Abstract
New insights into the distribution and biochemistry of the cyanotoxin cylindrospermopsin (CYN) have been provided by the recent determination of its biosynthesis gene cluster (cyr) in several cyanobacterial species. Raphidiopsis curvata CHAB1150 isolated from China was analyzed for CYN analogues. Only 7-deoxy-CYN was detected in the cell extracts. The cyr gene cluster of R. curvata CHAB1150 was sequenced, and the cyr genes of this strain were found to have extremely high similarities (96% to 100%) to those from other nostocalean species. These species include Cylindrospermopsis raciborskii AWT205, Aphanizomenon sp. strain 10E6, and Aphanizomenon ovalisporum ILC-146. Insertion mutation was identified within the cyrI gene, and transcripts of cyrI and another functional gene cyrJ were detected in R. curvata CHAB1150. General congruence between the phylogenetic trees based on both cyr and 16S rrn was displayed. Neutral evolution was found on the whole sequences of the cyr genes, and 0 to 89 negative selected codons were detected in each gene. Therefore, the function of CyrI is to catalyze the oxygenation of 7-deoxy-CYN in CYN biosynthesis. The transcripts of the mutated cyrI gene may result from polycistronic transcription. The high conservation of the cyr genes may be ascribed to purifying selection and horizontal gene transfer.
INTRODUCTION
Cyanotoxins are toxic compounds produced by cyanobacteria that are widespread in freshwater and marine ecosystems. The chemistry, toxicity, and biosynthesis of these toxins were fully documented (5, 6, 37). Specifically, the hepatotoxin cylindrospermopsin (CYN) is a zwitterionic alkaloid composed of a tricyclic guanidine group, a hydroxymethyluracil moiety, and a sulfonic acid group. The sulfonic acid group makes this toxin highly water soluble (36). CYN was first isolated from Cylindrospermopsis raciborskii and was proven to be the poison causing the Palm Island mystery disease, which is characterized by various symptoms of hepatitis and diarrhea (4, 10, 36). CYN can cause injury and cell necrosis in multiple organs, including the liver, thymus, kidneys, and heart (53), by inhibiting protein synthesis through a noncovalent linkage with a nonribosomal protein involved in the eukaryotic translation system (7) or by hindering the synthesis of glutathione (42). Extracellular accumulation and poor decomposition of CYN were found in lake water, which indicate a potential risk for human health (41, 54).
To date, only two natural analogues of CYN have been recorded, contrary to the prolific variants of microcystin. One of these analogues is 7-epi-CYN, a C-7 epimer of CYN with toxicity similar to CYN (2). The other is 7-deoxy-CYN, which lacks the hydroxyl group on C-7 and shows no toxicity to mice by intraperitoneal injection (35). In another study, however, 7-deoxy-CYN was proven to be a potent inhibitor of protein synthesis in vitro, and oxygenation at C7 was not essential for its toxicity (27). The three variants, CYN, 7-epi-CYN, and 7-deoxy-CYN, can be detected in the same cyanobacterial strain, Oscillatoria sp. strain PCC 6506 at different concentration ratios (28). CYN is a major part of the toxin pool in several species, including C. raciborskii (24), Aphanizomenon flos-aquae (40), Aphanizomenon ovalisporum (1), Anabaena lapponica (47), and Umezakia natans (9). In contrast, 7-deoxy-CYN is the major toxin in Raphidiopsis curvata (25) and Lyngbya wollei (45). The former only contains trace amounts of CYN. U. natans was originally classified under the Stigonematales. However, based on recent molecular evidence, U. natans has recently been reevaluated and results showed that it belongs to the Nostocales (34). Therefore, the known CYN producers belong to the two orders of cyanobacteria, namely, Nostocales and Oscillatoriales.
Feeding experiments on C. raciborskii have demonstrated that the amidination of glycine is most likely the first step in the biosynthesis of CYN. After this step, the polyketide chain of CYN is biosynthesized using the product guanidinoacetate and five units of acetate (3). Although the whole biochemical pathway for the synthesis of CYN has not been totally clarified, the genes involved in CYN production have been elucidated (44, 46). The amidination reaction is catalyzed by an enzyme encoded by the cyrA gene. This enzyme is the first reported cyanobacterial amidinotransferase gene, and arginine has been proven to be a donor of the amidino group (32). The CYN biosynthesis gene cluster (cyr) was first amplified from C. raciborskii AWT205, and a step-by-step catalytic pathway was proposed according to the putative function of each gene (31). Among the enzymes encoded by those genes, CyrB through CyrF participates in the five contiguous chain elongation reactions using acetate units. The sequences of CyrG and CyrH are highly similar and the two enzymes possibly transfer the second guanidino group to the polyketide chain, forming a uracil ring. A tailoring enzyme, CyrJ, may transfer a sulfonic acid group to the tricyclic ring. In the final step of the pathway, an iron oxygenase, CyrI, catalyzes the hydroxylation of C-7 binding to the uracil ring, and 7-deoxy-CYN was proven to be the substrate of CyrI (29). With respect to the transport of CYN, a transporter gene cyrK was also described in the cyr gene cluster (31). In addition, similar cyr gene clusters have been obtained from other cyanobacterial species, including C. raciborskii CS-505 (49), Aphanizomenon sp. strain 10E6 (50), Oscillatoria sp. strain PCC 6506 (28), and A. ovalisporum ILC-146 (46). The functions of these cyr genes in the CYN biosynthesis pathway are not conclusive, and additional evidence from in vivo gene interference experiments is needed.
Raphidiopsis is phylogenetically close to Cylindrospermopsis, and the two genera usually co-occur in freshwater bodies (55). Raphidiopsis species containing CYN and 7-deoxy-CYN have been reported from Asia and Australia (25, 30). However, no cyr genes of Raphidiopsis have been described up to the present. The characterization of the cyr gene cluster from Raphidiopsis is important in understanding the evolution and phylogenetic distribution of cyr genes within the whole realm of cyanobacteria. The present study aims to determine whether the cyr gene cluster in Raphidiopsis has close phylogenetic relations to that of Cylindrospermopsis and whether CYN chemotype correlates with cyr genotype.
MATERIALS AND METHODS
Cyanobacterial strains and DNA extraction.
Three cyanobacterial strains were used in the present study, including R. curvata CHAB1150 isolated from Chidonghu Lake (China), R. curvata HB1 (25), and C. raciborskii AWT205 (11). All strains were grown in liquid CT medium (14) at 25°C under constant white light intensity of 30 μmol photons · m−2 · s−1 and on a 12-h/12-h light/dark cycle. The total genomic DNA of the cyanobacterial cells was then extracted using the SDS-phenol method previously described (33). Finally, the purified DNA was dissolved in TE buffer and stored at −20°C.
PCR amplification, purification, and sequencing of the cyr genes and 16S rRNA gene.
Primer sets targeting cyr genes and intergenic spacers were designed using the Primer Premier 5.0 software based on cyr sequences of C. raciborskii AWT205. The primer sets were designed with overlaps to cover a complete cyr gene cluster. The 16S rRNA gene was then amplified using primer set 16SF/16SR (55). Afterward, PCRs were performed in 50-μl volumes containing 5 μl of 10× PCR Buffer (Takara, Japan), 500 μM each deoxynucleoside triphosphate (dNTP), 200 pmol of each primer, 1 U of LA Taq (Takara, Japan), and 98 ng genomic DNA. The cycling conditions applied were as follows: 94°C for 3 min, 35 cycles of 94°C for 30 s, 50°C to 60°C for 45 s, and 72°C for 1 min to 7 min, 72°C for 10 min, and a 4°C hold. The annealing temperature and extension time were adjusted for each primer set, respectively (see Table S1 in the supplemental material). The unknown intergenic sequences and the flanking regions of the cyr cluster were amplified by PCR using a Takara genome walking kit (Takara, Japan) in accordance to the procedures in the manual. Thermal cycling was then carried out in an MJ mini personal thermal cycler (Bio-Rad). The PCR products were purified using a PCR purification kit (Generay, China) and were used for TA cloning with the pMD18-T vector (Takara, Japan). Recombinant plasmids were extracted from the positive bacterial clones, and the target sequences were sequenced using the ABI 3730 automated sequencer (Applied Biosystems) in both directions.
Sequence analysis.
The similarity and conservation of the cyr sequences compared with the homologous sequences from GenBank were determined using BLASTN on the website of the National Center for Biotechnology Information (NCBI). Open reading frames were identified using the ORF Finder tool (NCBI). DNA contig assembly, amino acid sequence deduction, and sequence alignment were performed using the Bioedit V5.0.6 software. Transposase sequences were also analyzed using the IS Finder (http://www-is.biotoul.fr/is.html).
Detection of cyr gene transcripts.
Two milliliters of cyanobacterial culture at the exponential phase (optical density at 680 nm [OD680] of 1.0) was taken and centrifuged. The cell pellets were harvested and resuspended in 1 ml TRIzol reagent (Invitrogen) for total RNA preparation. The mixture was shaken using a Mini Beadbeater (Biospec) at 70 s−1 for 30 s and RNA was extracted according to procedures in the manual of the TRIzol reagent. The purified RNA was then dissolved in 30 μl diethylpyrocarbonate (DEPC)-treated water and stored at −80°C. Up to 10 μl of the RNA solution was digested by RNase-free DNase (Fermentas, Canada) prior to reverse transcription to obtain pure RNA extracts without genomic DNA contamination. Afterward, 5 μl of the products was taken for reverse transcription reactions using a PrimeScript 1st strand cDNA synthesis kit (Takara, Japan). The pure RNA extracts and total cDNA were used for PCR detection of transcripts of the two cyr genes, cyrI and cyrJ, using the primer sets RTcyrIF318/RTcyrIR588 and RTcyrJF295/RTcyrJR597 (see Table S1 in the supplemental material), respectively. Negative control was set by performing all the above-mentioned steps without cyanobacterial cells. Genomic DNA was used as the positive PCR control.
Phylogenetic analysis.
Cyanobacterial strains containing available cyr genes and 16S rRNA gene sequences were chosen in the phylogenetic analysis and four sequence data sets were constructed, including 16S rrn-5, multi-cyr, 16S rrn-10, and cyrA. The two data sets 16S rrn-5 and multi-cyr contained 16S rrn and cyr genes, respectively, from five cyanobacterial strains (see Table S2 in the supplemental material). The multi-cyr data set was a concatenation of the 11 cyr genes, cyrA to -K. Moreover, the data sets 16S rrn-10 and cyrA contained 16S rrn and cyrA genes, respectively, from nine cyanobacterial strains (see Table S2 in the supplemental material). The 16S rrn gene of Gloeobacter violaceus PCC 7421 and glycine amidinotransferase gene of Streptomyces bingchenggensis BCW-1 were used as outgroup sequences in these two data sets, respectively. Multiple sequence alignments were created using ClustalX v2.0 (23). The results were manually confirmed. The best substitution model for DNA sequence evolution was calculated by hierarchical likelihood ratio tests (HLRTS) using Modeltest v3.7 (38). Sequence alignments were used to construct maximum-likelihood (ML) trees using PHYML v3.0 (8) and PAUP v4.0b10 based on an optimal model and 1,000 bootstrap replicates. Bayes trees were created using MrBayes v3.1.2 (12). The parameters used were as follows: running, 5 million generations; sampling, every 1,000 generations, Nst, 2/6 (according to the model test results); rates, gamma; and burnin, 2,500. For each run, Markov chain Monte Carlo (MCMC) diagnostics were calculated in every 1,000 generations. The ML and Bayes trees were checked using TreeView software. Phylogenetic trees were reconstructed using the neighbor-joining (NJ) algorithm and the Kimura two-parameter model implemented within MEGA v4 (52). This method performed 1,000 bootstrap replicates. The GenBank accession numbers of DNA sequences are shown in Table S2 in the supplemental material.
Selection analysis.
The cyr gene sequences from R. curvata CHAB1150, C. raciborskii AWT205, Aphanizomenon sp. strain 10E6, Oscillatoria sp. strain PCC 6506, and cyrA from A. ovalisporum ILC-146 were used for the selection analysis of each cyr gene. These sequences were transformed into amino acid sequences using Bioedit V5.0.6. Protein sequence alignments were created using ClustalX v2.0 and then transformed into codon sequence alignments using the online tool PAL2NAL (51). The codon alignments were then submitted to a suite of phylogenetic analysis tools (http://www.datamonkey.org/) (19) for selection analysis. First, the best model for phylogenetic analysis was chosen using the Model Selection tool. Afterward, the recombination evidence of the sequence alignments was detected using the GARD program (21), and site-to-site rate variation was modeled by a beta-gamma distribution with four rate classes. The significance of the potential recombination breakpoints was examined using the Kishino Hasegawa test (18), which estimates the variance of the difference between log likelihood of different tree topologies. Thereafter, taking recombination events into account, selection was detected using three methods, namely, PARRIS (43), SLAC, and FEL (20). The level of significance was 0.05. In addition, Tajima's test of neutrality was performed using DnaSP v5 (26) for each alignment.
Toxin analysis.
Cyanobacterial cells were centrifuged, collected, and freeze-dried. Extraction and detection of the CYN analogues were then performed using the liquid chromatography-mass spectrometry (LC/MS) method according to a previous work (22). Modifications were made for the LC conditions applied, which were as follows: column, Amide-80, 2.0 mm by 150 mm (Toso Corporation, Japan); mobile phase, 75% CH3CN in 10 mM NH4HCOO–0.1% HCOOH (pH 3.0); injection volume, 5 μl; elution, isocratic; flow rate, 0.2 ml/min; column temperature, 40°C; and detection, photodiode array (PDA). Both positive (m/z 400.2 for 7-deoxy-CYN and m/z 416.2 for CYN and 7-epi-CYN) and negative modes (m/z 398.2 for 7-deoxy-CYN and m/z 414.2 for CYN and 7-epi-CYN) were monitored using the mass spectrometer. Identification of toxin-derived ions was performed according to standard toxins.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the current study are available under GenBank accession numbers JN873921, JN873922, JN873923, JN873924, JN873925, and JN873926.
RESULTS
Characterization of the cyr gene cluster in R. curvata CHAB1150.
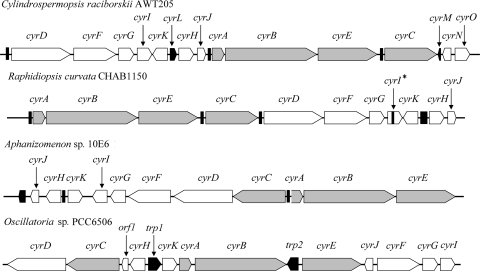
The cyr genes from R. curvata CHAB1150 were successfully amplified. Figure 1 shows the complete cyr gene cluster. The results showed that the organization of the cyr genes in R. curvata CHAB1150 is similar to that in the other two nostocalean species, C. raciborskii AWT205 and Aphanizomenon sp. strain 10E6, and divergent from that in Oscillatoria sp. strain PCC 6506. As displayed in Fig. 1, the cyr gene cluster of nostocalean species can be regarded as a rearrangement of the three conserved sections, namely, two polycistron sections called section 1 (cyrA-cyrB-cyrE) and section 3 (cyrD through cyrK) and the single gene section 2 (cyrC). The arrangement for these sections is as follows: section 3-1-2 for C. raciborskii AWT205, section 1-2-3 for R. curvata CHAB1150, and section (2-3) (reverse complement)-1 for Aphanizomenon sp. strain 10E6. Section 3 is divided into several parts located around sections 1 and 2 for the cyr cluster of Oscillatoria sp. strain PCC 6506.
Fig 1.
Structural organization of the cyr gene clusters from four cyanobacterial strains, including R. curvata CHAB1150, C. raciborskii AWT205 (31), Aphanizomenon sp. strain 10E6 (50), and Oscillatoria sp. strain PCC 6506 (28). Gray and white symbols, rearranged cyr genes; black symbols, transposase genes or vestiges thereof.
The similarities between the cyr genes of R. curvata CHAB1150 and the other cyanobacterial strains decrease in the following order: C. raciborskii AWT205 (99 to 100%), Aphanizomenon sp. strain 10E6 (96 to 99%), A. ovalisporum ILC-146 (96 to 97%), and Oscillatoria sp. strain PCC 6506 (86 to 93%) (Table 1). The similarities between 16S rrn of R. curvata CHAB1150 and the other cyanobacterial strains decrease in the following order: C. raciborskii AWT205 (99%), Aphanizomenon sp. strain 10E6 (95%), A. ovalisporum ILC-146 (94%), and Oscillatoria sp. strain PCC 6506 (91%).
Table 1.
Similarities of cyr genes and the 16S rRNA gene for R. curvata CHAB1150 and other CYN producers, including C. raciborskii AWT205, Aphanizomenon sp. strain 10E6, Oscillatoria sp. strain PCC 6506, and A. ovalisporum ILC-146
| CHAB1150 gene | % similarity to corresponding gene from strain: |
|||
|---|---|---|---|---|
| AWT205 | 10E6 | PCC 6506 | ILC-146 | |
| cyrA | 99 | 99 | 87 | 96 |
| cyrB | 99 | 99 | 89 | 97 |
| cyrC | 99 | 99 | 88 | 97 |
| cyrD | 100 | 99 | 86 | |
| cyrE | 99 | 99 | 87 | |
| cyrF | 99 | 98 | 87 | |
| cyrG | 99 | 96 | 86 | |
| cyrH | 99 | 97 | 88 | |
| cyrI | 100 | 98 | 88 | |
| cyrJ | 100 | 99 | 91 | |
| cyrK | 99 | 98 | 93 | |
| cyrN | 99 | |||
| cyrO | 89 | |||
| rrn (16S rRNA) | 99 | 95 | 91 | 94 |
Figure 1 shows the presence of the transposase sequences or vestiges thereof observed within or around the cyr gene clusters. The intergenic sequence between cyrK and cyrH from R. curvata CHAB1150 is similar (83%) to that of the transposase gene cyrL located between cyrK and cyrH from C. raciborskii AWT205. Partial sequence between cyrC and cyrD from R. curvata CHAB1150 has high similarities to the transposase vestiges located on both 3′-end flanking sequence of cyrC (98%) and 5′-end flanking sequence of cyrD (99%) from C. raciborskii AWT205. In addition, partial sequence flanking the 5′ end of cyrA from R. curvata CHAB1150 has high similarities to the transposase vestiges located between cyrC and cyrA from Aphanizomenon sp. strain 10E6 (100%) and between cyrJ and cyrA from C. raciborskii AWT205 (78%).
The cyrN and cyrO genes found in the cyr cluster of C. raciborskii AWT205 were also amplified from R. curvata CHAB1150. However, these genes are not adjacent to the cyr gene cluster. The identity of these cyrO sequences is only 89%, much lower than that of the other cyr genes. Therefore, it is likely that the two genes cyrN and cyrO do not belong to the cyr gene cluster, as described in Oscillatoria sp. strain PCC 6506 (28).
Mutation and transcription of cyrI gene.
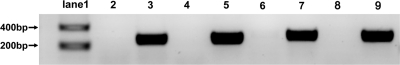
The alignment of cyrI sequences showed a 92-bp sequence insertion after the position of 85 bp (C. raciborskii AWT205 numbering) within the cyrI gene of R. curvata CHAB1150 (Fig. 2; see Fig. S1 in the supplemental material). This insertion causes a frameshift and several stop codons within the cyrI gene. As a result, the protein sequence of CyrI is truncated. In addition, the insertion sequence contains identical inverted terminal repeats (ITRs), implying a probable vestige of transposon. Moreover, a different insertion sequence (1,144 bp) within the cyrI gene of R. curvata HB1 (Fig. 3; see Fig. S2 in the supplemental material) was found. A frameshift and several stop codons (data not shown) also occur within this gene. Furthermore, the insertion sequence contains identical ITRs and a 921-bp open reading frame encoding 306 amino acids with 99% identity to the transposase family IS5 protein of C. raciborskii CS-505 (GenBank accession no. EFA68170). In an attempt to examine the transcription of cyrI in the two Raphidiopsis strains, a primer set targeting the 271-bp region after the insertion position was used. As displayed in Fig. 4, the two Raphidiopsis strains and C. raciborskii AWT205 showed identical results in which both cyrI and cyrJ were detected in the total cDNA. These results indicated that these two genes are both transcribed into the mRNA.
Fig 2.
Alignment of partial cyrI sequences and deduced protein sequences from R. curvata CHAB1150 and C. raciborskii AWT205. Asterisks, stop codons; dashed lines, gaps introduced into the alignment; bold lines, ITRs.
Fig 3.
Alignment of partial cyrI sequences from R. curvata HB1 and C. raciborskii AWT205. Dashed lines, gaps introduced into the alignment; bold lines, ITRs; rectangles, complementary nucleotides for the initial and stop codons; arrow, transcription direction of the transposase gene.
Fig 4.
PCR products of cyrI and cyrJ genes displayed on a 1% agarose gel. Lane 1, DNA marker. Lanes 2 to 5 represent the amplification products of cyrI (lanes 2 and 3) and cyrJ (lanes 4 and 5) from pure RNA extracts, and lanes 6 to 9 represent the amplification products of cyrI (lanes 6 and 7) and cyrJ (lanes 8 and 9) from total cDNA, shown in each case as negative and positive controls, respectively.
Phylogenetic analysis.
The phylogenetic analysis revealed general congruence between 16S rrn (Fig. 5A) and multi-cyr (Fig. 5B), implying a possible coevolution. R. curvata CHAB1150 and C. raciborskii (AWT205 and CS-505) were closely related in both trees and formed a Cylindrospermopsis-Raphidiopsis (C/R) group. In addition, Aphanizomenon sp. strain 10E6 was shown to have a short distance to the C/R group in the multi-cyr tree compared with the 16S rrn tree, implying a high conservation of the cyr genes between Aphanizomenon sp. strain 10E6 and the C/R group. The distant relationship between Oscillatoria sp. strain PCC 6506 and the nostocalean species in both trees revealed that cyr and 16S rrn have coevolved in Oscillatoria sp. strain PCC 6506. Moreover, the results coincided with different similarities between R. curvata CHAB1150 and the other species. A phylogenetic analysis was further performed using the first cyr gene, cyrA, and the corresponding 16S rrn from the same cyanobacterial species, including more available sequences. Similar to the results displayed in Fig. 5A and B, the 16S rrn (Fig. 5C) and cyrA (Fig. 5D) trees were congruent. Aphanizomenon sp. strains 10E6 and 22D11 were much closer to the C/R group in the cyrA tree than the 16S rrn tree. Moreover, U. natans TAC101 and A. ovalisporum ILC-146 were closely clustered in both trees and they were divergent from the C/R group.
Fig 5.
Phylogenetic trees of cyr genes and 16S rRNA genes. (A and B) Trees (topology based on NJ trees) constructed using data sets of 16S rrn-5 and multi-cyr genes. Trees in panels C and D (topology based on Bayesian trees) were constructed using data sets of 16S rrn-10 and cyrA. Bootstrap values above 50% were indicated in the order of ML, Bayesian, and NJ analysis at the nodes of the trees. Outgroup taxa and distantly related Oscillatoria sp. strain PCC 6506 were not shown in panels C and D. Vertical bar, C/R group, including Cylindrospermopsis and Raphidiopsis species. The dash in panel C means there are no data on this branch.
Selection analysis.
As described in Table 2, according to the improvement in Akaike information criterion (AICc) (39) score, the GARD program found 2 to 6 potential recombination breakpoints in the cyr genes. The Kishino Hasegawa test, however, found no significance for these breakpoints. Using the GARD-inferred phylogenetic tree, SLAC found only one negatively selected codon in cyrA and cyrC, respectively, and FEL found 4 to 89 negatively selected codons in each cyr gene. In contrast, no positively selected codon was identified using the SLAC and FEL methods. However, the PARRIS method detected significant evidence of positive selection in cyrK. In Tajima's test, no significant difference from neutral evolution was displayed for all of the cyr genes. To understand the selection stress acting on the cyrA gene of the nostocalean species, the cyrA sequences excluding that of Oscillatoria sp. strain PCC 6506 were analyzed independently. The analyses displayed results similar to those described above.
Table 2.
Number of potential recombination breakpoints and selection test results for the cyr genes
| Gene | No. of recombination breakpoints by GARD model | ΔAICca | Significance by KH test (P < 0.05)b | No. of codons found by negative selection with: |
Significance of positive selection by PARRIS (P < 0.05) | |
|---|---|---|---|---|---|---|
| SLAC | FEL | |||||
| cyrA | 2 | 8.7 | NSc | 1 | 10 | NS |
| cyrAd | 3 | 5.9 | NS | 0 | 1 | NS |
| cyrB | 6 | 182.9 | NS | 0 | 89 | NS |
| cyrC | 3 | 27.9 | NS | 1 | 29 | NS |
| cyrD | 5 | 46.5 | NS | 0 | 50 | NS |
| cyrE | 6 | 50.6 | NS | 0 | 43 | NS |
| cyrF | 6 | 90.6 | NS | 0 | 33 | NS |
| cyrG | 2 | 18.7 | NS | 0 | 26 | NS |
| cyrH | 2 | 29.7 | NS | 0 | 18 | NS |
| cyrJ | 2 | 4.4 | NS | 0 | 4 | NS |
| cyrK | 5 | 90.4 | NS | 0 | 12 | Se |
ΔAICc, improvement in the AICc score of the best-fitting GARD model compared to the single tree model.
KH test, Kishino Hasegawa test.
NS, no significance.
Sequences only from nostocalean species.
S, significant (P = 0).
Toxin analysis.
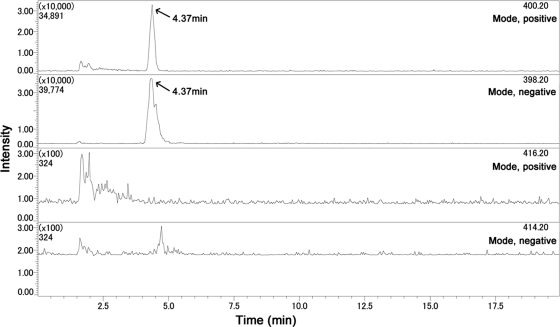
The cell extract of R. curvata CHAB1150 was analyzed using LC/MS. Figure 6 shows that only ions derived from 7-deoxy-CYN were detected in accordance with the ion peaks of the standard toxins (data not shown). The two peaks at m/z 400.20 and m/z 398.20 were both observed at 4.37 min. However, no ion peak for CYN or 7-epi-CYN was observed.
Fig 6.
LC-MS ion chromatogram for cell extracts of R. curvata CHAB1150. The top two graphs (m/z 400.20 and m/z 398.20) represent 7-deoxy-CYN, respectively, and the bottom two graphs (m/z 416.20 and m/z 414.20) represent CYN or 7-epi-CYN, respectively.
DISCUSSION
R. curvata CHAB1150 was detected to produce only 7-deoxy-CYN in the cell extracts. The cyr gene cluster of this Raphidiopsis strain was further characterized to understand whether this toxigenic phenotype results from the variation of the genetic basis. Phylogenetic analyses of the cyr genes were also carried out to explore the origin of these gene clusters.
Mazmouz et al. (29) demonstrated that a 2-oxoglutarate-dependent iron oxygenase encoded by the cyrI gene catalyzes 7-deoxy-CYN into CYN in vitro. The CyrI protein is most likely a tailoring enzyme controlling the synthesis of CYN from 7-deoxy-CYN in the cyanobacterial cells. The activity and variation of the cyrI gene may affect the production of CYN. Comparison of the cyrI gene sequences between R. curvata CHAB1150 and C. raciborskii AWT205 revealed an obvious insertion mutation in the cyrI gene of the former species. The mutation indicated that the cyrI gene is a natural mutant in R. curvata CHAB1150 that lacks CYN and 7-epi-CYN. This absence confirms the predicted function of CyrI in CYN biosynthesis. Similar results were also found for R. curvata HB1 with the production of a large amount of 7-deoxy-CYN but only a trace amount of CYN. The production of CYN in this strain is confusing because the cyrI gene of R. curvata HB1 is interrupted by a 1,144-bp insertion sequence, and the deduced protein sequence is truncated. It is likely that some other oxygenase catalyzes the oxygenation of 7-deoxy-CYN at low activity, thus replacing CyrI. The insertions containing ITRs and transposase sequences are probably mediated by the transposons. The natural mutant of toxin biosynthesis genes has been found in Lyngbya wollei in which the sxtI gene is truncated, and this species produces only decarbamoylated saxitoxin (17). The transcription of the mutated cyrI genes in Raphidiopsis strains may result from the cotranscription of polycistron (section 3).
The similarity of the cyr genes is equal to or higher than that of 16S rrn genes among the nostocalean species, indicating that cyr is even more conservative than 16S rrn. In contrast, cyr genes have lower similarities than 16S rrn between nostocalean species and Oscillatoria sp. strain PCC 6506. Purifying selection against nonsynonymous substitutions at the DNA level is often the mechanism for the conservative evolution of some important genes (15). This mechanism may have influenced the cyr genes from nostocalean species but not Oscillatoria sp. strain PCC 6506. As revealed by Tajima's test of neutrality and selection analysis, the whole sequences of the cyr genes are in neutral evolution, and only a few codons were found under purifying selection. Yilmaz and Phlips (56) indicated that the adenylation domain sequences of the cyrB gene are also under purifying selection. In addition, the PARRIS program detected evidence of positive selection in the cyrK gene. This evidence indicated that the cyr genes may not have identical or single evolution pathways. In general, purifying selection is not the only explanation for the conservation of the whole cyr gene clusters in nostocalean species.
Gene synteny can be observed for the cyr gene clusters, in which the gene organizations of nostocalean species are very similar to each other, but divergent from that of Oscillatoria sp. strain PCC 6506. Such gene synteny corresponds well to the 16S rrn-based phylogenetic trees. Additionally, the congruence of phylogenies indicated the coevolution of 16S rrn and cyr genes, as supported by previous results (16, 50). However, Aphanizomenon sp. strain 10E6 and Aphanizomenon sp. strain 22D11 were shown to be abnormally closer to the C/R group than the other Aphanizomenon species, A. ovalisporum ILC-146 in the cyrA tree (Fig. 5D) compared with the 16S rrn tree (Fig. 5C). This result may be ascribed to the recent horizontal gene transfer (HGT) between the C/R group and the two Aphanizomenon strains, 10E6 and 22D11. HGT has been proposed as the origin of cyr genes in Cylindrospermopsis species (16, 48) because the capability of producing CYN correlates with the possession of cyr genes and the distribution of CYN producers in the phylogenetic tree of cyanobacteria is sporadic. More evidence for HGT was provided by the atypical GC content of the cyr gene cluster (50).
The transposase vestiges between different sections indicated the presence of potential recombination breakpoints at these positions in the cyr gene clusters of the three nostocalean species. Compared with the cyr gene cluster of R. curvata CHAB1150 (section 1-2-3), C. raciborskii AWT205 showed that section 3 within the cyr gene cluster is translocated from downstream of section 1-2 to upstream in the same direction of transcription for section 3-1-2. Moreover, Aphanizomenon sp. strain 10E6 demonstrated that section (2-3) is translocated from downstream of section 1 to the upstream in the reverse direction of transcription for section (2-3) (reverse complement)-1. These translocations may have occurred intragenome independently or intergenome by HGT. The high similarity of transposase sequences at corresponding positions of the three cyr clusters suggested that there has been an original style of gene organization. As displayed in Fig. S3 in the supplemental material, the CyrK sequences of the nostocalean members are very similar but divergent from that of Oscillatoria sp. strain PCC 6506. However, Aphanizomenon sp. strain 10E6 has more identical amino acids to Oscillatoria sp. strain PCC 6506, indicating that the CyrK of Aphanizomenon sp. strain 10E6 is closer to the ancestral type than Raphidiopsis and Cylindrospermopsis.
At present, only few sequences from different CYN producers are available, so the phylogenetic analysis was limited in the present study. More CYN-producing species, multilocus sequence typing, and geographic divergence should be considered in further studies. The involvement of cytochrome P450 in the toxicity of CYN (13) indicates a probable common metabolic pathway and similar toxic metabolites for 7-deoxy-CYN. Therefore, 7-deoxy-CYN is of potential risk for human health and should be monitored together with CYN. The LC/MS method is better than the enzyme-linked immunosorbent assay (ELISA) method for quantifying both toxins simultaneously (28). Most of the known CYN producers belong to Nostocales, and the species of this order often form water blooms. Thus, more attention should be paid to them.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by State Key Basic Research and Development Plan of China (2008CB418002), the National Natural Science Foundation of China (31170189), the National Water Science and Technology Projects (2009ZX07101-013-02), and the Talent Scientist Program of the Chinese Academy of Sciences (082303-1-501).
Footnotes
Published ahead of print 27 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Banker R, et al. 1997. Identification of cylindrospermopsin in Aphanizomenon ovalisporum (Cyanophyceae) isolated from Lake Kinneret, Israel. J. Phycol. 33:613–616 [Google Scholar]
- 2. Banker R, et al. 2001. Uracil moiety is required for toxicity of the cyanobacterial hepatotoxin cylindrospermopsin. J. Toxicol. Environ. Health A 62:281–288 [DOI] [PubMed] [Google Scholar]
- 3. Burgoyne DL, Hemscheidt TK, Moore RE, Runnegar MTC. 2000. Biosynthesis of cylindrospermopsin. J. Org. Chem. 65:152–156 [DOI] [PubMed] [Google Scholar]
- 4. Byth S. 1980. Palm Island mystery disease. Med. J. Aust. 2:40–42 [DOI] [PubMed] [Google Scholar]
- 5. Carmichael WW. 1997. The cyanotoxins. Adv. Bot. Res. 27:211–256 [Google Scholar]
- 6. Dow CS, Swoboda UK. 2000. Cyanotoxins, p 613–632 In Whitton BA, Potts M. (ed), The ecology of cyanobacteria, their diversity in time and space. Kluwer Academic Press, Dordrecht, The Netherlands [Google Scholar]
- 7. Froscio SM, Humpage AR, Wickramasinghe W, Shaw G, Falconer IR. 2008. Interaction of the cyanobacterial toxin cylindrospermopsin with the eukaryotic protein synthesis system. Toxicon 51:191–198 [DOI] [PubMed] [Google Scholar]
- 8. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 9. Harada KI, et al. 1991. Isolation of cylindrospermopsin from a cyanobacterium Umezakia natans and its screening method. Toxicon 29:479–489 [DOI] [PubMed] [Google Scholar]
- 10. Hawkins PR, Runnegar MTC, Jackson ARB, Falconer IR. 1985. Severe hepatotoxicity caused by the tropical cyanobacterium (blue-green alga) Cylindrospermopsis raciborskii (Woloszynska) Seenaya and Subba Raju isolated from a domestic water supply reservoir. Appl. Environ. Microbiol. 50:1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hawkins PR, Chandrasena NR, Jones GJ, Humpage AR, Falconer IR. 1997. Isolation and toxicity of Cylindrospermopsis raciborskii from an ornamental lake. Toxicon 35:341–346 [DOI] [PubMed] [Google Scholar]
- 12. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 13. Humpage AR, Fontaine F, Froscio S, Burcham P, Falconer IR. 2005. Cylindrospermopsin genotoxicity and cytotoxicity: role of cytochrome P-450 and oxidative stress. J. Toxicol. Environ. Health Part A 68:739–753 [DOI] [PubMed] [Google Scholar]
- 14. Ichimura T. 1979. Media for the cultivation of algae, p 295–296 In Nishizawa K, Chihara M. (ed), Methods in phycological studies, Kyouritu Press, Tokyo, Japan: (In Japanese.) [Google Scholar]
- 15. Jordan IK, Rogozin IB, Wolf YI, Koonin EV. 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12:962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kellmann R, Mills T, Neilan BA. 2006. Functional modeling and phylogenetic distribution of putative cylindrospermopsin biosynthesis enzymes. J. Mol. Evol. 62:267–280 [DOI] [PubMed] [Google Scholar]
- 17. Kellmann R, Michali TK, Neilan BA. 2008. Identification of a saxitoxin biosynthesis gene with a history of frequent horizontal gene transfers. J. Mol. Evol. 67:526–538 [DOI] [PubMed] [Google Scholar]
- 18. Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29:170–179 [DOI] [PubMed] [Google Scholar]
- 19. Kosakovsky Pond, Frost SDW. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 20. Kosakovsky Pond SL, Frost SDW. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 21. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891–1901 [DOI] [PubMed] [Google Scholar]
- 22. Kubo T, Sano T, Hosoya K, Tanaka N, Kaya K. 2005. A new simply and effective fractionation method for cylindrospermopsin analyses. Toxicon 46:104–107 [DOI] [PubMed] [Google Scholar]
- 23. Larkin MA, Blackshields G. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 24. Li R, et al. 2001. Isolation and identification of the cyanotoxin cylindrospermopsin and deoxy-cylindrospermopsin from a Thailand strain of Cylindrospermopsis raciborskii (Cyanobacteria). Toxicon 39:973–980 [DOI] [PubMed] [Google Scholar]
- 25. Li R, et al. 2001. First report of the cyanotoxins cylindrospermopsin and deoxycylindrospermopsin from Rhaphidiopsis curvata (Cyanobacteria). J. Phycol. 37:1121–1126 [Google Scholar]
- 26. Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 27. Looper RE, Runnegar MTC, Williams RM. 2005. Synthesis of the putative structure of 7-deoxycylindrospermopsin: C7 oxygenation is not required for the inhibition of protein synthesis. Angew. Chem. Int. Edit. 44:3879–3881 [DOI] [PubMed] [Google Scholar]
- 28. Mazmouz R, et al. 2010. Biosynthesis of cylindrospermopsin and 7-epicylindrospermopsin in Oscillatoria sp. strain PCC 6506: identification of the cyr gene cluster and toxin analysis. Appl. Environ. Microbiol. 76:4943–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mazmouz R, Chapuis-Hugon F, Pichon V, Méjean A, Ploux O. 2011. The last step of the biosynthesis of the cyanotoxins cylindrospermopsin and 7-epi-cylindrospermopsin is catalysed by CyrI, a 2-oxoglutarate-dependent iron oxygenase. Chembiochem 12:858–862 [DOI] [PubMed] [Google Scholar]
- 30. McGregor GB, Sendall BC, Hunt LT, Eaglesham GK. 2010. Report of the cyanotoxins cylindrospermopsin and deoxy-cylindrospermopsin from Raphidiopsis mediterranea skuja (Cyanobacteria/Nostocales). Harmful Algae 10:402–410 [Google Scholar]
- 31. Mihali TK, Kellmann R, Muenchhoff J, Barrow KD, Neilan BA. 2008. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol. 74:716–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muenchhoff J, et al. 2010. A novel prokaryotic L-arginine:glycine amidinotransferase is involved in cylindrospermopsin biosynthesis. FEBS J. 277:3844–3860 [DOI] [PubMed] [Google Scholar]
- 33. Neilan BA, Jacobs D, Goodman AE. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 61:3327–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niiyama Y, Tuji A, Tsujimura S. 2011. Umezakia natans M. Watan. does not belong to Stigonemataceae but to Nostocaceae. Fottea 11:163–169 [Google Scholar]
- 35. Norris RL, et al. 1999. Deoxycylindrospermopsin, an analog of cylindrospermopsin from Cylindrospermopsis raciborskii. Environ. Toxicol. 14:163–166 [DOI] [PubMed] [Google Scholar]
- 36. Ohtani I, Moore RE, Runnegar MTC. 1992. Cylindrospermopsin: a potent hepatotoxin from the blue-green alga Cylindrospermopsis raciborskii. J. Am. Chem. Soc. 114:7942–7944 [Google Scholar]
- 37. Pearson L, Mihali T, Moffitt M, Kellmann R, Neilan B. 2010. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs 8:1650–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 39. Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over Likelihood Ratio Tests. Syst. Biol. 53:793–808 [DOI] [PubMed] [Google Scholar]
- 40. Preußel K, Stüken A, Wiedner C, Chorus I, Fastner J. 2006. First report on cylindrospermopsin producing Aphanizomenon flos-aquae (Cyanobacteria) isolated from two German lakes. Toxicon 47:156–162 [DOI] [PubMed] [Google Scholar]
- 41. Rücker J, et al. 2007. Concentrations of particulate and dissolved cylindrospermopsin in 21 Aphanizomenon-dominated temperate lakes. Toxicon 50:800–809 [DOI] [PubMed] [Google Scholar]
- 42. Runnegar MT, Kong SM, Zhong YZ, Lu SC. 1995. Inhibition of reduced glutathione synthesis by cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Pharmacol. 49:219–225 [DOI] [PubMed] [Google Scholar]
- 43. Scheffler K, Martin DP, Seoighe C. 2006. Robust inference of positive selection from recombining coding sequences. Bioinformatics 22:2493–2499 [DOI] [PubMed] [Google Scholar]
- 44. Schembri MA, Neilan BA, Saint CP. 2001. Identification of genes implicated in toxin production in the cyanobacterium Cylindrospermopsis raciborskii. Environ. Toxicol. 16:413–421 [DOI] [PubMed] [Google Scholar]
- 45. Seifert M, McGregor G, Eaglesham G, Wickramasinghe W, Shaw G. 2007. First evidence for the production of cylindrospermopsin and deoxy-cylindrospermopsin by the freshwater benthic cyanobacterium Lyngbya wollei (Farlow ex Gomont) Speziale and Dyck. Harmful Algae 6:73–80 [Google Scholar]
- 46. Shalev-Alon G, Sukenik A, Livnah O, Schwarz R, Kaplan A. 2002. A novel gene encoding amidinotransferase in the cylindrospermopsin producing cyanobacterium Aphanizomenon ovalisporum. FEMS Microbiol. Lett. 209:87–91 [DOI] [PubMed] [Google Scholar]
- 47. Spoof L, et al. 2006. First observation of cylindrospermopsin in Anabaena lapponica isolated from the boreal environment (Finland). Environ. Toxicol. 21:552–560 [DOI] [PubMed] [Google Scholar]
- 48. Stucken K, et al. 2009. Toxicity phenotype does not correlate with phylogeny of Cylindrospermopsis raciborskii strains. Syst. Appl. Microbiol. 32:37–48 [DOI] [PubMed] [Google Scholar]
- 49. Stucken K, et al. 2010. The smallest known genomes of multicellular and toxic cyanobacteria: comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS One 5:e9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stüken A, Jakobsen KS. 2010. The cylindrospermopsin gene cluster of Aphanizomenon sp. strain 10E6: organization and recombination. Microbiology 156:2438–2451 [DOI] [PubMed] [Google Scholar]
- 51. Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 53. Terao K, et al. 1994. Electron microscopic studies on experimental poisoning in mice induced by cylindrospermopsin isolated from blue-green alga Umezakia natans. Toxicon 32:833–843 [DOI] [PubMed] [Google Scholar]
- 54. Wiedner C, Rücker J, Fastner J, Chorus I, Nixdorf B. 2008. Seasonal dynamics of cylindrospermopsin and cyanobacteria in two German lakes. Toxicon 52:677–686 [DOI] [PubMed] [Google Scholar]
- 55. Wu Z, Shi J, Xiao P, Liu Y, Li R. 2011. Phylogenetic analysis of two cyanobacterial genera Cylindrospermopsis and Raphidiopsis based on multi-gene sequences. Harmful Algae 10:419–425 [Google Scholar]
- 56. Yilmaz M, Phlips EJ. 2011. Diversity of and selection acting on cylindrospermopsin cyrB gene adenylation domain sequences in Florida. Appl. Environ. Microbiol. 77:2502–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.