Abstract
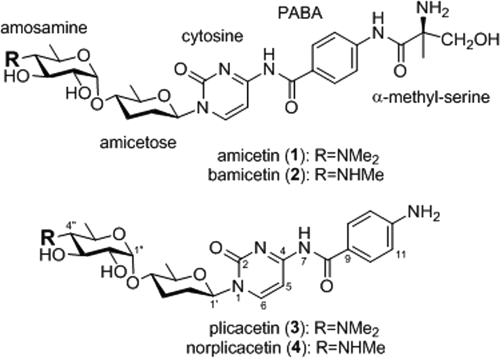
Amicetin, an antibacterial and antiviral agent, belongs to a group of disaccharide nucleoside antibiotics featuring an α-(1→4)-glycoside bond in the disaccharide moiety. In this study, the amicetin biosynthesis gene cluster was cloned from Streptomyces vinaceusdrappus NRRL 2363 and localized on a 37-kb contiguous DNA region. Heterologous expression of the amicetin biosynthesis gene cluster in Streptomyces lividans TK64 resulted in the production of amicetin and its analogues, thereby confirming the identity of the ami gene cluster. In silico sequence analysis revealed that 21 genes were putatively involved in amicetin biosynthesis, including 3 for regulation and transportation, 10 for disaccharide biosynthesis, and 8 for the formation of the amicetin skeleton by the linkage of cytosine, p-aminobenzoic acid (PABA), and the terminal (+)-α-methylserine moieties. The inactivation of the benzoate coenzyme A (benzoate-CoA) ligase gene amiL and the N-acetyltransferase gene amiF led to two mutants that accumulated the same two compounds, cytosamine and 4-acetamido-3-hydroxybenzoic acid. These data indicated that AmiF functioned as an amide synthethase to link cytosine and PABA. The inactivation of amiR, encoding an acyl-CoA-acyl carrier protein transacylase, resulted in the production of plicacetin and norplicacetin, indicating AmiR to be responsible for attachment of the terminal methylserine moiety to form another amide bond. These findings implicated two alternative strategies for amide bond formation in amicetin biosynthesis.
INTRODUCTION
Nucleoside antibiotics are a large family of microbial secondary metabolites exhibiting potent and diverse bioactive properties (36). Amicetin (compound 1 in Fig. 1) is a disaccharide pyrimidine nucleoside antibiotic produced by Streptomyces vinaceusdrappus and Streptomyces fasciculatis (22). Amicetin has been known for its activity against a number of both Gram-negative and Gram-positive bacteria, particularly Mycobacterium tuberculosis (16, 33), and was also found to have effects against herpesvirus 1 and poliovirus (5). Amicetin has been described as a universal antibiotic, exerting effects against microorganisms of different evolutionary origins, including archaea, bacteria, and eukarya (44), by functioning as a peptidyl transferase inhibitor to block protein biosynthesis (41). Although the binding of amicetin to a conserved structural motif of 23S rRNAs has been investigated by using site-directed mutations, chemical footprinting, and nuclear magnetic resonance (NMR) structure and molecular modeling studies (18, 44), the exact working mechanism for amicetin remains elusive.
Fig 1.
Chemical structures of amicetin (compound 1), bamicetin (compound 2), plicacetin (compound 3), and norplicacetin (compound 4). Me, methyl.
The chemical structure of amicetin was established in 1962 after extensive studies of its hydrolytic products (22, 56) and was finally confirmed by crystallization studies (55). From various actinobacterial strains, a group of 10 amicetin analogues had been isolated, including bamicetin (compound 2) and plicacetin (compound 3) (31), norplicacetin (compound 4) (21), oxamicetin (42), SF2457 (37), oxyplicacetin (11), and cytosaminomycins A to D (30). The presence of an α-(1→4)-glycoside bond between amosamine and amicetose is the most characteristic feature of the amicetin group antibiotics. A p-aminobenzoic acid (PABA) moiety is present in amicetin. PABA is an important precursor of folates in primary metabolism and is also incorporated into many secondary metabolites (61). PABA has been shown to serve as the starter unit initiating the biosynthesis of aureothin (32) and FR-008/candicidin (7) and contributes to the skeleton of chloramphenicol (3). PABA moieties in these secondary metabolites are derived from chorismate via two enzymatic steps catalyzed by a PABA synthase and a 4-amino-4-deoxychorismate (ADC) lyase (6, 32, 61). The primary amino group of cytosine is linked by PABA, to which a (+)-α-methylserine residue is attached, forming two amide bonds in the amicetin structure.
The biosynthesis gene clusters for a number of pyrimidine nucleoside antibiotics have been elucidated, including nikkomycin (4, 46), polyoxin (8), blasticidin S (13), A-500359s and A-503803s (25), caprazamycin (38), lipisidomycin (39) and its analogue A-90289 (24), pacidamycins (50, 60), muraymycin (12), and tunicamycin (9). However, biosynthesis studies for the amicetin group of disaccharide nucleoside antibiotics are not yet available. Herein we report the cloning and characterization of the amicetin biosynthesis gene cluster from S. vinaceusdrappus NRRL 2363. Heterologous expression of the ami gene cluster in Streptomyces lividans TK64 resulted in the production of amicetin and its analogues. The inactivation of 4 biosynthesis genes (amiI, amiF, amiL, and amiR) in the ami gene cluster led to mutants lacking amicetin production or accumulating biosynthesis intermediates, which provided experimental evidence for the functions of these genes in amicetin biosynthesis. This work paves the way to further study the unusual enzymology of amicetin biosynthesis, including the unusual retaining of glycosylation and the biochemical logic of forming two amide bonds.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
Bacterial strains and plasmids used and constructed in this study are listed in Table S1 in the supplemental material. The strain S. vinaceusdrappus NRRL 2363 was obtained from the National Center for Agricultural Utilization Research culture collection. Streptomyces strains were grown at 28°C, either in ISP2 medium (40) for growth and sporulation or in production medium [2.5% glucose, 0.7% soybean powder, 0.25% yeast extract, 0.5% (NH4)2SO4, 0.4% NaCl, 0.04% KH2PO4, 0.8% CaCO3] for fermentation. Chemicals, enzymes, and other molecular biological reagents were purchased from standard commercial sources and used according to the manufacturers' recommendations.
DNA isolation, manipulation, and sequencing.
DNA isolation from and manipulation in Escherichia coli and Streptomyces were carried out according to standard procedures (40, 53). Primers were synthesized at the Shanghai Invitrogen Biotech Co., Ltd. PCR amplifications were carried out on an authorized thermal cycler (Eppendorf AG). DNA sequencing was performed at the Invitrogen Biotech Co., Ltd. (Guangzhou, People's Republic of China), and the Chinese National Genome Center (Shanghai).
Genomic library construction and screening.
A pOJ446-based cosmid library of S. vinaceusdrappus NRRL 2363 genomic DNA was constructed using a Gigapack III XL packaging kit according to the manufacturer's specifications. For screening of the library, a ca. 0.55-kb conserved nucleoside diphosphate (NDP)-glucose 4,6-dehydratase gene fragment was obtained via a PCR approach using the following pair of degenerate primers: NGDH-F (5′-CSGGSGSSGCSGGSTTCATCGG-3′) and NGDH-R (5′-GGGWRCTGGYRSGGSCCGTAGTTG-3′) (17). Alternatively, a ca. 1.1-kb DNA fragment of a PABA synthase gene homologue was amplified using the degenerated primer pair PAPB-F (5′-GGGVGTSCAGTTCCMMCCSGAGTC-3′) and PABA-R (5′-CAGGTCGACGATCATSAGGTTCTCGGC-3′). The two PCR products were cloned, sequenced, and subsequently used as digoxigenin (DIG)-labeled probes for Southern blot colony hybridization (52).
Sequence analysis.
The sequence of the ami gene cluster was determined, and the open reading frames (ORFs) were deduced from the sequence by using the FramePlot 4.0 beta program. The corresponding deduced proteins were compared with known proteins by BLAST methods.
Construction of targeting vectors.
The subcloning strategy for pCSG3104 is depicted in Fig. S1 and the resulting subclones are summarized in Table S1 in the supplemental material. The gene inactivation experiments with S. vinaceusdrappus NRRL 2363 were carried out using lambda Red-mediated gene replacements according to standard procedures (29). Detailed procedures for disrupting and complementing individual ami genes are described in the supplemental material (see Fig. S1 to S8).
Heterologous production of amicetin and its analogues in S. lividans TK64.
The plasmid pCSG3104 was introduced into S. lividans TK64 by protoplast transformation (40). A single transformant was inoculated into 25 ml production medium [2.5% glucose, 0.7% soybean powder, 0.25% yeast extract, 0.5% (NH4)2SO4, 0.4% NaCl, 0.04% KH2PO4, 0.8% CaCO3] for 5 to 7 days, and production was monitored by high-performance liquid chromatography (HPLC). S. lividans TK64 transformed with the cosmid vector pOJ446 was used as a control.
Isolation and structural elucidation of accumulated novel analogues.
For the isolation of amicetin and accumulated analogues, each 400-ml culture was centrifuged at 4°C and 4,000 rpm for 10 min. After removal of the cell pellets, the resulting supernatant was extracted four to five times with 1-butanol at about one-third of the volume of the supernatant. The extracts were concentrated under vacuum conditions and dissolved in appropriate volumes of methanol for HPLC or HPLC-mass spectrometry (MS) analysis. HPLC analysis was carried out on a Luna C18 reversed-phase column (150 by 4.6 mm; particle size, 5 μm [Phenomenex]) with UV detection at 254 nm. The solvent system consisted of solvent A, 10% CH3CN in water supplemented with 0.08% trifluoroacetic acid (TFA), and solvent B, 90% CH3CN in water. The program was as follows: 0% to 40% solvent B (0 to 15 min), 40% to 80% solvent B (15 to 18 min), 80% to 0% solvent B (18 to 19 min), and 0% solvent B (19 to 25 min) at a flow rate of 1 ml/min. The targeted compounds from large-scale fermentation of corresponding mutants were isolated and purified by various chromatographic methods, including silica gel chromatography, thin-layer chromatography, medium-pressure column chromatography, reversed-phase silica gel column chromatography, and Sephadex LH-20 gel column chromatography (see the supplemental material for details). The purified compounds were analyzed by 1H and 13C NMR spectroscopy.
Nucleotide sequence accession number.
The sequence of the ami gene cluster was deposited in GenBank under accession number HM748814.
RESULTS AND DISCUSSION
Cloning, identification, and heterologous expression of the amicetin biosynthesis gene cluster.
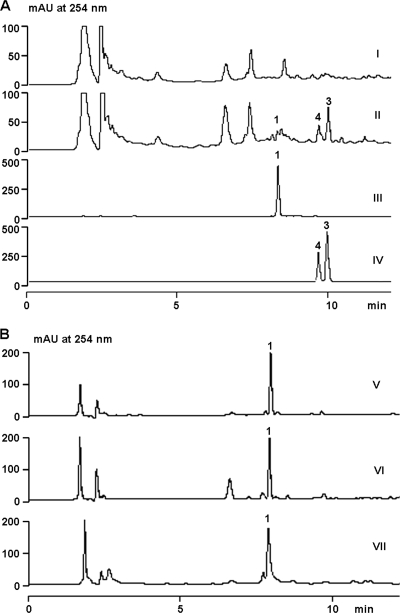
Structurally, amicetin contains two deoxysugar units and a PABA moiety. On the basis of these structural features, two complementary strategies were employed to screen for a putative amicetin biosynthesis gene cluster in a pOJ446-based genomic library from S. vinaceusdrappus NRRL 2363. Using a pair of previously reported degenerate primers targeting the conserved NDP-glucose 4,6-dehydratase gene, we were able to amplify a 550-bp PCR fragment, which was proven to be related to a dTDP-glucose 4,6-dehydratase gene (accession number YP_480774) from Frankia sp. strain CcI3. Alternatively, based on the universally conserved PABA synthase motifs, we designed a pair of degenerate primers (PABA-F and PABA-R) capable of amplifying a 1.1-kb fragment, which was sequenced and determined to be a homologue of aurG, a PABA synthase gene from the aureothin pathway in Streptomyces thioluteus (32). Among 1,920 clones in the genomic library from S. vinaceusdrappus NRRL 2363, 9 were positive with the NDP-glucose 4,6-dehydratase gene probe and 6 were positive with the PABA probe. A common positive clone, designated pCSG3104, was identified. Subsequently, pCSG3104 and pOJ446 were introduced into S. lividans TK64 by protoplast transformation and the resulting transformants were cultivated for 5 days. Interestingly, 3 metabolites were detected in the butanol extract of S. lividans TK64::pCSG3104 (Fig. 2A, trace II); however, none were present in the control, S. lividans TK64::pOJ446 (Fig. 2A, trace I). Upon HPLC-MS analyses, these metabolites were identified as amicetin (m/z 617.2 [M − H]−), norplicacetin (m/z 501.2 [M − H]−), and plicacetin (m/z 515.2 [M − H]−) by comparisons of their UV/visible spectra, retention times, and molecular masses with authentic standards (Fig. 2A, traces III and IV).
Fig 2.
Heterologous expression of the ami gene cluster in S. lividans TK64 and preliminary determination of the boundary. (A) HPLC analyses of the heterologous production of amicetin and its analogues in S. lividans TK64::pOJ446 (I) and TK64::pCSG3104 (II) in comparison to an amicetin (compound 1) standard (III) and plicacetin (compound 3) and norplicacetin (compound 4) standards (IV). (B) HPLC analyses of the metabolite profiles of wild type S. vinaceusdrappus NRRL 2363 (V), the Δorf(−1) mutant AM1001 (VI), and the Δorf1 mutant AM1011 (VII). mAU, milli-absorbance units.
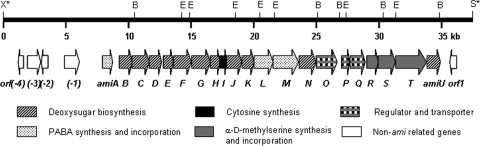
The insert in pCSG3104 was sequenced by a shotgun approach, yielding a 37,337-bp contiguous DNA sequence with an overall G+C content of 68.5% (GenBank accession number HM748814). Bioinformatic analysis of the sequence revealed the presence of 26 ORFs (Fig. 3), and the deduced functions of individual ORFs are summarized in Table 1. The four ORFs, orf(−4) to orf(−1), located upstream of amiA encode proteins whose functions are not relevant to amicetin biosynthesis (Table 1) and are probably outside the ami gene cluster. Indeed, the inactivation of orf(−1) (encoding an Mg- or Mn-dependent protein phosphatase) by replacement of an internal fragment with the aac(3)IV gene cassette (see Fig. S2 in the supplemental material) led to a mutant, AM1001, that produced amicetin at a yield comparable to that from the wild-type strain (Fig. 2B, traces V and VI). This indicated that orf(−1) was not involved in amicetin biosynthesis. The inactivation of the orf1 gene (see Fig. S3 in the supplemental material), encoding a GCN5-related N-acetyltransferase (GNAT) family protein, led to a mutant, AM1011, displaying no effects on amicetin production (Fig. 2B, trace VII), excluding a functional role for orf1 in amicetin biosynthesis. Based on these experimental results, we preliminarily outlined the ami gene cluster in S. vinaceusdrappus NRRL 2363 to contain 21 genes, from amiA to amiU, spanning a DNA region of approximate 27 kb. Interestingly, these 21 genes are transcribed in the same direction (Fig. 3).
Fig 3.
Restriction map of the DNA insert in pCSG3104 and organization of the ami gene cluster for amicetin biosynthesis. Restriction sites: E, EcoRI site; B, BamHI site; X*, XbaI site originating from the cosmid vector pOJ446; S*, SpeI site originating from the cosmid vector pOJ446. Proposed functions of individual ORFs are identified and are summarized in Table 1.
Table 1.
Deduced ORFs and their predicted functions in the ami gene cluster
| Gene | Product size (aa)a | Proposed function of gene productb | Closest protein homologue (origin) | Accession no. | % similarity/% identity |
|---|---|---|---|---|---|
| orf(−4) | 137 | Putative NAD(P)H-dependent FMN reductase | SSQG_07138 (Streptomyces viridochromogenes DSM 4073) | EFL36620 | 90/84 |
| orf(−3) | 346 | Putative transcription antitermination regulator | DSMT0073 (Streptomyces ambofaciens) | CAK50916 | 90/84 |
| orf(−2) | 173 | Putative regulator | SAMT0056 (Streptomyces ambofaciens ATCC 23877) | CAI78259 | 78/70 |
| orf(−1) | 396 | Mg- or Mn-dependent protein phosphatase | SSLG_00983 (Streptomyces sp. strain SPB78) | EFK98924 | 83/75 |
| amiA | 264 | Putative 4-amino-4-deoxychorismate lyase | PabC (Streptomyces sp. FR-008) | AAQ82550 | 61/49 |
| amiB | 372 | NDP-sugar aminotransferase | SULAZ_0355 (Sulfurihydrogenibium azorense) | YP_002728349 | 53/35 |
| amiC | 458 | NDP-hexose 2,3-dehydratase | SchS2 (Streptomyces sp. strain SCC 2136) | CAF31368 | 51/36 |
| amiD | 322 | NDP-hexose 3-ketoreductase | SpnN (Saccharopolyspora spinosa) | AAG23275 | 44/29 |
| amiE | 257 | Glucose-1-phosphate thymidylyltransferase | Thal_1465 (Thermocrinis albus DSM 14484) | YP_003474221 | 55/37 |
| amiF | 468 | GCN5-related N-acetyltransferase | Dbac_1649 (Desulfomicrobium baculatum DSM 4028) | YP_003158157 | 50/32 |
| amiG | 495 | Glycosyltransferase, group 1 | DAMO_2983 (NC10 bacterium) | CBE70056 | 46/30 |
| amiH | 249 | NDP-aminohexose N-dimethyltransferase | Veis_4849 (Verminephrobacter eiseniae) | YP_999558 | 51/34 |
| amiI | 182 | (Deoxy)cytidine deoxyribosyltransferase | MilB (“Streptomyces rimofaciens”) | ACA14349 | 46/27 |
| amiJ | 403 | Cytosylglucuronic acid synthase/glycosyltransferase | BlsD (Streptomyces griseochromogenes) | AAP03118 | 51/39 |
| amiK | 315 | NDP-hexose 4-ketoreductase | PokS6 (Streptomyces diastatochromogenes) | ACN64824 | 44/35 |
| amiL | 498 | Benzoate-CoA synthase | SBI_01640 (Streptomyces bingchenggensis) | ADI04761 | 50/38 |
| amiM | 677 | 4-Amino-4-deoxychorismate synthase | PapA (Streptomyces venezuelae) | BAD21140 | 61/49 |
| amiN | 439 | NDP-4-keto-6-deoxyglucose-3-dehydratase | Gra-orf23 (Streptomyces violaceoruber) | CAA09644 | 86/75 |
| amiO | 559 | ABC transporter | Kfla_6094 (Kribbella flavida DSM 17836) | YP_003383896 | 83/74 |
| amiP | 212 | TetR family transcriptional regulator | HMPREF9621_00706 (Propionibacterium acnes) | EFS75074 | 75/59 |
| amiQ | 397 | Putative major facilitator superfamily transporter | Svir_26710 (Saccharomonospora viridis DSM 43017) | YP_003134482 | 76/65 |
| amiR | 299 | Malonyl CoA-acyl carrier protein transacylase | ECA2695 (Pectobacterium atrosepticum) | YP_050786 | 48/29 |
| amiS | 452 | Putative glycine/serine hydroxymethyltransferase | GlyA (Azospirillum sp. strain B510) | YP_003450671 | 64/50 |
| amiT | 821 | Nonribosomal peptide synthetase (A-PCP) | McnA (“Microcystis” sp. strain NIVA-CYA 172/5) | AAZ03550 | 54/36 |
| amiU | 354 | dTDP-glucose 4,6-dehydratase | AaLAA1DRAFT_2011 (Alicyclobacillus acidocaldarius LAA1) | EED06826 | 65/49 |
| orf1 | 179 | GCN5-related N-acetyltransferase (GNAT) family protein | Sros_7636 (Streptosporangium roseum DSM 43021) | ACZ90310 | 75/65 |
aa, amino acids.
FMN, flavin mononucleotide; A-PCP, adenylation domain and peptidyl carrier protein.
Genes involved in transport and regulation.
Two putative transporter-encoding genes, amiO and amiQ, are identified in the ami gene cluster. AmiO shows 73% sequence identity to a putative ABC transporter, Kfla_6094, from Kribbella flavida DSM 17836. AmiO has two typical ABC-cobalt-CbiO domains, one at the N terminus and one at the C terminus. Each domain contains motifs characteristic of an ATP binding site, Walker A and Walker B motifs, and Q-loop, D-loop, and H-loop motifs (27). AmiQ shares 64% sequence identity with an arabinose efflux permease family protein (ACU97655), belonging to the major facilitator superfamily (MFS), from Saccharomonospora viridis DSM 43017. MFS proteins constitute a large and diverse group of secondary transporters that facilitate the transport of a variety of substrates across cytoplasmic or internal membranes (43). AmiP exhibits 59% sequence identity to a putative TetR family transcriptional regulator (accession no. EFS75074) from Propionibacterium acnes. Functions regulated by TetR family members have been clustered into 10 groups, of which the most frequent function is the regulation of efflux pumps and transporters involved in antibiotic resistance and tolerance to toxic chemicals (51). Thus, AmiP may play a regulatory role in amicetin biosynthesis.
Genes involved in deoxysugar biosynthesis.
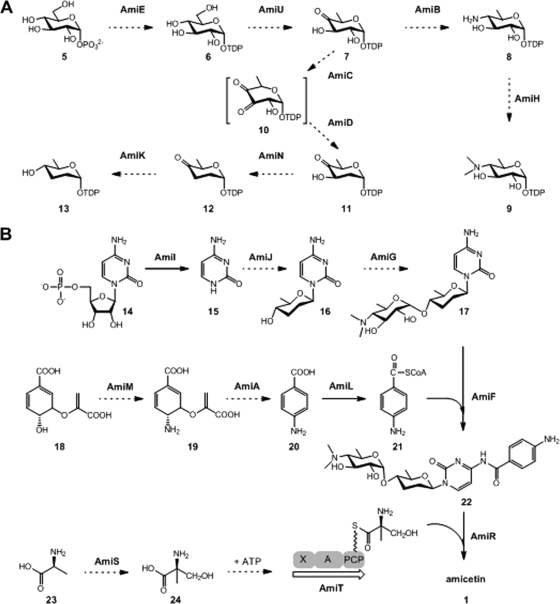
Eight genes, amiB to amiE, amiH, amiK, amiN, and amiU, are putatively involved in the biosynthesis of two deoxysugar moieties, d-amosamine and d-amicetose. The sequence alignment of the products of these genes with known deoxysugar biosynthesis enzymes leads to a proposal of the biosynthesis pathway for both sugar units (Table 1; Fig. 4A). The d-glucose-1-phosphate (compound 5) thymidylyltransferase AmiE may catalyze the formation of TDP-d-glucose (compound 6), which is further converted to TDP-4,6-dideoxy-4-keto-d-glucose (compound 7) by the TDP-glucose 4,6-dehydratase AmiU. The biosynthesis of amosamine and amicetose may branch at a common intermediate, compound 7, which is universally found in many deoxysugar pathways (57). The amiB gene, encoding a pyridoxal phosphate (PLP)-dependent aminotransferase, may be responsible for the introduction of an amino group at the C-4 position of compound 7 to form TDP-d-viosamine (compound 8). AmiH carries a type 12 methyltransferase domain (pfam08242) and is probably involved in the dimethylation of compound 8 to form TDP-d-amosamine (compound 9).
Fig 4.
Proposed amicetin biosynthesis pathway. (A) Proposed biosynthesis of two deoxysugars. (B) Two proposed alternative strategies for amide bond formation in amicetin biosynthesis. The dashed-line arrows denote biosynthesis steps predicted solely from bioinformatic analyses, and the solid-line arrows represent biosynthesis steps based on data from in vivo gene inactivation experiments. The orders for sugar attachments are deduced from the accumulation of compound 17 in the ΔamiF mutant AM1003 and the ΔamiL mutant AM1007 (Fig. 5). Sugar attachments can also occur after the formation of the two amide bonds. X, domain with unknown function; A, adenylation domain; PCP, peptidyl carrier protein.
Four enzymes, AmiC, AmiD, AmiN, and AmiK, are expected to catalyze the conversion of compound 7 to TDP-d-amicetose (compound 13) (Fig. 4A). The C-2 deoxygenation converting compound 7 to TDP-3,4-diketo-2,6-dideoxy-d-glucose (compound 10) is putatively catalyzed by AmiC, which shows the highest homology to SchS2 (36% identity), a predicted NDP-hexose 2,3-dehydratase in the biosynthesis of amicetose in angucyclines Sch 47554 and Sch 47555 (2). AmiD displays 29% identity to SpnN, a characterized TDP-hexose-3-ketoreductase which catalyzes the formation of TDP-4-keto-2,6-dideoxy-d-glucose in the TDP-d-forosamine biosynthesis pathway for spinosyn in Saccharopolyspora spinosa (35). Thus, AmiD may reduce the C-3 keto in compound 10 to form the equatorial hydroxyl group in TDP-4-keto-2,6-dideoxy-d-glucose (compound 11). The subsequent C-3 deoxygenation of compound 11, leading to compound 12, is probably performed by AmiN in a way similar to that of its homologue SpnQ (72% identity), a mechanistically characterized TDP-4-keto-6-deoxyglucose-3-dehydratase in the spinosyn pathway (34). Finally, the C-4 reduction on compound 12 to form TDP-d-amicetose (compound 13) is putatively catalyzed by AmiK, which is homologous to a predicted NDP-hexose 4-ketoreductase, PokS6 (35% identity), from the polyketomycin pathway (15).
Genes involved in glycosylation.
Genes for two putative glycosyltransferases (GTs), AmiJ and AmiG, are found in the ami gene cluster. By using a BLAST search of the GenBank database, AmiJ is found to exhibit 39% sequence identity to the cytosylglucuronic acid synthase BlsD from the blasticidin S pathway, which catalyzes the attachment of glucuronic acid to cytosine to form cytosylglucuronic acid (13). Thus, AmiJ is thought to catalyze the coupling of amicetose and cytosine to form an N-glycosidic bond in the amicetin pathway. AmiG is closely related to the GT1 family of GTs, with the highest similarity (30% identity) to a putative GT from an anaerobic bacterium (20). Distinct from most natural-product GTs in secondary metabolic pathways, which often catalyze glycosyl group transfer with inversion of the anomeric stereochemistry with respect to the donor sugar (14), AmiG may be responsible for the formation of the α-(1→4)-glycosidic bond between amosamine and amicetose, proceeding with retention of the anomeric configuration of amosamine. Consistent with this unique property, AmiG displays little similarity to a number of natural-product inverting GTs that have been well studied by in vivo inactivation or by in vitro biochemical characterization (57, 58).
Genes involved in cytosine and PABA biosynthesis.
Similar to the biosynthesis of blasticidin S and mildiomycin (13, 45), the biosynthesis of amicetin may originate from CMP (compound 14) (Fig. 4B). AmiI shows sequence similarity to various nucleoside 2-deoxyribosyltransferases that catalyze the cleavage of the glycosidic bond of 2-deoxyribonucleosides, among which BlsM (34% identity) from the blasticidin S pathway and MilB (27% identity) from the mildiomycin pathway have been functionally characterized (28, 45). The amiI gene was inactivated to probe its functional role in amicetin biosynthesis, resulting in the ΔamiI mutant AM1006. AM1006 lost the ability to produce amicetin or detectable intermediates (Fig. 5A, trace I). The production of amicetin could be partially restored by in trans complementation (see Fig. S8 in the supplemental material). These data supported the possibility that AmiI might participate in an early step of amicetin biosynthesis, providing free cytosine (compound 15) from hydrolysis of CMP (Fig. 4B), in analogy to the well-studied enzymes BlsM and MilB.
Fig 5.
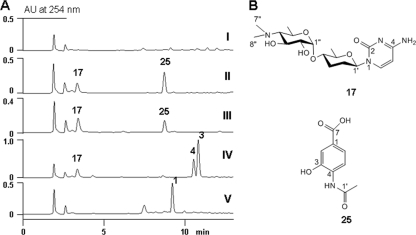
Production analyses of ami gene inactivation mutants. (A) HPLC analyses of the metabolite profiles of the ΔamiI mutant AM1006 (I), the ΔamiL mutant AM1007 (II), the ΔamiF mutant AM1003 (III), the ΔamiR mutant AM1009 (IV), and the wild type S. vinaceusdrappus NRRL 2363 (V). (B) Chemical structures of compounds isolated from the mutant AM1007.
Four genes in the ami gene cluster, amiM, amiA, amiL, and amiF, may encode enzymes responsible for the incorporation of the compound 20 moiety into amicetin. AmiM contains both type I glutamine amidotransferase (cd01653) and chorismate binding (pfam00425) motifs and exhibits high levels of sequence identity to various putative PABA synthases, such as AurG in aureothin biosynthesis (32) and PabAB for chloramphenicol in S. venezuelae (47% identity), which was functionally characterized (3). AmiA encodes an ADC lyase with 48% identity to PabC-1 for FR-008/candicidin biosynthesis in Streptomyces sp. strain FR-008 (61). Thus, AmiM may act to transfer the amide nitrogen of glutamine to chorismate (compound 18), forming ADC (compound 19), and subsequently, AmiA carries out the elimination of pyruvate from ADC to produce PABA (compound 20), which is further modified by AmiL, a benzoate coenzyme A (benzoate-CoA) ligase, to form PABA-CoA (compound 21) (Fig. 4B). Bioinformatic analysis shows that the amiF gene encodes a protein of 468 amino acids. Interestingly, the N terminus of AmiF contains a conserved NAT_SF (cd04301) domain for N-acetyltransferase superfamily enzymes. However, the N terminus of AmiF exhibits similarities to only two GCN5-related N-acetyltransferases, from Segniliparus rotundus DSM 44985 (ACU8974; 32% identity) and Desulfomicrobium baculatum DSM 4028 (ADG97057; 34% identity). Given the capability of N-acetyltransferases to catalyze the transfer of the acetyl group from acetyl-CoA to a primary amine (19), we hypothesized that AmiF may be responsible for the formation of an amide bond between cytosine and PABA (Fig. 4B).
To probe the specific functions of amiF and amiL in amicetin biosynthesis, both genes were inactivated, yielding two mutants, AM1003 (ΔamiF) and AM1007 (ΔamiL). HPLC analyses of the butanol extracts from both mutants showed that they lost amicetin-producing ability and accumulated the same two products (Fig. 5A, traces II and III), and in trans complementation of AM1003 (ΔamiF) and AM1007 (ΔamiL) restored amicetin production (see Fig. S8 in the supplemental material). Upon large-scale (9-liter) fermentation of AM1003 and subsequent product isolation efforts, one product was revealed to be cytosamine (compound 17) by comparison of its 1H NMR data (see Table S3 in the supplemental material) with those reported previously (42). Another product was elucidated as 4-acetamido-3-hydroxybenzoic acid (compound 25) (Fig. 5B). Careful analyses of the 1H 13C distortionless enhancement by polarization transfer (DEPT)-NMR spectra of compound 25 (see Table S3 in the supplemental material) indicated that there was a 4-amino-3-hydroxybenzoic acid moiety [δH 8.72 (d, H-5), 8.30 (s, H-2), 8.08 (d, J = 8.5 Hz, H-6); δC 169.3 (s, C-7), 148.2 (s, C-3), 132.5 (s, C-4), 128.2 (s, C-1), 122.2 (d, C-6), 121.1 (d, C-5), 117.4 (d, C-2)]. Additionally, an acetyl moiety [δH 2.34 (s, Me-2′); δC 170.2 (s, C-1′), 24.5 (q, Me-2′)] was present in compound 25. The heteronuclear multiple-bond correlation (HMBC) data from the proton [δH 10.02 (s)] of the amino group to C-3, C-5, and C-1′ of the acetyl moiety indicated that the C-4 amino group was acetylated. Based on these NMR data, compound 25 was elucidated as 4-acetamido-3-hydroxybenzoic acid (Fig. 5B). However, it is hard to explain the role of compound 25 in the amicetin biosynthesis pathway. We hypothesized that PABA was accumulated as a biosynthesis intermediate in both AM1003 (ΔamiF) and AM1007 (ΔamiL). PABA was converted to compound 25 by a hydroxylase and an N-acetyltransferase encoded outside the ami gene cluster. This hypothesis could be inferred from the biotransformation of exogenously supplemented PABA (compound 20) into compound 25 by the ΔamiI mutant AM1006 (see Fig. S9 in the supplemental material). Given the predicted role of AmiL as a PABA-CoA synthase, production of cytosamine and compound 25 in both AM1003 and AM1007 indicated that AmiF probably functioned to coordinate amide bond formation, linking PABA-CoA (compound 21) and cytosamine (compound 17) (Fig. 4B). Additionally, these data indicated that the glycosyltransferase AmiG was capable of utilizing amicetosyl-cytosine (compound 16) as a substrate (Fig. 4B). However, the timing for AmiG in the compound 1 biosynthesis pathway was still not clear; AmiG glycosylation could also occur as a tailoring step after amide bond formation.
Genes involved in the biosynthesis and incorporation of (+)-α-d-methylserine.
Three genes, amiS, amiT, and amiR, were predicted to be involved in the biosynthesis and incorporation of (+)-α-d-methylserine into amicetin. AmiS shows high similarity to glycine/serine hydroxymethyltransferase (SHMT), belonging to the PLP-dependent aspartate aminotransferase superfamily (fold I), which catalyzes the transfer of the hydroxymethyl group of N5,N10-methylene tetrahydrofolate to glycine, resulting in the formation of serine and tetrahydrofolate (54). Therefore, AmiS may catalyze the conversion of d-alanine (compound 23) to α-d-methylserine (compound 24) in the amicetin pathway (Fig. 4B). Subsequently, α-d-methylserine may be activated by AmiT, a protein displaying significant similarity to nonribosomal peptide synthetases (Fig. 4B). Bioinformatic analysis shows that AmiT contains an adenylation domain (A domain), with predicted substrate specificity for serine, and a putative thiolation domain (T domain). Interestingly, no domains are predicted in the first 200 amino acid residues at the N terminus of AmiT, and a required condensation domain (C domain) is missing in AmiT. AmiR exhibits weak similarity (less than 30% identity) to putative acyl-CoA-acyl carrier protein transacylases, which transfer an acyl group to an acyl carrier protein, resulting in the formation of a carbon-sulfur bond. Recently, this kind of enzyme has also been shown to be capable of forming a carbon-nitrogen bond (1).
To determine the function of AmiR in amicetin biosynthesis, the amiR gene was inactivated to give the double-crossover mutant AM1009. The mutant was devoid of amicetin production (Fig. 5A, trace IV), which could be restored by in trans complemention (see Fig. S8 in the supplemental material). Two compounds distinct from amicetin were produced by the ΔamiR mutant AM1009 (Fig. 5A, trace IV). The two compounds were then isolated and determined to be plicacetin (compound 3) and norplicacetin (compound 4) by 1H 13C correlation spectroscopy (COSY), heteronuclear single-quantum correlation (HSQC), and HMBC NMR characterizations (see Tables S3 and S4 in the supplemental material). Comparing the 1H and 13C NMR spectra of compounds 3 and 4 with those of amicetin revealed that the signals of the (+)-α-methyl serine moiety were absent in the spectra of both compounds. The only difference between compounds 3 and 4 was that the double N-methyl signals [δH 2.50 (6H, s, 7″-Me), δH 42.4 (q, 7″-Me)] in compound 3 were replaced by a single N-methyl signal [δH 2.51 (3H, s, 7″-Me), δC 33.0 (q, 7″-Me)] in compound 4. Thus, compounds 3 and 4 were confirmed to be plicacetin and norplicacetin. Interestingly, compound 17 was also found to be a minor product in the ΔamiR mutant AM1009. These data indicated that AmiR played an essential role in amide bond formation between PABA and the terminal (+)-α-d-methylserine (Fig. 4B). However, its exact function remains unclear.
Implications for amide bond formation in amicetin biosynthesis.
Commonly occurring amide bonds in natural products are most often formed by the condensation domain of nonribosomal peptide synthetases. Nature also develops diverse ways to form amide bonds. For instance, amide bonds in aminocoumarins and fredericamycins are introduced by amide synthethases (10, 47), and thioesterase domains at the C termini of polyketide synthases (PKS) are proposed to catalyze amide bond formation in macrolactams (48). Transglutaminase homologue AdmF-catalyzed amide bond formation in andrimid represents a novel condensation strategy (23). Unusual strategies for amide bond formation in the biological assembly of peptidyl nucleoside antibiotics have been demonstrated recently. For instance, an ATP-independent strategy for amide bond formation in the A-503803 pathway was revealed by Funabashi et al. (26), and tRNA-dependent peptide bond formation in pacidamycin biosynthesis was validated by Zhang et al. (59). In amicetin biosynthesis, the amide linkage between PABA and (+)-α-d-methylserine moieties is likely to be formed by a common strategy through a nonribosomal peptide synthetase, AmiT. However, bioinformatic analysis shows that AmiT lacks a crucial condensation domain. We propose that AmiR, an acyltransferase-like protein, may act as a free-standing C domain in compensation for AmiT, to furnish the amide bond formation between PABA and (+)-α-d-methylserine. This hypothesis is supported by the fact that the amiR inactivation mutant accumulated two biosynthesis intermediates, plicacetin (compound 3) and norplicacetin (compound 4), both lacking the terminal (+)-α-d-methylserine moiety. Interestingly, the inactivation of amiF, an N-acetyltransferase-encoding gene, led to a mutant that produced the same two compounds (17 and 25) as the mutant with the inactivation of amiL, a PABA-CoA synthase gene, indicating that AmiF was essential for amide bond formation between the cytosine and PABA moieties. In rifamycin biosynthesis, an arylamine N-acetyltransferase, RifF, was also proposed as an amide synthase (49). Taken together, the data indicate that two alternative strategies, involving a nonribosomal peptide synthetase complex (comprising AmiT and AmiR) and an N-acetyltransferase (AmiF), were utilized to form the two amide bonds present in the amicetin structure.
Conclusion.
In this study, we cloned and characterized the amicetin biosynthesis gene cluster from S. vinaceusdrappus NRRL 2363. Additionally, the ami gene cluster was expressed in a heterologous host, S. lividans TK64, to produce amicetin and its analogues plicacetin and norplicacetin. The identity of the ami gene cluster was further confirmed by in vivo inactivation experiments with 4 putative biosynthesis genes (amiI, amiF, amiL, and amiR) and subsequent isolation of metabolites from the gene knockout mutants, thereby providing hints of two alternative strategies for amide bond formation in amicetin biosynthesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank U. F. Wehmeier and W. Piepersberg (Bergische University, Wuppertal, Germany) for generous gifts of the cloning vector pUCPU21. We are grateful to the analytical facility of the South China Sea Institute of Oceanology for recording NMR data.
This work was supported in part by grants from the National Science Foundation of China (30870060 and 31125001), the Funds of the Chinese Academy of Sciences for Key Topics in Innovation Engineering (KSCX2-YW-G-065, KZCX2-YW-JC202, LYQY200805, and KSCX2-EW-G-12), and the National Basic Research Program of China (2010CB833805). Changsheng Zhang is a scholar of the “100 Talents Project” of the Chinese Academy of Sciences (08SL111002). Gaiyun Zhang is a recipient of a fellowship from the China Postdoctoral Science Foundation (20090460837).
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abe T, Hashimoto Y, Hosaka H, Tomita-Yokotani K, Kobayashi M. 2008. Discovery of amide (peptide) bond synthetic activity in acyl-CoA synthetase. J. Biol. Chem. 283: 11312–11321 [DOI] [PubMed] [Google Scholar]
- 2. Basnet DB, et al. 2006. Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC-2136: cloning, sequencing, and characterization. Mol. Cells 22: 154–162 [PubMed] [Google Scholar]
- 3. Brown MP, Aidoo KA, Vining LC. 1996. A role for pabAB, a p-aminobenzoate synthase gene of Streptomyces venezuelae ISP5230, in chloramphenicol biosynthesis. Microbiology 142 (Pt. 6): 1345–1355 [DOI] [PubMed] [Google Scholar]
- 4. Bruntner C, Lauer B, Schwarz W, Mohrle V, Bormann C. 1999. Molecular characterization of co-transcribed genes from Streptomyces tendae Tu901 involved in the biosynthesis of the peptidyl moiety of the peptidyl nucleoside antibiotic nikkomycin. Mol. Gen. Genet. 262: 102–114 [DOI] [PubMed] [Google Scholar]
- 5. Carrasco L, Vazquez D. 1984. Molecular bases for the action and selectivity of nucleoside antibiotics. Med. Res. Rev. 4: 471–512 [DOI] [PubMed] [Google Scholar]
- 6. Chang Z, Sun Y, He J, Vining LC. 2001. p-Aminobenzoic acid and chloramphenicol biosynthesis in Streptomyces venezuelae: gene sets for a key enzyme, 4-amino-4-deoxychorismate synthase. Microbiology 147: 2113–2126 [DOI] [PubMed] [Google Scholar]
- 7. Chen S, et al. 2003. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem. Biol. 10: 1065–1076 [DOI] [PubMed] [Google Scholar]
- 8. Chen W, et al. 2009. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J. Biol. Chem. 284: 10627–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen W, et al. 2010. Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1: 1093–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, et al. 2010. Characterization of FdmV as an amide synthetase for fredericamycin A biosynthesis in Streptomyces griseus ATCC 43944. J. Biol. Chem. 285: 38853–38860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y, Zeeck A, Chen Z, Zahner H. 1985. Studies on metabolites produced by Streptomyces ramuloses Tue-34. II. The structural elucidation of oxyplicacetin, a new member of amicetin group. Kangshengsu 10: 285–295 [Google Scholar]
- 12. Cheng L, et al. 2011. Identification of the gene cluster involved in muraymycin biosynthesis from Streptomyces sp. NRRL 30471. Mol. Biosyst. 7: 920–927 [DOI] [PubMed] [Google Scholar]
- 13. Cone MC, Yin X, Grochowski LL, Parker MR, Zabriskie TM. 2003. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. Chembiochem 4: 821–828 [DOI] [PubMed] [Google Scholar]
- 14. Coutinho PM, Deleury E, Davies GJ, Henrissat B. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328: 307–317 [DOI] [PubMed] [Google Scholar]
- 15. Daum M, et al. 2009. Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. Chembiochem 10: 1073–1083 [DOI] [PubMed] [Google Scholar]
- 16. DeBoer C, Caron EL, Hinman JW. 1953. Amicetin, a new streptomyces antibiotic. J. Am. Chem. Soc. 75: 499–500 [Google Scholar]
- 17. Decker H, et al. 1996. A general approach for cloning and characterizing dNDP-glucose dehydratase genes from actinomycetes. FEMS Microbiol. Lett. 141: 195–201 [DOI] [PubMed] [Google Scholar]
- 18. Donarski J, Shammas C, Banks R, Ramesh V. 2006. NMR and molecular modelling studies of the binding of amicetin antibiotic to conserved secondary structural motifs of 23S ribosomal RNAs. J. Antibiot. 59: 177–183 [DOI] [PubMed] [Google Scholar]
- 19. Dyda F, Klein DC, Hickman AB. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29: 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ettwig KF, et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548 [DOI] [PubMed] [Google Scholar]
- 21. Evans JR, Weare G. 1977. Norplicacetin, a new antibiotic from Streptomyces plicatus. J. Antibiot. 30: 604–606 [DOI] [PubMed] [Google Scholar]
- 22. Flynn EH, Hinman JW, Caron EL, Woolf DO. 1953. The chemistry of amicetin, a new antibiotic. J. Am. Chem. Soc. 75: 5867–5871 [Google Scholar]
- 23. Fortin PD, Walsh CT, Magarvey NA. 2007. A transglutaminase homologue as a condensation catalyst in antibiotic assembly lines. Nature 448: 824–827 [DOI] [PubMed] [Google Scholar]
- 24. Funabashi M, et al. 2010. The biosynthesis of liposidomycin-like A-90289 antibiotics featuring a new type of sulfotransferase. Chembiochem 11: 184–190 [DOI] [PubMed] [Google Scholar]
- 25. Funabashi M, et al. 2009. Identification of the biosynthetic gene cluster of A-500359s in Streptomyces griseus SANK60196. J. Antibiot. 62: 325–332 [DOI] [PubMed] [Google Scholar]
- 26. Funabashi M, et al. 2010. An ATP-independent strategy for amide bond formation in antibiotic biosynthesis. Nat. Chem. Biol. 6: 581–586 [DOI] [PubMed] [Google Scholar]
- 27. Gaudet R, Wiley DC. 2001. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 20: 4964–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grochowski LL, Zabriskie TM. 2006. Characterization of BlsM, a nucleotide hydrolase involved in cytosine production for the biosynthesis of blasticidin S. Chembiochem 7: 957–964 [DOI] [PubMed] [Google Scholar]
- 29. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100: 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haneda K, et al. 1994. Cytosaminomycins, new anticoccidial agents produced by Streptomyces sp. KO-8119. I. Taxonomy, production, isolation and physico-chemical and biological properties. J. Antibiot. 47: 774–781 [DOI] [PubMed] [Google Scholar]
- 31. Haskell TH, et al. 1958. The isolation and characterization of three crystalline antibiotics from Streptomyces plicatus. J. Am. Chem. Soc. 80: 743–747 [Google Scholar]
- 32. He J, Hertweck C. 2003. Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster. Chem. Biol. 10: 1225–1232 [DOI] [PubMed] [Google Scholar]
- 33. Hinman JW, Caron EL, DeBoer C. 1953. The isolation and purification of amicetin. J. Am. Chem. Soc. 75: 5864–5866 [Google Scholar]
- 34. Hong L, Zhao Z, Liu HW. 2006. Characterization of SpnQ from the spinosyn biosynthetic pathway of Saccharopolyspora spinosa: mechanistic and evolutionary implications for C-3 deoxygenation in deoxysugar biosynthesis. J. Am. Chem. Soc. 128: 14262–14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hong L, Zhao Z, Melancon CE, III, Zhang H, Liu HW. 2008. In vitro characterization of the enzymes involved in TDP-d-forosamine biosynthesis in the spinosyn pathway of Saccharopolyspora spinosa. J. Am. Chem. Soc. 130: 4954–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Isono K. 1988. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J. Antibiot. 41: 1711–1739 [DOI] [PubMed] [Google Scholar]
- 37. Itoh J, Miyadoh S. 1992. SF2457, a new antibiotic related to amicetin. J. Antibiot. 45: 846–853 [DOI] [PubMed] [Google Scholar]
- 38. Kaysser L, et al. 2009. Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J. Biol. Chem. 284: 14987–14996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaysser L, Siebenberg S, Kammerer B, Gust B. 2010. Analysis of the liposidomycin gene cluster leads to the identification of new caprazamycin derivatives. Chembiochem 11: 191–196 [DOI] [PubMed] [Google Scholar]
- 40. Kieser T, Bibb M, Butter M, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 41. Kirillov S, Porse BT, Vester B, Woolley P, Garrett RA. 1997. Movement of the 3′-end of tRNA through the peptidyl transferase centre and its inhibition by antibiotics. FEBS Lett. 406: 223–233 [DOI] [PubMed] [Google Scholar]
- 42. Konishi M, Naruishi M, Tsuno T, Tsukiura H, Kawaguchi H. 1973. Oxamicetin, a new antibiotic of bacterial origin. II. Structure of oxamicetin. J. Antibiot. 26: 757–764 [DOI] [PubMed] [Google Scholar]
- 43. Law CJ, Maloney PC, Wang DN. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62: 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leviev IG, et al. 1994. A conserved secondary structural motif in 23S rRNA defines the site of interaction of amicetin, a universal inhibitor of peptide bond formation. EMBO J. 13: 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li L, et al. 2008. The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5′-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. Chembiochem 9: 1286–1294 [DOI] [PubMed] [Google Scholar]
- 46. Liu G, Tian Y, Yang H, Tan H. 2005. A pathway-specific transcriptional regulatory gene for nikkomycin biosynthesis in Streptomyces ansochromogenes that also influences colony development. Mol. Microbiol. 55: 1855–1866 [DOI] [PubMed] [Google Scholar]
- 47. Luft T, Li SM, Scheible H, Kammerer B, Heide L. 2005. Overexpression, purification and characterization of SimL, an amide synthetase involved in simocyclinone biosynthesis. Arch. Microbiol. 183: 277–285 [DOI] [PubMed] [Google Scholar]
- 48. Ogasawara Y, et al. 2004. Cloning, sequencing, and functional analysis of the biosynthetic gene cluster of macrolactam antibiotic vicenistatin in Streptomyces halstedii. Chem. Biol. 11: 79–86 [DOI] [PubMed] [Google Scholar]
- 49. Pompeo F, Mushtaq A, Sim E. 2002. Expression and purification of the rifamycin amide synthase, RifF, an enzyme homologous to the prokaryotic arylamine N-acetyltransferases. Protein Expr. Purif. 24: 138–151 [DOI] [PubMed] [Google Scholar]
- 50. Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ. 2010. Pacidamycin biosynthesis: identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. Chembiochem 11: 1700–1709 [DOI] [PubMed] [Google Scholar]
- 51. Ramos JL, et al. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69: 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Riley LK, Caffrey CJ. 1990. Identification of enterotoxigenic Escherichia coli by colony hybridization with nonradioactive digoxigenin-labeled DNA probes. J. Clin. Microbiol. 28: 1465–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 54. Schirch L, Jenkins WT. 1964. Serine transhydroxymethylase. Transamination of d-alanine. J. Biol. Chem. 239: 3797–3800 [PubMed] [Google Scholar]
- 55. Smith JL, Sundaralingam M. 1981. The structure of the antibiotic amicetin consisting of nucleobase, disaccharide and amino acid moieties. Acta Crystallogr. B 37: 1095–1101 [Google Scholar]
- 56. Stevens CL, Nagarajan K, Haskell TH. 1962. The structure of amicetin. J. Org. Chem. 2: 2991–3005 [Google Scholar]
- 57. Thibodeaux CJ, Melancon CE, III, Liu HW. 2008. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. Engl. 47: 9814–9859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Williams GJ, Thorson JS. 2009. Natural product glycosyltransferases: properties and applications. Adv. Enzymol. Relat. Areas Mol. Biol. 76: 55–119 [DOI] [PubMed] [Google Scholar]
- 59. Zhang W, Ntai I, Kelleher NL, Walsh CT. 11 July 2011, posting date. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc. Natl. Acad. Sci. U. S. A. doi: 10.1073/pnas.1109539108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang W, Ostash B, Walsh CT. 2010. Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc. Natl. Acad. Sci. U. S. A. 107: 16828–16833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y, Bai L, Deng Z. 2009. Functional characterization of the first two actinomycete 4-amino-4-deoxychorismate lyase genes. Microbiology 155: 2450–2459 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.