Abstract
In the present study, bacterial communities in 200-liter biogas reactors containing liquid manure consecutively fed with casein, starch, and cream were investigated over a period of up to 33 days. A 16S rRNA gene clone library identified Bacteroidetes and Firmicutes as the most abundant bacterial groups in the starting material, at 58.9% and 30.1% of sequences, respectively. The community development of both groups was monitored by real-time PCR and single-strand conformation polymorphism (SSCP) analysis. The Firmicutes and Bacteroidetes communities were unexpectedly stable and hardly influenced by batch-feeding events. The continuous feeding of starch led to community shifts that nevertheless contributed to a stable reactor performance. A longer starving period and a change in the pH value resulted in further community shifts within the Bacteroidetes but did not influence the Firmicutes. Predominant DNA bands from SSCP gels were cloned and sequenced. Sequences related to Peptococcaceae, Cytophagales, and Petrimonas sulfuriphila were found in all samples from all experiments. Real-time PCR demonstrated the abundance of members of the phylum Bacteroidetes and also reflected changes in gene copy numbers in conjunction with a changing pH value and acetate accumulation.
INTRODUCTION
The demand for energy is growing worldwide. At the same time, the availability of fossil fuels is decreasing, and methods for their extraction, for example, the extraction of crude oil from the deep sea, are becoming more and more expensive and risky for both humans and the environment. Global climate change caused by the increase of greenhouse gases in the atmosphere is another problem caused by the consumption of fossil fuels. Therefore, finding sustainable and cost-effective energy sources is a basic necessity for the future. One important part of the future's energy mixture will be the production of biogas, which is an energy-efficient and ecologically friendly way of producing bioenergy (40).
The process of anaerobic digestion leading to methane formation is well known, while knowledge about the compositions and the changes in the microbial communities involved is still limited but may help to improve biogas production and avoid process failures (16, 40). Methane-forming processes take place under strictly anoxic conditions and can be divided into three steps. First, primary fermenting bacteria hydrolyze complex organic compounds and convert them to oligomers and monomers. While hydrogen, acetate, and carbon dioxide or other C-1 compounds like formate can be utilized directly by methanogenic Archaea, other fermentation products have to be further degraded by so-called secondary fermenting bacteria. Additionally, C-1 compounds and hydrogen are transformed to acetate by syntrophic bacteria. These bacteria live in mutual dependence with methanogenic Archaea, performing an interspecies hydrogen transfer (1, 32). For a better understanding of the complex microbial interactions leading to biogas formation, a better knowledge of the involved microorganisms is required.
In the last few years, much research work has been conducted on the microbial communities in biogas plants. In most studies, culture-independent methods like clone library analyses, real-time PCR, or molecular fingerprinting methods have been applied. Archaeal communities, among others, have been investigated by using anoxic reactors that utilized liquid manure and different agricultural products as substrates (17, 23, 26). Many studies that analyzed both the archaeal and bacterial communities (10, 16, 18, 19, 39, 42) or only bacterial communities (7) were accomplished, but the majority of the microbially diverse populations in anaerobic digesters has still not yet been sampled, as revealed by a recent meta-analysis (25). Additionally, the majority of the sequences found could not be assigned to an established genus (25). In most studies of microbial biogas communities, biogas reactors digesting manure or anaerobic sludge supplemented with complex substrates like fodder beet silage (16), grass silage (39), maize silage and green rye (18), or different agricultural and municipal solid wastes (41) were examined. As we ascertained, little research has been conducted on anaerobic microbial communities in biogas reactors digesting liquid manure supplemented with different pure or defined substrates in order to identify the most important key players in the degradation of the basic components of organic material. In addition, a whole fermentation time course instead of a single sampling event should be taken into account. Lue et al. (21) previously investigated bacterial communities during the anaerobic degradation of different complex organic wastes at different pH values and also glucose and glutamic acid in 100-ml bottles containing anaerobic sludge from a sewage treatment plant. Samples were taken at four different time points. Their results indicated that the pH value exerted a greater influence on microbial communities than did the biochemical components. The dominant bacterial groups were identified as lactic acid bacteria at pH 5 and Clostridia at pH 7. In another study, the bacterial community in anaerobic reactors during continuous fermentation with starch as the sole carbon source in a nutrient medium inoculated with pig slurry was examined in final volumes of 700 ml over a time period of 14 days (30). During that time, the bacterial diversity was significantly reduced. Here, Clostridium cluster XIVa was the dominant bacterial group (30). However, analyses of anaerobic microbial communities in volumes smaller than 1 liter will not be able to completely reflect the processes in a full-scale bioreactor, but similar experiments with biogas-producing reactors with a higher volume had not yet been accomplished.
In this study, we describe the development of bacterial communities in 200-liter biogas reactors operating under mesophilic conditions. Casein, starch, and cream with a fat percentage of 33% represented some of the basic components of organic material and were successively added to the biogas reactors in concentrations in which they occur in common biogas substrates like maize silage. The community composition of the most abundant bacterial groups was monitored over a time period of up to 1 month, depending on substrate degradation and methane formation. The study combines for the first time the use of pilot-scale reactors and the degradation of defined substrates, long observation periods, and frequent sampling.
MATERIALS AND METHODS
Reactor operation and sampling.
Anaerobic reactor experiments were carried out at the Eichhof in Bad Hersfeld, Germany. All liquid manure for laboratory experiments was taken from a biogas reactor with a volume of 600 m3 and consisted of liquid cattle manure (70%) and liquid pig manure (30%) with a dry mass of 3 to 4%. This reactor was regularly fed with maize silage and bruised grain and was kept at a temperature of 36°C to 38°C. For all experiments, laboratory-scale reactors of 200 liters were used, with anchor stirrers that reached the bottom of the tanks. Stirring was conducted for 15 min per h. The temperature was maintained at 39°C. Gas production was measured with a drum-type gas meter (type 1/6; Ritter, Bochum, Germany), and gas was collected into bags. Methane contents were determined with an infrared measuring cell (type GS IRM 100; GS Messtechnik GmbH, Ratingen, Germany) once an hour. Concentrations of short-chain fatty acids were determined by gas chromatography using an Agilent 6890N chromatograph (Agilent Technologies, Böblingen, Germany) with a flame ionization detector and a Zebron ZB-WAX-Plus column (30 m by 0.25 mm by 0.25 μm) (Phenomenex, Torrance, CA). After each of the five experiments, the manure samples from the parallel reactors were interchanged and mixed among each other to make sure that potential community changes in the following experiments resulted only from the substrate added. Before the laboratory experiments were started, liquid manure was incubated until no further gas formation could be observed, and new experiments were also started when no further gas formation from the last substrate could be detected. This resulted in rest periods of up to 6 weeks between the experiments. During this time, heating and stirring of the reactors were conducted as described above. The following defined substances were added as substrates: casein (95%; Merck KGaA, Darmstadt, Germany), starch (Toffena potato starch; Südstärke GmbH, Schrobenhausen, Germany), and Rama cream (33% fat; Unilever, Rotterdam, Netherlands). In the first experiment (experiment A1), 2.45 g liter−1 casein was added to two parallel-running 200-liter reactors. In a second experiment (experiment A2), after 2 weeks of rest, 8 g liter−1 casein was added. This was followed by a third fermentation (experiment B), after 6 weeks of rest, with 4.8 g liter−1 starch and, at a second feeding time point after 184 h, 6.6 g liter−1 starch. In this case and in the following experiments, a third reactor that was not fed with any substrate was set up as a control. After another rest period of 1 week, a fourth experiment (experiment C) with continuous starch feeding was carried out (0.6 g liter−1 day−1), and finally, after 1 week of rest, a fermentation (experiment D) with cream containing 33% fat (2.2 g liter−1 fat) was conducted. Here, the feeding of the reactors was also conducted twice, with a second feeding step after 382 h with the same amount of cream. The substrate concentrations chosen reflect the approximate concentrations of protein (in the case of 8 g liter−1 casein), starch, and fat as those typically found in biogas substrates like maize silage. In this way, the experimental conditions were supposed to be similar to the actual conditions in full-scale agricultural biogas reactors.
Samples were taken in sterile 50-ml plastic centrifugation tubes (Falcon; Greiner Bio-One, Frickenhausen, Germany) via a valve that was situated at half the height of the tank reactor and immediately frozen at −20°C. These samples were transported to Gießen, Germany, in a freezing box to prevent thawing and stored again at −20°C. Samples that were taken at distinctive points in the course of substrate degradation were chosen for molecular analyses, always including one sample taken directly before the addition of substrate.
DNA extraction.
Manure samples were thawed and homogenized for 1 min in a Stomacher80 Biomaster laboratory homogenizer (Seward Laboratory Systems Inc.). Two hundred milligrams from each sample was used for DNA extraction with the QIAamp stool minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, including the following modifications. After the addition of ASL buffer (provided with the Qiagen QIAamp stool minikit), 0.6 g sterile zirconium beads (0.1 mm; Carl Roth GmbH, Karlsruhe, Germany), and four sterile glass beads (2.5 mm; Carl Roth GmbH), the samples were mixed thoroughly. Cells were then disrupted for 3 min at maximum speed in an MM200 cell mill (Retsch, Haan, Germany). Further treatments of the samples were carried out according to Qiagen protocols. The purity and amount of DNA were checked by measurements at 260 nm with a photometer (Thermo Scientific Genesys 20; Thermo Fisher Scientific Inc., Waltham, MA) and by agarose gel electrophoresis (1% [wt/vol] agarose; staining of DNA with 0.5 μg ml−1 ethidium bromide).
PCR.
All primers used in this study are listed in Table 1 and were purchased from Eurofins MWG Operon (Ebersberg, Germany). PCR was performed with total volumes of 50 μl in 200-μl reaction tubes (Life Science Products) with a MyCycler instrument (Bio-Rad, Munich, Germany). Chemicals and enzymes for PCR were purchased from Fermentas, St. Leon-Rot, Germany. Reaction mixtures contained 1× Dream Taq PCR buffer with 2 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), 8 μg bovine serum albumin (BSA), 1 U Dream Taq polymerase, 0.2 to 0.4 μM forward primer and reverse primer, and 10 to 30 ng of DNA. DNase/RNase-free distilled water (UltraPure; Invitrogen, Life Technologies, Darmstadt, Germany) was added to obtain the final reaction mixture volume. PCRs with Firmicutes primers Firm350 forward and Firm814 reverse (Table 1) (22) started with an initial denaturation step for 4 min at 96°C, followed by 30 cycles that included denaturation for 30 s at 96°C, annealing for 30 s at 57°C, and extension for 2 min at 74°C. A final extension step of 30 min at 74°C was added. The PCR profile for Bacteroidetes primers CFB555 forward and CFB968 reverse (Table 1) (22) included an initial denaturation step for 4 min at 96°C followed by 8 touchdown cycles for 30 s at 96°C, 30 s at 65°C to 61°C (0.5°C temperature decrease per cycle), and 30 s at 74°C. After this, 22 cycles for 30 s at 96°C, 30 s at 61°C, and 30 s at 74°C were added, followed by a final step for 30 min at 74°C. PCR with primers 9bfm and 1512uR (Table 1) (22) followed a profile described previously by Mühling et al. (22), except that the annealing temperature was changed to 53°C and the extension step was prolonged to 90 s. The PCR profile for primers M13 forward and M13 reverse (Table 1) (Promega GmbH, Mannheim, Germany) consisted of an initial denaturation step for 5 min at 95°C, followed by 25 cycles of 30 s at 95°C, 50 s at 55°C, and 2 min at 72°C and a final extension step for 10 min at 72°C. With primers Com1 forward and Com2 reverse (Table 1) (34), PCR was carried out starting with 3 min at 95°C, followed by 16 touchdown cycles for 30 s at 95°C, 30 s at 65°C to 57°C (0.5°C temperature decrease per cycle), and 30 s at 72°C. The profile continued with 9 cycles for 30 s at 95°C, 30 s at 57°C, and 30 s at 72°C, followed by a final step for 30 min at 72°C. For PCR products that were used for single-strand digestion, phosphorylated reverse primers for the corresponding PCRs were used. PCR products were checked for amounts and quality by agarose gel electrophoresis and measurement at 260 nm with a photometer.
Table 1.
Summary of PCR primers used in this study
| Primer | Sequence (5′→3′) | Target | Reference or source |
|---|---|---|---|
| Firm350 forward | GGCAGCAGTRGGGAATCTTC | Firmicutes | 22 |
| Firm814 reverse | ACACYTAGYACTCATCGTTT | Firmicutes | 22 |
| CFB555 forward | CCGGAWTYATTGGGTTTAAAGGG | Bacteroidetes | 22 |
| CFB968 reverse | GGTAAGGTTCCTCGCGTA | Bacteroidetes | 22 |
| 9bfm | GAGTTTGATYHTGGCTCAG | Bacteria | 22 |
| 1512uR | ACGGHTACCTTGTTACGACTT | Bacteria | 22 |
| M13 forward | CGCCAGGGTTTTCCCAGTCACGAC | Vector pGEM-T | Promega GmbH |
| M13 reverse | TCACACAGGAAACAGCTATGAC | Vector pGEM-T | Promega GmbH |
| Com1 forward | CAGCAGCCGCGGTAATAC | Bacteria | 34 |
| Com2 reverse | CCGTCAATTCCTTTGAGTTT | Bacteria | 34 |
| 907 reverse | CCGTCAATTCCTTTGAGTTT | Bacteria | 24 |
Real-time PCR.
Quantitative real-time PCR analyses were performed with a SYBR green assay. Real-time PCRs were carried out with four replicates in 10-μl reaction mixture volumes using a 72-well rotor in a Rotor-Gene 3000 cycler (Corbett Research, Sydney, Australia). Reaction mixtures contained a 1× concentration of ABsolute QPCR SYBR green mix from Thermo Fisher Scientific Inc. (including Thermo-Start DNA polymerase and a final MgCl2 concentration of 3 mM), 2 μg bovine serum albumin, a 0.07 μM concentration (each) of the corresponding forward primer and reverse primer, and 5 to 15 ng of DNA. DNase/RNase-free distilled water (Invitrogen) was added to obtain the final reaction mixture volume. All real-time PCRs started with an initial denaturation step at 95°C for 15 min and ended with a final elongation step at 72°C for 4 min. For the cytophaga-Flavobacterium-Bacteroides (CFB) primer pair (Table 1) (22), 30 cycles were performed for 60 s at 95°C, 60 s at 64°C, 30 s at 72°C, and 20 s at 80°C for the fluorescence read. For real-time PCR with the Firm primer pair (Table 1) (22), 40 cycles were performed, including 30 s at 95°C, 30 s at 60°C, 20 s at 72°C, and 20 s at 80°C. For every real-time PCR run, a melt curve analysis at a range from 60°C to 95°C was done. Standards for real-time PCR were generated by cloning the corresponding genes from Bacteroides intestinalis (CFB primer pair, DSM 17393; DSMZ, Braunschweig, Germany) and Clostridium acetobutylicum (Firm primer pair, DSM 792), as described below. Recombinant plasmids were isolated by using the Roti-Prep plasmid minikit (Carl Roth GmbH). PCR with M13 primers (Table 1) (Promega) was performed as described above, and PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The amount of double-stranded DNA in the purified PCR product solution was determined by using the fluorescent nucleic acid stain PicoGreen (Quant-iT PicoGreen dsDNA reagent; Invitrogen) in a Tecan GENios FL fluorescence reader (Tecan Group Ltd., Männedorf, Switzerland), and copy numbers of standard fragments were calculated according to the following equation: (Cw × NA)/(N × MN) = Cs (where Cs is the target number [μl−1]; Cw is the DNA concentration [ng μl−1]; N is the number of bases in the amplicon; MN is 649.5 [ng nmol−1], the average molecular weight of a base pair; and NA is 6.23 × 1023 × 10−9 [nmol−1], the Avogadro constant). Univariate analysis of variance (ANOVA) was used for testing differences in copy numbers between different reactors and substrates with the SPSS software program, version 19 (IBM, New York, NY).
Preparation of single-stranded DNA.
PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA was eluted with 50 μl Tris-HCl (pH 8.0). To remove the phosphorylated strand, purified PCR products were digested with 6 U Lambda exonuclease (Fermentas) per 1,000 ng DNA in 1× Lambda exonuclease reaction buffer in a total volume of 50 μl. For each digestion, 2,000 ng of double-stranded DNA was used. Reaction mixtures were incubated for 2.5 h at 37°C. Single-stranded DNA was purified by using the MinElute PCR purification kit (Qiagen) according to the manufacturer's instructions, eluted in 20 μl Tris-HCl (pH 8.0), and subsequently mixed with 8 μl of denaturing loading buffer (98% formamide, 20 mM EDTA, 0.05% [wt/vol] bromophenol blue, 0.05% [wt/vol] xylene cyanol). Single-stranded DNA was denatured for 4 min at 95°C and cooled on ice immediately afterwards. After this procedure, samples were directly loaded onto a polyacrylamide gel for single-strand conformation polymorphism (SSCP) analysis or stored at −20°C.
Single-strand conformation polymorphism analysis.
Electrophoresis was carried out with a nondenaturing polyacrylamide gel consisting of 0.6× MDE solution (Biozym Scientific GmbH, Hessisch Oldendorf, Germany) and 1× TBE buffer (0.89 M Tris, 0.89 M boric acid, and 20 mM EDTA [pH 8.0]) with an INGENYphorU system (Ingeny International BV, Goes, Netherlands). Gels were run at 450 V at 19.5°C for 18 h. One of the glass plates was treated with 2 ml PlusOneRepel Silane ES (GE Healthcare, Chalfont St. Giles, Great Britain) prior to electrophoresis to avoid the gel sticking to both plates. Gels were stained by using the PageSilverSilver staining kit (Fermentas) according to the manufacturer's instructions for maximum sensitivity. Samples were applied completely onto the gel. As a migration marker, a mixture of single-stranded rRNA genes from different pure cultures was used, including the following bacterial strains: Geobacter sp. strain RA6, Bacillus licheniformis, Bacillus aquamaris, Pseudomonas putida, Lactococcus lactis (DSM 20481), and Agrobacterium vitis. All strains except for L. lactis were unpublished isolates from the working group of Sylvia Schnell. The gene fragments were amplified via PCR with the Com primer pair (Table 1) (34) and processed for SSCP analysis in the same way as the other samples. Processing and normalization of gel scans were conducted with GelComparII, version 5.1 (Applied Maths BVBA, Sint-Martens-Latem, Belgium).
Isolation of DNA fragments from polyacrylamide gels.
DNA bands visible after silver staining were cut out with a scalpel and transferred into 1.5-ml reaction tubes, and 100 μl of “crush-and-soak” buffer (34), consisting of 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA (pH 8.0), and 0.1% (wt/vol) sodium dodecyl sulfate, was added. The tubes were incubated at room temperature for several minutes, and the swollen gel slices were then cut into smaller pieces with a sterile pipette tip. This step was followed by an incubation at 37°C at 800 rpm for 3 h in a heating block (HLC BioTech, Bovenden, Germany). The samples were centrifuged at 10,000 × g for 5 min in a microcentrifuge (Heraeus Fresco; Thermo Fisher Scientific Inc.). Supernatants were transferred into new 1.5-ml reaction tubes, and 2 volumes of ethanol (98%) were added for overnight precipitation at −20°C. After the removal of ethanol, samples were dried at 37°C in a heating block (HLC BioTech), and DNA was resolved in 20 μl TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
Cloning.
PCR products of the DNA bands were purified with the Qiaquick PCR purification kit (Qiagen) according to the manufacturer's instructions. DNA was eluted with 30 μl DNase/RNase-free distilled water (Invitrogen). Fragments were ligated into vector pGEM-T (Promega, Mannheim, Germany) via TA cloning. Competent cells of Escherichia coli JM109 (Promega) were transformed with the generated plasmids according to the manufacturer's instructions. Transformed cells were selected by blue-white screening. Clones harboring plasmids with correct inserts were identified by colony PCR with primers M13 forward and M13 reverse (Promega).
Cloning of the bacterial 16S rRNA genes for the construction of a bacterial clone library of the manure without the addition of substrate was carried out by LGC Genomics GmbH (Berlin, Germany). For this purpose, the 16S rRNA gene was amplified with primers 9bfm and 1512uR (30), and the PCR product was sent to LGC Genomics.
Sequencing.
All sequencing was carried out by LGC Genomics. For sequencing with primer M13 forward, clones to be sequenced were sent to LGC Genomics in a 96-well microtiter plate filled with LB (lysogeny broth) agar with 50 μg ml−1 ampicillin. For the sequencing of the bacterial clone library, primer 907 reverse (Table 1) (24) was used.
Phylogenetic analyses.
Quality checks and cutting of sequences were performed by using the software package MEGA, version 4.0 (36). Sequences were analyzed for chimeras with the Bellerophon program (version 3) (13) on the greengenes website (6). Alignments were done with the SILVA Web aligner (27), and phylogenetic trees were constructed with the ARB software package (20), using the maximum likelihood algorithm for the nearly full-length sequences (>1,400 bp). Clone sequences (>400 bp) were added by using the ARB parsimony tool. Similarity values for operational taxonomic unit (OTU) classifications were calculated by using the neighbor-joining algorithm (PHYLIP [8] implemented in the ARB software package). For sequence comparisons, the SILVA SSU 106 Ref database (released April 2011) was used.
Nucleotide sequence accession numbers.
All sequences were deposited in the NCBI GenBank database under the following accession numbers: JN254924 to JN255110 (Bacteroidetes and Firmicutes clone sequences) and JN399099 to JN399171 (bacterial clone sequences from the clone library).
RESULTS
Bacterial clone library.
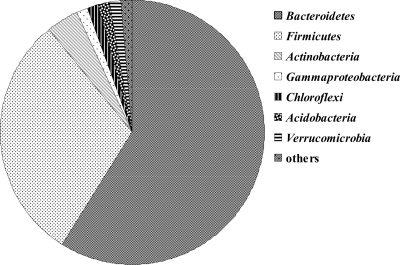
Before the experiments were started, a 50-ml sample from the 600-m3 reactor from the Eichhof in Bad Hersfeld, Germany, was taken, and DNA was isolated. In order to gain an overview of the most abundant bacterial groups, a clone library of the complete 16S rRNA genes was constructed, and 73 clone insert sequences were obtained (Fig. 1). The phyla Bacteroidetes and Firmicutes dominated, with 58.9% and 30.1% of the sequences, respectively. Other groups detected were Actinobacteria (4.1%), Gammaproteobacteria (1.4%), Chloroflexi (1.4%), Acidobacteria (1.4%), and Verrucomicrobia (1.4%). Based on these results, Firmicutes primers Firm350 forward and Firm814 reverse as well as Bacteroidetes primers CFB555 forward and CFB968 reverse, described previously by Mühling et al. (22), were chosen for further molecular experiments.
Fig 1.
Results of sequence analysis of a bacterial clone library. In order to gain an overview of abundant bacterial groups in the biogas plant, a sample was taken from the main reactor, from which all manure for further experiments originated. With DNA from this sample, PCR with universal primers 9bfm and 1512uR (30), cloning, and subsequent transformation were performed. Seventy-three clones were used for sequencing. Dominant groups and their percentages of the whole number of sequences are shown. Results were used to guide the choice of bacterial groups to be investigated.
Acetate and methane production.
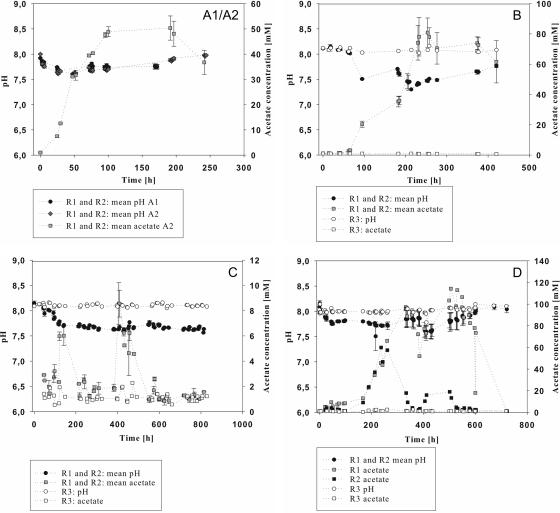
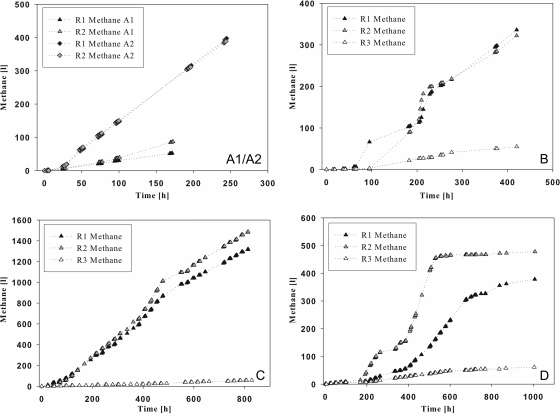
Data for acetate production are shown in Fig. 2. Acetate concentrations from up to 8.5 mM during the continuous feeding of starch (Fig. 2C) to up to 120 mM during cream fermentation were achieved. Here, acetate concentrations in reactors 1 and 2 showed large variations (Fig. 2D). Methane (Fig. 3) accumulated in amounts of up to 1,500 liters (Fig. 3C). Methane formation proceeded differently in reactors 1 and 2 during cream fermentation. In reactor 3 (no substrate), acetate and methane were produced in only minor amounts (Fig. 2 and 3).
Fig 2.
Development of acetate concentrations and pH values in 200-liter biogas reactors fed with casein at 2.45 g liter−1 (A1) and 8 g liter−1 (A2), starch (4.8 g liter −1 after 0 h and 6.6 g liter−1 after 184 h) (B), starch (continuous feeding of 0.6 g liter−1 day−1) (C), and cream (generating a fat concentration of 2.2 g liter−1, with feeding steps after 0 h and 382 h) (D). Reactors 1, 2, and 3 were operated under the same conditions, but no substrate was added to reactor 3. The x axis refers to the incubation time after the addition of substrate, and the y axis refers to pH values and acetate concentrations (mM).
Fig 3.
Development of methane accumulation in 200-liter biogas reactors fed with casein at 2.45 g liter−1 (A1) and 8 g liter−1 (A2), starch (4.8 g liter−1 after 0 h and 6.6 g liter−1 after 184 h) (B), starch (continuous feeding of 0.6 g liter−1 day−1) (C), and cream (generating a fat concentration of 2.2 g liter−1, with feeding steps after 0 h and 382 h) (D). Reactors 1, 2, and 3 were operated under the same conditions, but no substrate was added to reactor 3. The x axis refers to the incubation time after the addition of substrate, and the y axis refers to methane accumulation (liters).
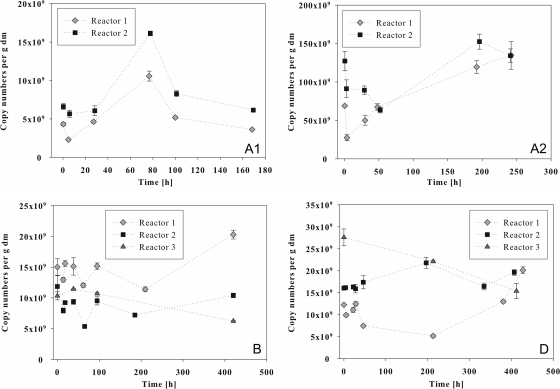
Real-time PCR.
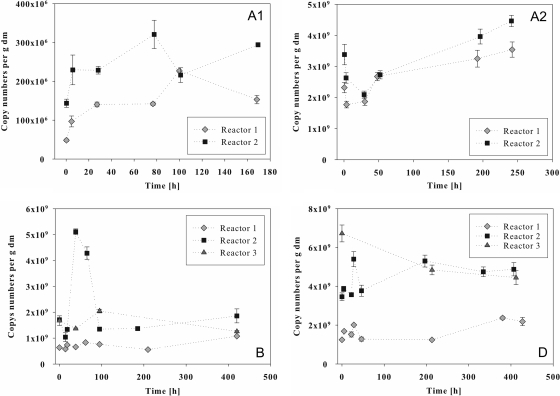
Real-time PCR analyses were accomplished for the batch experiments to detect changes and the development of the corresponding gene copy numbers per g dry mass during the fermentations. Real-time PCR analyses with the CFB primer pair (Fig. 4) reflected the important role of the Bacteroidetes that had already been observed with the clone library. Before the addition of 2.45 g liter−1 casein, about 6 × 109 copies per g dry mass of the corresponding 16S rRNA gene fragment were detected (Fig. 4A1). A maximum of 1.0 × 1010 to 1.6 × 1010 copies was found after ca. 80 h, with decreasing copy numbers per g dry mass at the end of the fermentation. The second fermentation with 8 g liter−1 casein (Fig. 4A2) started with copy numbers of about 7 × 1010 copies per g dry mass in reactor 1 and 13 × 1010 copies per g dry mass in reactor 2. After 3 h of fermentation, the copy numbers had decreased and then significantly (P = 0.003) increased, reaching maxima of ca. 1.2 × 1011 and 1.5 × 1011 copies per g dry mass, respectively, at 200 h of fermentation. At this point, acetate formation also reached a maximum of 50 mM (Fig. 2A). During the fermentation of starch (Fig. 4B), copy numbers per g dry mass ranged from 5 × 109 to 2.0 × 1010. After 95 h, a decrease of the pH value from ca. 8.1 to ca. 7.5 occurred, reaching a minimum of pH 7.3 after 200 h (Fig. 2B). This value was correlated with an increasing acetate concentration, reaching a maximum of 70 mM after 257 h (Fig. 2B), and slightly decreasing Bacteroidetes copy numbers per g dry mass. Reactor 3, which was not fed with starch, showed a constant pH value of 8.1 and no acid formation (Fig. 2B). While reactors 1 and 2 exhibited increasing Bacteroidetes copy numbers per g dry mass after the second feeding step with 6.6 g liter−1 starch after 184 h of incubation, reactor 3 showed a constant decrease. The fermentation of cream (Fig. 4D) at 5 × 109 to 2.8 × 1010 copies per g dry mass led to copy numbers similar to those with the fermentation of starch. In this case, data for reactor 1 and reactor 2 did not run as parallel as in the previous experiments, while reactor 3 again showed constantly decreasing copy numbers per g dry mass. In both reactors, feeding events correlated with increasing Bacteroidetes copy numbers, while no increase and no acetate formation were observed for reactor 3. A pairwise comparison of the three reactors resulted in significant differences (P ≤ 0.018), determined by the calculation of the average deviations resulting from the comparison of the pairs of copy number of the corresponding sampling points. The acetate concentrations also developed differently in reactors 1 and 2, reaching maxima of 97 mM after 332 h and 114 mM after 506 h in reactor 1 but only 59 mM after 238 h and 20 mM after 443 h in reactor 2 (Fig. 2D and 3D). A pairwise comparison of the four fermentations by calculating the average deviations between copy numbers of reactors 1 and 2 at all time points showed that during the fermentation of 8 g liter−1 casein, copy numbers were significantly higher than during the other three fermentations (P = 0.000).
Fig 4.
Real-time PCR analyses of Bacteroidetes communities in 200-liter biogas reactors fed with casein at 2.45 g liter−1 (A1) and 8 g liter−1 (A2), starch (4.8 g liter−1 after 0 h and 6.6 g liter−1 after 184 h) (B), and cream (generating a fat concentration of 2.2 g liter−1, with feeding steps after 0 h and 382 h) (D). Reactors 1, 2, and 3 were operated under the same conditions, but no substrate was added to reactor 3. The x axis refers to the incubation time after the addition of substrate, and the y axis reflects the 16S rRNA gene copy number of the fragment detected by primer pair CFB555f/CFB968r per g of dry mass (dm).
Real-time PCR analyses with the Firm primer pair during casein fermentation (Fig. 5A1) resulted in 5.0 × 107 to 1.5 × 108 gene copies per g dry mass before the addition of substrate, reaching up to 3 × 108 gene copies per g dry mass after 80 h with 2.45 g liter−1 casein. The second fermentation, with 8 g liter−1 casein (Fig. 5A2), started with 1.8 × 109 to 3.4 × 109 copies per g dry mass and reached 4.5 × 109 copies per g dry mass at the end of the experiment. Increasing Firmicutes copy numbers (P = 0.001) from 30 h to 50 h and from 50 h to 195 h of fermentation correlated with increasing acetate concentrations (Fig. 2A2). During the batch fermentation of starch (Fig. 5B), copy numbers per g dry mass ranged from 1 × 109 to 5 × 109 copies in reactor 2 and almost remained constant in reactor 1, with 5.6 × 108 to 1 × 109 copies per g dry mass, and in reactor 3, with 1.2 × 109 to 2.0 × 109 copies per g dry mass. Copy numbers per g dry mass for cream fermentation (Fig. 5D) ranged from 1.2 × 109 to 2.4 × 109 in reactor 1, from 3.5 × 109 to 5.4 × 109 in reactor 2, and from 4.4 × 109 to 6.7 × 109 in reactor 3.
Fig 5.
Real-time PCR analyses of Firmicutes communities in 200-liter biogas reactors fed with casein at 2.45 g liter−1 (A1) and 8 g liter−1 (A2), starch (4.8 g liter−1 after 0 h and 6.6 g liter−1 after 184 h) (B), and cream (generating a fat concentration of 2.2 g liter−1, with feeding steps after 0 h and 382 h) (D). Reactors 1, 2, and 3 were operated under the same conditions, but no substrate was added to reactor 3. The x axis refers to the incubation time after the addition of substrate, and the y axis reflects the 16S rRNA gene copy number of the fragment detected by primer pair Firm350f/Firm814r per g of dry mass (dm).
SSCP analyses.
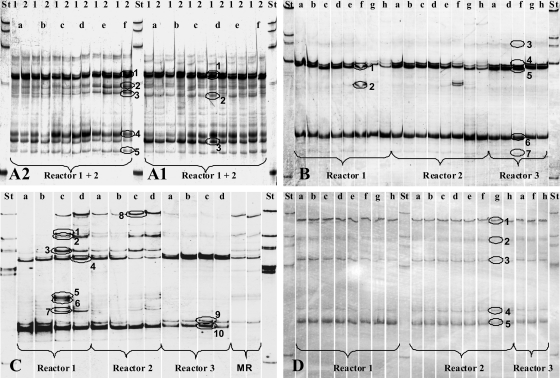
During the two fermentations with casein, the Bacteroidetes (Fig. 6A1 and A2) showed a high level of diversity, and no substantial changes in community composition occurred over the complete fermentation period. Diversity was considerably reduced after 6 weeks of rest until no further gas was produced. The addition of starch did not change the community composition (Fig. 6B). The above-mentioned pH decrease from pH 8.1 to 7.5 in reactors 1 and 2 (Fig. 2B) led to the appearance of additional DNA bands after 95 h. During the continuous feeding of starch (Fig. 6C), reactor 3 showed a band pattern similar to that for the batch fermentation of starch. Additional DNA bands appeared in reactors 1 and 2, especially after 409 h and 812 h. Two replicate samples from the 600-m3 main reactor (MR) showed a community similar to those of reactors 1 and 2. After the feeding of cream, the fingerprints for all 3 reactors were identical and did not change during the course of fermentation (Fig. 6D).
Fig 6.
SSCP analyses of Bacteroidetes communities in 200-liter biogas reactors fed with casein at 2.45 g liter−1 (A1) and 8 g liter−1 (A2), starch (4.8 g liter−1 after 0 h and 6.6 g liter−1 after 184 h) (B), starch (continuous feeding with 0.6 g liter−1 day−1) (C), and cream (generating a fat concentration of 2.2 g liter−1, with feeding steps after 0 h and 382 h) (D). Reactors 1, 2, and 3 were operated under the same conditions, but no substrates were added to reactor 3. The two samples from the main reactor (MR) were taken at the same time. All manure for the experiments originated from this reactor. Circled bands were cut out, and DNA was isolated for cloning and subsequent sequencing. For each experiment, one sample was taken before the first feeding step (0 h). Feeding experiments were carried out in the following order: A1, A2, B, C, and D. Between the experiments, reactors were incubated for several days or weeks until no further gas formation could be observed. Lowercase letters refer to the following incubation times: for panel A1, a is 0 h, b is 5 h, c is 28 h, d is 77 h, e is 100 h, and f is 169 h; for panel A2, a is 0 h, b is 3 h, c is 30 h, d is 50 h, e is 195 h, and f is 241 h; for panel B, a is 0 h, b is 14 h, c is 18 h, d is 38 h, e is 63.5 h, f is 95 h, g is 200 h, and h is 420 h; for panel C, a is 0 h, b is 49 h, c is 409 h, and d is 812 h; and for panel D, a is 0 h, b is 4.5 h, c is 22 h, d is 28 h, e is 46 h, f is 213 h, g is 360 h, and h is 415 h. St, standard; 1, reactor 1; 2, reactor 2.
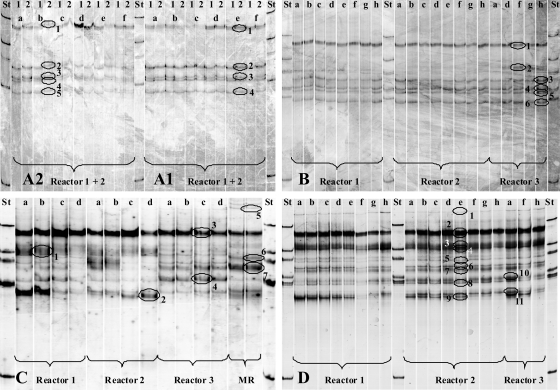
The SSCP fingerprints of Firmicutes communities of the two casein fermentations (Fig. 7A1 and A2) and the batch fermentation of starch (Fig. 7B) were identical, showing 5 predominant DNA bands. No changes in community composition were observed during these experiments. In the samples from the continuous starch fermentation (Fig. 7C), the DNA band patterns indicated a higher level of diversity, and the DNA band patterns of reactors 1 and 2 showed changes in community composition during the course of fermentation. Reactor 3, to which no starch was added, displayed a constant DNA band composition. During the fermentation of cream (Fig. 7D), there was still a higher level of diversity in all samples, with 12 distinguishable bands.
Fig 7.
SSCP analyses of Firmicutes communities in 200-liter biogas reactors fed with casein at 2.45 g liter−1 (A1) and 8 g liter−1 (A2), starch (4.8 g liter−1 after 0 h and 6.6 g liter−1 after 184 h) (B), starch (continuous feeding of 0.6 g liter−1 day−1) (C), and cream (generating a fat concentration of 2.2 g liter−1, with feeding steps after 0 h and 382 h) (D). Reactors 1, 2, and 3 were operated under the same conditions, but no substrates were added to reactor 3. The two samples from the main reactor (MR) were taken at the same time. All manure for the experiments originated from this reactor. Circled bands were cut out, and DNA was isolated for cloning and subsequent sequencing. For each experiment, one sample was taken before the first feeding step (0 h). Feeding experiments were carried out in the following order: A1, A2, B, C, and D. Between the experiments, reactors were incubated for several days or weeks until no further gas formation could be observed. Lowercase letters refer to the following incubation times: for panel A1, a is 0 h, b is 5 h, c is 28 h, d is 77 h, e is 100 h, and f is 169 h; for panel A2, a is 0 h, b is 3 h, c is 30 h, d is 50 h, e is 195 h, and f is 241 h; for panel B, a is 0 h, b is 14 h, c is 18 h, d is 38 h, e is 63.5 h, f is 95 h, g is 200 h, and h is 420 h; for panel C, a is 0 h, b is 49 h, c is 409 h, and d is 812 h; and for panel D, a is 0 h, b is 4.5 h, c is 22 h, d is 28 h, e is 46 h, f is 213 h, g is 360 h, and h is 415 h. St, standard; 1, reactor 1; 2, reactor 2.
Phylogenetic analyses.
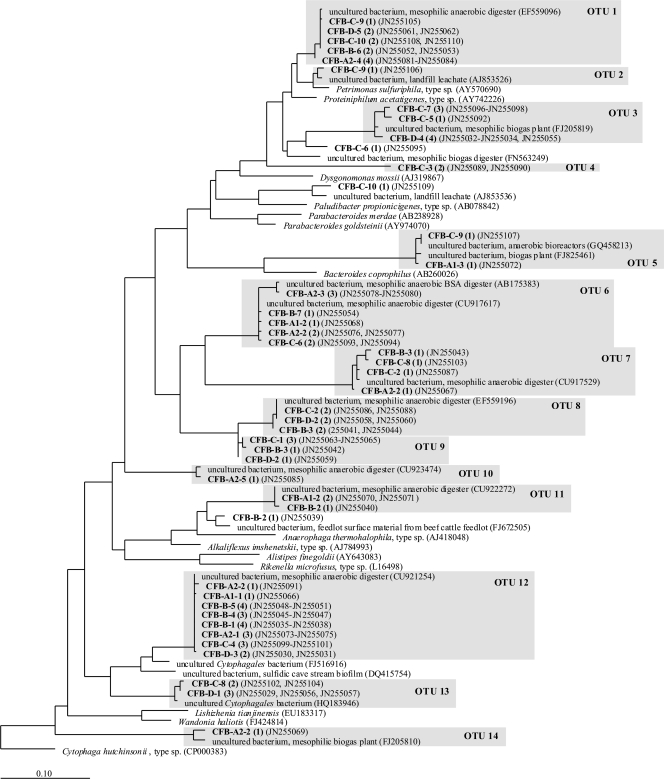
Predominant DNA bands from SSCP gels were excised, and DNA was isolated. After reamplification via PCR and cloning, 3 to 4 clones per DNA band were sequenced. Sequences identified as putative chimeras were deleted. A total of 105 16S rRNA partial gene sequences for Firmicutes and 82 sequences for Bacteroidetes were obtained. For Bacteroidetes, phylogenetic analyses resulted in a phylogenetic tree with 14 operational taxonomic units (OTUs) with ≥98% sequence identity among each OTU (Fig. 8). None of these OTUs included any hitherto-cultured species. OTUs containing species that occurred in all samples and all five fermentation experiments were OTU 1 and OTU 12 (see Fig. S1 in the supplemental material). The sequences in OTU 1 were obtained from DNA bands that were detected at the same position in all SSCP gels. All these clone sequences showed more than 98.8% sequence identity to a sequence from an uncultured bacterium found in a mesophilic anaerobic digester (GenBank accession number EF559096). The next cultured relative of OTU 1 was Petrimonas sulfuriphila (accession number AY570690), with sequence identity values of 92.5 to 96.3%. The sequences in OTU 12, which was the largest OTU, were closely related to an uncultured Cytophagales bacterium (95.3 to 96.2% sequence identity) (accession number FJ516916). OTU 6 was closely related to an uncultured bacterium from a mesophilic anaerobic BSA digester (98.7 to 99.8% sequence identity) (accession number AB175383). Sequences from OTU 11 were related to Alkaliflexus imshenetskii, with 88.5 to 95.8% sequence identity. This OTU appeared in the “f” samples during the batch fermentation of starch when the pH value decreased (Fig. 6B). In OTU 13, sequences which occurred for the first time during the continuous feeding of starch were found. They were situated in equal positions in the SSCP gels and were closely related to an uncultured Cytophagales bacterium from leachate sediment (minimum of 98.6% sequence identity) (accession number HQ183946).
Fig 8.
Phylogenetic tree of Bacteroidetes 16S rRNA partial gene sequences from 200-liter biogas reactors. The tree was constructed by using the maximum likelihood algorithm for the nearly full-length sequences (>1,400 bp). Clone sequences (in boldface type) were added by using the ARB parsimony tool. CFB, Bacteroidetes. A1, A2, B, C, and D refer to the fermentation experiments (Fig. 6). The first number following refers to the DNA band number (Fig. 6), and the second number following in parentheses refers to the number of sequences from the same DNA band represented by this position in the tree. GenBank accession numbers are also in parentheses.
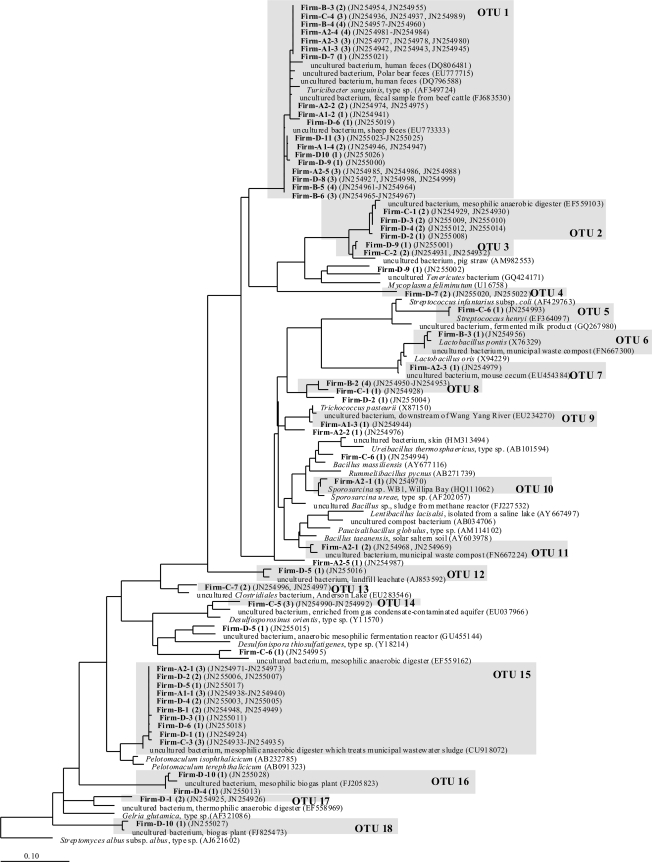
Phylogenetic analyses of 105 Firmicutes sequences (Fig. 9) resulted in 18 OTUs with a minimum of 98% sequence identity, except for OTU 1, which contained 44 clone sequences, and among these were a minority of sequences with lower identity values. Only 31 of the sequences belonged to the class Clostridia (groups 13 to 19), and 7 clone sequences could not be assigned to any of the OTUs. OTUs 1 and 15 contained the majority of the sequences and were present in all experiments (see Fig. S2 in the supplemental material). In OTU 1, 37 of the 44 clone sequences were closely related to Turicibacter sanguinis (GenBank accession number AF349724), with an identity value of at least 98%. Closely related uncultured bacteria were found in feces of humans (accession numbers DQ796588 and DQ806481), cattle (accession number FJ683530), polar bears (accession number EU777715), and sheep (accession number EU773333). OTU 8 contained four sequences from a species that marked the only difference between the samples from reactor 3 and those from reactors 1 and 2 during the batch experiment with starch. The next cultured relative of OTU 8 was Paucisalibacillus globulus (accession number AM114102) (92.1 to 93% sequence identity). OTU 15 was the OTU with the second most sequences and contained only members of the family Peptococcaceae. Its next cultured relative was Pelotomaculum isophthalicicum (accession number AB232785), with sequence identity values of 94 to 95.7%.
Fig 9.
Phylogenetic tree of Firmicutes 16S rRNA partial gene sequences from 200-liter biogas reactors. The tree was constructed by using the maximum likelihood algorithm for the nearly full-length sequences (>1,400 bp). Clone sequences (in boldface type) were added by using the ARB parsimony tool. Firm, Firmicutes. A1, A2, B, C, and D refer to the fermentation experiments (Fig. 7). The first number following refers to the DNA band number (Fig. 7), and the second number following in parentheses refers to the number of sequences from the same DNA band represented by this position in the tree. GenBank accession numbers are also in parentheses.
The highest number of OTUs within the Bacteroidetes was present during the continuous feeding of starch, whereas the highest number of Firmicutes OTUs could be found during cream fermentation.
DISCUSSION
We constructed a bacterial clone library that revealed the dominance of the phyla Bacteroidetes (58.9%) and Firmicutes (30.1%) in the full-scale biogas reactor from which all manure for our experiments was taken. The dominance of these two phyla in biogas reactors was also demonstrated previously by Klocke et al. (16), among others, who constructed a 16S rRNA gene clone library of Bacteria and Archaea. Of all sequences obtained, 44% belonged to the phylum Firmicutes, and 21% belonged to the phylum Bacteroidetes. The third major bacterial group found by Klocke et al. was the phylum Proteobacteria (24%), which constituted only 1.4% of the sequences in our clone library. A very small fraction of Proteobacteria, 0.5%, was also found by Liu et al. (19), who constructed a 16S rRNA gene clone library with 96 bacterial OTUs containing 47.2% sequences belonging to the Firmicutes and 35.4% sequences belonging to the Bacteroidetes. The differences between the results of the clone libraries are most probably due to the different substrates used, as Klocke et al. (16) investigated reactors treating fodder beet silage and cattle manure and Liu et al. (19) investigated a reactor fed with pig manure. Phyla that were represented by only few clones in our clone library, like Actinobacteria (4.1%), Gammaproteobacteria (1.4%), and Verrucomicrobia (1.4%), were also found in small numbers in rRNA gene amplicon libraries from bovine feces (35). Metagenomic studies also identified the Firmicutes and Bacteroidetes as dominant groups. A sample from a production-scale biogas reactor fed with maize silage, green rye, and chicken manure which was operated under mesophilic conditions was analyzed (33). Here, 52% of the contigs were allocated to Clostridia, 12% were allocated to Bacilli, and 16% were allocated to Bacteroidetes.
The phylum Bacteroidetes is a group of bacteria found in many different habitats, living in soil or aquatic environments as well as being associated with humans and animals, and displays a high level of phenotypic and metabolic diversity (12, 38). Members of the Bacteroidetes are, together with members of the Firmicutes, the major inhabitants of the rumen (15) and of the human gut (14). A high abundance of these bacteria was also found by real-time PCR (Fig. 4 and 5), which successfully showed the correlation between chemical data, such as acetate formation, and gene copy numbers. During most of the fermentations, a decrease of gene copy numbers of Bacteroidetes and Firmicutes detected by real-time PCR comparing the first sample (taken before the addition of substrate) and the second sample (taken after the addition of substrate) was observed (e.g., see Fig. 4A1, A2, and B and 5A2), followed by increasing gene copy numbers. This was probably due to the disturbance caused by the contact with oxygen during the opening of the reactors for the addition of substrate. This is reinforced by the fact that this did not occur in reactor 3, which served as a negative-control reactor and was not opened. Moreover, the copy numbers of Bacteroidetes and Firmicutes during cream fermentation showed large variations between reactors 1 and 2 (Fig. 4D and 5D). Regarding acetate and methane formation (Fig. 2D and 3D), variation between the replicate reactors was also observed. The reason for this could have been the fact that cream is a microemulsion that is not as good soluble as casein or starch. Therefore, the cream might have dissolved better in reactor 2, which also produced more methane (Fig. 3D). The highest Bacteroidetes gene copy numbers, 1.5 × 1011 copies per g dry mass, were found during the fermentation of 8 g liter−1 casein. Therefore, Bacteroidetes might play an important role in protein degradation in biogas reactors treating manure supplemented with other organic materials. A large number of Bacteroidetes OTUs was found in association with casein fermentation (see Fig. S1 in the supplemental material). Tang et al. (37) also demonstrated previously that the phylum Bacteroidetes was a dominating phylum during the digestion of bovine serum albumin (BSA) as the sole carbon source in anaerobic reactors with synthetic wastewater. One of the sequences found by Tang et al. was also found to be closely related to OTU 6 (Fig. 8). The initially high level of diversity of the Bacteroidetes was significantly reduced after a 6-week rest period without feeding. This reduced but stable Bacteroidetes community showed changes only in association with alterations of the pH value or the continuous feeding of a certain substrate. This finding indicated that a stable reactor performance was still maintained with a reduced set of dominating Bacteroidetes species. Sequences from OTU 1 and OTU 12 could be found in all Bacteroidetes SSCP gels. The sequences from OTU 12 were related to Cytophagales. Bacteria of this order are found mainly in habitats rich in organic material, especially soil, decaying plant material, or the feces of herbivores (29). These bacteria can also adapt to rather low nutrient levels (8), which occurred during the rest periods between the fermentation experiments. Sequences belonging to OTU 12 were present in all SSCP gels (see Fig. S1 in the supplemental material), so it can be assumed that the corresponding bacteria belong to a group capable of enduring phases of nutrient limitation. This was supported by the fact that the corresponding DNA band was weak in samples from the main reactor (Fig. 6C). P. sulfuriphila, which was related to OTU 1, is a strictly anaerobic bacterium that ferments carbohydrates and organic acids (11). Many bacteria identified as being closely related to Bacteroidetes OTUs from this study, like Proteiniphilum acetatigenes, P. sulfuriphila, and A. imshenetskii, were also found previously in other biogas reactors (18, 19).
In contrast to the Bacteroidetes community, the decrease of the pH value after 95 h did not correlate with changes in the Firmicutes community composition (Fig. 7B). As already observed by SSCP analyses of Bacteroidetes, the continuous feeding of starch had an influence on the community and also led to different developments in reactors 1 and 2, whereas reactor 3 was harboring a constant community (Fig. 7C). The reason for this was probably an insufficient mixing of the manure before the new experiment was started, as the Firmicutes communities in all three reactors already differed from each other in the first sample (0 h). The addition of cream to the reactors supported a high level of diversity of Firmicutes. This is evident in Fig. S2 in the supplemental material, which shows that the highest number of OTUs was found during cream fermentation, indicating that the most abundant degraders of this substrate can be found within the Firmicutes. A phylogenetic analysis of Firmicutes revealed that the sequences from OTU 1, containing the majority of the Firmicutes clone sequences, were most closely related to T. sanguinis, a strictly anaerobic bacterium that can ferment only two carbohydrates, maltose and 5-ketogluconate (2). As maltose is a degradation product of starch, the fact that the manure used for the experiments was taken from a reactor fed with maize silage might have led to a selection of starch-degrading Firmicutes. T. sanguinis branched deeply in the Firmicutes group, being only distantly related to other bacteria (2). These results show that still, an enormous number of unexpected and hitherto uncultured bacteria was found in the biogas reactors. As the second largest OTU, OTU 15 contained only sequences belonging to the family Peptococcaceae and was predominant in all samples of all fingerprint experiments (see Fig. S2 in the supplemental material). Peptococcaceae are anaerobic, chemoorganotrophic bacteria that ferment proteins or carbohydrates, with lower fatty acids as the main fermentation products (31). The closest relative of OTU 15 was P. isophthalicicum, an anaerobic bacterium that only grows in a syntrophic relationship with a methanogen (28). It was able to degrade phthalate isomers in coculture with Methanospirillum hungatei, a methanogen that we also found in our samples (data not shown). This can be an indication of syntrophic relationships between methanogens and bacteria from OTU 15, as these microbial interactions are crucial for anaerobic degradation (32). The genus Pelotomaculum was also frequently detected in a previous metagenomic study of a mesophilic biogas reactor (33). The surprisingly low Firmicutes diversity observed is probably, as Clostridia are already known to play a key role in the biogas-producing process (16, 18, 19), partly due to the fact that many of these species were not detected by the primers. Using the Probe Match tool provided by the Ribosomal Database Project (RDP) (5), we found that Firmicutes primers Firm350f and Firm814r (Table 1) (22) match about 54.1% (forward primer) and 49.3% (reverse primer) of the Firmicutes sequences in the RDP database, and among these sequences, 95.5% and 89%, respectively, were from the class Bacilli, but only 10% and 5.9% were from the class Clostridia. However, better-matching primers for the whole phylum Firmicutes or for the whole class Clostridia have not been designed so far. Nevertheless, this study shows that Firmicutes bacteria not belonging to the Clostridia contribute to anaerobic digestion in the biogas process. In the above-mentioned metagenomic study (33), 12% of the contigs were allocated to the Bacilli, which showed that nonclostridial Firmicutes are also abundant in biogas reactors. Regarding the phylum Bacteroidetes, primers CFB555f and CFB968r (Table 1) (22) match 86.5% and 93.8% of the Bacteroidetes sequences, respectively, so it is most likely that the majority of the abundant species could be detected. In different studies, the bacterial community during the digestion of cattle manure or glucose showed rapid changes over short periods (3, 9) and different bacterial populations with different substrates (4), in contrast to our results. However, in those three studies and in others (21, 39), bioreactors with volumes of at most 1.5 liters were used. Laboratory reactors with higher volumes may be more similar to full-scale biogas reactors and less susceptible to external disturbances like oxygen contamination. This was supported by the fact that in the full-scale main reactor (MR) (Fig. 6 and 7), SSCP band patterns similar to those for the laboratory-scale reactors were found. Predominant DNA bands that were found in all samples on the laboratory scale were also present in the full-scale reactor. Other reasons for the observed stability include the fact that the substrates used in this study are easily accessible for microorganisms, in contrast to plant raw material, and that an introduction of additional microorganisms along with fermented substrates like maize silage was very unlikely.
In conclusion, it was demonstrated for the first time that Bacteroidetes and Firmicutes communities in 200-liter biogas reactors fed with defined substrates like casein, starch, and cream were very stable during the digestion of the substrates. Both communities were hardly influenced by batch-feeding events. Only starving periods, changes of pH values, or long-time continuous feeding correlated with community shifts. Additionally, it should be considered that analyses at the DNA level reflect the presence of microorganisms but not their activity. Studies using RNA-based analyses should be accomplished to complete the understanding of complex bacterial communities.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded and supported by the Hessisches Ministerium für Umwelt, Energie, Landwirtschaft und Verbraucherschutz, Referat VIII7, Förderung der Nutzung von Biorohstoffen.
We thank Hubertus Brunn and Enno Janßen for their continuous support during this study. We also thank the anonymous reviewers for their critical and constructive advice.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bagi Z, et al. 2007. Biotechnological intensification of biogas production. Appl. Microbiol. Biotechnol. 76: 473–482 [DOI] [PubMed] [Google Scholar]
- 2. Bosshard PP, Zbinden R, Altwegg M. 2002. Turicibacter sanguinis gen. nov., sp nov., a novel anaerobic, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52: 1263–1266 [DOI] [PubMed] [Google Scholar]
- 3. Chackhiani M, et al. 2004. 16S rDNA characterisation of bacterial and archaeal communities during start-up of anaerobic thermophilic digestion of cattle manure. Bioresour. Technol. 93: 227–232 [DOI] [PubMed] [Google Scholar]
- 4. Cirne DG, Lehtomaki A, Bjornsson L, Blackall LL. 2007. Hydrolysis and microbial community analyses in two-stage anaerobic digestion of energy crops. J. Appl. Microbiol. 103: 516–527 [DOI] [PubMed] [Google Scholar]
- 5. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72: 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dohrmann AB, Baumert S, Klingebiel L, Weiland P, Tebbe CC. 2011. Bacterial community structure in experimental methanogenic bioreactors and search for pathogenic clostridia as community members. Appl. Microbiol. Biotechnol. 89: 1991–2004 [DOI] [PubMed] [Google Scholar]
- 8. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- 9. Fernandez A, et al. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65: 3697–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goberna M, Insam H, Franke-Whittle IH. 2009. Effect of biowaste sludge maturation on the diversity of thermophilic bacteria and archaea in an anaerobic reactor. Appl. Environ. Microbiol. 75: 2566–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grabowski A, Tindall BJ, Bardin V, Blanchet D, Jeanthon C. 2005. Petrimonas sulfuriphila gen. nov., sp. nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int. J. Syst. Evol. Microbiol. 55: 1113–1121 [DOI] [PubMed] [Google Scholar]
- 12. Gupta RS. 2004. The phylogeny and signature sequences characteristics of Fibrobacteres, Chlorobi, and Bacteroidetes. Crit. Rev. Microbiol. 30: 123–143 [DOI] [PubMed] [Google Scholar]
- 13. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319 [DOI] [PubMed] [Google Scholar]
- 14. Karlsson FH, Ussery DW, Nielsen J, Nookaew I. 2011. A closer look at Bacteroides: phylogenetic relationship and genomic implications of a life in the human gut. Microb. Ecol. 61: 473–485 [DOI] [PubMed] [Google Scholar]
- 15. Kim M, Morrison M, Yu ZT. 2011. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 76: 49–63 [DOI] [PubMed] [Google Scholar]
- 16. Klocke M, Mahnert P, Mundt K, Souidi K, Linke B. 2007. Microbial community analysis of a biogas-producing completely stirred tank reactor fed continuously with fodder beet silage as mono-substrate. Syst. Appl. Microbiol. 30: 139–151 [DOI] [PubMed] [Google Scholar]
- 17. Klocke M, et al. 2008. Characterization of the methanogenic archaea within two-phase biogas reactor systems operated with plant biomass. Syst. Appl. Microbiol. 31: 190–205 [DOI] [PubMed] [Google Scholar]
- 18. Krause L, et al. 2008. Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotechnol. 136: 91–101 [DOI] [PubMed] [Google Scholar]
- 19. Liu FH, et al. 2009. The structure of the bacterial and archaeal community in a biogas digester as revealed by denaturing gradient gel electrophoresis and 16S rDNA sequencing analysis. J. Appl. Microbiol. 106: 952–966 [DOI] [PubMed] [Google Scholar]
- 20. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32: 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lue F, Shao LM, Bru V, Godon JJ, He PJ. 2009. Synergetic effect of pH and biochemical components on bacterial diversity during mesophilic anaerobic fermentation of biomass-origin waste. J. Appl. Microbiol. 106: 580–591 [DOI] [PubMed] [Google Scholar]
- 22. Mühling M, Woolven-Allen J, Murrell JC, Joint I. 2008. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2: 379–392 [DOI] [PubMed] [Google Scholar]
- 23. Munk B, Bauer C, Gronauer A, Lebuhn M. 2010. Population dynamics of methanogens during acidification of biogas fermenters fed with maize silage. Eng. Life Sci. 10: 496–508 [Google Scholar]
- 24. Muyzer G, Teske A, Wirsen CO, Jannasch HW. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel-electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164: 165–172 [DOI] [PubMed] [Google Scholar]
- 25. Nelson MC, Morrison M, Yu ZT. 2011. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour. Technol. 102: 3730–3739 [DOI] [PubMed] [Google Scholar]
- 26. Nettmann E, Bergmann I, Mundt K, Linke B, Klocke M. 2008. Archaea diversity within a commercial biogas plant utilizing herbal biomass determined by 16S rDNA and mcrA analysis. J. Appl. Microbiol. 105: 1835–1850 [DOI] [PubMed] [Google Scholar]
- 27. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu YL, et al. 2006. Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185: 172–182 [DOI] [PubMed] [Google Scholar]
- 29. Reichenbach H. 2006. The order Cytophagales, p 549–590 In Dworkin M, et al. (ed), The prokaryotes, 3rd ed, vol 7 Springer, New York, NY [Google Scholar]
- 30. Ricca DM, Ziemer CJ, Kerr BJ. 2010. Changes in bacterial communities from swine feces during continuous culture with starch. Anaerobe 16: 516–521 [DOI] [PubMed] [Google Scholar]
- 31. Rogosa M. 1971. Peptococcaceae, a new family to include the Gram-positive, anaerobic cocci of the genera Peptococcus, Peptostreptococcus, and Ruminococcus. Int. J. Syst. Bacteriol. 21: 234–237 [Google Scholar]
- 32. Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61: 262–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schlüter A, et al. 2008. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J. Biotechnol. 136: 77–90 [DOI] [PubMed] [Google Scholar]
- 34. Schwieger F, Tebbe CC. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64: 4870–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shanks OC, et al. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 77: 2992–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 37. Tang YQ, Shigematsu T, Morimura S, Kida K. 2005. Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J. Biosci. Bioeng. 99: 150–164 [DOI] [PubMed] [Google Scholar]
- 38. Vingadassalom D, et al. 2005. An unusual primary sigma factor in the Bacteroidetes phylum. Mol. Microbiol. 56: 888–902 [DOI] [PubMed] [Google Scholar]
- 39. Wang H, et al. 2010. Development of microbial populations in the anaerobic hydrolysis of grass silage for methane production. FEMS Microbiol. Ecol. 72: 496–506 [DOI] [PubMed] [Google Scholar]
- 40. Weiland P. 2010. Biogas production: current state and perspectives. Appl. Microbiol. Biotechnol. 85: 849–860 [DOI] [PubMed] [Google Scholar]
- 41. Weiss A, Jerome V, Freitag R, Mayer HK. 2008. Diversity of the resident microbiota in a thermophilic municipal biogas plant. Appl. Microbiol. Biotechnol. 81: 163–173 [DOI] [PubMed] [Google Scholar]
- 42. Ziganshin AM, et al. 2011. Bacteria and archaea involved in anaerobic digestion of distillers grains with solubles. Appl. Microbiol. Biotechnol. 89: 2039–2052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.