Abstract
We took advantage of a plant-root enrichment culture system to characterize mesophilic soil archaea selected through the use of organic and inorganic amendments. Comparative analysis of 16S rRNA and amoA genes indicated that specific archaeal clades were selected under different conditions. Three amoA sequence clades were identified, while for a fourth group, identified by 16S rRNA gene analysis alone and referred to as the “root” clade, we detected no corresponding amoA gene. The amoA-containing archaea were present in media with either organic or inorganic amendments, whereas archaea representing the root clade were present only when organic amendment was used. Analysis of amoA gene abundance and expression, together with nitrification-coupled growth assays, indicated potential growth by autotrophic ammonia oxidation for members of two group 1.1b clades. Increased abundance of one of these clades, however, also occurred upon the addition of organic amendment. Finally, although amoA-containing group 1.1a archaea were present in enrichments, we detected neither expression of amoA genes nor evidence for nitrification-coupled growth of these organisms. These data support a model of a diverse metabolic community in mesophilic soil archaea that is just beginning to be characterized.
INTRODUCTION
The realization that newly discovered and apparently ubiquitous members of the domain Archaea carry out the process of ammonia oxidation (15, 63) is one of the most remarkable recent findings from research in environmental microbiology. The ammonia oxidation reaction is the first, and rate-limiting, step in the two-step process of the microbial oxidation of ammonia to nitrate (nitrification). Beginning in 1992 with two reports on the recovery of archaeal 16S rRNA gene sequences from marine plankton (10, 17), numerous studies in conventional marine and terrestrial habitats established high abundance and incidence of a novel taxon of archaea, considered at the time to place within the division Crenarchaeota. This, in itself, was a remarkable finding because the division was comprised exclusively of thermophilic and hyperthermophilic archaea. Just as unexpected was the recovery from temperate, oxygenated environments, more than a decade later, of genes encoding the putative archaeal ammonia monooxygenase enzyme that were linked to 16S rRNA genes of these mesophilic Crenarchaeota (75, 80). Previously, aerobic ammonia oxidation was known for only a few genera of bacteria from the Beta- and Gammaproteobacteria lineages (56).
There have been several reports of enrichments of mesophilic Crenarchaeota and ammonia-oxidizing archaea (AOA) (3, 9, 23, 24, 65, 83), and a large number of surveys have shown widespread distribution and high copy numbers of the gene (amoA) putatively encoding the active subunit of the archaeal ammonia monooxygenase enzyme (16, 41, 44, 83). In fact, AOA amoA gene copies were found to outnumber those of bacterial ammonia oxidizers (AOB) in many marine and soil habitats (15, 63). Two isolates (one marine and one soil) of AOA have been reported (36, 74), with some notable differences in their growth requirements. While the marine “Candidatus Nitrosopumilus maritimus” strain SCM1 achieves 107 cells ml−1 growing autotrophically on bicarbonate and NH4+ (36, 42), the soil “Ca. Nitrososphaera viennensis” strain EN76 requires supplementation with organic carbon, i.e., pyruvate, to achieve continuous growth and reasonably high cell numbers (74). Strain EN76 tolerates higher concentrations of NH4+ than SCM1 and can use urea in place of NH4+ as the sole energy source. Kinetic studies on strain SCM1 indicated that the organism has a very high specific affinity for its substrate and high specific oxidation rates at low ammonium concentrations found in open oceans (42). Together with results from studies quantifying the uptake of inorganic carbon using 13C (84) and 14C (25, 35) bicarbonate tracer experiments and the natural distribution of radiocarbon in archaeal membrane lipids (30), these results suggest autotrophic ammonia oxidation by marine archaea is substantial.
Studies in terrestrial ecosystems, however, have been less conclusive. Although archaeal amoA genes are ubiquitous in soils and, similar to marine habitats, frequently outnumber bacterial amoA genes (12, 32, 41), relating the abundance of AOA to ammonia oxidation activity has been challenging. In some studies, AOA appear to be active in natural samples, based on quantification of amoA gene expression in soil microcosms (48, 75), while in other studies AOA gene abundance did not increase with soil nitrification activity (12, 32, 43, 86). Furthermore, archaeal members of the clade referred to as Crenarchaeota group 1.1b or group 1.1b (11), also referred to as “soil lineage,” are by far the most abundant and ubiquitous in soil (50), but linking the actual growth of AOA to nitrification activity in soil has been reported (40, 51, 73, 87) only for group 1.1a archaea (11), also referred to as the “marine lineage,” occurring there at much lower abundance. There is also evidence suggesting that AOA may have a selective advantage at low ammonium concentrations, possibly exploiting mineralization as a major source of ammonium (42, 69).
Our previous work (65) showed the selection and growth of group 1.1b archaea in enrichment cultures using root extract as substrate, and others have shown that marine (30, 35, 52, 72, 79) and soil (74) archaea can take up and utilize organic carbon or low amounts of CO2 (46) independent of ammonium addition. These results suggest that the capacity for mixotrophic and/or heterotrophic metabolism provides a possible explanation for their abundance in soils worldwide (1). We hypothesized that some of these archaea may be classical heterotrophs, others may be heterotrophic ammonia oxidizers, which do not derive energy from the reaction and oxidize ammonia at low rates that are difficult to detect, and still others might be capable of mixotrophic ammonia oxidation. We previously used 16S rRNA gene analysis to document selection of a specific group 1.1b clade from the roots of Lycopersicon esculentum (tomato) plants grown in Wisconsin soil, using root extract as a growth substrate in culture. Clones from this clade were designated TRC, for tomato root Crenarchaeota (64), and TREC, for tomato root enrichment Crenarchaeota (65). With this report, we further explore the diversity and metabolic potential of root-colonizing soil archaea by examining the AOA assemblage selected in enrichment culture. We document selection of different clades of group 1.1 archaea, including putative AOA, in incubations with organic and inorganic amendments. Our data suggest that as a whole, the group 1.1b archaea are metabolically diverse, with additional strategies besides autotrophic ammonia oxidation. These results are consistent (i) with previous studies in finding that the growth of members of group 1.1b may be uncoupled from ammonia oxidation activity (13, 32, 43, 46, 51, 53, 87) and (ii) with the newly established requirement of organic carbon for growth of “Ca. Nitrososphaera viennensis” strain EN76 (74).
MATERIALS AND METHODS
Inoculum and root extract preparation.
Tomato plants were grown and root inoculum was prepared as described previously (65), except that the plants were grown in an outdoor plot instead of in a growth chamber. Briefly, plants were grown in Oregon silt loam soil (24.9% sand, 60.8% silt, 14.3% clay) containing 4.8% organic matter. Root inoculum was prepared from tomato plants by rinsing roots thoroughly in sterile-distilled H2O to remove soil particles prior to bath sonication for 30 s in sterile Milli-Q H2O. The roots were rinsed in a similar manner for root extract preparation, but instead of sonication, they were ground with a mortar and pestle under liquid N2.
Culture conditions.
The recyclostat was operated on intermittent chemostat/batch cycles such that medium replacement in the 3-liter vessel occurred approximately every 2 months. Temperature was maintained at 22°C by water circulation through the jacket of the reactor. Oxygen levels were maintained under 10% by sparging the culture vessel intermittently (twice weekly) with a gas mixture containing 2% O2 and 0.035% CO2, with the balance as N2. The “SC batch” subculture was inoculated from the recyclostat with a 1:10 dilution into 500 ml of synthetic freshwater Crenarchaeota medium containing 500 μM NH4Cl, 1 mM NaHCO3, 10 mM HEPES, 1 g of NaCl liter−1, 0.4 g of MgCl2·6H2O liter−1, 0.1 g CaCl2·2H2O liter−1, and 0.5 g of KCl liter−1 (with other medium components as described in reference 36). Before inoculation into the flask, the supernatant was removed, and the cells were washed gently three times with fresh medium. The culture was maintained in the dark at 22°C, in semicontinuous batch mode in a 1-liter spinner flask (Bellco Glass Inc., Vineland, NJ) with ports for transport of medium into and out of the vessel. The measured pH was 6.8 and did not change during experiments.
DNA/RNA extraction, purification, and quantification.
Genomic DNA was extracted using the Fast DNA spin kit for soil (Thermo Fisher Scientific, Waltham, MA). For gene expression analysis, total DNA and RNA were extracted from the same homogenized sample in a 1:9 volume ratio, respectively. Total RNA was isolated using a modified phenol-chloroform method (67). RNA was treated with DNase (Promega, Southampton, United Kingdom) and purified with a Qiagen RNeasy minikit (Qiagen, Valencia, CA). Purified RNA was examined by PCR to ensure no DNA contamination was present. DNA and RNA concentrations were quantified using PicoGreen and RiboGreen dye (Invitrogen Corp., Carlsbad, CA), respectively, on a Nanodrop fluorometer (Thermo Scientific, Wilmington, DE), according to the manufacturer's protocol.
Reverse transcription-PCR.
Total RNA was converted to cDNA with random hexamers using a SuperScript III First-Strand Synthesis Supermix for qRT-PCR (Invitrogen Corp.) according to the manufacturer's protocol.
PCR, rRNA gene cloning, and sequencing.
Total extracted DNA from enrichments was amplified by PCR using 16S rRNA gene-specific primers as previously described (65). PCR was performed in separate reactions with the archaea-biased forward primer 109fF (5′-ACKGCTCAGTAACACGT-3′) (19) or 133F (5′-TGTTGACTACGTGTTACTGAG-3′) (64) and the multidomain reverse primer 1492R (5′-GGYTACCTTGTTACGACTT-3′) (39). According to the method of Francis et al. (16), the amoA gene-specific primers Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) were used to generate amoA gene fragments with the following PCR protocol: 95°C for 5 min; followed by 30 cycles consisting of 94°C for 45 s, 53°C for 60 s, and 72°C for 60 s; and finally 72°C for 15 min. PCR products were cloned by ligation using the pGEM-T vector system (Promega Corp., Madison, WI), employing blue-white colony screening. Inserts were confirmed by PCR amplification, sequencing, and sequence analysis. Partial sequences were generated by bidirectional sequencing of cloned products using the vector primers SP6 and T7. Sequencing reactions were carried out according to the OHSU DNA Services Core standardized protocols (http://www.ohsu.edu/xd/research/research-cores/index.cfm) and were read on an ABI 3130xl capillary fluorescence instrument.
Phylogenetic analysis.
16S rRNA gene sequences were aligned in ARB by using the Silva website (55). The number of added neighbors was limited to 40 while generating the alignment, and the auto-reverse/complement option was selected. Regions of ambiguous positional homology were removed, and additional manual corrections were made using MEGA version 4.0 (71) to create an alignment of 79 sequences containing 1,183 homologous nucleotide positions (the alignment of OREC sequences by themselves consisted of 1,299 homologous nucleotide positions). The CLC Genomics Workbench program (CLC Bio, Aarhus, Denmark) was used to align amoA gene sequences, using the MUSCLE algorithm with default settings, which yielded an alignment of 49 sequences containing 590 homologous nucleotide positions, and a translated amino acid alignment of 49 sequences containing 196 homologous amino acid positions.
Phylogenetic analysis was carried out for all data sets in MEGA 4.0, CLC Genomics Workbench, and PhyML (version 2.4.5) (20). Nonparametric bootstrapping was carried out with 1,000 replicates in all cases. Neighbor-joining (NJ) trees were constructed in CLC Genomics Workbench. Minimum-evolution (ME) trees were constructed in MEGA using close neighbor interchange (CNI) with a search level of 2. For ME, nucleotide trees were generated using the maximum-composite-likelihood substitution model, including transitions and transversions, and amino acid trees were generated using the JTT matrix substitution model. Maximum-parsimony (MP) trees were constructed in MEGA using CNI with a search level of 3, and initial trees were constructed by random addition (10 repetitions). Maximum-likelihood (ML) trees were constructed in PhyML. For ML, nucleotide trees were made using the GTR model with base frequency estimates set to ML and estimating for the transition-to-transversion (ts/tv) ratio, the proportion of invariable sites, and gamma distribution; amino acid trees were constructed with the Blosum62 model estimating for the proportion of invariable sites and gamma, using the JTT matrix substitution model. Trees were rooted with multiple outgroups and beautified in Dendroscope (29) for visual clarity.
Primer design for qPCR.
The primers used for quantitative PCR (qPCR) with 16S rRNA genes were 669F (5′-CGACGGTGAGGGATGAAAG-3′) (65) and 88611GR (5′-CCAGGCGGCAGACTTAAC-3′) (88611GR contains an A-to-G modification at position 11 in 886R [65]). Three gene-specific primer sets targeting individual amoA clades were also designed using CLC Genomics workbench software. Primer specificity was tested by PCR with sequenced clones, and the primers were found to amplify only the targeted clade. Primer sequences and cycling parameters are listed in Table S3 in the supplemental material.
qPCR.
qPCR was performed as described for 16S rRNA genes (65), with modifications for amoA genes as described below. In each experiment, three independent master mixes were prepared for each culture, and two independent master mixes were prepared for standard DNA and tested in duplicate within a single microtiter plate. Control reactions (with water in place of template) were also included in each experiment. PCR was conducted in a MyiQ Real-Time qPCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA). Reactions were performed in a volume of 25 μl containing 1× iQ SYBR green Supermix (Bio-Rad) and a 0.2 μM concentration of each primer at optimized annealing temperature (see Table S3 in the supplemental material). Each template DNA or cDNA was amplified using the following cycling conditions: 95.0°C for 4 min; 40 cycles of denaturation at 95.0°C for 30 s, annealing at 59.0°C for 45 s, and elongation at 72.0°C for 45 s; followed by a final extension step at 72°C for 4 min. A melting-curve protocol began after amplification and consisted of 1 min at 95°C, followed by 1 min at 55°C and 80 10-s steps with a 0.5°C increase in temperature at each step. The PCR threshold value and the amplification efficiency of individual wells (82.4 to 98.6%) were calculated using the LinReg PCR program (57). Inhibition was tested for by serial dilution of templates and was not observed. Standard curves were generated by serial dilutions of linearized plasmids. The linear ranges were from 107 to 100 template copies with R2 values of the standard curves of ≥0.99 for all assays. The results were normalized for starting template concentration. Mean values and standard deviations were calculated using Microsoft Excel, version 12.2.8.
PCR-SSCP.
PCR–single-strand conformation polymorphism analysis (PCR-SSCP) was conducted by using the Crenarchaeota-biased primers 133F and 248R5P (66). Template DNA was PCR amplified in a 75-μl reaction mixture containing 37.5 μl of IQ Supermix (Bio-Rad), 33 μl of PCR water, 0.75 μl of each primer (200 nM final concentration), and 3 μl of template DNA. The reaction mixtures were subjected to touchdown PCR as follows: 1 min at 94°C, followed by 20 touchdown cycles consisting of 30 s of denaturation at 94°C, 1.5 min of annealing at 65°C, and 1.5 min of elongation at 72°C. During touchdown, the annealing temperature was reduced every two cycles by 0.5°C, until the final annealing temperature of 55°C was reached. This was followed by 15 cycles of 30 s at 94°C, 1.5 min at 55°C, and 1.5 min at 72°C, with a final extension of 5 min at 72°C. After amplification, the PCR products were purified using a Promega Wizard SV gel and a PCR purification kit (Promega Corp., Madison, WI) according to the manufacturer's protocol. PCR products were then incubated with 15 U of lambda exonuclease in 1× buffer (New England BioLabs, Beverly, MA) at 37°C for 1.5 h, followed by heat inactivation at 65°C for 10 min. The recovered products were desalted using Sephadex G50 microspin columns (Aldrich Chemical Co., Milwaukee, WI). After purification, each PCR product was combined with 15 μl of SSCP stop solution (Lonza, Basel, Switzerland), denatured at 95°C for 3 min, and immediately transferred onto ice. The products were electrophoresed on 1× MDE (Lonza Group, Ltd., Basel, Switzerland) polyacrylamide gels at 300 V for 22 h at 17°C. The gels were scanned on a Typhoon variable mode imager (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), and images were analyzed using GelCompar software II (Applied Maths, Austin, TX). Peak height cutoffs ranged from 5 to 20 relative fluorescence units, depending on the signal intensity.
TOC and TKN measurements.
Total Kjeldahl nitrogen (TKN) and total organic carbon (TOC) were measured at TestAmerica Laboratories, Inc. (Portland, OR) using standard U.S. Environmental Protection Agency methods (76, 77). TKN was determined by semiautomated colorimetry with sulfuric acid digestion. TOC determination was based on the method of UV-promoted persulfate oxidation, converting organic carbon to carbon dioxide.
Nitrification activity assays.
Recyclostat biomass was resuspended 1:10 in 150 ml of synthetic freshwater Crenarchaeota medium as described for SC batch cultures, except 250 μM NH4Cl and 2 mM NaHCO3 were used. Assays were incubated aerobically in the dark at 22°C. Potential ammonia oxidation/nitrification activity was monitored by measuring changes in the concentrations of ammonium, nitrite, and nitrate. Ammonium concentrations were determined by colorimetric measurements of absorbance at 340 nm (45) using a Sigma-Aldrich ammonia assay kit (St. Louis, MO). Alternatively, for low concentrations of ammonium present in root extract, a fluorimetric method (27, 33) was used. Nitrite and nitrate concentrations were determined by photometric measurement of absorbance at 540 nm, using a nitrite/nitrate colorimetric assay kit (Cayman Chemical, Ann Arbor, MI) in a two-step process (18, 49). Colorimetric and fluorimetric measurements were made using a Spectramax M2 plate reader (Molecular Devices, Sunnyvale, CA). The detection limits for nitrite and nitrate were 2 and 2.5 μM, respectively. The detection limits for ammonium were 1.5 nM and 10 μM for the fluorescence and colorimetric assays, respectively.
Nucleotide sequence accession numbers.
All sequences have been deposited in the GenBank database with accession numbers JF799588 to JF799670 (16S rRNA genes) and JF799671 to JF799755 (amoA genes).
RESULTS
Enrichment culture was used to examine the selection of rhizosphere archaea in response to different substrates and growth conditions. Sonicate prepared from the surface of tomato roots grown in Oregon silt loam soil was used to simultaneously inoculate duplicate batch cultures and a semicontinuous biomass recycle reactor, referred to here as a recyclostat (37, 78). The cultures were incubated in parallel in a defined medium containing 1 mM NH4Cl, to which 1× root extract (RE; 5 g liter−1) was added. 1× RE contained 32.6 ± 10.1 mg organic carbon liter−1, 3 μM ammonium, and no detectable nitrate or organic nitrogen (detection limits of 2.5 μM and 1.25 mg liter−1, respectively). The proportion of archaea to bacteria was estimated to range from 10 to 30% using a semiquantitative PCR method, with analysis by fluorescence in situ hybridization and microscopy supporting this assessment (data not shown).
16S rRNA gene phylogeny.
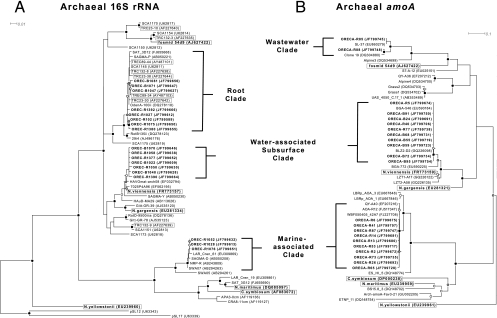
DNA isolated from batch and recyclostat cultures was used in PCR with a forward primer biased broadly toward archaea (109F [19]), or more specifically toward mesophilic soil Crenarchaeota (133F [64]), and the reverse primer 1492R (39) hybridizing broadly to archaeal and bacterial 16S rRNA genes to generate gene fragments for the construction of libraries. Sequence analysis was performed on ≥30 randomly chosen clones from libraries constructed separately with DNA from the recyclostat and from one batch culture. Similar results were obtained with both forward primers (data not shown). Recovered clones were designated “OREC,” for ORegon Enrichment Culture. The clones recovered from recyclostat were more diverse than those from the batch culture, clustering into 3 versus 2 clades, respectively. The majority of OREC sequences recovered from both cultures clustered in the same two major groups, one containing TRC and TREC clones, referred to herein as the “root” clade, and another designated as the “water-associated subsurface” clade (WaS; Fig. 1A). In all, from batch and recyclostat cultures, 46 (35 nonidentical) sequences grouped within the root clade and 32 (30 nonidentical) sequences grouped within the WaS clade (see Table S1 in the supplemental material). Root clade sequences clustered closely (98.1 to 100% sequence identity over 1,183 nucleotides [nt]) with TRC and TREC clones recovered directly from tomato roots grown in Wisconsin (WI) soil (e.g., accession no. AF227642 [64]), and from enrichment cultures inoculated with WI-grown tomato roots (e.g., accession no. AY487103 [65]), respectively. This clade also contained sequences recovered from sediments dominated by the marsh grass Spartina alterniflora (e.g., accession no. FJ655668 [47]) and soils from a fallow agricultural site in Wisconsin (e.g., accession no. U62811 [2]) and the Austrian Central Alps (e.g., accession no. DQ278118).
Fig 1.
Inferred phylogenetic (ML) tree of archaeal 16S rRNA (A) and amoA (B) gene sequences recovered from plant root enrichment cultures. OREC and ORECA clones from the present study (see the text) are shown in boldface type, as are other clones anchoring phylogenetic clades, e.g., “Ca. Nitrososphaera gargensis.” clones anchoring phylogenetic clades, including tomato root clones TRC and TREC, from previous studies, are boxed. Clone number prefixes “B” and “R” indicate recovery from batch and recyclostat enrichments, respectively. GenBank accession numbers are listed parenthetically. Branch points supported by bootstrap values of ≥90%, ≥75%, and ≥50% in all MP and ED methods are indicated by filled, gray, and open circles, respectively. Scale bars: 0.01 (A) and 0.1 (B) change per nucleotide.
WaS clade sequences from recyclostat and batch cultures shared high sequence identity (see Table S1 in the supplemental material) and were most closely related (98.0 to 99.0% identity over 1,183 nt) to biofilm samples from an underground lava cave in Hawaii (e.g., accession no. EF032794), deep subsurface water samples (e.g., accession no. EF562195 [61]) and “Ca. Nitrososphaera viennensis” strain EN76 (74) (Fig. 1A). These sequences were also closely related to clones recovered from moderate and high-temperature subsurface water samples (e.g., accession no. AB113626, ≥97% identity over 1,183 nt) and the archaeal ammonia oxidizer, “Ca. Nitrososphaera gargensis,” enriched from a moderately thermophilic hot spring (≥96% sequence identity [24]).
Six (five nonidentical) OREC clones grouped within a separate cluster (Fig. 1A) most closely related (≥99.6% sequence identity over 1,183 nt) to clones recovered from rice paddy fields (e.g., accession no. AB243809 [62]), deep subsurface waters (e.g., accession no. AB050208 [70]), and the rhizosphere of the freshwater macrophyte Littorella uniflora (e.g., accession no. EU309869 [26]). This sequence group was recovered only from the recyclostat and clustered with group 1.1a, in a clade associated (88.8% identity over 1,183 homologous nucleotides) with the “marine” group (11, 50). Others (40, 48, 86) have referred to this clade, which contains sequences retrieved from freshwater, soils, sediments, and the subsurface, as 1.1a associated. In keeping with this idea, we refer to it here as the “marine-associated” (Ma) clade. The marine clade itself contains sequences of the archaeal ammonia oxidizer “Ca. Nitrosopumilus maritimus,” SCM1 isolated from aquarium sediments (36), the sponge symbiont “Ca. Cenarchaeum symbiosum” (54), and “Ca. Nitrosoarchaeum limnia” SFB1, enriched from low-salinity sediments of San Francisco Bay (3). Upon comparing the three groups recovered from enrichment cultures, the root and WaS clade sequences shared higher identity, both placing into group 1.1b, whereas the Ma clade sequences were more divergent from the others and, as mentioned above, clustered with group 1.1a (see Table S1 in the supplemental material).
Relative abundance of 16S rRNA gene sequence clades under different growth conditions.
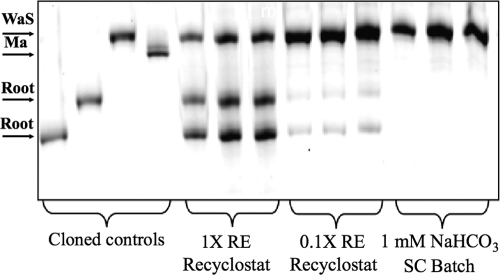
The relative abundance of the different clades was determined in the recyclostat in the presence of 1× RE, or a 10-fold lower concentration (0.5 g liter−1, referred to here as 0.1× RE), using PCR-SSCP analysis. Bands corresponding to individual clades were identified by comigration on polyacrylamide gels with sequenced controls. For the root clade, two different sequence types (sharing 98% identity over the 147-bp amplified region) were identified and corresponded to separate bands on PCR-SSCP gels, whereas sequences from the other clades each produced a single band (Fig. 2). Incubation with 0.1× RE resulted in dominance of the WaS clade (represented by a single band on PCR-SSCP gels) over the root clade (Fig. 2 and Table 1). The root clade (represented by two bands on PCR-SSCP gels) was dominant, however, when 1× RE was used as a substrate. The WaS clade alone was detected in a semicontinuous batch (SC batch) culture inoculated with biomass from the recyclostat and incubated for 4 months with NaHCO3 as the sole carbon source (Fig. 2 and Table 1). The Ma clade was not detected under any conditions using this approach.
Fig 2.
Representative PCR-SSCP gels of the archaeal assemblage in cultures with organic and inorganic amendments. DNA fragments (bands) correspond to root, WaS, and Ma clades, as indicated by comigration with cloned and sequenced controls. Culture conditions are indicated across the bottom of the gel.
Table 1.
Relative abundance of 16S rRNA gene sequence clades in cultures with organic and inorganic amendments
| Clade | % peak abundance (SE)a |
||
|---|---|---|---|
| 1× Recyclostat | 0.1× Recyclostat | SC batch | |
| Root | 65.9 (1.3) | 15.6 (3.4) | ND |
| WaS | 34.1 (1.4) | 84.4 (3.4) | 100 (0) |
| Ma | ND | ND | ND |
Values represent the mean relative percentage of each individual clade's peak area to total peak area for three replicates per treatment in PCR-SSCP assays. ND, not detected.
Selection and growth of the root clade with root extract incubations.
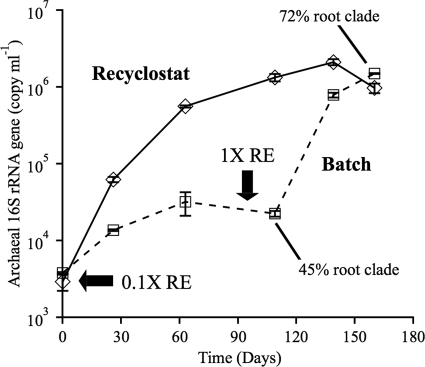
To determine whether root-extract-dependent growth of archaea occurred, abundance was measured by qPCR, using the 669F/886-11GR primer set biased broadly toward the 16S rRNA genes of soil crenarchaeotes (65). Total archaeal abundance (inferred from 16S rRNA gene copies and based on an estimation of one copy per genome) was 1 to 2 orders of magnitude higher in the recyclostat than the batch cultures when both were incubated with 0.1× RE for ∼3.5 months, at which time the batch cultures were supplemented with 1× RE. Over ∼1 month after the addition, archaeal abundance in the batch culture increased 2 orders of magnitude and reached levels similar to those present in the recyclostat (Fig. 3). Similar to the results for the recyclostat (Fig. 2), strong selection for the root clade was observed in batch culture after supplementation with 1× RE (relative abundance of the clade increased from 45 to 72%, Fig. 3), determined by PCR-SSCP analysis (data not shown).
Fig 3.
Change in 16S rRNA gene copy number for total archaea in the recyclostat (diamonds) and batch (squares) cultures over time. The horizontal arrow indicates the time both cultures were supplemented with 0.1× RE; the vertical arrow indicates the time at which batch cultures were supplemented with 1× RE. Representative data are shown. Boxed numbers indicate the change in relative abundance of the root clade in batch culture over the time period indicated (see the text). Point values indicate the means and error bars indicate the standard deviations from three replicate PCRs (tested in duplicate).
amoA gene phylogeny.
The Arch-amoAF and Arch-amoAR primers, biased toward AOA amoA genes, were used to generate clone libraries from the enrichment cultures as described previously (16). Recovered clones were designated “ORECA,” for ORegon Enrichment Culture amoA. Similar to results from the 16S rRNA gene libraries, amoA sequences recovered from the recyclostat were more diverse, clustering into three clades, while those from the batch culture grouped into a single clade. All 43 ORECA sequences retrieved from batch culture clustered with the amoA gene from “Ca. Nitrososphaera viennensis” strain EN76 (74) (Fig. 1B; see also Table S2 in the supplemental material) in the WaS amoA clade, corresponding to the WaS 16S rRNA gene sequence clade. Sequences recovered from the recyclostat clustered mainly into two groups (Fig. 1B): (i) the WaS clade (12 clones) and (ii) a clade of 28 sequences most closely related (97.6% identity over 590 nt) to amoA sequences recovered from marine and estuarine sediments (e.g., accession no. FJ227708), and acidic soils (e.g., accession no. FJ517347, 86). Analogous to the Ma 16S rRNA gene sequence clade, sequences in this amoA clade were highly identical (93.6% identity over 590 nt) to amoA sequences recovered from the roots and rhizosphere of the macrophyte L. uniflora and were associated with group 1.1a. Thus, we refer to this as the Ma amoA clade (see Table S2 in the supplemental material). Two additional sequences were recovered from the recyclostat that clustered most closely with amoA gene sequences recovered from activated sludge (e.g., accession no. EU860279 [88]) and a wastewater treatment plant (e.g., accession no. DQ304880), and therefore these are referred to as the “wastewater (WW)” amoA clade (Fig. 1B; see Table S2 in the supplemental material). Amplification was not detected for AOB amoA genes with the amoA-1F and amoA-2R primers (60) in multiple tests by either endpoint or qPCR (data not shown).
Abundance and expression of amoA genes under different growth conditions.
Primers were designed to differentiate the three amoA clades detected by sequence analysis. The primers were used in qPCR and reverse transcription-qPCR analyses to quantify the abundance of amoA genes and gene transcripts, respectively, representing each clade. WaS clade amoA genes were approximately 3- to 5-fold more abundant in 1× RE incubations compared to incubations with 0.1× RE, while amoA gene copies were 2- to 4-fold more abundant in 0.1× RE incubations for both Ma and WW clades compared to cultures with 1× RE (Table 2).
Table 2.
Comparison of amoA gene copy numbers in cultures with 1× and 0.1× root extract
| Clade | 1× REa |
0.1× RE |
||
|---|---|---|---|---|
| Copy no. | SD | Copy no. | SD | |
| Per ml | ||||
| WaS | 6.2 × 105 | 2.2 × 104 | 3.7 × 105 | 5.1 × 103 |
| Ma | 3.0 × 102 | 2.8 × 101 | 2.0 × 103 | 1.5 × 101 |
| WW | 5.5 × 103 | 4.0 × 102 | 4.1 × 104 | 1.3 × 103 |
| Per ng | ||||
| WaS | 3.7 × 104 | 1.3 × 103 | 1.1 × 104 | 1.7 × 103 |
| Ma | 1.8 × 101 | 1.6 × 100 | 5.8 × 101 | 5.2 × 100 |
| WW | 3.3 × 102 | 2.4 × 101 | 1.2 × 103 | 4.4 × 102 |
RE, root extract.
WaS clade amoA genes were the most abundant under all conditions tested, and the difference was greatest for incubations with 1× RE (2 to 3 orders of magnitude more WaS clade gene copies compared to WW and Ma clades, respectively [Tables 2 and 3]). Overall, amoA gene copies decreased by more than 2 orders of magnitude in the SC batch culture (with NaHCO3 as sole carbon source) but were still 10-fold higher for the WaS clade than for the WW clade (Table 3). The Ma clade dropped below the level of detection in the SC batch culture. Expression of amoA genes was observed only for the WaS clade in both recyclostat and SC batch cultures (Table 3). Due to observations of cell clumping during incubation in the SC batch culture, gene and transcript abundance was estimated per ng of total DNA or RNA, respectively.
Table 3.
Abundance of amoA genes and gene transcripts in cultures with organic (recyclostat) and inorganic (SC batch) amendments
| Clade | Recyclostata |
SC batcha |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene abundance |
Gene expression |
Gene abundance |
Gene expression |
|||||
| Copy no. | SD | Copy no. | SD | Copy no. | SD | Copy no. | SD | |
| WaS | 1.1 × 104 | 1.7 × 103 | 1.4 × 105 | 2.7 × 104 | 5.7 × 101 | 1.6 × 100 | 5.1 × 102 | 1.3 ×102 |
| Ma | 5.8 × 101 | 5.2 × 100 | ND | ND | ND | ND | ND | ND |
| WW | 1.2 × 103 | 4.4 × 102 | ND | ND | 3.4 × 100 | 2.7 × 100 | ND | ND |
Values are means and standard deviations per ng of total DNA or RNA for three replicate PCRs (tested in duplicate). ND, not detected or detection only after 35 PCR cycles.
Nitrification activity and change in abundance of amoA clades.
Potential nitrification activity was measured by incubating resuspended recyclostat biomass in a defined medium containing 260 μM NH4Cl and 2 mM NaHCO3 and determining the loss of ammonium and production of nitrite and nitrate over time (Fig. 4). Although a small amount of nitrite and nitrate was produced and accumulated in these experiments, it was never equivalent to the ammonium lost, except during the first few days of incubation (e.g., after 3 days, 25.5 ± 4.0 μM NH4Cl lost versus 28.8 ± 11.3 μM nitrite/nitrate accumulation). Incubations carried out for longer periods resulted in greater losses of ammonium and decreasing concentrations of measured nitrite and nitrate, indicating assimilation by the enrichment culture consortium (e.g., after 6 days of incubation, a decrease of ∼100 μM ammonium was observed, but the accumulation of nitrite and nitrate was only ∼30 μM). Longer incubations always resulted in lower accumulations of nitrite and nitrate, and the addition of root extract to the assay yielded the same general trend but increased the variability (data not shown). Comparison of amoA gene copy numbers in the starting inoculum to gene copies after 4 days of incubation showed small increases for two of the three clades (Table 4).
Fig 4.
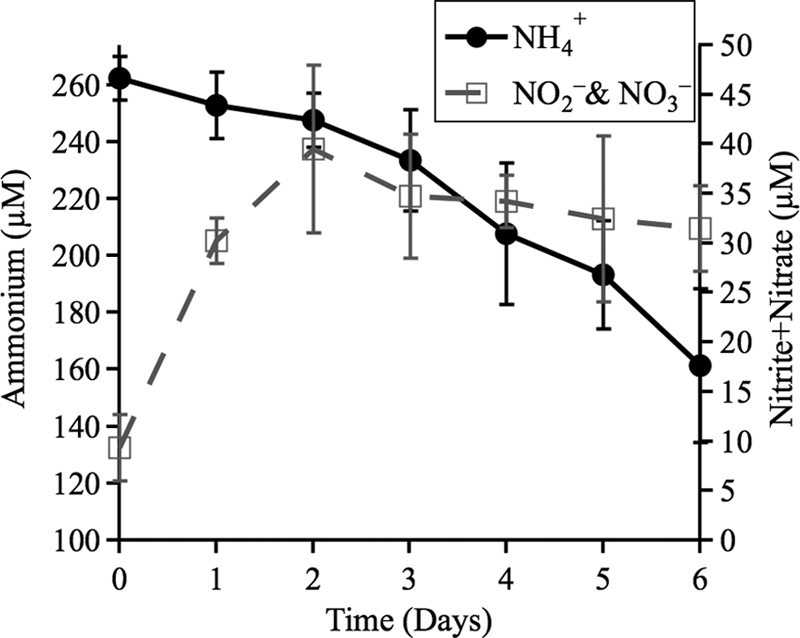
Changes in the concentrations of ammonium and nitrite plus nitrate in incubations for nitrification activity. Means and standard deviations from two representative experiments (with four replications each) are shown.
Table 4.
Change in archaeal amoA gene copy number during nitrification activity assaysa
| Clade | Day 0 |
Day 4 |
Fold changeb | ||
|---|---|---|---|---|---|
| Copy no. | SD | Copy no. | SD | ||
| WaS | 2.0 × 103 | 1.1 × 102 | 4.3 × 103 | 2.5 × 102 | 2.2 |
| Ma | 2.2 × 100 | 5.2 × 10−1 | ND | ND | |
| WW | 2.2 × 101 | 1.4 × 100 | 1.2 × 102 | 1.7 × 101 | 5.5 |
Means and standard deviations per ml are shown; values were estimated from three replicate PCRs (tested in duplicate) for each clade. ND, not detected or detection of signal only after 35 PCR cycles.
That is, the fold change in abundance over 4 days of incubation.
DISCUSSION
Recent discoveries of novel, mesophilic Crenarchaeota and the ability of at least some members of this group to oxidize ammonia have transformed our understanding of microbial nitrogen cycling. These organisms are so different from other cultivated archaea, in fact, that comparative analysis with the few available genomes led to a proposal (4) and support (5, 21, 68) for a new archaeal phylum, the Thaumarchaeota.
Metabolic diversity revealed by selection and growth of specific clades with organic and inorganic amendments.
Similar to an earlier study on archaea from Wisconsin soil (65), we showed here the root extract-dependent selection and growth, in culture, of members of the root clade from Oregon soil. Root clade sequences alone were recovered from previous batch enrichments (65), while sequences recovered in the present study were more diverse; however, more clones were also analyzed. For the first time, putative AOA were also identified in root extract enrichments by recovery of amoA gene sequences. Two different types of enrichments were compared in the recyclostat (semicontinuous replenishment of medium components) and batch (medium components depleted over time) cultures fed with the same root extract-containing medium. The root clade was dominant in the recyclostat culture supplied with 1× RE and was highly enriched in batch culture upon amendment with fresh 1× RE after 3 months of incubation. The WaS clade was dominant in the recyclostat at the lower RE concentration and in batch cultures after depletion of RE or upon supplementation with bicarbonate as the sole carbon source. However, the WaS clade was also more abundant in cultures containing RE than in those without. These results and others (24, 74) suggest a potential mixotrophic lifestyle for members of the WaS clade. There was no apparent selection by root extract for the Ma clade. In fact, the Ma clade was not detected at all in the cultures by PCR-SSCP, probably due to a combination of low abundance (revealed by qPCR) and a single-nucleotide mismatch to Ma clade sequences in both forward and reverse SSCP primers (data not shown). While the mismatch did not affect detection of the control (Fig. 2), relatively low representation in a complex DNA mixture most likely made detection by this method more difficult (66).
Autotrophic and/or mixotrophic ammonia oxidation indicated for the WaS clade.
Expression analysis revealed transcriptional activity from the WaS clade amoA genes only, regardless of culture conditions. These results are consistent with results showing autotrophic ammonia oxidation by the terrestrial hot springs thaumarchaeon “Ca. Nitrososphaera gargensis” (24), possessing genes with high identity to WaS clade sequences. The WaS clade's increase in abundance when incubated with 1× RE, however, is consistent with genomic evidence suggesting that AOA may have the ability to take up organic carbon compounds to supplement growth (22, 23, 82) and with direct evidence for an organic carbon requirement for growth of “Ca. Nitrososphaera viennensis” strain EN76 (74), also closely related to members of the WaS clade. Furthermore, Chen et al. (7) reported a higher abundance of AOA in the rice paddy rhizosphere compared to nonrhizosphere soil, presumably due to root exudates.
Results from experiments examining potential nitrification activity suggested that ammonia oxidation occurred with concomitant growth of archaea from the WaS clade (Fig. 4, Table 4). Isolation in pure culture or 13C-CO2 labeling studies will be necessary, however, to determine whether these organisms actually grow by oxidizing ammonia. Such a result would be consistent with reports showing growth of AOA isolates under nitrifying conditions for similar time periods (42, 74). Documentation of autotrophic and/or mixotrophic ammonia oxidation and growth of group 1.1b archaea in culture (24, 74) raises the question of why it has been so difficult to link ammonia oxidation activity to growth of members of this clade in soil. A possible explanation could be that although these organisms are selected for in cultivation experiments, they are not representative of the most abundant archaeal clades in soil (1), making it difficult to detect their activity due to their low numbers. Consistent with this idea, a recent study on nitrification in soil microcosms (85) found that relatively low numbers of “Ca. Nitrososphaera gargensis”-like amoA and 16S rRNA genes were labeled, compared to AOB genes, in 13C-CO2 uptake experiments. If the majority of AOA in soil are heterotrophic nitrifiers, correspondingly lower rates of ammonia oxidation may also be harder to detect.
Although their numbers were smaller, the fold increase for the WW clade was even greater than for the WaS clade during the nitrification assays (Table 4). Taken together with reduced numbers of this clade in the presence of higher concentrations of root extract, this result may indicate an ability to oxidize ammonia autotrophically, although we did not detect amoA gene transcripts from this group. Their relatively low numbers may have made such detection more difficult, however. Further research using 13C-CO2 labeling experiments may shed more light on the metabolic capabilities of this group.
Are members of the root clade heterotrophs?
Phylogenetic analysis of clone libraries indicated correspondence between 16S rRNA and amoA genes of both WaS and Ma clades but not between the 16S rRNA gene of the root clade and the amoA gene of the WW clade. Furthermore, the WW clade amoA genes were detected in the SC batch culture, while the root clade 16S rRNA genes were not. In total, our data suggest that either our primers did not detect an existing amoA gene for the root clade or one does not exist. In our reading of the literature, we found no direct evidence for an amoA gene corresponding to this clade, although results from one study comparing 16S rRNA and amoA clones retrieved from the same soil samples were somewhat suggestive (48). It is possible that such a gene(s) may be revealed by additional sequence analysis in the future. In fact, the amoA primer set used in the present study was shown to have mismatches with amoA sequences from, for example, “Ca. Nitrosocaldus yellowstonii,” “Ca. Nitrosopumilus maritimus,” and “Ca. Cenarchaeum symbiosum” (38). Our failure to recover the corresponding 16S rRNA gene for the WW amoA clade may have been due to a primer mismatch(es) or to the fact that at the time of cloning we appeared to be near the limit of detection for this clade, recovering only two clones.
Given the preferential selection of the root clade by root extract and its absence in incubations with bicarbonate as sole carbon source, it is tempting to speculate that the root clade is heterotrophic and that members of this group utilized organic matter in the root extract for growth. Although we did not discover one, it is also possible that they possess an as-yet-undetected amoA gene and are, in fact, capable of heterotrophic ammonia oxidation. Some heterotrophic bacteria, for instance, perform ammonia oxidation by a process that is analogous to that used by chemoautotrophic AOB (8). Organic nitrogen was not detected in the root extract (detection limit, 1.25 mg/liter), and while it is formally possible that the root clade responded to inorganic ammonium in the root extract in batch culture experiments, this is unlikely to have been the case in the recyclostat because the concentration of ammonium in the incubation medium was much higher (1 mM) than the ammonium in root extract (3 μM), making organic carbon seem more likely as a driver of the selection. Of course, it is possible that members of the root clade responded to some component of root extract other than organic carbon and/or nitrogen or to detritus from increased biomass and turnover of the enrichment consortium as a whole. It is interesting that the response was similar to root extract produced from plants grown in different soils and locations. Experiments are planned to determine directly whether root-derived carbon is taken up by archaea.
Based on somewhat controversial data (14, 81), heterotrophic nitrification is thought to be less important than autotrophic nitrification in the environment. However, many widespread and abundant heterotrophic bacteria and fungi have the ability to oxidize a variety of nitrogenous compounds (6, 14). In addition, several heterotrophic species of bacteria previously considered to be “poor” nitrifiers actually nitrify and denitrify simultaneously, giving a falsely low impression of their nitrification potential (58, 59). In some cases, the heterotrophic nitrification rates are less than a factor of 10 lower than rates for autotrophs (31). Heterotrophic nitrifiers do not appear to derive energy from ammonia oxidation but may instead use nitrification for the dissipation of excess reductant (58). The fact that growth rates of heterotrophs tend to be higher than those of autotrophs and that heterotrophic nitrifiers can be present in considerably higher numbers than autotrophic nitrifiers (14) suggests that the ecological significance of heterotrophic nitrification is not well understood. Together with the fact that the high abundance of AOA in many soil environments has not been linked to (generally higher rates of) autotrophic ammonia oxidation, the idea that heterotrophic (and/or mixotrophic) nitrification is carried out by archaea appears to be a viable hypothesis and is supported by the requirement for organic carbon by “Ca. Nitrososphaera viennensis” strain EN76 and its ability to oxidize organic nitrogen (74).
The hypothesis is additionally compelling in light of evidence that marine Thaumarchaeota take up some forms of organic carbon (30, 35, 52, 72, 79). Although the focus on archaeal nitrification has largely been on autotrophic ammonia oxidation, analysis of mesophilic Thaumarchaeota and AOA in natural environmental samples and microcosms indicates potential for mixotrophy or heterotrophy. Isotopic analysis of archaeal lipid biomarkers suggested that ca. 17% of these organisms in mesopelagic zones of the ocean may assimilate organic carbon (30). Research also indicated that Thaumarchaeota take up amino acids in deep ocean waters (52, 72, 79), and one report described their assimilation of protein and diatom extracellular polymers in both surface and deeper waters of the Western Arctic Ocean (35). Furthermore, the genome sequences of “Ca. Nitrosopumilus maritimus” SCM1 (82) and “Ca. Cenarchaeum symbiosum” revealed several putative ABC-type transporter systems for organic carbon acquisition (22, 23). Genes predicted to encode a nearly complete oxidative tricarboxylic acid cycle were identified, which is consistent with the consumption of organic carbon and the production of intermediates for amino acid and cofactor biosynthesis. Evidence for organic carbon consumption was also uncovered in the genome sequences of “Ca. Nitrosoarchaeum limnia” SFB1 (3) and “Ca. Nitrosoarchaeum koreensis” MY1, enriched from rhizosphere soil (34). Finally, the well-studied autotrophic ammonia oxidizing AOB, Nitrosomonas europaea, was recently shown to be a facultative mixotroph, utilizing fructose and pyruvate in the absence of CO2, its preferred carbon source (28). In conclusion, our results suggest that mesophilic soil archaea harbor a diverse set of metabolisms. Although autotrophic ammonia oxidation by AOA seems to be clearly important in marine environments and potentially so in soil habitats, it may turn out that the ability of these organisms to carry out mixotrophic and/or heterotrophic forms of metabolism largely explains their high abundance in soils.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Science Foundation grants MCB-0644468 and EF-0644468.
We are thankful to Rachel Henson and Adam Bonin for technical assistance.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bates ST, et al. 2010. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bintrim SB, Donahue TJ, Handelsman J, Roberts GP, Goodman RM. 1997. Molecular phylogeny of archaea from soil. Proc. Natl. Acad. Sci. U. S. A. 94: 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blainey PC, Mosier AC, Potanina AP, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6: e16626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6: 245–252 [DOI] [PubMed] [Google Scholar]
- 5. Brochier-Armanet C, Gribaldo S, Forterre P. 2011. Spotlight on the Thaumarchaeota. ISME J. 6: 227–230 (Commentary) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castignettii D, Hollocher TC. 1984. Heterotrophic nitrification among denitrifiers. Appl. Environ. Microbiol. 47: 620–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X-P, Zhu Y-G, Xia Y, Shen J-P, He J-Z. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10: 1978–1987 [DOI] [PubMed] [Google Scholar]
- 8. Crossman LC, Moir JWB, Enticknap JJ, Richardson DJ, Spiro S. 1997. Heterologous expression of heterotrophic nitrification genes. Microbiology 143: 3775–3783 [DOI] [PubMed] [Google Scholar]
- 9. de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10: 810–818 [DOI] [PubMed] [Google Scholar]
- 10. DeLong EF. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89: 5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeLong EF. 1998. Everything in moderation: archaea as “non-extremophiles.” Curr. Opin. Genet. Dev. 8: 649–654 [DOI] [PubMed] [Google Scholar]
- 12. Di H, et al. 2009. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2: 621–624 [Google Scholar]
- 13. Di H, et al. 2010. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72: 386–394 [DOI] [PubMed] [Google Scholar]
- 14. Focht DD, Verstraete W. 1977. Biochemical ecology of nitrification and denitrification [Soils]. Adv. Microb. Ecol. 1: 135–214 [Google Scholar]
- 15. Francis CA, Beman JM, Kuypers MMM. 2007. Minireview: new processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1: 19–27 [DOI] [PubMed] [Google Scholar]
- 16. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102: 14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuhrman JA, McCallum K, Davis AA. 1992. Novel major archaebacterial group from marine plankton. Nature 36: 148–149 [DOI] [PubMed] [Google Scholar]
- 18. Green LC, et al. 1982. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids Anal. Biochem. 126: 131–138 [DOI] [PubMed] [Google Scholar]
- 19. Grobkopf R, Stubner S, Liesack W. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64: 4983–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- 21. Gupta R, Shami A. 2011. Molecular signatures for the Crenarchaeota and the Thaumarchaeota. Antonie Van Leeuwenhoek 99: 133–157 [DOI] [PubMed] [Google Scholar]
- 22. Hallam SJ, et al. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U. S. A. 103: 18296–18301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hallam SJ, et al. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4: e95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hatzenpichler R, et al. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herndl G, et al. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71: 2303–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herrmann M, Saunders AM, Schramm A. 2008. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 74: 3279–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holmes RM, Aminot A, Kérouel R, Hooker BA, Peterson BJ. 1999. A simple and precise method for measuring ammonium in marine and freshwater ecosystems Can. J. Fish. Aquat. Sci. 56: 1801–1808 [Google Scholar]
- 28. Hommes NG, Sayavedra-Soto LA, Arp DJ. 2003. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J. Bacteriol. 185: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huson DH, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 8: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ingalls AE, et al. 2006. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. U. S. A. 103: 6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jetten MSM, et al. 1997. Novel principles in the microbial conversion of nitrogen compounds. Antonie Van Leeuwenhoek 71: 75–93 [DOI] [PubMed] [Google Scholar]
- 32. Jia Z, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11: 1658–1671 [DOI] [PubMed] [Google Scholar]
- 33. Kérouel R, Aminot A. 1997. Fluorometric determination of ammonia in sea and estuarine waters by direct segmented flow analysis. Mar. Chem. 57: 265–275 [Google Scholar]
- 34. Kim BK, et al. 2011. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J. Bacteriol. 193: 5539–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kirchman DL, Elifantz H, Dittel AI, Malmstrom RR, Cottrell MT. 2007. Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnol. Oceanogr. 52: 495–507 [Google Scholar]
- 36. Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546 [DOI] [PubMed] [Google Scholar]
- 37. Konopka A, Zakharova T, Oliver L, Camp D, Turco RF. 1996. Biodegradation of organic wastes containing surfactants in a biomass recycle reactor. Appl. Environ. Microbiol. 62: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konstantinidis K, Braff J, Karl D, DeLong E. 2009. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at Station ALOHA in the North Pacific subtropical gyre. Appl. Environ. Microbiol. 75: 5345–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Goodfellow ES. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc, New York, NY: [Google Scholar]
- 40. Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U. S. A. 108: 15892–15897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leininger S, et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809 [DOI] [PubMed] [Google Scholar]
- 42. Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979 [DOI] [PubMed] [Google Scholar]
- 43. Mertens J, et al. 2009. Bacteria, not archaea, restore nitrification in a zinc-contaminated soil. ISME J. 3: 1–8 [DOI] [PubMed] [Google Scholar]
- 44. Mincer TJ, et al. 2007. Quantitative distribution of presumptive archaeal bacterial nitrifiers in Monterey Bay the North Pacific subtropical gyre. Environ. Microbiol. 9: 1162–1175 [DOI] [PubMed] [Google Scholar]
- 45. Mondzac A, Ehrlich GE, Seegmiller JE. 1965. An enzymatic determination of ammonia in biological fluids. J. Lab. Clin. Med. 66: 526–531 [PubMed] [Google Scholar]
- 46. Mussmann M, et al. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108: 16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nelson KA, Moin NS, Bernhard AE. 2009. Archaeal diversity and the prevalence of Crenarchaeota in salt marsh sediments. Appl. Environ. Microbiol. 75: 4211–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10: 2966–2978 [DOI] [PubMed] [Google Scholar]
- 49. Nims RW, et al. 1995. Colorimetric methods for the determination of nitric oxide concentration in neutral aqueous solutions. Methods Companion Methods Enzymol. 7: 48–54 [Google Scholar]
- 50. Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. 2004. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real-time PCR. Environ. Microbiol. 5: 787–797 [DOI] [PubMed] [Google Scholar]
- 51. Offre P, Prosser JI, Nicol GW. 2009. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 70: 99–108 [DOI] [PubMed] [Google Scholar]
- 52. Pérez MT, Pausz C, Herndl GJ. 2003. Major shift in bacterioplankton utilization of enantiomeric amino acids between surface waters and the ocean's interior. Limnol. Oceanogr. 48: 755–763 [Google Scholar]
- 53. Pratscher J, Dumont M, Conrad R. 2011. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U. S. A. 108: 4170–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Preston CM, Wu KY, Molinski TF, Delong EF. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. U. S. A. 93: 6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned rRNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Purkhold U, et al. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66: 5368–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66 [DOI] [PubMed] [Google Scholar]
- 58. Robertson LA, Kuenen JG. 1990. Combined heterotrophic nitrification and aerobic denitrification in Thiosphaera pantotropha and other bacteria. Antonie Van Leeuwenhoek 57: 139–152 [DOI] [PubMed] [Google Scholar]
- 59. Robertson LA, Van Niel EWJ, Torremans RAM, Kuenen JG. 1988. Simultaneous nitrification and denitrification in aerobic chemostat cultures of Thiosphaera pantotropha. Appl. Environ. Microbiol. 54: 2812–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rotthauwe J-H, Witzel K-P, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63: 4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sahl JW, et al. 2008. Subsurface microbial diversity in deep-granitic-fracture water in Colorado. Appl. Environ. Microbiol. 74: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sakai S, et al. 2007. Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster Rice Cluster I. Appl. Environ. Microbiol. 73: 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schleper C, Nicol G. 2010. Ammonia-oxidizing archaea: physiology, ecology and evolution. Adv. Microb. Phys. 57: 1–41 [DOI] [PubMed] [Google Scholar]
- 64. Simon HM, Dodsworth JA, Goodman RM. 2000. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2: 495–505 [DOI] [PubMed] [Google Scholar]
- 65. Simon HM, et al. 2005. Cultivation of mesophilic crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71: 4751–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sliwinski MK, Goodman RM. 2004. Spatial heterogeneity of crenarchaeal assemblages within mesophilic soil ecosystems as revealed by PCR–single-stranded conformation polymorphism profiling. Appl. Environ. Microbiol. 70: 1811–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smith M, et al. 2010. Seasonal changes in bacterial and archaeal gene expression patterns across salinity gradients of the Columbia River coastal margin. PLoS One 5: e13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Spang A, et al. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18: 332–340 [DOI] [PubMed] [Google Scholar]
- 69. Stopnisek N, et al. 2010. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 76: 7626–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67: 5750–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 72. Teira E, Aken HMV, Veth C, Herndl GJ. 2006. Archaeal uptake of enantiomeric amino acids in meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51: 60–69 [Google Scholar]
- 73. Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10: 1357–1364 [DOI] [PubMed] [Google Scholar]
- 74. Tourna M, et al. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108: 8420–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Treusch AH, et al. 2005. Novel genes for nitrite reductase and Amo-related proteins indicated a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7: 1985–1995 [DOI] [PubMed] [Google Scholar]
- 76. U.S. Environmental Protection Agency 1983. Methods for chemical analysis of water and wastes, vol EPA-600/4-79-020, p 351.2. Environmental Monitoring and Support Laboratory, Cincinnati, OH: [Google Scholar]
- 77. U.S. Environmental Protection Agency 1983. Methods for chemical analysis of water and wastes, vol EPA-600/4-79-020, p 415.2. Environmental Monitoring and Support Laboratory, Cincinnati, OH: [Google Scholar]
- 78. van Verseveld HW, Arbige M, Chesbro WR. 1984. Continuous culture of bacteria with biomass retention. Trends Biotechnol. 2: 8–12 [Google Scholar]
- 79. Varela MM, van Aken HM, Sintes E, Herndl GJ. 2008. Latitudinal trends of Crenarchaeota and Bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ. Microbiol. 10: 110–124 [DOI] [PubMed] [Google Scholar]
- 80. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74 [DOI] [PubMed] [Google Scholar]
- 81. Verstraete W. 1975. Heterotrophic nitrification in soils and aqueous media. Izv. Akad. Nauk. SSSR Ser. Biol. 4: 541–558 (Translation) [Google Scholar]
- 82. Walker CB, et al. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107: 8818–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wuchter C, et al. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103: 12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wuchter C, Schouten S, Boschker HTS, Damste JSS. 2003. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol. Lett. 219: 203–207 [DOI] [PubMed] [Google Scholar]
- 85. Xia W, et al. 2011. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 6: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ying J-Y, Zhang L-M, He J-Z. 2010. Putative ammonia-oxidizing bacteria and archaea in an acidic red soil with different land utilization patterns. Environ. Microbiol. Rep. 2: 304–312 [DOI] [PubMed] [Google Scholar]
- 87. Zhang L-M, et al. 2010. Autotrophic ammonia oxidation by soil thaumarchaea. Proc. Natl. Acad. Sci. U. S. A. 107: 17240–17245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang T, et al. 2009. Occurrence of ammonia-oxidizing Archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J. Appl. Microbiol. 107: 970–977 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.