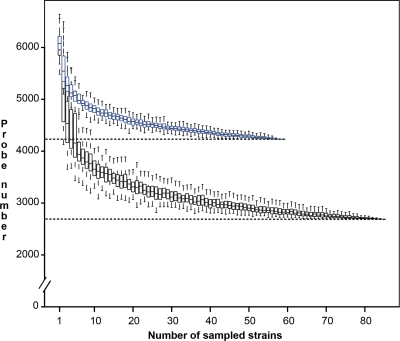
Depletion curves (Fig. 1) represent the relation between the number of strains hybridized on the pangenomic array and the number of positive probes. Curves were computed and plotted for all R. solanacearum strains (black curve) and phylotype IIB strains (blue curve). In both curves, after a massive decrease in the number of positive probe responses for the first 10 strains sampled, the number of positive probes tended to stabilize with an increasing number of strains tested and reached a plateau at 2,690 probes for all strains and 4,228 probes for the IIB strains.
Fig 1.
Depletion curves of the core probe set. The depletion of the core probe set was analyzed for phylotype IIB strains (n = 59) (blue whisker plots) and for the four Ralstonia solanacearum phylotypes (n = 85) (black whisker plots). Each whisker plot represents the distribution of the number of positive probes for a strain of the tested group (size given on the horizontal axis) after 100 resamplings.
The array core probes (ArCP) revealed by the pangenomic microarray represented 24.7% (n = 2,690) of the biological probes and targeted mainly chromosomal genes (83.0%). By focusing on the minimal gene set described by Gil et al. (16), which includes well-conserved housekeeping genes for basic metabolism and macromolecular synthesis, 199 out of 205 genes were found to be targeted by at least one probe from the ArCP group.
Phylogeny in the Ralstonia solanacearum species complex.
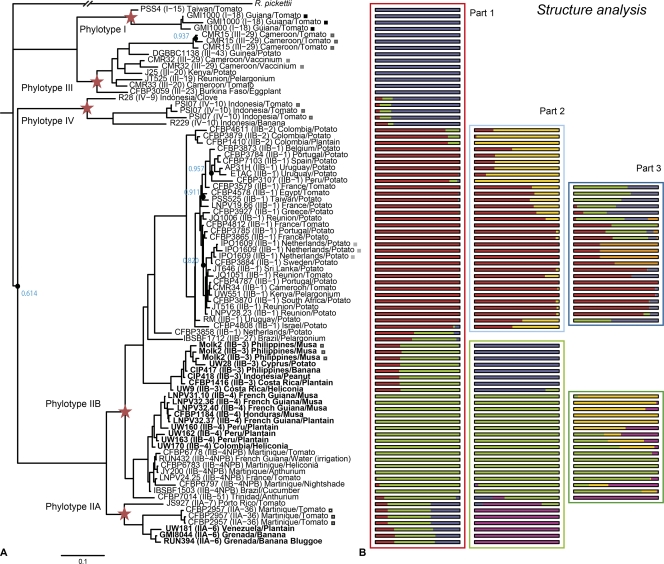
Phylogenetic reconstruction showed with high statistical support that strains are distributed into three major clusters and many subdivisions, reflecting the revised scheme for division into phylotypes and sequevars (30) (Fig. 2A). All the phylotypes were characterized as monophyletic in the tree, and all replicated hybridizations were clustered together. R. pickettii strain LMG5942T clearly constituted an outlier from the R. solanacearum species complex and was assigned as an outgroup. At the phylotype level, three distinct clusters were described: phylotype IV on one side, phylotypes I and III grouping together, and phylotype II constituting the last cluster. A closer look within phylotypes reveals, with high statistical support, that three strains clearly constituted outliers from their sequevars: strain CMR15 (III-29), strain CFBP6797 (IIB-4NPB), and strain CFBP3858 (IIB-1). Reconstruction of the population structure, performed with STRUCTURE software, also revealed three clusters (Fig. 2B, part 1) but grouped phylotypes I, III, and IV together in a first cluster (blue bars), phylotypes IIB-1 and IIB-2 in a second cluster (red bar), and the other sequevars in a last cluster (green bar). The analysis also revealed admixed signatures of individuals (i.e., their interbreed origins) by showing a mixed proportion of estimated membership probability for at least two groups. The two strains CFBP3858 (IIB-1) and IBSBF1712 (IIB-27) displayed this hybrid profile between the second cluster, to which they belong, and the third cluster, composed of other phylotype II strains. Phylotype IIA strains also showed signatures with the three clusters admixed.
Fig 2.
(A) Phylogenetic tree of 72 Ralstonia solanacearum strains, computed by MrBayes, version 3.2. Blue numbers and black circles represent statistical support levels below 0.99; red stars indicate the main node for every phylotype. For each strain, the phylotype and sequevar (in parentheses) are given, along with the country and host of isolation. Moko disease strains are shown in boldface. Brown rot strains belong to phylotypes IIB-1 and IIB-2. Squares indicate repetitions of genomic DNA hybridization. (B) Population structure analysis. Each box represents an independent analysis of all R. solanacearum strains (box outlined in red), brown rot strains from phylotypes IIB-1 and IIB-2 (boxes outlined in light or dark blue), or Moko disease-causing strains from phylotypes IIA-6, IIB-3, and IIB-4 (boxes outlined in light or dark green). Within an analysis, each solid bar represents the proportions of ancestral nucleotides inherited from each of the inferred ancestral populations.
Phylogeny of brown rot strains.
The diversity and structure of ecological groups, such as brown rot- and Moko disease-causing strains, were further investigated using the same approach. Brown rot strains were distributed into sequevars 1 and 2 (Fig. 2A; Table 1), which are both monophyletic. Analysis revealed a first partition into two groups, where sequevar 2 strains grouped with sequevar 1 strains (Table 1; Fig. 2B, part 2, top box, yellow bars). Those strains originated mainly in the Andean region. The other cluster, composed only of sequevar 1 strains (Fig. 2B, part 2, top box, red bars), was found to be subdivided into at least five clusters reflecting their geographical areas of isolation (Table 1; Fig. 2B, part 3, top box). One group was associated with the African and Indian Ocean regions (Table 1, subcluster A; Fig. 2B, part 3, top box, red bars); two groups were associated with the European and Mediterranean regions (Table 1, subclusters B and C; Fig. 2B, part 3, top box, green and blue bars); and one group was associated with Northern Europe (Table 1, subcluster D; Fig. 2B, part 3, top box, orange bars).
Table 1.
Bayesian inference of phylogeny and structure reconstruction analysis for brown rot strains from phylotypes IIB-1 and IIB-2
| Strain | Phylotypea | Sequevara | Country | Host | Phylogenyb | Structurec |
|
|---|---|---|---|---|---|---|---|
| Major cluster | Subcluster | ||||||
| CFBP4787 | IIB | 1 | Portugal | Potato | AIO | 1 | A |
| UW551 | IIB | 1 | Kenya | Pelargonium | AIO | 1 | A |
| CMR34 | IIB | 1 | Cameroon | Tomato | AIO | 1 | A |
| JT516 | IIB | 1 | Réunion | Potato | AIO | 1 | A |
| CFBP3870 | IIB | 1 | South Africa | Potato | AIO | 1 | A |
| JT646 | IIB | 1 | Sri Lanka | Potato | AIO | 1 | A |
| LNPV28.23 | IIB | 1 | Réunion | Potato | AIO | 1 | A |
| JQ1051 | IIB | 1 | Réunion | Tomato | AIO | 1 | A |
| CFBP3865 | IIB | 1 | France | Potato | EuMr | 1 | A |
| CFBP3785 | IIB | 1 | Portugal | Potato | EuMr | 1 | B |
| CFBP4812 | IIB | 1 | France | Tomato | EuMr | 1 | B |
| JQ1006 | IIB | 1 | Réunion | Potato | EuMr | 1 | B |
| CFBP3579 | IIB | 1 | France | Tomato | EuMr | 1 | B |
| CFBP4578 | IIB | 1 | Egypt | Tomato | EuMr | 1 | B |
| PSS525 | IIB | 1 | Taiwan | Potato | EuMr | 1 | B |
| LNPV19.66 | IIB | 1 | France | Potato | EuMr | 1 | C |
| RM | IIB | 1 | Uruguay | Potato | EuMr | 1 | C |
| CFBP3927 | IIB | 1 | Greece | Potato | EuMr | 1 | C |
| CFBP3884 | IIB | 1 | Sweden | Potato | North | 1 | D |
| IPO1609 | IIB | 1 | Netherlands | Potato | North | 1 | D |
| AP31H | IIB | 1 | Uruguay | Potato | Andean | 2 | |
| CFBP3784 | IIB | 1 | Portugal | Potato | Andean | 2 | |
| CFBP3873 | IIB | 1 | Belgium | Potato | Andean | 2 | |
| CFBP7103 | IIB | 1 | Spain | Potato | Andean | 2 | |
| ETAC | IIB | 1 | Uruguay | Potato | Andean | 2 | |
| CFBP3107 | IIB | 1 | Peru | Potato | Andean | 2 | |
| CFBP4808 | IIB | 1 | Israel | Potato | EuMr | 2 | |
| CFBP1410 | IIB | 2 | Colombia | Banana, plantain | 2 | ||
| CFBP3879 | IIB | 2 | Colombia | Potato | 2 | ||
| CFBP4611 | IIB | 2 | Colombia | Potato | 2 | ||
| CFBP3858 | IIB | 1 | Netherlands | Potato | Out | ND | |
Determined by sequencing of the partial endoglucanase (egl) gene (13).
Cluster determined from the phylogenetic reconstruction performed using MrBayes, version 3.2 (Fig. 2). Designations reflect the main origin of strains within each cluster: AIO, African–Indian Ocean regions; EuMr, European and Mediterranean regions; North, Northern European region; Andean, Latin American region. “Out” refers to the outer phylogenetic position of the strain compared to its sequevar.
Statistical groups were obtained by a Bayesian population structure assessment analysis with STRUCTURE software for numbers of clusters (K) ranging from 1 to 10. The major population was assessed as having 2 clusters, according to the likelihood of assignment decreasing from a ΔK value of 74 for 1 cluster to 1.44 for 6 clusters. Cluster 1 was assigned with an average inference of 0.872, and cluster 2 was assigned with an average inference of 0.799. The subpopulation was assessed as having 4 clusters, according to the likelihood of assignment increasing from a ΔK value of 11.210 for 1 cluster to 25.814 for 3 clusters, and then decreasing until K reached a value of 10. Subcluster A was assigned with an average inference of 0.837; subcluster B, with an average inference of 0.576; subcluster C, with an average inference of 0.554; and subcluster D, with an average inference of 0.693. ND, not done.
Moko disease-causing strains and phylogeny of emerging strains.
Moko disease-causing strains are partitioned into phylotypes IIA-6, IIB-3, and IIB-4; thus, this ecotype is not considered monophyletic. Analysis of this ecotype, along with emergent strains from phylotype IIB-4NPB and IIB-51, and other phylotype IIA strains, revealed a partition into three clusters (Table 2; Fig. 2B, part 2, bottom box), where phylotype IIB-3 strains constituted a first cluster (blue bars); phylotype IIB-4, IIB-4NPB, and IIB-51 strains constituted a second cluster (green bars); and phylotype IIA strains constituted a third cluster (purple bars). Focusing on the second cluster revealed a partition into three groups (Table 2; Fig. 2B, part 3, bottom box), where Moko disease-causing strains from sequevar 4 constituted a first cluster (yellow bars), distinct from emerging strains from sequevar 4NPB, grouped into a second cluster (green bar). Finally, strains UW162 (IIB-4), CFBP6797 (IIB-4NPB), and CFBP7014 (IIB-51) constituted a third cluster (Fig. 2B, part 3, bottom box, purple bars). Surprisingly, the Moko disease-causing sequevar 4 was split into two groups: strains UW162, UW163, and UW170 (Table 2, phylogeny 4A) were phylogenetically closer to the emerging sequevar 4NPB than the other sequevar 4 strains (Table 2, phylogeny 4B).
Table 2.
Bayesian inference of phylogeny and structure reconstruction analysis for Moko disease-causing strains from phylotypes IIA-6, IIB-3, IIB-4, IIB-4NPB, and IIB-51
| Strain | Phylotypea | Sequevara | Country | Host | Phylogenyb | Structurec |
|
|---|---|---|---|---|---|---|---|
| Major cluster | Subcluster | ||||||
| CFBP1416 | IIB | 3 | Costa Rica | Banana, plantain | 1 | ||
| CIP417 | IIB | 3 | Philippines | Banana | 1 | ||
| CIP418 | IIB | 3 | Indonesia | Peanut | 1 | ||
| Molk2 | IIB | 3 | Philippines | Musa | 1 | ||
| UW28 | IIB | 3 | Cyprus | Potato | 1 | ||
| UW9 | IIB | 3 | Costa Rica | Heliconia | 1 | ||
| UW170 | IIB | 4 | Colombia | Heliconia | 4A | 2 | a |
| CFBP6778 | IIB | 4NPB | Martinique | Tomato | NPB | 2 | a |
| CFBP6783 | IIB | 4NPB | Martinique | Heliconia | NPB | 2 | a |
| RUN432 | IIB | 4NPB | French Guiana | Water (irrigation) | NPB | 2 | a |
| IBSBF1503 | IIB | 4NPB | Brazil | Cucumber | NPB | 2 | a |
| LNPV24.25 | IIB | 4NPB | France | Tomato | NPB | 2 | a |
| JY200 | IIB | 4NPB | Martinique | Anthurium | NPB | 2 | a |
| CFBP1184 | IIB | 4 | Honduras | Musa | 4B | 2 | b |
| LNPV31.10 | IIB | 4 | French Guiana | Musa | 4B | 2 | b |
| LNPV32.36 | IIB | 4 | French Guiana | Musa | 4B | 2 | b |
| LNPV32.37 | IIB | 4 | French Guiana | Banana, plantain | 4B | 2 | b |
| LNPV32.40 | IIB | 4 | French Guiana | Musa | 4B | 2 | b |
| UW160 | IIB | 4 | Peru | Banana, plantain | 4B | 2 | b |
| UW163 | IIB | 4 | Peru | Banana, plantain | 4A | 2 | b |
| UW162 | IIB | 4 | Peru | Banana, plantain | 4A | 2 | c |
| CFBP6797 | IIB | 4NPB | Martinique | American nightshade | NPB | 2 | c |
| CFBP7014 | IIB | 51 | Trinidad | Anthurium | 2 | c | |
| GMI8044 | IIA | 6 | Grenada | Banana | 3 | ||
| RUN394 | IIA | 6 | Grenada | Banana, bluggoe | 3 | ||
| UW181 | IIA | 6 | Venezuela | Banana, plantain | 3 | ||
| JS927 | IIA | 7 | Porto Rico | Tomato | 3 | ||
| CFBP2957 | IIA | 36 | Martinique | Tomato | 3 | ||
| CFBP2957 | IIA | 36 | Martinique | Tomato | 3 | ||
| CFBP2957 | IIA | 36 | Martinique | Tomato | 3 | ||
Determined by sequencing of the partial endoglucanase (egl) gene (13).
Cluster determined from the phylogenetic reconstruction performed using MrBayes, version 3.2.
Statistical groups were obtained by a Bayesian population structure assessment analysis with STRUCTURE software for numbers of clusters (K) ranging from 1 to 10. The major population was assessed as having 3 clusters, according to the likelihood of assignment increasing from a ΔK value of 1.858 for 2 clusters to 1,401.232 for 3 clusters, and then decreasing until K reached a value of 10. Cluster 1 was assigned with an average inference of 0.980; cluster 2, with an average inference of 0.976; and cluster 3, with an average inference of 0.956. The subpopulation was assessed as having 3 clusters, according to the likelihood of assignment increasing from a ΔK value of 0.948 for 2 clusters to 98.391 for 3 clusters, and then decreasing until K reached a value of 10. Subcluster a was assigned with an average inference of 0.881; subcluster b, with an average inference of 0.827; and subcluster c, with an average inference of 0.677.
Comparative genomic hybridizations.
Analysis of the gene content revealed by the pangenomic microarray highlighted differences among phylotype II ecotypes, but no gene repertories were found to be associated with the pathogenicity traits of the different ecotypes. Nonetheless, two particular groups of genes were observed. A zonula occludens toxin that increases cell permeability, leading to the disassembly of intercellular tight junctions, was found in every brown rot strain from phylotype IB-1. This toxin gene was also found in strains CMR15 (III-29) and K60 (IIA-7). A total of three out of eight genes of the rhi-type antimitotic toxin operon were found in every emerging strain from phylotype IIB-4NPB. Variable numbers of genes in this operon were found in phylotype IIB-4 and IIA strains PSI07 (IV-10), CFBP3059 (III-23), and DGBBC1138 (III-43).
DISCUSSION
Exploration of the diversity within almost clonal strains of R. solanacearum was necessary in order to understand their relationships and the evolutionary mechanisms that shaped this successful plant pathogen. Hence, we analyzed the relationships of the 72 R. solanacearum strains, along with R. pickettii strain LMG5942T as the outgroup strain, using statistical methods for phylogeny reconstruction and population structure inference. It is important here to point out one of the particular features of this second analysis. The STRUCTURE software aims to infer groups of individuals that are under the Hardy-Weinberg equilibrium and linkage equilibrium. While this assumption seems clearly valid for phylotypes I, III, and IV, concerns can be raised regarding phylotype II. This phylotype was recently estimated to be partly clonal, which may induce a bias into the population structure inference. The effect of complete population clonality would be that the inferred clustering of individuals would reflect subdivisions of the diversity, as a tree would do, rather than actual populations (12, 24). It remains difficult to estimate the degree of disruption caused in our inference, and one must be cautious when interpreting those particular results.
Unsurprisingly, major divisions and subdivisions of the phylogeny were congruent with the revised phylotype/sequevar scheme obtained from genome analysis and with previous CGH microarray or genomic data analyses (19, 30, 44) that support the subdivision of the R. solanacearum species complex into three different groups: phylotypes I and III, phylotype II, and phylotype IV. In the population structure analysis, phylotype IV appeared to be associated with phylotypes I and III, while phylotype II was divided into two groups. This result was quite surprising in that a separation between phylotypes I and III, on the one hand, and phylotype IV, on the other hand, was expected. In fact, phylotype IV presents some admixed signals until K reaches 5, when phylotype IV splits from phylotypes I and III (data not shown).
Topologies were also congruent regardless of the data set targeted by the probes used: the whole genome, the chromosome (see Fig. S2 in the supplemental material), the megaplasmid (see Fig. S3), and type III effectors (see Fig. S4) or, more generally, genes recognized to be involved into pathogenicity (see Fig. S5). This is consistent with the hypothesis of a long coevolution of the two replicons (6, 15, 19) and the ancestral character of R. solanacearum as a plant pathogen (15). Finally, the results confirmed the main repartition of core genes on the chromosome (19, 30, 31), but no major gene repertories explained the known phylogeny of R. solanacearum, other than the zonula occludens toxin in brown rot IIB-1 strains and the rhi-type toxin operon in emerging IIB-4NPB strains. This suggests, rather than a gene content difference, a fine-tuning of gene expression and regulation; nonetheless, many genes were associated with unknown and uncharacterized proteins.
Brown rot ecotype strains.
The pangenomic microarray approach developed in this study was successful in distinguishing between phylogenetic positions relative to brown rot strains from phylotype IIB-1, Moko disease-causing strains from phylotype IIB-4, and emerging strains from phylotype IIB-4NPB. The investigation first focused on cold-tolerant phylotype IIB-1 strains, which represent a serious threat for potato production in Europe and temperate regions of the world. These strains were formerly known to present a clonal structure (20, 23, 27, 38), and considerable diversity associated with the geographical distribution of strains was inferred in this study. Analysis showed two major clusters, but surprisingly, whereas the inference of phylogeny reflected a separation between sequevar 1 and 2 strains (phylotype II), the Bayesian population structure assessment method grouped strains from the Andean region together. Since the potato (S. tuberosum) originated in the highlands of Central America, this finding suggests a common evolutionary past for brown rot IIB-1 and IIB-2 strains, before their evolution as two distinct phylogenetic clusters. Strains in this “Andean” cluster could be referred to as the Andean brown rot strains, distinct from the 20 other strains constituting the other major cluster in phylotype IIB-1. However, strains CFBP3873 (Belgium) and CFBP4808 (Israel), assigned to this “Andean” cluster, displayed high levels of membership in both of those clusters, suggesting a hybrid profile and close paths of evolution.
A focus on the major cluster composed only of phylotype IIB-1 strains revealed at least four subclusters correlated with the geographical origins of strains and named after these locations. The group names must nevertheless be taken with caution, since the location of isolation may not represent the real center of origin. A first cluster, named “EuMr,” contained strains mainly originating from Europe and the Mediterranean and was phylogenetically heterogeneous, partitioning into two structure clusters: B and C. The hybrid profile revealed by the estimated membership probability values among strains from clusters B and C and among strains within each cluster suggests high gene flows between those two populations. To our knowledge, assignment of strain PSS525, isolated in Taiwan from potato, to the European cluster B supports the hypothesis of its introduction into Taiwan through Pelargonium material (P. Prior, unpublished data; J.-F. Wang, personal communication). Similarly, it is anticipated that strain JQ1006, isolated from wilted potato in Réunion Island, may also have been introduced, since it also belongs to the European cluster B. Such data provide epidemiological evidence of the introduction and spread of strains across countries, supposedly carried by infected material.
Whereas European strains were previously described as heterogeneous, the AIO phylogenetic cluster, comprising strains with a geographical origin mainly in Africa or the Indian Ocean, was consistent with structure cluster A and was characterized by a high estimated membership. We could then hypothesize that those strains were exposed to limited gene flow and may be characterized as endemic to the African and Indian Ocean regions, since they were limited to these regions. However, three strains present with possible gene flow events between the endemic African brown rot strains and European strains. Strains CFBP3865 (isolated in France from potato), CFBP3785 (Portugal, potato), and CFBP4812 (France, tomato) were assigned to the EuMr phylogenetic cluster but also to the African structure cluster, showing a hybrid profile with a European structure cluster. This suggests that these strains carry a hybrid profile between African–Indian Ocean and European populations and thus may represent a bridge between these two major brown rot subclusters. Hence, two distinct events may explain the diversity of brown rot strains observed in Europe: a former Andean origin that may have spread worldwide along with a massive movement of potato material and a more recent African–Indian Ocean origin. The African lineage for brown rot strains is thus of major interest as a model for studying microevolution events at a continental level.
The last subcluster found within the brown rot strains from phylotype I was referred to as the “North” phylogenetic cluster and structure cluster D, corresponding to strains isolated in Netherlands and Sweden. The reference strain IPO1609 was isolated by Janse in 1995, and strain CFBP3884 was isolated by Ollson and received by Janse in 1980 (J. Janse, personal communication). As revealed by the pangenomic microarray, these two strains were clearly distinct from other IIB-1 clusters. Strain IPO1609 was recently reported to carry a 77-kb DNA deletion (17) compared to other R. solanacearum genomes, especially IIB-1 strain UW551. This chromosomal region included 43 genes, from which 2 encoded proteins related to pathogenicity traits. The high similarity between IPO1609 and CFBP3884 and the negative probe signal targeting this particular 68-kb region (confirmed by replicates [data not shown]) strongly indicate that this deletion is present in both strains. This deletion event should not be considered an exception in R. solanacearum strains. Both strains IPO1609 and CFBP3884 are nonpathogenic to potato (5), and IPO1609 was proved to show limited aggressiveness to Solanaceae (25). A strain UW551 mutant with this large DNA fragment deleted showed a dramatic reduction of virulence (17). This particular deletion was not the only difference in gene content between the “North” phylogenetic cluster and those distributed in structure clusters A and B; the microarray data flagged at least 91 additional genes, 8 of which were reported to be related to pathogenicity. Nevertheless, the lack of pathogenicity for potato and the generally low virulence traits of strains IPO1609 and CFBP3884 remain to be further investigated.
Moko disease-causing strains.
Insect-transmitted and Moko disease-causing strains that were distributed into phylotypes IIA-6, IIB-3, and IIB-4 are devastating to banana production. A major phylogenetic issue was that Moko IIB-4 strains were phylogenetically undistinguishable from emergent strains and new pathological variants assigned to IIB-4NPB by use of a neutral marker approach or partial egl sequencing. These emerging strains were surveyed in the French West Indies (42) and clustered with the Moko sequevar 4 strains, though they showed a completely different host range: they were highly pathogenic to Solanaceae, along with brown rot phylotype IIB-1 strains (5), but nonpathogenic to banana (NPB). This study resolved that disputed phylogenetic position, since the microarray data clearly showed distinct lineages. However, Moko IIB-4 strains were distributed into two phylogenetic clusters, 4A and 4B; cluster 4A is closely related to the emerging IIB-4NPB strains, in contrast to cluster 4B. Although emerging strains and Moko disease-causing sequevar 4 strains showed a close phylogenetic relationship, it is difficult to explain how virulence traits evolved over time among those two ecotypes. Emerging strains from sequevar 4NPB are not pathogenic to banana and are highly aggressive on Solanaceae, whereas all Moko IIB-4 strains are highly pathogenic to banana, and some retained pathogenicity to Solanaceae and could overcome genetic resistance resources (5). These data are consistent with the assumption that strains IIB-4NPB emerged from the IIB-4 lineage as a new ecotype (43). This hypothesis suggests that a loss of particular pathogenicity traits could trigger an emergence of highly host adapted strains. The phylogenetic position of strain CFBP6797 (4NPB) outside the emerging cluster, but within cluster 4B, confirms the close relationship between the Moko strains and the emerging ecotypes. The particular phylogenetic status of strain CFBP7014, of the newly described sequevar 51, is also a matter of interest, since this sequevar differed phylogenetically from Moko disease-causing strains and emergent strains. Strain CFBP7014, previously characterized as sequevar 4NPB by PCR (42), was assigned as a close outlier of 4NPB sequevars. This strain was shown to be highly pathogenic on sensitive Solanaceae and could establish latent infections on resistant Solanaceae but did not penetrate into banana plant tissues (5). Thus, like sequevar 4NPB strains in the French West Indies, strain CFBP7014 might be considered an emergent pathological variant in Trinidad.
Combining the natural competence of R. solanacearum for transformation and its wide phylogenetic diversity, it is easier to understand the success of R. solanacearum in extending its host range, its phenotype diversity, and its geographical distribution. Gene content explains the phylogenetic diversity, but not the pathogenic profile: a study focusing on gene expression and on fine-tuning of alleles within the pangenome should be carried out. Accessing those data by questioning the full genome content on a large strain collection remains problematic; nevertheless, the use of the pangenomic microarray developed in this study provides a resolution never before reached for R. solanacearum diversity studies.
This study brought new insights into aspects of the diversity of R. solanacearum, especially with regard to the epidemiology of three ecotypes within phylotype II of this plant pathogen: brown rot-causing strains, Moko disease-causing strains, and emerging strains. However, more research needs to be done on the evolutionary past in order to fully understand the relationships of ecotypes and phylotypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank the institutions cited in the text for their courtesy in sharing Ralstonia solanacearum strains; the staff of the supercomputer TITAN, Saint Denis, Université de la Réunion, for statistical analysis computation; and J. J. Cheron for microbiological laboratory support.
This work was funded by the Fédération Nationale des Producteurs de Plants de Pommes de Terre, Mission-DAR, grant 7124 of the French Ministry of Food, Agriculture, and Fisheries. The European Regional Development Fund (ERDF) of the European Union, Conseil Régional de La Réunion, also provided financial support as part of a Biorisk program developed at CIRAD. We thank INRA for funding the “PARASOL” project for the development of the pangenomic DNA microarray.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buddenhagen I, Kelman A. 1964. Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 2: 203–230 [Google Scholar]
- 3. Buddenhagen I, Sequeira L, Kelman A. 1962. Designation of races in Pseudomonas solanacearum. Phytopathology 52: 726 [Google Scholar]
- 4. Buddenhagen IW. 1986. Bacterial wilt revisited. In Persley GJ, et al. (ed), Bacterial wilt disease in Asia and the South Pacific: proceedings of an international workshop held at PCARRD, Los Baños, Philippines, 8 to 10 October 1985, p. 126–143 ACIAR proceedings no. 13 Australian Centre for International Agricultural Research, Canberra, Australia [Google Scholar]
- 5. Cellier G, Prior P. 2010. Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology 100: 1250–1261 [DOI] [PubMed] [Google Scholar]
- 6. Coenye T, Vandamme P. 2003. Simple sequence repeats and compositional bias in the bipartite Ralstonia solanacearum GMI1000 genome. BMC Genomics 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohan FM, Perry EB. 2007. A systematics for discovering the fundamental units of bacterial diversity. Curr. Biol. 17: R373–R386 [DOI] [PubMed] [Google Scholar]
- 8. Cook D, Barlow E, Sequeira L. 1989. Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment polymorphisms with DNA probes that specify virulence and hypersensitive response. Mol. Plant Microbe Interact. 2: 113–121 [Google Scholar]
- 9. Cook D, Sequeira L. 1994. Strain differentiation of Pseudomonas solanacearum by molecular genetic methods, p 77–93 In Hayward AC, Hartman GL. (ed), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom [Google Scholar]
- 10. Eden-Green SJ. 1994. Diversity of Pseudomonas solanacearum and related bacteria in South East Asia: new direction for Moko disease, p 25–34 In Hayward AC, Hartman GL. (ed), Bacterial wilt: the disease and its causative organism, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom [Google Scholar]
- 11. Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- 12. Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fegan M, Prior P. 2005. How complex is the “Ralstonia solanacearum species complex,” p 449–461 In Allen C, Prior P, Hayward AC. (ed), Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, St. Paul, MN [Google Scholar]
- 14. Fegan M, Taghavi M, Sly LI, Hayward AC. 1998. Phylogeny, diversity and molecular diagnostics of Ralstonia solanacearum, p 19–33 In Prior P, Allen C, Elphinstone J. (ed), Bacterial wilt disease: molecular and ecological aspects. INRA Editions, Paris, France [Google Scholar]
- 15. Genin S, Boucher C. 2004. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42: 107–134 [DOI] [PubMed] [Google Scholar]
- 16. Gil R, Silva FJ, Pereto J, Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68: 518–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez A, Plener L, Restrepo S, Boucher C, Genin S. 2011. Detection and functional characterization of a large genomic deletion resulting in decreased pathogenicity in Ralstonia solanacearum race 3 biovar 2 strains. Environ. Microbiol. 13: 3172–3185 [DOI] [PubMed] [Google Scholar]
- 18. Guidot A, Coupat B, Fall S, Prior P, Bertolla F. 2009. Horizontal gene transfer between Ralstonia solanacearum strains detected by comparative genomic hybridization on microarrays. ISME J. 3: 549–562 [DOI] [PubMed] [Google Scholar]
- 19. Guidot A, et al. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayward AC. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29: 67–87 [DOI] [PubMed] [Google Scholar]
- 21. Hayward AC. 1994. The hosts of Pseudomonas solanacearum, p 9–24 In Hayward AC, Hartman GL. (ed), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom [Google Scholar]
- 22. Hudson ME. 2008. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Res. 8: 3–17 [DOI] [PubMed] [Google Scholar]
- 23. Janse JD. 1996. Potato brown rot in western Europe—history, present occurrence and some remarks on possible origin. EPPO Bull. 26: 17 [Google Scholar]
- 24. Kaeuffer R, Reale D, Coltman DW, Pontier D. 2007. Detecting population structure using STRUCTURE software: effect of background linkage disequilibrium. Heredity (Edinb.) 99: 374–380 [DOI] [PubMed] [Google Scholar]
- 25. Mahbou Somo Toukam G, et al. 2009. Broad diversity of Ralstonia solanacearum strains in Cameroon. Plant Dis. 93: 1123–1130 [DOI] [PubMed] [Google Scholar]
- 26. Poussier S, Prior P, Luisetti J, Hayward C, Fegan M. 2000. Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 23: 479–486 [DOI] [PubMed] [Google Scholar]
- 27. Poussier S, et al. 2000. Genetic diversity of Ralstonia solanacearum as assessed by PCR-RFLP of the hrp gene region, AFLP and 16S rRNA sequence analysis, and identification of an African subdivision. Microbiology 146(Pt 7): 1679–1692 [DOI] [PubMed] [Google Scholar]
- 28. Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. R Development Core Team 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 30. Remenant B, et al. 2010. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salanoubat M, et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415: 497–502 [DOI] [PubMed] [Google Scholar]
- 32. Sheather SJ, Jones MC. 1991. A reliable data-based bandwidth selection method for kernel density estimation. J. R. Stat. Soc. Ser. B Stat. Methodol. 53: 683–690 [Google Scholar]
- 33. Silverman BW. 1986. Density estimation. Chapman and Hall, London, United Kingdom [Google Scholar]
- 34. Smith JJ, et al. 1998. Genetic diversity amongst Ralstonia solanacearum isolates of potato in Europe. EPPO Bull. 28: 83–84 [Google Scholar]
- 35. Staley JT. 2006. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361: 1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taghavi M, Hayward C, Sly LI, Fegan M. 1996. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 46: 10–15 [DOI] [PubMed] [Google Scholar]
- 37. Thwaites R, Mansfield J, Eden-Green S, Seal S. 1999. RAPD and rep PCR-based fingerprinting of vascular bacterial pathogens of Musa spp. Plant Pathol. 48: 121–128 [Google Scholar]
- 38. Timms-Wilson TM, Bryant K, Bailey MJ. 2001. Strain characterization and 16S-23S probe development for differentiating geographically dispersed isolates of the phytopathogen Ralstonia solanacearum. Environ. Microbiol. 3: 785–797 [DOI] [PubMed] [Google Scholar]
- 39. van der Wolf JM, et al. 1998. Genetic diversity of Ralstonia solanacearum race 3 in Western Europe determined by AFLP, RC-PFGE and Rep-PCR, p 44–49 In Prior P, Allen C, Elphinstone J. (ed), Bacterial wilt disease: molecular and ecological aspects. Springer-Verlag, Berlin, Germany [Google Scholar]
- 40. Vaneechoutte M, Kämpfer P, De Baere T, Falsen E, Verschraegen G. 2004. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int. J. Syst. Evol. Microbiol. 54: 317–327 [DOI] [PubMed] [Google Scholar]
- 41. Villa JE, et al. 2005. Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase, and hrpB gene sequences. J. Gen. Plant Pathol. 71: 39–46 [Google Scholar]
- 42. Wicker E, et al. 2007. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 73: 6790–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Prior P. 2009. Epidemiological evidence for the emergence of a new pathogenic variant of Ralstonia solanacearum in Martinique (French West Indies). Plant Pathol. 58: 853–861 [Google Scholar]
- 44. Wicker E, et al. 17 November 2011. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. [Epub ahead of print.] doi: 10.1038/ismej.2011.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39: 897–904 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.