Abstract
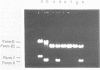
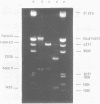
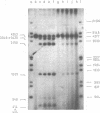
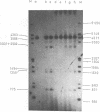
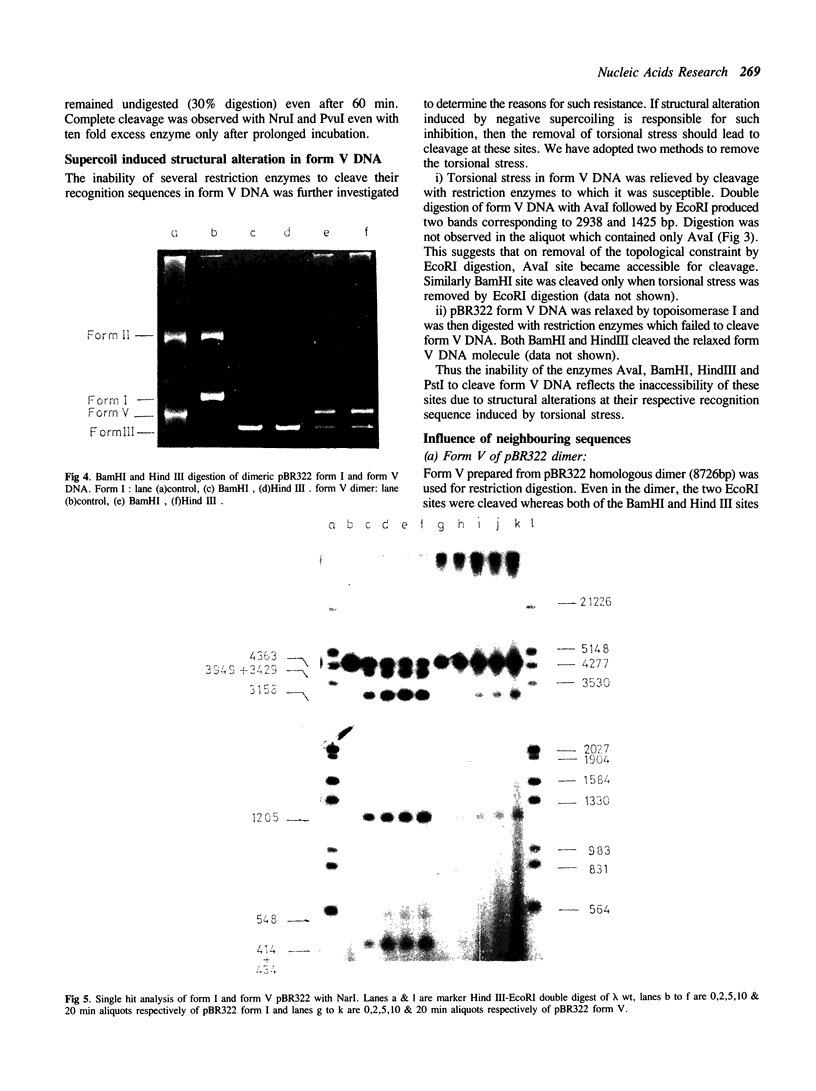
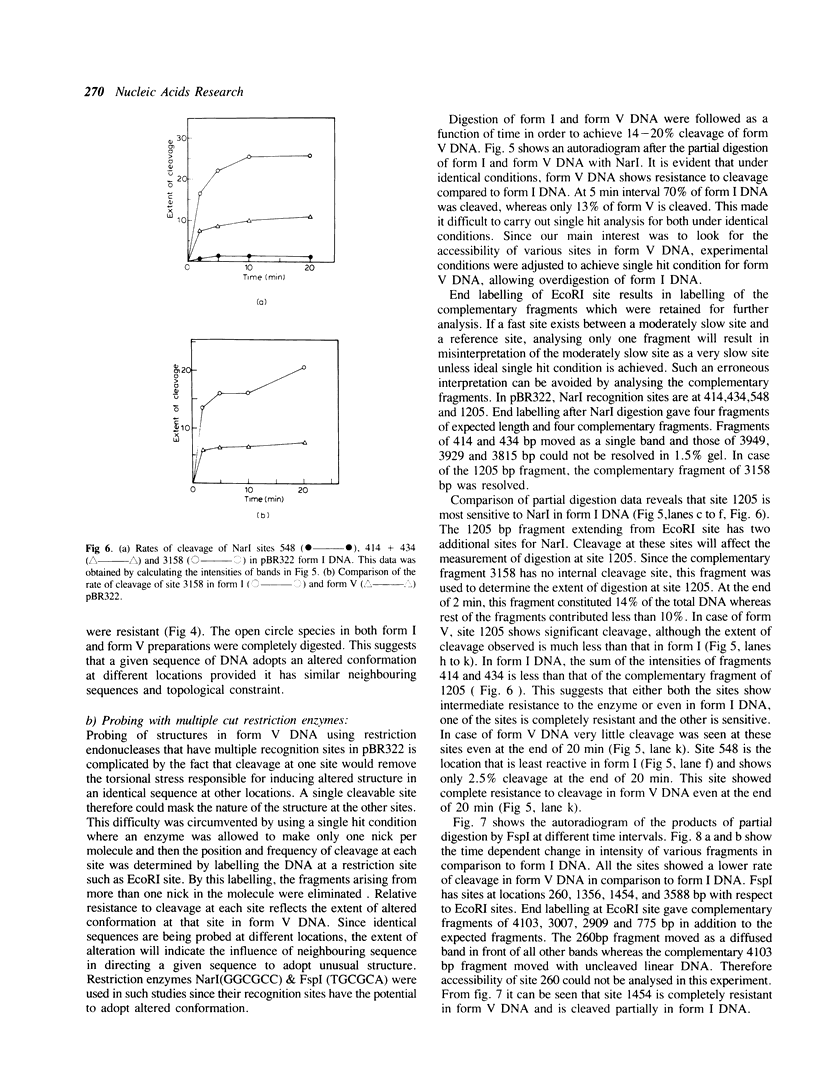
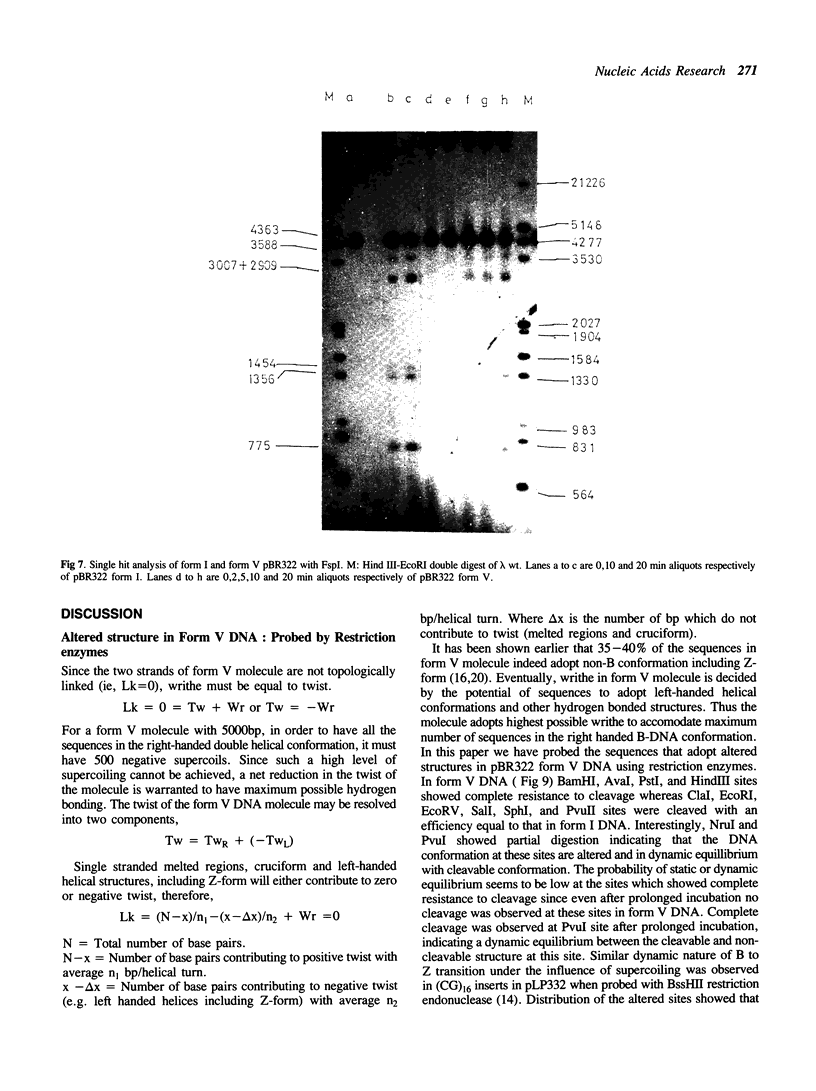
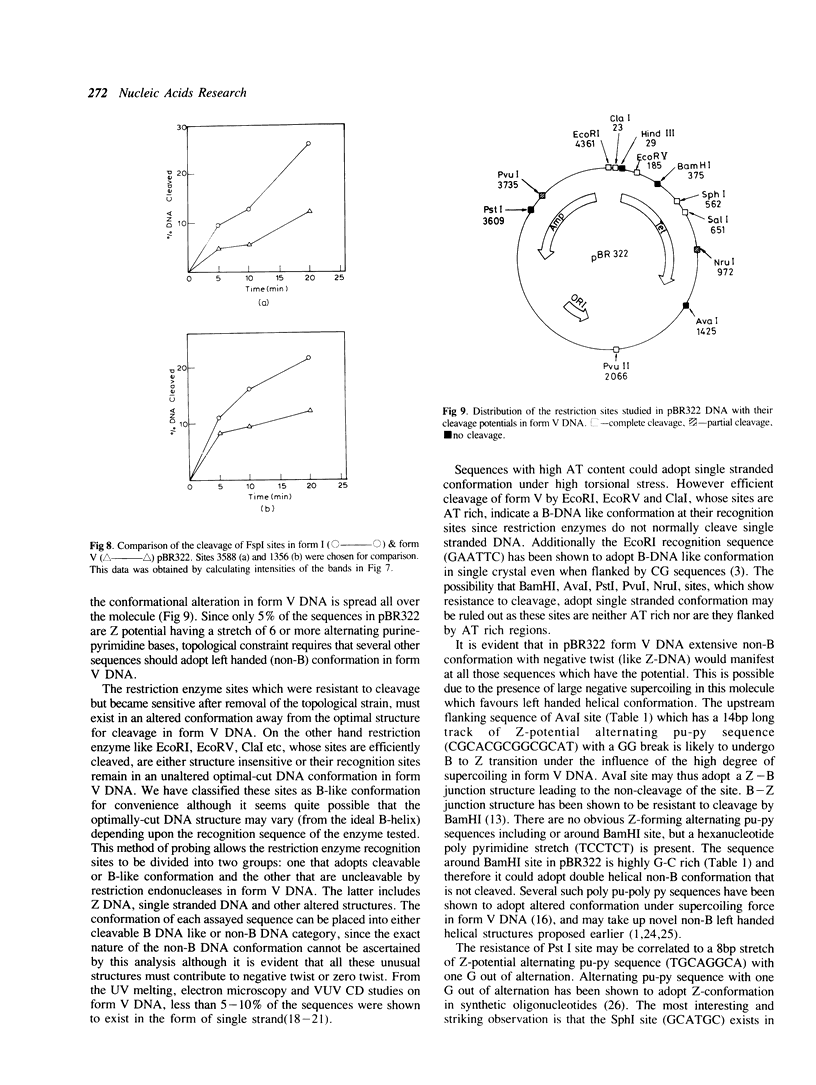
The ability of DNA sequences to adopt unusual structures under the superhelical torsional stress has been studied. Sequences that are forced to adopt unusual conformation in topologically constrained pBR322 form V DNA (Lk = 0) were mapped using restriction enzymes as probes. Restriction enzymes such as BamHI, PstI, AvaI and HindIII could not cleave their recognition sequences. The removal of topological constraint relieved this inhibition. The influence of neighbouring sequences on the ability of a given sequence to adopt unusual DNA structure, presumably left handed Z conformation, was studied through single hit analysis. Using multiple cut restriction enzymes such as NarI and FspI, it could be shown that under identical topological strain, the extent of structural alteration is greatly influenced by the neighbouring sequences. In the light of the variety of sequences and locations that could be mapped to adopt non-B conformation in pBR322 form V DNA, restriction enzymes appear as potential structural probes for natural DNA sequences.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S. K., Shouche Y. S., Cantor C. R., McClelland M. Sequences that adopt non-B-DNA conformation in form V DNA as probed by enzymic methylation. J Mol Biol. 1987 Jan 5;193(1):201–211. doi: 10.1016/0022-2836(87)90637-1. [DOI] [PubMed] [Google Scholar]
- Brahms S., Vergne J., Brahms J. G., Di Capua E., Bucher P., Koller T. Natural DNA sequences can form left-handed helices in low salt solution under conditions of topological constraint. J Mol Biol. 1982 Dec 5;162(2):473–493. doi: 10.1016/0022-2836(82)90539-3. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985 Jun 25;13(12):4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Jendrisak J. J., Hager D. A., Burgess R. R. Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme). J Biol Chem. 1981 Jun 10;256(11):5860–5865. [PubMed] [Google Scholar]
- Feigon J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Z-DNA forms without an alternating purine-pyrimidine sequence in solution. Science. 1985 Oct 4;230(4721):82–84. doi: 10.1126/science.4035359. [DOI] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of sequence in the stabilization of left-handed DNA helices in vitro and in vivo. Biochim Biophys Acta. 1988 Sep 7;950(3):243–254. doi: 10.1016/0167-4781(88)90120-0. [DOI] [PubMed] [Google Scholar]
- Mishra R. K., Latha P. K., Brahmachari S. K. Interruptions of (CG)n sequences by GG, TG and CA need not prevent B to Z transition in solution. Nucleic Acids Res. 1988 May 25;16(10):4651–4665. doi: 10.1093/nar/16.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982 Dec;31(2 Pt 1):309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Vasser M., Dinkelspiel K., Crea R. Conformational stability of alternating d (CG) oligomers in high salt solution. Nucleic Acids Res. 1981 May 11;9(9):2195–2206. doi: 10.1093/nar/9.9.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N., Brahmachari S. K. Critical cation balance in B leads to Z transition: role of Li+. FEBS Lett. 1983 Nov 28;164(1):33–37. doi: 10.1016/0014-5793(83)80013-1. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Brahmachari S. K. Dynamic nature of B to Z transition: role of DNA supercoiling and Z-DNA binding protein. Indian J Biochem Biophys. 1988 Dec;25(6):542–547. [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Sasisekharan V., Pattabiraman N., Gupta G. Some implications of an alternative structure for DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4092–4096. doi: 10.1073/pnas.75.9.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Wells R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 May;80(9):2447–2451. doi: 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]