Abstract
Lactobacillus rhamnosus GG is a human intestinal isolate that has been studied intensively because of its probiotic properties. We have previously shown that L. rhamnosus GG produces proteinaceous pili that earlier had been observed only in Gram-positive pathogens (M. Kankainen et al., Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198, 2009). These pili were found to be encoded by the spaCBA gene cluster, and the pilus-associated SpaC pilin was shown to confer on the cells a mucus-binding ability. In addition to the spaCBA cluster, another putative pilus cluster, spaFED, was predicted from the L. rhamnosus GG genome sequence. Herein, we show that only SpaCBA pili are produced by L. rhamnosus, and we describe a detailed analysis of cell wall-associated and affinity-purified SpaCBA pili by Western blotting and immunogold electron microscopy. Our results indicate that SpaCBA pili are heterotrimeric protrusions with a SpaA subunit as the shaft-forming major pilin. Only a few SpaB subunits could be observed in pilus fibers. Instead, SpaB pilins were found at pilus bases, as assessed by immunogold double labeling of thin sections of cells, suggesting that SpaB is involved in the termination of pilus assembly. The SpaC adhesin was present along the whole pilus length at numbers nearly equaling those of SpaA. The relative amount and uniform distribution of SpaC within pili not only makes it possible to exert both long-distance and intimate contact with host tissue but also provides mucus-binding strength, which explains the prolonged intestinal residency times observed for L. rhamnosus GG compared to that of nonpiliated lactobacilli.
INTRODUCTION
Pili or fimbriae are proteinaceous surface appendages found in both Gram-negative and Gram-positive bacteria. In Gram-positive bacteria, pili have been studied intensively for the last decade and typically are very thin in these organisms, ranging from 1 to 10 nm in diameter (27). Pili thus far have been identified in a few pathogenic Gram-positive species, such as Corynebacterium diphtheriae (5, 26, 28), Actinomyces naeslundii (17, 31), Bacillus cereus (3), Enterococcus faecalis (19), Enterococcus faecium (7, 9), and in several members of the genus Streptococcus, such as Streptococcus agalactiae (14), Streptococcus pneumoniae (2), and Streptococcus pyogenes (18). In Gram-negative bacteria, where the pili have been studied for decades, the pilus backbone generally is composed of homopolymers of noncovalently bound subunits (21). In contrast, the pili in Gram-positive bacteria are composed of strings of major pilin subunits that are covalently linked to each other by a series of sortase-mediated transpeptidation reactions (8).
The monomeric pilins are secreted across the plasma membrane of Gram-positive bacteria by the Sec-dependent pathway (24). During secretion the nascent pilins are sequestered to the membrane by a hydrophobic tail preceded by a conserved LPXTG motif. The individual pilus fibers are formed by the polymerization of major subunits, in which a lysine (K) in the proposed WxxxVxYPKN-like pilin motif of the added subunit forms an isopeptide bond with a threonine (T) residue in the LPXTG motif of the pilin subunit already attached to the growing pilus chain (27). This reaction, which is catalyzed by a pilus-specific sortase (also referred to as class C sortase), is repeated many times, as there are some 100 or more pilin subunits in a single pilus fiber. Therefore, the biosynthesis of growing pilus chains follows the classical head-to-tail growth mode found in the formation of many biological polymers (10). In the last step the mature pilus fiber is joined to the peptidoglycan precursor lipid II by another sortase enzyme, the housekeeping sortase (termed the class A sortase), ultimately leading to the covalent attachment of the pilus to the cell wall (25, 29).
Pili in Gram-positive bacteria generally are composed of two or three different subunits (8, 16). In addition to the above-mentioned major pilin, individual pili also contain one or two minor pilins. It can be envisioned that the minor subunits are linked to pilus fibers in three different ways. A minor pilin lacking the pilin motif but containing the LPXTG motif could serve as an initiator of pilus polymerization and therefore become located at the pilus tip. Alternatively, a minor pilin could be added to the chain similarly to the major pilin, i.e., through the pilin motif in the minor subunit. The third, albeit less defined, attachment mechanism is assumed to involve the conserved residues in the E box motif (YxLxETxAPxGY) usually located between the pilin motif and the LPXTG sequence; the replacement of the glutamate in the E box of the SpaA major subunit has been shown to abolish the incorporation of the SpaB minor pilin to the SpaABC pili in C. diphtheriae (29). Of note, in the latter mechanism the minor subunit is not added by the head-to-tail mechanism and therefore will not be an integral part of the pilus fiber; rather, it is attached to the pilus from the outside in a decorative manner.
Recently, we identified pili in the probiotic Lactobacillus rhamnosus GG, challenging the dogma that pili are pathogenicity factors. L. rhamnosus GG is a human commensal intestinal isolate with a well-documented record of safe consumption and health-promoting effects. Based on the genome sequence, L. rhamnosus GG was predicted to possess two pilus gene clusters, spaCBA and spaFED (12). By using Western blotting and immunogold transmission electron microscopy (TEM), we demonstrated that L. rhamnosus GG harbors SpaC-containing pili. In addition, L. rhamnosus GG cells and recombinant SpaC pilin were shown to bind to human mucus. This binding could be inhibited with SpaC antiserum; also, an insertional mutant strain, L. rhamnosus GG-ΩspaC, completely lacked the mucus-binding ability (12). Taking these data together, L. rhamnosus GG (and possibly other probiotic lactobacilli) produce pili that are able to bind to the mucus layer of the intestinal tract, thereby potentially allowing L. rhamnosus GG to exert its probiotic actions. In a further detailed analysis, we produced the remaining pilins (SpaA, SpaB, SpaD, SpaE, and SpaF) as histidine-tagged recombinant proteins in Escherichia coli and assessed their mucus-binding properties (30). In this work it was noted that the SpaB and SpaF pilins also possess a mucus-binding ability. The finding that SpaB, SpaC, and SpaF show adhesive characteristics is in line with other studies, since minor pilin genes in various pilus clusters have frequently been shown to encode pilus adhesins; in fact, thus far no major subunits of pili have been assigned adhesive functions (13). Moreover, as predicted from the amino acid sequence, the SpaC adhesin was shown to contain a von Willebrand factor-like domain with possible lectin-type binding. However, the mucus-binding property of SpaB, the smallest subunit encoded by the spaCBA gene cluster, could be explained simply by electrostatic interactions with the negatively charged glycan chains present in mucus, as it has a net positive charge (isoelectric point, 8.0) (30).
Aside from findings from adhesion studies, there are questions about pilus structures and subunits involved in pilus biosynthesis; notably, in L. rhamnosus GG there are two pilus gene clusters, spaCBA and spaFED, which are located distantly from each other as individual islands in the GG genome. Since different types of pili have been reported to be simultaneously present on the surfaces of C. diphtheriae, E. faecium, and S. pneumoniae (1, 5, 7, 26, 28), we performed a detailed study utilizing Western blotting and immunogold TEM in combination with antibodies (Ab) raised against recombinant SpaCBA and SpaFED pilins to reveal the individual pilins making up pili in L. rhamnosus GG and to assess the spatial organization of different subunits within the fiber. Here we report the presence of heterotrimeric SpaCBA pili and the absence of SpaFED pili in L. rhamnosus GG. Based on our findings about the multisubunit organization of the SpaCBA pili, we present a refined structural model of heterotrimeric pili in Gram-positive bacteria with multiple copies of the adhesive minor subunit in pilus fiber.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Lactobacillus rhamnosus GG (ATCC 53103) strain was kindly provided by Valio Ltd. (Finland) and was routinely cultivated in static de Man-Rogosa-Sharpe (MRS) broth (Difco) at 37°C. In certain cases L. rhamnosus GG cells also were grown in MRS without glucose, MRS supplemented with bile (0.6% oxgall; Sigma), or modified tryptic soy broth (TSB) at 30 and 37°C.
Cell fractionation and isolation of cell wall proteins.
L. rhamnosus GG cell fractionation and the isolation of cell wall-associated proteins were performed as described previously (12). Briefly, L. rhamnosus GG cells (1 × 1010 CFU) grown overnight were harvested by centrifugation, washed once with phosphate-buffered saline (PBS) (pH 7.2), and then broken mechanically three times for 120 s with glass beads in a milling machine (Bühler Vibrogen-Zellmühle). A 0.5-ml volume of PBS was added to the disrupted cell suspension, and the unbroken cells and cellular debris were removed by low-speed centrifugation (1,000 × g for 1 min). The cell extract then was collected and fractionated by centrifugation at 16,000 × g for 30 min at 4°C to separate cytosolic and cell wall-bound components. The supernatant represented the cell-free cytosolic fraction and was retained for further use. To recover proteins associated with the cell wall fraction, the pellet was resuspended in 50 μl of digestion buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 5 mM CaCl2, 10 mg/ml lysozyme, and 150 U/ml mutanolysin), incubated at 37°C for 3 h, and then analyzed by SDS-PAGE and immunoblotting.
Immunoaffinity purification of native L. rhamnosus GG SpaCBA pili.
L. rhamnosus GG cells were grown to early log phase (optical density at 600 nm [OD600] of 0.5) at 37°C in 1 liter of MRS broth and then were harvested by centrifugation at 5,000 × g for 10 min. The cell pellet was washed once with PBS, resuspended to a 100-ml final volume in protoplasting buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 5 mM CaCl2, 20% sucrose, 10 mg/ml lysozyme, and 450 U/ml mutanolysin), and then incubated at 37°C for 2 h. The protoplast suspension was centrifuged at 5,000 × g for 10 min to remove cellular debris, and the supernatant containing the proteins enzymatically released from the cell wall was collected for use in the purification of SpaCBA pili. The affinity isolation of SpaCBA pili was carried out using a previously produced antiserum directed against the recombinant form of the SpaA major pilin constituent of the pilus structure (30). Polyclonal Ab were first purified from anti-SpaA rabbit serum by using a Hi-Trap protein G HP column (GE Healthcare Bio-Sciences, Uppsala, Sweden) according to the protocol recommended by the manufacturer. Briefly, a 1-ml column preequilibrated with binding buffer (20 mM sodium phosphate, pH 7.0) was loaded with filtered (0.45-μm-pore-size filter) SpaA antiserum and washed with at least 10 bed volumes of the same buffer. The Ab were eluted from the column with 0.1 M glycine-HCl (pH 2.7), and the elution fractions were immediately mixed with neutralization buffer (1 M Tris-HCl, pH 9.0). The Ab-containing fractions judged to be pure by SDS-PAGE analysis were pooled, buffer exchanged with coupling buffer (0.2 M NaHCO3–0.5 M NaCl, pH 8.3) by using an EconoPac 10 DG desalting column (Bio-Rad), and then passed through a 0.45-μm-pore-size membrane filter. These affinity-purified polyclonal Ab (∼3.75 mg) then were coupled covalently to the resin in a 1-ml Hi-Trap N-hydroxysuccinimide (NHS)-activated column (GE Healthcare Bio-Sciences, Uppsala, Sweden) by following the manufacturer's protocol. The SpaA antibody-bound column was used to isolate SpaCBA pili from the cell wall-associated protein extract using the purification procedure described above. The elution fractions containing SpaCBA pili were assessed for purity by SDS-PAGE analysis and immunoblotting with SpaCBA pilin subunit antisera, and the protein concentration was estimated by A280 measurements.
Detection of L. rhamnosus GG proteins by immunoblotting.
Cell wall-associated proteins, cell-free extract, and affinity-purified SpaCBA pili were separated by SDS-PAGE under denaturing conditions on 4 to 15% gradient gels (Bio-Rad) and then electroblotted onto polyvinylidene difluoride (PVDF) Immobilon P membranes (Millipore). Membranes were probed with anti-SpaA (diluted 1:25,000), anti-SpaB (diluted 1:5,000), anti-SpaC (diluted 1:25,000), anti-SpaF (diluted 1:10,000), or anti-SpaD (diluted 1:10,000) rabbit sera produced previously (12, 30, and international patent application no. PCT/FI2010/050059) and then incubated with goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad) diluted 1:100,000. The pilin-related proteins were visualized by enhanced chemiluminescence using the Amersham ECL Advance Western blotting detection kit (GE Healthcare Bio-Sciences, Uppsala, Sweden) according to the manufacturer's instructions. The cross-reactivity and detection limits of the SpaA-, SpaB-, and SpaC-specific antisera were each assessed with the previously produced recombinant hexahistidine-tagged SpaCBA pilin proteins (12, 30) under denaturing conditions by Western blot analysis. Besides the SpaB antiserum, which cross-reacted slightly with SpaC, no cross-reactivity could be observed in Western blotting (data not shown).
Immunogold labeling of whole cells and affinity-purified pili.
L. rhamnosus GG cells were grown overnight, washed once with PBS, and then diluted (OD600 of 1.0) in the same buffer. Formvar-carbon-coated copper grids first were floated for 1 h on droplets of the diluted L. rhamnosus GG cells or affinity-purified SpaCBA pili diluted 1:20 in PBS, washed several times with 0.02 M glycine in PBS, and then treated with a blocking solution of 1% bovine serum albumin (BSA) in PBS. The grids then were floated for 1 h on droplets of blocking solution containing anti-SpaA (diluted 1:100), anti-SpaB (diluted 1:200), anti-SpaC (diluted 1:100), anti-SpaF (diluted 1:100), or anti-SpaD (diluted 1:100) serum, washed several times with 0.1% BSA in PBS to remove unbound antibodies, and incubated for 20 min with protein A conjugated to 5- or 10-nm-diameter gold particles (pAg) diluted 1:55 in blocking solution. After one wash in PBS, the grids were fixed with 1% glutaraldehyde, washed with distilled water, and then negatively stained with a mixture of 1.8% methylcellulose–0.4% uranyl acetate. The steps of the procedure beginning with antiserum treatment and ending before negative staining were repeated once for double-labeling experiments with various combinations of the different pilin-specific antisera. The dilutions of the antibodies in the second staining cycle were 1:100 for SpaA and SpaC and 1:200 for SpaB. The cross-reactivities of the antibodies under native (nondenaturing) conditions were tested by immuno-dot blot, which revealed that neither SpaB nor SpaC Ab cross-reacted, whereas SpaA Ab bound to recombinant SpaB (data not shown). To avoid bias generated by the SpaA Ab cross-reactivity, in all double-labeling immunogold TEM experiments the first antiserum added was the SpaB or the SpaC antiserum.
Preparation of immunogold-labeled GG thin sections.
L. rhamnosus GG cells grown overnight were washed once with phosphate buffer (0.1 M Na-phosphate, pH 7.4) and fixed for 4 h at room temperature in 4% paraformaldehyde and 0.1% glutaraldehyde in phosphate buffer. After fixation the cells were collected by centrifugation and resuspended into 2% paraformaldehyde in phosphate buffer. The cells next were embedded in Lowicryl HM20 resin at cold temperatures using a freeze substitution method. Ultrathin plastic sections cut from polymerized Lowicryl were mounted on nickel grids and incubated for 20 min in blocking buffer (1% BSA, 0.5% fish skin gelatinase [FSG], and 1% fetal calf serum in phosphate buffer) followed by floating overnight at 4°C on a droplet of SpaB antiserum diluted 1:200 in antibody buffer (2% BSA, 0.1% Tween 20, and 0.1% FSG in phosphate buffer). The grids then were washed several times with pAg buffer (0.2% BSA, 0.01% Tween 20, and 0.01% FSG in phosphate buffer) and subsequently incubated for 20 min at room temperature with 10-nm pAg diluted 1:55 in pAg buffer. The grids then were washed several times with phosphate buffer followed by extensive washing with water. All steps of the procedure beginning with antiserum treatment were repeated by using either SpaA or SpaC antiserum (diluted 1:100) and 5-nm pAg. Finally, the grids were poststained by uranyl acetate and lead citrate using an UltroStain apparatus (Leica).
TEM.
The grids were examined and images were obtained using a JEOL 1200 EX II transmission electron microscope. To estimate the average number of different pilin subunits in the SpaCBA pilus structure, the pAg content from a minimum of 20 randomly selected pili per immunogold-labeling experiment was determined.
RESULTS
Immunoblotting.
A standard way to identify pilus proteins in Gram-positive bacteria is to analyze cell wall (CW) extract by Western blotting. The subunits involved in a pilus structure are visualized on Western blots as high-molecular-weight (HMW) ladders or smears, since pili in Gram-positive bacteria are composed of consecutive major pilins covalently linked to each other to form a fiber in which the individual shaft pilins are much like beads on a string. Therefore, in this study we first analyzed the presence of different molecular species corresponding to either monomeric or HMW molecules in cell wall and cell-free extracts (cytosolic and membrane-associated proteins) as well as in affinity-purified pili isolated from L. rhamnosus GG cells by Western blotting using SpaCBA Ab (Fig. 1). SpaF and SpaD antibodies were used as well, but no signal could be detected on membranes treated with these antibodies (data not shown). To test the possibility that SpaFED pilus expression is environmentally controlled, we grew L. rhamnosus GG in different media and at various temperatures (as specified in Materials and Methods) prior to Western analysis but still were unable to obtain any signal using SpaF or SpaD Ab (data not shown).
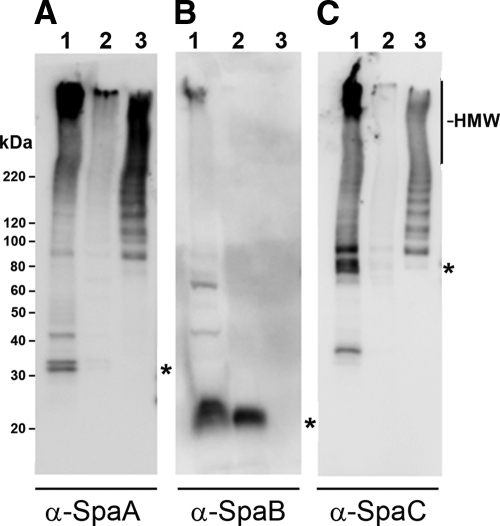
Fig 1.
Western analysis of fractionated GG cells and affinity-purified pili. Membranes were probed with polyclonal rabbit antisera against recombinant SpaA (A), SpaB (B), or SpaC (C). The wells were loaded with cell wall-associated extract (lane 1), cell-free extract (lane 2), or affinity-purified pili (lane 3). The calculated position of the corresponding monomeric pilin is indicated with an asterisk on the right side of each panel.
As expected for subunits building up a pilus structure, HMW species were identified in CW extract of whole cells with every antibody, indicating that all three SpaCBA subunits participate in the mature pilus fiber (Fig. 1A to C, lanes 1). The signal intensities were highest for SpaA and SpaC, whereas with SpaB Ab only weak staining was observed. With SpaA Ab we observed HMW banding in the cell-free fraction, representing growing pili still being attached to the membrane (Fig. 1A, lane 2). To a lesser extent this was also observed with SpaC Ab (Fig. 1C, lane 2), while none could be detected with SpaB Ab (Fig. 1B, lane 2). However, monomeric SpaB was found in high concentrations in both cell walls and cell-free extracts (Fig. 1B, lanes 1 and 2). This was remarkable, since SpaB was only sparsely detected in the HMW area of CW extract. One possibility is that SpaB is only rarely incorporated into assembling pili, resulting into the accumulation of monomeric SpaB pilins to the membrane and the cell wall. With SpaA Ab a doublet band was observed close to the 30-kDa standard band in the CW preparation, whereas this doublet was absent from the cell extract (Fig. 1A, lanes 1 and 2). Recombinant SpaA has previously been shown to migrate as a doublet on SDS-PAGE gels (30), and this aberrant migration has been assumed to represent different isoforms resulting from alternative intramolecular isopeptide bonding (4, 10, 11) that also has been observed for pilins from species other than L. rhamnosus (22). As observed for SpaA Ab, with SpaC Ab the monomeric form was evident in CW extract but was absent from the cell-free preparation (Fig. 1C, lanes 1 and 2). Interestingly, bands probably representing dimeric and trimeric SpaB also could be found in the cell wall extract (Fig. 1B, lane 1). This indicates that the SpaB subunits can polymerize independently of the SpaA or SpaC subunit.
With affinity-purified pili, HMW proteins were evident with SpaA (Fig. 1A, lane 3) and SpaC Ab (Fig. 1C, lane 3), whereas no HMW protein could be identified with the SpaB Ab (Fig. 1B, lane 3). The absence of HMW signal in affinity-purified pili using SpaB Ab most likely is related to the detection limit in the Western system used, since the HMW staining is only barely detectable in the CW extract. In contrast to the results from the L. rhamnosus GG culture in the stationary phase, intermediate-sized molecules (starting from 90 kDa) were clearly present in affinity-purified pilus preparations obtained from a logarithmic-growth-phase culture (Fig. 1A and C, lanes 3). This dissimilarity between these two pilus-containing samples most likely results from different growth phases used to obtain them, as the intermediate-sized molecular species obviously represent nascent pilus fibers still being assembled by the pilus-specific sortase in metabolically active L. rhamnosus GG cells. This assumption is further supported by the observation that whereas SpaA and SpaC monomers were present in the CW extract (Fig. 1A and C, lanes 1), they could not be found in the affinity-purified pilus preparations (Fig. 1A and C, lanes 3), indicating that these pilins are added to growing pilus fibers in actively dividing L. rhamnosus GG cells once they are secreted from the cytosol.
Immunogold TEM.
The SpaCBA pili were visualized using rabbit antisera raised against recombinant SpaA, SpaB, and SpaC pilin subunits in combination with protein A conjugated to 5- or 10-nm gold particles. The same approach was used to study the presence of the putative SpaFED pili that was not detected by Western blotting. The comparative amounts of individual pilin subunits in both cell wall-bound and affinity-purified pilus fibers were analyzed using single-labeling immunogold TEM by counting pAg from labeled pili and normalizing the obtained values with pilus lengths (Fig. 2 and 3). The relative positions of the different subunits within pilus fibers were revealed by the double-labeling TEM (Fig. 4 and 5) of whole and thin-sectioned cells as well as of affinity-purified pili.
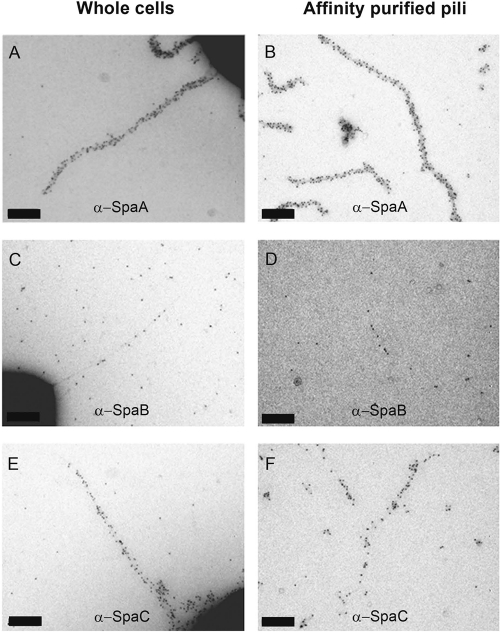
Fig 2.
TEM image of Lactobacillus rhamnosus GG cells labeled with SpaA antiserum and 10-nm protein A gold particles (pAg). The image shows numerous pili with lengths of up to 1 μm. Scale bar, 200 nm.
Fig 3.
TEM images of single-labeled GG cells and affinity-purified SpaCBA pili. Representative images of single-labeled cell wall-associated pili (A, C, and E) or affinity-purified pili (B, D, and F). The pili were labeled with SpaA (A and B), SpaB (C and D), or SpaC antiserum (E and F), followed by the addition of 5-nm pAg. Scale bars indicate 100 nm.
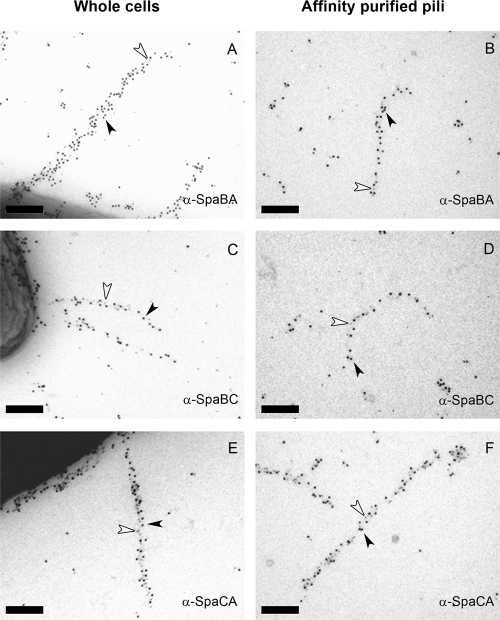
Fig 4.
Double-labeled immunogold images of GG cells and affinity-purified SpaCBA pili. Whole cells (A, C, and E) and affinity-purified intact pili (B, D, and F) were first labeled with SpaB (A to D) or SpaC Ab (E and F), followed by 5-nm protein A gold particles (arrows). The pili next were labeled with SpaA (A and B), SpaC (C and D), or SpaA Ab (E and F) and 10-nm pAg (arrowheads). Scale bars, 200 nm.
Fig 5.
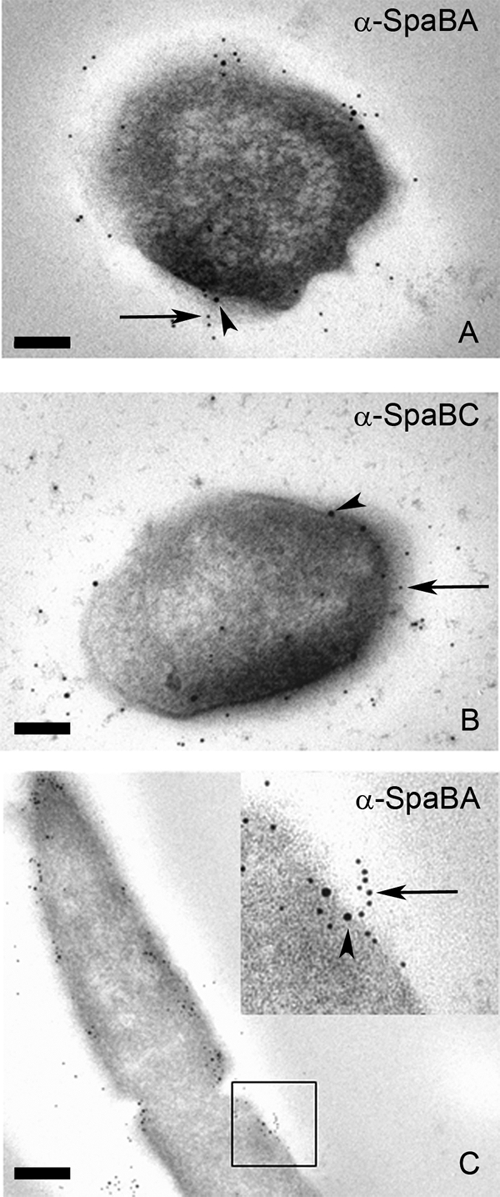
TEM images of immunogold-labeled GG thin sections. Whole GG cells first were labeled with SpaB antibodies followed by 10-nm protein A gold particles (arrowheads). The pili then were labeled with SpaA (A and C) or SpaC antiserum (B) and 5-nm pAg (arrow). The area enclosed in the black square in panel C is highlighted in the upper right corner of the panel. Scale bars, 100 (A and B) or 200 nm (C).
We demonstrate that the same pili reacting with the SpaC Ab in our previous study (12) also contain the SpaA and SpaB pilins, whereas with SpaF and SpaD Ab no pili could be visualized. Multiple pili in each L. rhamnosus GG cell with lengths of up to 1 μm and each harboring a few dozen pilin subunits could be observed with antisera raised against SpaA and SpaC subunits (Fig. 2 and 3). In contrast, the visualization of pili using SpaB Ab was challenging, since only a few protein A gold particles could be seen in each pilus fiber (Fig. 3). The relative numbers of the SpaCBA subunits obtained by the quantification of pAg from single-labeled pili revealed a SpaC/SpaA ratio of 1:2. SpaB numbers could not reliably be compared to those of SpaA or SpaC, as only a few SpaB molecules could be detected in individual pili (Fig. 3). In addition, the L. rhamnosus GG SpaCBA pili were proven to be heterotrimeric in a series of double-labeling immunogold TEM experiments, since all three SpaCBA Ab combinations used showed the double labeling of individual pilus fibers (Fig. 4).
The small number of SpaB in mature pili revealed by immunoelectron microscopy together with large amounts of SpaB monomers observed in cell extracts by Western blotting prompted us to study the intracellular location of SpaB and compare that to those of SpaA and SpaC. Remarkably, in double-labeled GG thin sections, a clear colocalization between SpaA and SpaB could be seen near the cell membrane and cell wall, where in most cases a single SpaB molecule was surrounded by SpaA subunits. In contrast, SpaC was distributed apparently independently of SpaB (Fig. 5). Moreover, SpaB subunits were frequently detected at the base of protruding pili (Fig. 5C, inset). It worth nothing that in thin sections the pili are embedded in a plastic resin together with cells, and due to the flexible nature of Gram-positive pili they only rarely exist in thin sections even as short (a few pilins long) elongated fibers; rather, they exist as clumps close to the cell surface. The frequent clustering of multiple SpaA pilins around singular SpaB molecules together with the observed short elongated SpaA stretches with single SpaB molecules at the base suggests that intracellular SpaB pools serve as a reservoir of molecular switches that, when incorporated into the growing pili, occasionally terminate pilus elongation and ultimately lead to the covalent anchoring of the mature pilus fiber to the peptidoglycan chain.
DISCUSSION
The genome of the probiotic Lactobacillus rhamnosus GG is known to contain two pilus gene clusters, spaCBA and spaFED (12). Despite the presence of the spaFED genes in the genome, the SpaCBA pili appear to be the only pili produced by L. rhamnosus GG under the experimental conditions tested in this study. The absence of SpaFED pili is likely to result from the lack of appropriate environmental stimuli to activate the expression of the spaFED genes, as there was no notable transcription of spaFED genes under any tested laboratory conditions (Kati Laakso and Tuomas Salusjärvi, personal communication). It cannot be excluded, however, that the SpaFED pili are produced in the intestinal tract, similarly to the Tad pilus genes in Bifidobacterium breve that are expressed only in vivo in mouse intestine (20).
The present knowledge about subunit organization of pili in Gram-positive bacteria is, to a large extent, based on pioneering work with C. diphtheriae pili (5, 8, 26, 28). This resulted in a proposal for a heterotrimeric prototype pilus in which the pilus shaft is formed mainly of consecutive major pilin subunits and in which each pilus fiber has a single adhesin (larger-sized minor subunit) molecule on the pilus tip (5, 6, 28). In addition, a second minor pilin (smaller minor subunit) might be incorporated into the pilus shaft via the pilin motif (6) or might be linked to the pilus fiber in a decorative manner (5) through a yet-to-be clarified reaction, possibly involving the E box. Furthermore, as shown for a number of different Gram-positive species, more than one type of pili can coexist in the same cell (1, 5, 7, 26, 28). Based on the data presented here, the SpaCBA pili follow the prototype pilus model only partially. The L. rhamnosus GG SpaCBA pili are heterotrimeric, i.e., they contain all three different subunits, and we found the larger minor pilin, SpaC, to be located at the pilus tips. The tip position of SpaC is in line with the genetic organization of the spaCBA operon, since, being the first pilin subunit translated from the spaCBA pilus gene cluster, the SpaC would also be the first one to be transported through the membrane. Once secreted, SpaC might initiate pilus assembly and therefore would be situated at the pilus tip. However, a major exception to the canonical Gram-positive pilus model is that the position of the SpaC subunit is not restricted to the pilus tip. Instead, SpaC can be found all along the pilus fiber, and furthermore, having a 1:2 ratio in molecular numbers with SpaA, the amount of the SpaC subunit is almost comparable to that of the SpaA major pilin. The presence of larger minor pilins along the pilus shaft has been reported with GBS67 and GBS104 subunits of Streptococcus agalactiae type PI-2a and PI-1 pili, respectively (23). Although no stoichiometric calculations of numbers of different subunits were made in S. agalactiae, the relative amounts of these minor pilins compared to those of the corresponding major pilins were substantially lower than that of SpaC in L. rhamnosus GG SpaCBA pili, as seen in double-labeling immunogold TEM experiments (23). It is unlikely, however, that SpaC would be incorporated into the pilus shaft, since it lacks any recognizable pilin motif but harbors two putative E box elements instead. We therefore infer that in addition to being a tip adhesin, the SpaC subunit is attached to the pilus polymer in a decorative manner, possibly through its E box motif or by a yet-undetermined mechanism. This has two major implications: first, it allows L. rhamnosus GG cells to make both long-distance contacts with host mucosal tissue, using fully extended pili involving SpaC molecules present at pilus tips, as well as intimate short-distance contacts with SpaC subunits situated closer to pilus bases. Second, since there are multiple SpaC adhesins in individual pili and several pili in individual cells, the overall mucus-binding avidity (i.e., sum of the affinities of individual SpaC molecules) provided by the pilus-associated SpaC molecules to the cells likely substantially promotes the competitive properties of L. rhamnosus GG cells in the ecological niche of mucosal surfaces in the human gastrointestinal tract. This assumption is further justified by the results from a human intervention study that showed the intestinal residency time of Lactobacillus rhamnosus GG to be considerably longer than that of the related pilus-deficient Lactobacillus rhamnosus LC705 strain (12). A similar difference in colonization ability was recently observed with the Tad pilus-expressing Bifidobacterium breve UCC2003PK1 and its pilus-deficient mutant UCC2003-tadAPK1: when germ-free Swiss Webster mice were inoculated with these two strains, the amounts of bacterial cells shed into feces remained equally high (109 CFU/g feces) for both strains as long as the mice were kept under germ-free conditions; when the mice were introduced to competitive gut microbiota by transferring them into a non-germ-free area and supplying their cages with fecal pellets from conventional mice, the amount of the wild-type strain dropped to a level of 106 CFU/g feces within 7 days after recolonization, and that of the mutant strain went down to a scarcely detectable level. Furthermore, for the following 2 weeks the amount of the wild-type strain UCC2003PK1 remained constant at 106 CFU/g feces, whereas no sign of the pilus-deficient strain UCC2003-tadAPK1 could be detected (20). As Lactobacillus rhamnosus GG is currently the only known probiotic strain with mucus-binding pili, it can be speculated that the SpaCBA pilus-mediated mucus adherence and the resulting competitive advantage in the mucosal environment provides Lactobacillus rhamnosus GG with advanced probiotic properties compared to those of other probiotic strains. As Lactobacillus rhamnosus GG is currently the only known probiotic strain with mucus-binding pili, it can be speculated that the SpaCBA pilus-mediated mucus adherence and the resulting competitive advantage in the mucosal environment provide Lactobacillus rhamnosus GG with advanced probiotic properties compared to those of other probiotic strains.
The smaller minor pilin SpaB in L. rhamnosus GG is also somewhat distinctive among Gram-positive pilins described to date, because it has dualistic characteristics. On the one hand, based on our observation that the SpaB pilin is clustered together with SpaA close to the cell wall, we favor the model presented for the SpaB pilin in corynebacteria, in which SpaB is assumed to act as a molecular switch that terminates pilus polymerization and predestines mature pili to be covalently attached to the peptidoglycan by the housekeeping sortase (15). On the other hand, we have previously shown by using an in vitro mucus-binding assay and recombinant SpaB that the L. rhamnosus GG SpaB also is an adhesin that binds human intestinal mucus, probably via electrostatic interactions (30). Therefore, SpaB can be viewed as a multipotent molecule exerting functions in both cell wall attachment of pili and mucus adhesion of L. rhamnosus GG cells. The overall impact of SpaB in human mucus binding, however, is likely to be minor compared to that of SpaC, as SpaC greatly outcompetes SpaB in terms of molecular numbers in the cell envelope. Since SpaB contains both the pilin motif and the E box, it remains to be elucidated whether the association of SpaB into pilus fibers follows the in-chain type, a decorative type, or a combination of both. This same rationale applies to the SpaA subunit of L. rhamnosus GG pili, and therefore the possibility for SpaA decoration cannot be exclusively ruled out at this point.
The overall findings presented in this study lead us to propose a novel pilus structure among Gram-positive bacteria. In our scheme (Fig. 6), the major rearrangement deals with the larger-sized minor pilin, SpaC, the occurrence of which has been widened from a sole tip location to cover the full pilus length. This observation, together with the presence of yet another mucus-binding adhesin, SpaB, in Lactobacillus rhamnosus GG SpaCBA pili makes these surface protrusions unique among the Gram-positive pili studied thus far.
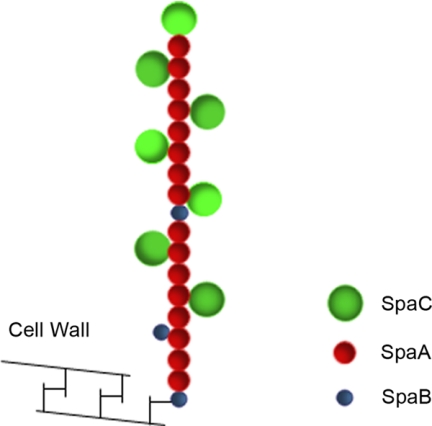
Fig 6.
Schematic model of Lactobacillus rhamnosus GG SpaCBA pili. In the proposed model, the heterotrimeric SpaCBA pili in GG are composed of shaft-forming SpaA major pilins together with SpaB and SpaC minor pilins. The SpaC adhesin can be found not only at the pilus tip but also throughout the pilus at a ratio with SpaA of approximately 1:2. The SpaB minor pilin is located at the pilus base, and few SpaB subunits can be found on the pilus fibers.
ACKNOWLEDGMENTS
This work was supported by the Academy of Finland (research grants 118165 and 117877) and was part of the Center of Excellence in Microbial Food Safety Research (MiFoSa) and the Research Program on Nutrition, Foods, and Health (ELVIRA). W.M.D.V. is funded as a Finland Distinguished Professor by the Finnish Funding Agency for Technology and Innovation (TEKES) and is also funded by grant 25.0.1.7 from the European Research Council (MicrobesInside). The research of J.R. was partially supported by a special grant from Valio Ltd., Helsinki, Finland.
We thank Ilkka Palva for his invaluable guidance and discussions throughout this study. We thank the Electron Microscopy Unit of the Institute of Biotechnology–University of Helsinki for providing laboratory facilities.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Bagnoli F, et al. 2008. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J. Bacteriol. 190: 5480–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barocchi MA, et al. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103: 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Budzik JM, Marraffini LA, Schneewind O. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66: 495–510 [DOI] [PubMed] [Google Scholar]
- 4. Budzik JM, et al. 2009. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. U. S. A. 106: 19992–19997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaspar AH, Ton-That H. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 188: 1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guttilla IK, et al. 2009. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J. Bacteriol. 191: 5603–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hendrickx AP, et al. 2008. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 154: 3212–3223 [DOI] [PubMed] [Google Scholar]
- 8. Hendrickx AP, Budzik JM, Oh SY, Schneewind O. 2011. Architects at the bacterial surface-sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 9: 166–176 [DOI] [PubMed] [Google Scholar]
- 9. Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. 2009. LPxTG surface proteins of enterococci. Trends Microbiol. 17: 423–430 [DOI] [PubMed] [Google Scholar]
- 10. Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. 2007. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 318: 1625–1628 [DOI] [PubMed] [Google Scholar]
- 11. Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. 2009. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. U. S. A. 106: 16967–16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kankainen M, et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106: 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5: 580–592 [DOI] [PubMed] [Google Scholar]
- 14. Lauer P, et al. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309: 105. [DOI] [PubMed] [Google Scholar]
- 15. Mandlik A, Das A, Ton-That H. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 105: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16: 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mishra A, Das A, Cisar JO, Ton-That H. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 189: 3156–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mora M, et al. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 102: 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nallapareddy SR, et al. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 116: 2799–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Connell Motherway M, et al. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108: 11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria–structure, assembly and their role in disease. Cell. Mol. Life Sci. 66: 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quigley BR, Zahner D, Hatkoff M, Thanassi DG, Scott JR. 2009. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol. Microbiol. 72: 1379–1394 [DOI] [PubMed] [Google Scholar]
- 23. Rosini R, et al. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61: 126–141 [DOI] [PubMed] [Google Scholar]
- 24. Scott JR, Zahner D. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62: 320–330 [DOI] [PubMed] [Google Scholar]
- 25. Swaminathan A, et al. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swierczynski A, Ton-That H. 2006. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J. Bacteriol. 188: 6318–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4: 509–519 [DOI] [PubMed] [Google Scholar]
- 28. Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50: 1429–1438 [DOI] [PubMed] [Google Scholar]
- 29. Ton-That H, Schneewind O. 2004. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12: 228–234 [DOI] [PubMed] [Google Scholar]
- 30. von Ossowski I, et al. 2010. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 76: 2049–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeung MK, Donkersloot JA, Cisar JO, Ragsdale PA. 1998. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect. Immun. 66: 1482–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]