Abstract
The cyclic AMP (cAMP) pathway represents a central signaling cascade with crucial functions in all organisms. Previous studies of Trichoderma reesei (anamorph of Hypocrea jecorina) suggested a function of cAMP signaling in regulation of cellulase gene expression. We were therefore interested in how the crucial components of this pathway, adenylate cyclase (ACY1) and cAMP-dependent protein kinase A (PKA), would affect cellulase gene expression. We found that both ACY1 and PKA catalytic subunit 1 (PKAC1) are involved in regulation of vegetative growth but are not essential for sexual development. Interestingly, our results showed considerably increased transcript abundance of cellulase genes in darkness compared to light (light responsiveness) upon growth on lactose. This effect is strongly enhanced in mutant strains lacking PKAC1 or ACY1. Comparison to the wild type showed that ACY1 has a consistently positive effect on cellulase gene expression in light and darkness, while PKAC1 influences transcript levels of cellulase genes positively in light but negatively in darkness. A function of PKAC1 in light-modulated cellulase gene regulation is also reflected by altered complex formation within the cel6a/cbh2 promoter in light and darkness and in the absence of pkac1. Analysis of transcript levels of cellulase regulator genes indicates that the regulatory output of the cAMP pathway may be established via adjustment of XYR1 abundance. Consequently, both adenylate cyclase and protein kinase A are involved in light-modulated cellulase gene expression in T. reesei and have a dampening effect on the light responsiveness of this process.
INTRODUCTION
The habitat of fungi is a diversified environment, and successful competition requires adaptation to changing conditions in an appropriate way. After sensing of the extracellular signal through a receptor, the signaling cascade starts and the signal is transduced by, for example, secondary messengers such as cyclic AMP (cAMP). As a secondary messenger, cAMP is involved in stress response, carbon and lipid metabolism, sporulation, development, virulence, mating, mycoparasitism, and other responses to extracellular signals (1, 9, 14, 28, 39, 45, 68).
The main target of cAMP in this signaling cascade is cAMP-dependent protein kinase A (PKA). After binding of cAMP to a regulatory subunit, one of the catalytic kinase subunits dissociates and in turn phosphorylates other target proteins (12, 17, 41, 67). The cAMP signaling pathway is highly conserved in fungi. The pathway of heterotrimeric G-protein signaling, and especially that of subgroup I and III G-protein alpha subunits, is linked to the cAMP pathway, predominantly via its effect on adenylate cyclase, which generates cAMP (7, 73).
The potential to efficiently degrade lignocellulose caused Trichoderma reesei (anamorph of Hypocrea jecorina) to become one of the most important industrial producers of cellulases. These important hydrolytic enzymes are produced on various carbon sources. Not only cellulose but also the soluble carbon sources cellobiose (33), lactose (34), and l-sorbose and several more (36) can cause induction of cellulase gene expression. Lactose, a by-product of the cheese manufacture and whey processing industries (46), does not occur in the natural habitat of T. reesei. However, the fact that this carbon source efficiently induces cellulase gene expression makes it an important renewable raw material in biotechnology. If cultivation in a medium without solids is preferred, lactose represents a valuable alternative to cellulose (59). In recent years, considerable progress has been made in understanding the mechanism of cellulase gene regulation on lactose (24).
Regulation of the major cellulolytic genes of T. reesei is governed by numerous extracellular signals and intracellular regulatory factors (24, 52), including transcription factors such as ACE1, ACE2, and XYR1 (2, 3, 44, 47). ACE1 is a repressor of cellulase gene expression (2), but ACE2 and XYR1 act positively on cellulase gene transcript abundance (3, 62). Transcription of ace1, ace2, and xyr1 is induced upon growth on lactose. While ace1 is carbon catabolite repressed, the carbon catabolite repressor CRE1 is necessary for full induction of ace2 and xyr1 (43).
Analysis of the genome of T. reesei revealed, surprisingly, that this industrial workhorse has an unexpectedly small number of genes for cellulases, hemicellulases, and pectinases (37). Consequently, this small inventory of hydrolytic enzymes must be efficiently and specifically adjusted to environmental conditions in order to enable successful competition in the natural habitat. One possible way to accomplish this aim is to precisely adapt enzyme biosynthesis to environmental signals by means of a tightly regulated signal transduction cascade. The regulatory mechanisms underlying this adaptation can be exploited for biotechnological applications.
It has been known for quite some time that the formation of endoglucanases can be altered by addition of cAMP in Trichoderma (61). A relatively small amount of cAMP (up to 10 μM dibutyryl cAMP [dbcAMP]) stimulates endoglucanase activity, whereas 1,000-fold-higher concentrations of cAMP inhibit the formation of endoglucanases. Nevertheless, neither the mechanisms nor the roles of intracellular regulation and its central components were characterized with respect to their impact on cellulase gene expression.
Besides the response to a given substrate composition, cellulase gene transcript levels are modulated by environmental factors such as light and by ENV1, BLR1, and BLR2, the central components of the light signaling machinery in T. reesei (50, 51). The pathway of heterotrimeric G-protein signaling and especially two G-protein alpha subunits, GNA1 and GNA3 (belonging to subgroups I and III), which are likely to transmit nutrient signals are also involved in this mechanism (53, 58). Interestingly, in Neurospora crassa, PKAC-1, the major catalytic subunit of this fungus (6), is a crucial element of the circadian clock and stabilizes both the white collar complex WCC and FREQUENCY (encoded by frq1) (20, 38). In Trichoderma atroviride, a connection between the two blue light receptors BLR-1 and BLR-2 and the cAMP signaling pathway also has been observed (9). Recently, we showed a clear effect of ENV1 on intracellular cAMP levels, which is likely mediated via regulation of a phosphodiesterase (69). These data suggest a close connection between the cAMP signaling pathway, light response, and cellulase gene expression.
Consequently, we sought to gain insight into the mechanisms of the two major components in cAMP-mediated signal transduction, adenylate cyclase and cAMP-dependent protein kinase A. We investigated both the light responsiveness of cellulase gene expression (difference between light and darkness in one strain) and the effect of lack of functional adenylate cyclase or protein kinase A (by comparison of the wild type and a deletion mutant in light or darkness) in order to evaluate the contributions of these two components to cross talk between light response and nutrient signaling.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
T. reesei wild-type strain QM9414 (ATCC 26921) and Hypocrea jecorina CBS999.97 MAT1-1 (60) were used in the present study. For quantitative reverse transcription-PCR (qRT-PCR) analysis and biomass determination, T. reesei was grown in liquid culture in 200 ml Mandels-Andreotti minimal medium (32) supplemented with 0.1% (wt/vol) peptone to induce germination and with 1% (wt/vol) lactose (Merck, Darmstadt, Germany) as a carbon source. For evaluation of the effect of cAMP supplementation, T. reesei QM9414 along with recombinant strains was grown on 3% (wt/vol) malt extract agar plates for 10 days at 28°C. Exogenous dbcAMP (Sigma-Aldrich, St. Louis, MO) was added to the medium to a final concentration of 5 mM. Mating experiments were performed on 3% (wt/vol) malt extract medium with 2% (wt/vol) agar. Plates were incubated at 20 to 25°C in daylight for 10 to 14 days. Escherichia coli JM109 was used for the propagation of vector molecules and DNA manipulations (74).
Construction of T. reesei Δpkac1 and Δacy1 strains and copy number determination.
To construct a mutant strain in which PKAC1 is nonfunctional, the protein kinase A catalytic domain was disrupted. A 4,276-bp fragment which contained the entire gene, including flanking sequences, was PCR amplified using primers pkac1F and pkac1R (the sequences of all oligonucleotides used in this study are listed in Table 1). The 3′ sequence to be used for construction of a deletion vector was excised from the 4,276-bp fragment by digestion with PstI and NruI (resulting in a 1,424-bp fragment) and cloned into a PstI/SmaI-digested pBluescript SK(+) (Stratagene, La Jolla, CA) to obtain pBpkac3. For the 5′ sequence, the 4,276-bp fragment was digested with SalI and EcoRV (resulting in a 1,004-bp fragment) and cloned into the corresponding sites in pBpkac3 to obtain pBpkac35. The hph (pkiP:hph:cbh2T) marker cassette (30) was digested with XhoI and HindIII. Blunt ends were created by Klenow fill-in, and then the fragment was inserted into the EcoRV site of pBpkac35 to obtain pDELpkac1. The deletion cassette was excised from pDELpkac1 by restriction digestion with NotI, and this linear fragment was used for the transformation of protoplasts of the wild-type strain QM9414 (18). Transformants were selected on plates containing 50 μg/ml hygromycin B (Carl Roth, Karlsruhe, Germany). Fungal DNAs were isolated from transformants and subjected to PCR using primers pkac1MF and pkac1MR to verify the replacement of the pkac1 gene with the hph gene. These primers bind within the 3′ and 5′ regions of pkac1. Successful deletion of pkac1 results in an amplification of the whole deletion construct (3,097 bp) and lack of the original open reading frame, which would be represented by a smaller fragment (1,974 bp) (data not shown).
Table 1.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| qRT-cbh1F | 5′-GAAGAACTGCTGTCTGGA-3′ |
| qRT-cbh1R | 5′-AATGGAGAGGCTGTTACC-3′ |
| qRT-cbh2F | 5′-CCAACGCCTTCTTCATCAC-3′ |
| qRT-cbh2R | 5′-CCAGACAAACGAATCCAGC-3′ |
| qRT-ace1F | 5′-GACAAGACGGATGTGTTCCAG-3′ |
| qRT-ace1R | 5′-GTTGAAGATGTCGGGCTGTG-3′ |
| qRT-ace2F | 5′-TCAACATCCTCCACCACCAGTC-3′ |
| qRT-ace2R | 5′-TGTCGGCGTACTCTCTCAGC-3′ |
| qRT-xyr1F | 5′-CTTCCTCCTCCTGCTCATCG-3′ |
| qRT-xyr1R | 5′-TCGTGTGCCCTAACAATGGTC-3′ |
| pkac1F | 5′-AACCACAGCGCCTTTCCAG-3′ |
| pkac1R | 5′-AATCGCGACCGGATCAGTC-3′ |
| pkac1MF | 5′-TTCCTCATCACCCTGTGG-3′ |
| pkac1MR | 5′-CGCCAATCTACCTAATACCG-3′ |
| acy1D3F | 5′-ATCTCGAGGCTCAGCTGTACATTACGAAC-3′ |
| acy1D3R | 5′-ATGGTACCTCTAGACGCGCTTCATCTTCTATG-3′ |
| acy1D5R | 5′-ATGGATCCGTGAGAGGCTCCTACACAC-3′ |
| acy1D5F | 5′-ATGCGGCCGCATTAAGTGACTTGTAACACCC-3′ |
| acy1MF | 5′-CTCTGGCAACGAGGAAGCAATG-3′ |
| acy1MR | 5′-GATCCCTCCCGACCATGATTG-3′ |
| acy1R1 | 5′-CGACCCGGTGGATACATTTC-3′ |
| acy1F1 | 5′-CGCCGCATGGACTACTTTG-3′ |
| CKT057 | 5′-GATCGCTTCTTTATTGGGTAATATACAGCCAGGCGGGG-3′ |
| CKT058 | 5′-GATCCCCCGCCTGGCTGTATATTACCCAATAAAGAAGC-3′ |
| CKT083 | 5′-GATCGCTTCTTTAAAGGGTTTTATACAGCCAGGCGGGG-3′ |
| CKT084 | 5′-GATCCCCCGCCTGGCTGTATAAAACCCTTTAAAGAAGC-3′ |
| cbh2T_F1 | 5′-GGCTTGCTCGCTGACTGATAC-3′ |
| cbh2T_F2 | 5′-GAGGGAGACGAGGTTGTGATG-3′ |
| HPH_F1 | 5′-GATGTAGGAGGGCGTGGATATG-3′ |
| HPH_R1 | 5′-GGGAGATGCAATAGGTCAGG-3′ |
Underlining indicates restriction sites introduced to facilitate cloning.
For acy1 gene deletion, the 3′ flanking sequence was PCR amplified using primers acy1D3F and acy1D3R to obtain a 1,672-bp fragment. This product was digested with XhoI and Acc65I and cloned into the XhoI-Acc65I site of pBSXH (51) to obtain pDelacy3h. The 5′ flanking sequence was PCR amplified using primers acy1D5R and acy1D5F to obtain a 1,575-bp PCR product. This fragment was digested with NotI and BamHI and cloned into the NotI-BamHI site of pDelacy3h to obtain pDELacy1. For transformation, pDelacy1 was linearized with NotI and purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany).
The linear plasmid was used for electroporation of spores of T. reesei QM9414 (see below for details). Fungal DNAs were isolated from transformants and used for diagnostic PCR. Deletion of acy1 was verified by PCR using primers acy1MF and acy1MR. Both primers bind within the 3′ and 5′ flanking regions of the open reading frame of acy1. Successful integration of the deletion vector in the genome resulted in amplification of a 3,154-bp fragment. Since in this case the wild-type fragment amplified with this primer combination is considerably larger (7,019 bp) and thus unlikely to be detected even in case of ectopic integration of the cassette, we confirmed deletion of acy1 using qRT-acy1R1 and qRT-acy1F1 as primers. The deletion mutant showed no amplification product, while when wild-type acy1 was present, a 170-bp fragment was amplified (data not shown).
The number of copies of the cassette integrated in the genome was analyzed by quantitative PCR on genomic DNA of wild-type and mutant strains as described previously (70). This method is preferable to Southern blotting because of the linearity of qPCR over several orders of magnitude and elimination of problems with limited resolution in the case of larger fragments to be evaluated in Southern blots. Briefly, we used primers which bind within the hph gene (HPH_F1 and HPH_R1 [Table 1]) to check for presence of the deletion cassette. Additionally we used primers binding to the cbh2 terminator (cbh2T_F1 and cbh2T_F2 [Table 1]), which is comprised within the hph cassette that we used (30). A strain for which single integration had been proven by Southern blotting (69) was used as a control. For both the Δpkac1 and Δacy1 strains, only single integration at the locus was detected. The parental strain QM9414 was used as a control. This analysis confirmed that in both strains only one copy of the deletion cassette was present and no ectopic integration had occurred (see Fig. S1 in the supplemental material).
Transformation of T. reesei by electroporation.
Electroporation was performed as described elsewhere (54). Briefly, spores of T. reesei QM9414 were harvested from a freshly sporulated 90-mm malt extract agar plate and suspended in 1.2 M sorbitol. Spores were washed twice, resuspended in 100 μl 1.2 M sorbitol, and cooled on ice. A 75-μl portion of cold spore suspension was mixed with 20 μg of vector of interest and electroporated with a Micro Pulser electroporator (Bio-Rad Laboratories, Hercules, CA) at 1.8 kV with preset values of 10 μF for capacitance and 600 Ω for parallel resistance. A Gene Pulser cuvette (Bio-Rad) with a 0.2-cm gap was used. Thereafter, 1 volume of the reaction mixture (consisting of spore suspension plus vector DNA solution) of YEPD (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 1% [wt/vol] glucose) and 4 reaction volumes 1.2 M sorbitol were added and mixed. For regeneration, the whole mixture was incubated overnight at room temperature. Thereafter, spores were streaked out on plates containing selection medium.
Phylogenetic analysis.
Clustal X 2.0.7 (25) was used for alignment of amino acid sequences. MEGA 4.0.2 (66) was used for phylogenetic analysis, using the minimum-evolution method. Amino acid sequences were obtained from the T. reesei, Trichoderma viride, or T atroviride genome database (JGI webpage: http://genome.jgi-psf.org/). For N. crassa, Aspergillus spp., Fusarium spp., and Magnaporthe grisea, amino acid sequences were retrieved from the BROAD genome database (http://www.broadinstitute.org/science/data).
DNA isolation.
A 0.3-g portion of glass beads (no. A554.1; Carl Roth, Karlsruhe, Germany) was placed in a 2-ml reaction tube, and 600 μl of puffer PB (QIAquick PCR purification kit; Qiagen, Hilden, Germany) was added. T. reesei strains were grown for 3 to 4 days on plates containing 3% (wt/vol) malt extract medium covered with cellophane, and then a small amount of mycelia was harvested, added to the 2-ml tube, and mixed thoroughly. The mixture was centrifuged briefly, and purification was performed using the QIAquick PCR purification kit (Qiagen).
RNA isolation and cDNA synthesis.
Fungal mycelia were harvested by filtration, briefly washed with tap water, and frozen in liquid nitrogen. Total RNA was isolated with the RNeasy plant minikit (Qiagen). RNA was treated with DNase I (Fermentas, Vilnius, Lithuania) and purified with the RNeasy MinElute Cleanup kit (Qiagen). A 5-μg portion of each sample was then reverse transcribed using the RevertAID H minus first-strand cDNA synthesis kit (Fermentas).
qRT-PCR.
Quantitative RT-PCR was performed as described previously (69). All PCRs were performed in an iCycler iQ5 Bio-Rad system (Bio-Rad Laboratories). The Bio-Rad iQ5 software was used to compile PCR protocols and to define the plate setups. All reactions were performed in triplicates. The final mixture for each sample contained 12.5 μl iQ SYBR green mix (Bio-Rad Laboratories), 9.5 μl sterile water, 6.25 μM forward and reverse primers, and 1 μl cDNA (diluted 1:20). Each run included a nontemplate control. Primer and probe sequences are given in Table 1. The following PCR program was used: 3 min of initial denaturation at 95°C and 40 cycles of 15 s at 95°C, 20 s at the gene-specific temperature, and 20 s at 72°C. PCR efficiencies were calculated by linear-regression-of-efficiency analysis using a dilution series and the Bio-Rad iQ5 software (69). All qPCR efficiencies were above 90%. As a reference gene we used the gene encoding ribosomal protein L6e of T. reesei (69). A homologue of this gene has been shown to be suitable for qRT-PCR in N. crassa (16, 26, 40).
Biomass determination.
T. reesei QM9414 and the Δacy1 and Δpkac1 mutants were grown in 200 ml Mandels-Andreotti minimal medium liquid culture supplemented with 0.1% (wt/vol) peptone to induce germination and with 1% (wt/vol) lactose as a carbon source in constant light or constant darkness. Fungal mycelia were harvested after 20, 30, and 40 h to increase statistical significance. Mycelia were collected on a Whatman glass fiber filter (Whatman, Maidstone, England) by filtration. After drying overnight at 80°C, the mycelium was weighed. The biomass formation was calculated from two independent experiments with two biological replicates each.
EMSA and preparation of cell extracts.
T. reesei wild-type strain QM9414 and the Δpkac1 mutant were grown in Mandels-Andreotti minimal medium with 1% (wt/vol) lactose as a carbon source. After 30 h, mycelia were harvested and frozen in liquid nitrogen. Cell extracts were prepared as described previously (75). For electrophoretic mobility shift assay (EMSA), radioactively labeled complementary oligonucleotides CKT057/CKT058 (75) were used for binding studies. Five nanograms of probe and 30 μg of protein extracts were applied per lane. Complementation with CKT057/CKT058 in a 100-fold molar excess showed that the complexes seen indeed resulted from protein binding on the fragment. Complementation with a 100-fold molar excess of oligonucleotides CKT083/CKT084 (75), which comprise a deleted version of the binding motif CAE, was done in order to confirm that the complexes seen in our experiment were indeed specific for binding within CAE. EMSA was performed as described previously (57, 75). Densitometric scanning of band intensity was performed using the Bio-Rad GS800 calibrated densitometer with the Bio-Rad Quantity One software package (Bio-Rad). Three different exposures were analyzed, and after background subtraction, band densities were normalized to the intensity of the uppermost band of QM9414 in darkness for comparison between examples.
RESULTS
Characterization of T. reesei acy1 and pkac1.
The aim of this study was to evaluate the role of the central components of the cAMP pathway in regulation of cellulase gene expression. We therefore investigated the influence of the T. reesei homologues of adenylate cyclase and cAMP-dependent protein kinase A in this process.
The genome of T. reesei (http://genome.jgi-psf.org/Trire2/Trire2.home.html) contains one putative adenylate cyclase gene (acy1 [TR_124340]) and two genes encoding catalytic subunits of cAMP-dependent protein kinase A (pkac1 [TR_57399] and pkac2 [TR_65873]) (49). pkac1 (protein kinase A catalytic subunit 1) of T. reesei consists of a predicted 1,593-bp open reading frame interrupted by four introns and encodes a protein of 449 amino acids. Phylogenetic analysis of PKAC1 and PKAC2 along with their orthologues in other fungi confirms their classification (see Fig. S2 in the supplemental material). In contrast to the phenotype of mutants with mutations in the catalytic subunit 1 of protein kinase A, deletion of pkac2-like genes in M. grisea and Ustilago maydis does not affect the cell morphology (27). Also, a recent study of N. crassa did not reveal any phenotype upon deletion of the pkac2 homologue (42). We therefore focused on pkac1 in our study.
acy1 (adenylate cyclase 1) of T. reesei consists of a predicted 6,649-bp open reading frame interrupted by four introns, which encodes a protein of 2,037 amino acids. ACY1 was checked for conserved domains using the NCBI Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (35). The structure of fungal adenylate cyclases is conserved between species. We found the domains characteristic for fungal adenylate cyclases (5) to be similar to those of T. reesei ACY1. Furthermore, phylogenetic analysis of adenylate cyclases in various species shows that ACY1 corresponds to the fungal adenylate cyclases of class III (5) (data not shown).
ACY1 and PKAC1 are involved in regulation of vegetative growth.
For detailed analysis of the roles of ACY1 and PKAC1 in regulation of cellulase gene expression, we constructed mutant strains lacking these proteins. Inoculation of the Δpkac1 and Δacy1 mutants on malt extract plates (3%, wt/vol) and incubation at 28°C in constant darkness or constant light for 4 days showed that the hyphal extension rates of these strains are strongly decreased compared to that of the parental strain QM9414, which is in accordance with data from other fungi (Fig. 1A). Deletion of the corresponding PKAC genes in different fungi shows a typical phenotype, with dramatically decreased growth rate on solid media (6), (13). Lack of acy1 leads to a significantly reduced growth rate, which is similar to the growth of the pkac1 mutant strain, and abolished intracellular cAMP production. Similar phenotypes were obtained for Aspergillus fumigatus (29), Aspergillus nidulans (13), Magnaporthe grisea (11), and Trichoderma virens (39). Microscopic inspection of growth of the Δpkac1 and Δacy1 mutants revealed very dense mycelium and loss of hyphal avoidance (19, 72) in these strains (Fig. 1A). Supplementation with dbcAMP enhanced growth of the Δacy1 mutant, although not to wild-type levels (Fig. 1B), as was also shown for A. fumigatus (29).
Fig 1.
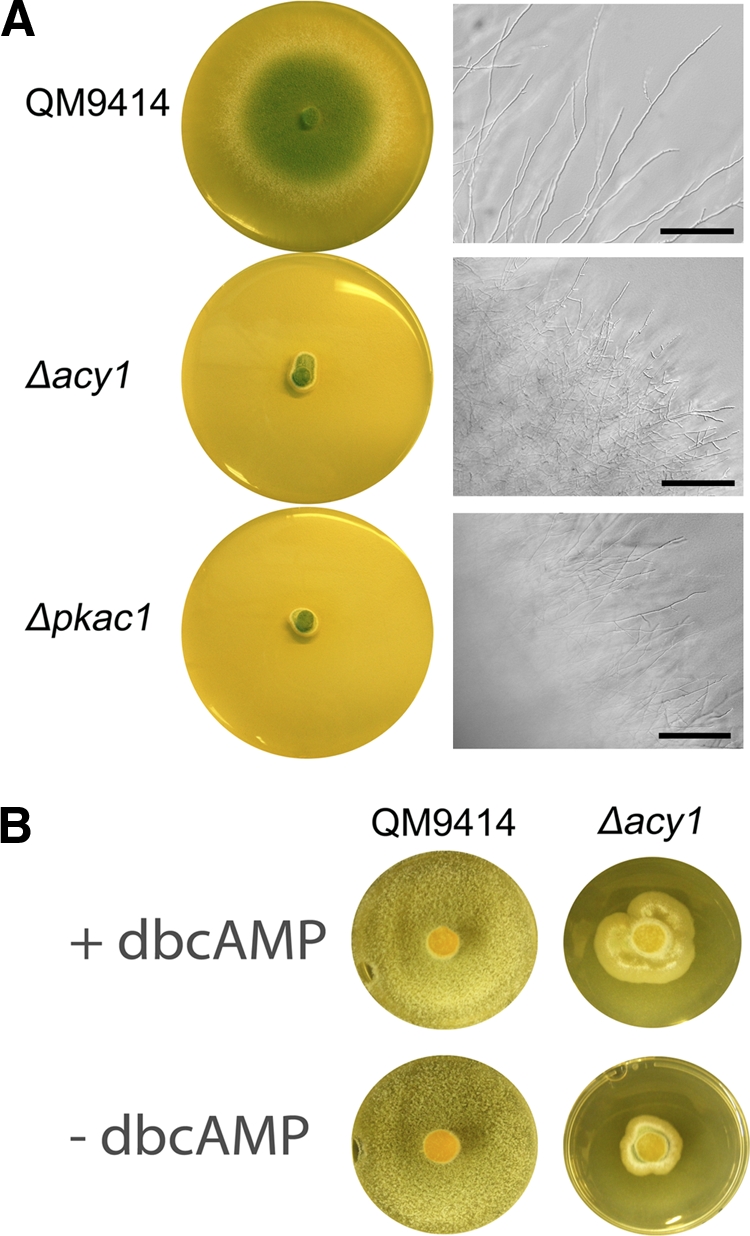
Phenotypes of wild-type QM9414 and the Δacy1 and Δpkac1 mutants. (A) Strains were grown on malt extract agar plates for 4 days at 28°C. Scale bars are equivalent to 100 μm. (B) Supplementation of dbcAMP to the Δacy1 mutant alleviates the severe growth phenotype but does not rescue wild-type growth. QM9414 and the Δacy1 mutant were grown on malt extract agar plates supplemented with 5 mM dbcAMP for 4 days at 28°C.
Investigation of growth on the soluble carbon source lactose in liquid cultures in constant light and constant darkness for 20, 30, and 40 h revealed decreased biomass formation, predominantly at earlier time points, in the Δpkac1 and Δacy1 mutants, although the growth defects in liquid minimal medium were clearly less severe than those on solid malt extract medium. Only minor differences between growth in darkness and in light were observed (see Fig. S3 in the supplemental material).
Deletion of pkac1 and acy1 does not perturb fruiting body formation.
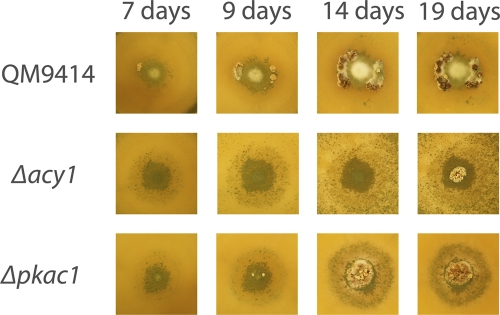
The recently discovered possibility of crossing in T. reesei (60) prompted us to investigate whether this tool would still be available in strains lacking ACY1 or PKAC1. Conidia of the sexually competent wild-type strain CBS999.97 (MAT1-1) (60) were inoculated with equal amounts of the respective mutant strain and with QM9414 (MAT1-2) as a control. This method eliminates the influence of growth rate on evaluation of fruiting body formation (Fig. 2). In both the Δpkac1 and Δacy1 mutants delayed fruiting body formation was observed, with the most severe effect occurring upon lack of acy1. Confrontation assays with these strains inoculated opposite from each other confirmed this result (see Fig. S4 in the supplemental material). Fruiting bodies formed with both mutants were of a size comparable to those of the wild type.
Fig 2.
Effect of pkac1 and acy1 on T. reesei on fruiting body formation with H. jecorina CBS999.97. Fruiting body formation upon coinoculation of H. jecorina CBS999.97 (MAT1-1) and wild-type QM9414 (MAT1-2), the Δpkac1 mutant (MAT1-2), or the Δacy1 mutant (MAT1-2) is shown. Plates were inoculated with equal amounts of spores of both mating partners in the center of the plate to prevent interference of different growth rates of wild-type and mutants Strains were grown on malt extract agar plates at 25°C in daylight. Pictures were taken at 7, 9, 14, and 19 days after inoculation.
These results are in accordance with data on N. crassa cr-1 (adenylate cyclase), deletion of which causes delayed fruiting body formation (21). However, we did not observe a reduction in fruiting body size as reported for Sordaria macrospora (22). We conclude that the cAMP pathway provides a positive input to the regulation of sexual development but is not essential for this process in T. reesei.
Analysis of progeny from crosses revealed that the severe growth phenotype on solid media clearly segregates with resistance to hygromycin B and consequently confirms that the defects of the Δpkac1 and Δacy1 mutants are not caused by random changes elsewhere in the genome.
Light responsiveness of cellulase gene expression is dampened by PKAC1 and ACY1.
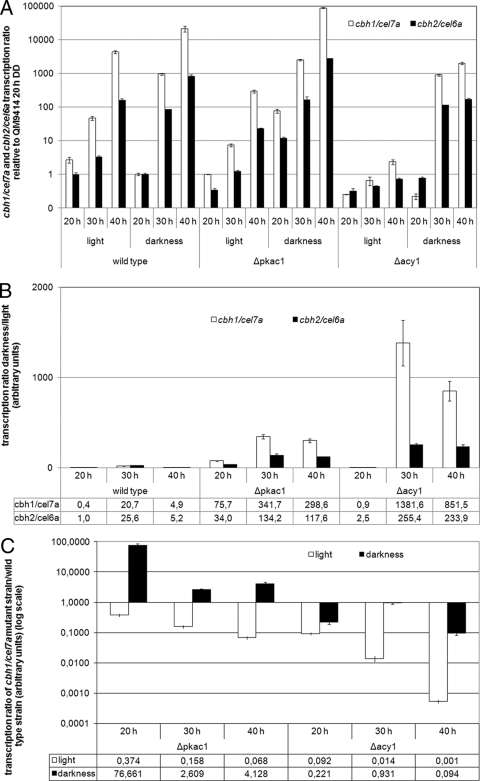
Earlier studies indicated an involvement of cAMP in regulation of cellulase gene expression (53, 61). Additionally, we had found in preliminary Northern blot experiments that cellulase gene expression on lactose is regulated differently in response to light than on cellulose (A. Schuster and M. Schmoll, unpublished data). We thus analyzed the roles of ACY1 and PKAC1 in regulating the transcript abundances of the two major cellulase genes cel7a/cbh1 and cel6a/cbh2 in light and darkness (Fig. 3A).
Fig 3.
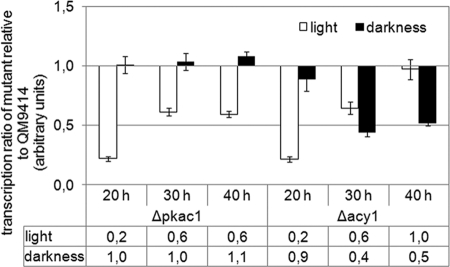
Transcript levels of the major cellulase genes cbh1/cel7a and cbh2/cel6a as influenced by the absence of PKAC1 or ACY1. Transcript abundance was analyzed by qRT-PCR after 20, 30, or 40 h of growth in Mandels-Andreotti minimal medium with 1% (wt/vol) lactose as a carbon source in constant light or constant darkness. (A) Transcript levels of cbh1/cel7a and cbh2/cel6a in the wild-type QM9414 and the Δacy1 and Δpkac1 mutants. Values are shown as fold changes relative to that for QM9414 after 20 h in darkness. (B) Transcript ratios in darkness versus light of cbh1/cel7a and cbh2/cel6a in wild-type QM9414 and the Δacy1 and Δpkac1 mutants. Relative mean values of fold regulation for cbh1/cel7a and cbh2/cel6a are given below the diagram. (C) cbh1/cel7a transcript ratio in mutant versus wild-type strains in constant light or constant darkness. Relative mean values of fold regulation are given below the diagram.
As expected, cel7a/cbh1 and cel6a/cbh2 are coregulated, with cel7a/cbh1 showing higher transcript levels than cel6a/cbh2. Interestingly, in contrast to data on cellulose (51), light does not stimulate cellulase gene expression on lactose but has a negative effect, hence confirming our earlier Northern blot data. We found considerably decreased transcript levels in darkness compared to in light for both genes, reaching more than 25-fold-enhanced transcript abundance in darkness at 30 h (Fig. 3B). This differential expression of cellulase genes between light and darkness is even more apparent in the Δpkac1 and Δacy1 mutants, with more than 1,000-fold upregulation of cel7a/cbh1 transcript abundance in the Δacy1 mutant in darkness compared to light (Fig. 3B). Alterations in light-dependent gene expression on different carbon sources are not without precedent, since the influence of light on growth is also dependent on the carbon source (8, 14, 15, 55). A recent study reported considerable differences in light-dependent gene expression even on different concentrations of the same carbon source (4). Nevertheless, deletion of pkac1 or acy1 affects only the extent of differential expression of cel7a/cbh1 and cel6a/cbh2 in light and darkness and does not abolish or reverse light-dependent regulation. Therefore, pkac1 and acy1 are not assumed to determine the relevance of the received nutrient signals in light and darkness. Instead, these genes appear to balance cross talk between nutrient and light signaling.
Transcript levels of the major cellulase genes are decreased by light on lactose.
In addition to the effect of the cAMP pathway on light responsiveness of cellulase gene expression, we also investigated how PKAC1 and ACY1 would alter cellulase gene expression compared to that in the wild type. Comparison of cel7a/cbh1 transcript levels in the Δpkac1 and Δacy1 mutants with that in QM9414 showed that in light both PKAC1 and ACY1 exert a positive effect, with ACY1 being more effective (Fig. 3C). In darkness, however, the positive effect of ACY1 is less pronounced, and PKAC1 even acts negatively on transcript levels of cel7a/cbh1. Despite the severely perturbed growth of the two mutants on solid medium, growth in liquid medium was only slightly decreased in both mutants (see Fig. S3 in the supplemental material). Hence, a direct correlation between biomass formation and transcription of the major cellulase genes is not obvious in darkness, and the assumption that growth rate would be correlated with cellulase expression is not supported. Rather, delayed germination of the Δpkac1 and Δacy1 mutants is likely to cause their lower biomass formation.
We conclude that both PKAC1 and ACY1 are involved in adjustment of cellulase gene transcript levels to light or darkness and consequently are components of the integrated signaling cascades of light and nutrient signaling.
Complex formation within the promoter of cel6a/cbh2 is altered in light and in the Δpkac1 mutant.
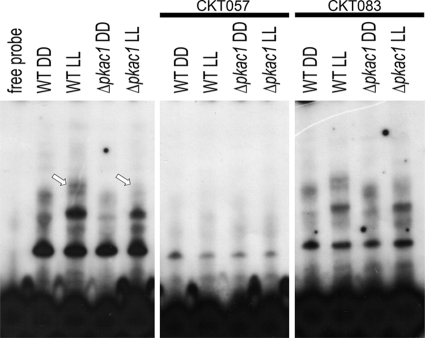
Because of the effect of PKAC1 on transcript levels of cellulase genes, we were interested in whether this effect would be mediated directly or indirectly via phosphorylation of transcription factors by EMSA. This method reveals binding of protein complexes to a given regulatory promoter motif by retardation of a radioactively labeled DNA fragment due to complex formation. In the case of an indirect effect, such as phosphorylation of upstream factors regulating the abundances of important cellulase transcription factors such as XYR1, ACE1, or ACE2, we should observe differences in complex formation of regulatory factors binding within cellulase promoters in light and darkness and/or in the Δpkac1 mutant. Direct effects, i.e., phosphorylation of XYR1, ACE1, or ACE2 itself, would not necessarily cause detectable alterations in complex formation of proteins within the respective promoter. Nevertheless, enhanced binding efficiency due to phosphorylation cannot be excluded.
As a probe, we used a fragment comprising the well-studied cellulase-activating element CAE of the cel6a/cbh2 promoter (CKT057) (75), which is coregulated with cel7a/cbh1, as a test case for EMSA. Our results clearly show different complex formation within the CAE in light and darkness upon growth on lactose in the wild-type (Fig. 4). Lack of PKAC1, however, results in weaker complex formation within the CAE, which is most obvious in light. Densitometric analysis of band intensities revealed that the binding strength of the two uppermost complexes formed in the Δpkac1 mutant is decreased by 50% ± 11% in darkness and by 74% ± 18% in light (indicated by the arrows in Fig. 4). We conclude that the influence of PKAC1 on transcript levels of cellulase genes is mediated via alteration of the abundance and/or binding efficiency of positive or negative regulating factors which bind within the cellulase promoters or the promoters of cellulase-regulating upstream factors.
Fig 4.
Characterization of protein complex formation in cell extracts from QM9414 and the Δpkac1 mutant. Strains were grown in Mandels-Andreotti minimal medium with 1% (wt/vol) lactose as a carbon source. CKT057, comprising the cellulase-activating element (CAE), was used as a probe. Thirty micrograms of T. reesei cell extract was loaded. The most clearly altered protein complex in light is indicated by arrows. Competition experiments with 100-fold excesses of CKT057 (wild type) and CKT083 (oligonucleotide with deleted CAE) as indicated confirmed specific binding of complexes by abolishment of complex formation (CKT057; wild-type) or unaltered complex formation (CKT083; deleted binding site).
Transcription factors of cbh1 and cbh2 are influenced by the presence of light.
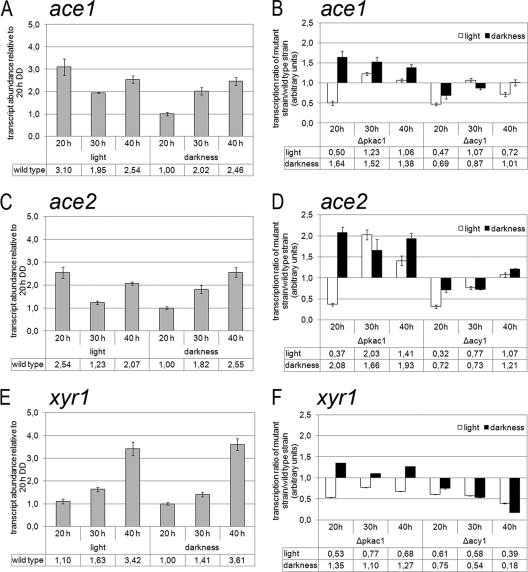
The striking effects of ACY1 and PKAC1 on cellulase gene transcription led to the question how this regulatory impact would be mediated. Moreover, EMSA indicated an influence on transcription factors to be responsible for regulation of cellulase genes. We therefore analyzed the transcript abundances of three of the most important transcription factor genes with respect to regulation of cellulase gene expression, ace1, ace2, and xyr1, in light and darkness. Interestingly, all three transcription factors have cAMP-dependent protein kinase phosphorylation sites and, additionally, several casein kinase II and protein kinase C phosphorylation sites (71).
In QM9414, we detected only minor differences in transcript levels of these transcription factor genes in light and darkness (Fig. 5A, C, and E). The mutant strains, however, did show differences in the transcript abundances of ace1, ace2, and xyr1. While PKAC1 seems to have a negative effect on transcript levels of all three transcription factor genes in darkness, ACY1 has only small effects on the transcript abundances of ace1 and ace2 but a clearly positive effect on that of xyr1 in both light and darkness (Fig. 5B, D, and F). Since the transcript pattern of xyr1 in the Δpkac1 and Δacy1 mutants bears a striking similarity to that of cel7a/cbh1 in these mutants, we assume the major effect of PKAC1 and ACY1 to be exerted via regulation of the transcript abundance of xyr1 by phosphorylation of an upstream factor. Therefore, the (negative) effect of lack of pkac1 on complex formation within the cbh2-promoter also is likely to be indirect and may involve this upstream component. Nevertheless, we cannot rule out a contribution to cel7a/cbh1 regulation by ACE1 or ACE2, which may be phosphorylated directly or regulated indirectly by PKAC1.
Fig 5.
Analysis of transcript abundances of the transcription factor-encoding genes ace1 (A and B), ace2 (C and D), and xyr1 (E and F) by qRT-PCR. (A, C, and E) Transcript abundances of ace1 (A), ace2 (C), and xyr1 (E) in QM9414. Values are shown as fold changes relative to the value for QM9414 in constant darkness after 20 h of growth. (B, D, and F) Transcript ratios of ace1 (B), ace2 (D), and xyr1 (F) between QM9414 and mutant strains. Transcript abundance was analyzed by qRT-PCR during cultivation of wild-type QM9414 and the Δpkac1 and Δacy1 mutants in liquid Mandels-Andreotti medium with 1% (wt/vol) lactose as a carbon source after 20, 30, or 40 h of growth in constant light or constant darkness.
Transcription of env1 is induced by the photoreceptor BLR1, the homologue of which was shown to be phosphorylated by protein kinase A in N. crassa (20). The light-dependent function of ACY1 and PKAC1 together with the recently described influence of ENV1 on cAMP levels (66) led us to investigate this regulatory interrelationship in more detail. Indeed, lack of either pkac1 or acy1 causes decreased transcript levels of env1 in light (Fig. 6). Consequently, we assume that the influence of protein kinase A on the activity of the photoreceptor complex as shown in N. crassa (20) may also be operative in T. reesei, due to the decreased transcript levels of env1. This feedback cycle may serve as fine-tuning mechanism for integration of light and nutrient signaling.
Fig 6.
Analysis of transcript abundance of the light regulatory protein-encoding gene env1. The transcript ratios of env1 between QM9414 and mutant strains are shown. Transcript levels were analyzed by qRT-PCR during cultivation of wild-type QM9414 and the Δpkac1 and Δacy1 mutants in liquid Mandels-Andreotti medium with 1% (wt/vol) lactose as a carbon source after 20, 30, or 40 h of growth in constant light or constant darkness.
DISCUSSION
The relevance of the secondary messenger cAMP for physiology and development has been subject to extensive research with organisms of all kingdoms of life. In fungi, perturbation of the central components of cAMP signaling, adenylate cyclase or cAMP-dependent protein kinase A, consistently leads to severe growth defects on solid media and defects in numerous physiological processes (6, 11, 13, 29, 39, 76). In accordance with these findings, we also observed such defects for deletion of acy1 or pkac1 in T. reesei.
For Trichoderma spp., the function of cAMP with respect to cellulase gene expression was studied more than a decade ago (61), although the influence of adenylate cyclase or protein kinase A remained unknown. Early studies on the cAMP pathway of Trichoderma spp. also revealed an involvement of light in regulation of cAMP levels (50). Positive effects of light on activity have been shown for both adenylate cyclase (23) and protein kinase A (9). This increased activity of PKA occurs even in the absence of one of the photoreceptors, BLR-1 or BLR-2, in T. atroviride. In this fungus, expression of blue light-regulated genes also is influenced by alterations in activity of PKA (9). Finally, a connection between the regulatory impact of cAMP and light was discovered by studying the adenylate cyclase-activating G-protein alpha subunit GNA3, which enhances both cAMP levels and cellulase gene expression in light (53). We now provide insights into the mechanism of cAMP-dependent, light-modulated cellulase gene expression in T. reesei. All reports available so far on light-modulated cellulase gene expression concentrate on cellulose as a carbon source. Interestingly, we also found light-modulated cellulase gene expression on lactose, but we were surprised that the effect of light is reversed compared to that with cellulose, since on lactose darkness is beneficial for high levels of cellulase gene transcripts. A similar phenomenon was seen for induction of cellulase gene transcription by sophorose (56). Consequently the assessment of the significance of a given nutrient signal with respect to its relevance in light and darkness can also be observed on lactose. In other words, the presence of lactose has a different significance for T. reesei in light and darkness, which is reflected in the light responsiveness of cellulase gene expression. Therefore, cellulase gene expression on this carbon source is adjusted to the light status. However, the importance of cellulase gene expression on lactose must be rated differently than that on cellulose. This may be due to the fact that cellulases are not essential for growth on lactose (59), yet they are on cellulose.
Analysis of transcript abundances of cellulase genes in the wild type and the Δpkac1 and Δacy1 mutants revealed intriguing insights into the mechanism of cellulase regulation by the cAMP pathway (Fig. 7). Although differential gene expression in light and darkness (light responsiveness) was also observed in the wild type (roughly 20-fold), these relative differences were considerably higher in the mutant strains. In the Δpkac1 mutant transcript abundance is 300-fold higher in darkness than in light, and in the Δacy1 mutant the difference is even above 1,000-fold. Despite these huge differences in transcript abundance, no-template controls confirmed that even the very low levels of transcripts are not artifacts. Hence, the cAMP pathway obviously plays an important role in adaptation of metabolism (to which extracellular hydrolytic enzymes are of crucial importance) to the altered environmental conditions in light and darkness. Both PKAC1 and ACY1 are important for balanced cellulase gene expression in light and darkness and dampen light responsiveness (Fig. 7).
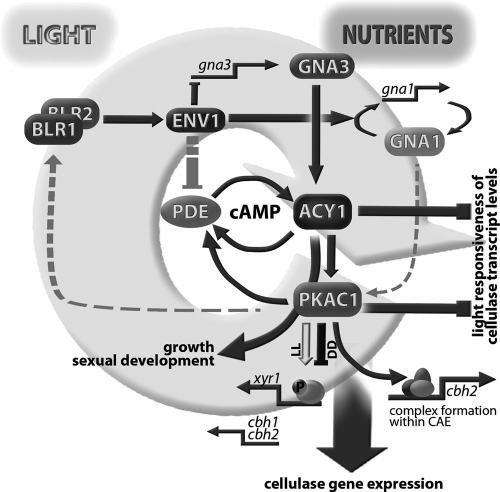
Fig 7.
Schematic representation of the functions of ACY1 and PKAC1 in integration of nutrient and light signals. Nutrient signals are transmitted by GNA1 and GNA3, activation of which leads to increased cAMP levels (53, 58). ENV1 enhances feedback regulation of gna1, acts negatively on transcription of gna3, and is assumed to dampen phosphodiesterase (PDE) function (69). ACY1 and PKAC1 decrease light responsiveness of cellulase transcript levels. Their effect on cellulase gene expression is mediated mainly by PKAC1, which alters protein complex formation within the cbh2 promoter and likely phosphorylates an upstream factor regulating xyr1 gene expression. Comparable effects of deletion of gna1 (58) and pkac1 suggest an additional level of regulation for the function of PKAC1 by GNA1, as proposed earlier (13). Fine-tuning of cross talk between light and nutrient signaling is likely to be accomplished by feedback of PKAC1 via the BLR1-BLR2 complex.
Interestingly, comparison to wild-type levels of cellulase transcripts shows that in darkness, PKAC1 and ACY1 have contrary effects, while in light both factors act positively. The presence of ACY1 has a positive influence on transcript levels of cellulase genes under both conditions. However, the effect of PKAC1 is clearly light dependent. We assume that while ACY1 provides an output signal in terms of cAMP levels, PKAC1 does not simply transmit this signal but is a further checkpoint for integration of different signals.
In this respect it is interesting that in N. crassa, cAMP-dependent protein kinase A is a key component of the circadian clock. PKA phosphorylates FREQUENCY (20) and acts as a priming kinase for phosphorylation of the white collar complex (20). In T. reesei, the corresponding photoreceptors BLR1 and BLR2 are involved in regulation of cellulase gene expression (10) and may consequently be part of an integrated feedback loop connecting light response and nutrient signaling, presumably exerted by heterotrimeric G-proteins (53, 58) and in part via adjustment of cAMP levels (Fig. 7). Further support for this hypothesis comes from a recent study which clearly showed an interconnection between the light response pathway and cAMP signaling. This pathway is assumed to work via adjustment of cAMP levels by ENVOY acting on a phosphodiesterase (69). The influence of ACY1 and PKAC1 on the transcript abundance of env1 confirms this regulatory interrelationship of ENVOY with the cAMP pathway. These results suggest a feedback circuit that is operative between the cAMP pathway and the light response pathway. This interpretation would be consistent with results on T. atroviride (9) showing that BLR1 activity is increased when PKA activity is enhanced. However, since in N. crassa PKA inhibits the light function of the WCC (20), the regulatory network connecting PKA, the BLR complex, and ENV1 in T. reesei requires additional investigation. Decreased transcript levels of env1 should lead to increased phosphodiesterase activity (69), causing lower levels of cAMP, which in turn should lead to decreased cellulase levels (61). Such a negative feedback cycle would be consistent with the decreased cellulase levels observed in light in this study.
In darkness, ACY1 but not PKAC1 positively influences transcript levels of env1, and PKAC1 acts negatively on cellulase transcription, in contrast to ACY1. Two alternative explanations for this phenomenon are conceivable. First, PKA-independent functions and alternative targets of cAMP (48) may bypass PKAC1-mediated gene regulation and may also feed back on env1 transcription. However, our recent findings that ENV1 overrules G-protein alpha function in light but in darkness only the G-protein-related phenotype is observed (69) provide a more intriguing interpretation. According to this mechanism, the function of GNA1 is dominant over that of ENV1 in darkness. If the GNA1 pathway acts at least in part via direct regulation of PKAC1, as suggested earlier (13), the higher levels of cellulase transcripts upon lack of pkac1 would be consistent with results for lack of gna1. The proposed feedback of PKAC1 on env1 transcription via BLR1 is abolished in darkness, and hence no alteration in transcript levels is observed. However, in the absence of ACY1, the feedback of PKAC1 on env1 transcription due to GNA1 activation of PKAC1 is still operative, which is consistent with the negative effect of GNA1 on env1 transcription as shown earlier (69).
XYR1 is an essential transcription factor of cellulase gene expression not only on cellulose but also on lactose (43, 62–64). It was shown that XYR1 binds to one of its target promoters (xyn1) not only under inducing conditions but also under repressing conditions (44). Our data raise the question of whether this is also true for different light conditions, which were not considered in previous studies. Hence, this topic warrants further investigation.
Transcript levels of xyr1 are influenced negatively by ACE2, which is assumed to require phosphorylation to contact DNA (65). xyr1 transcript abundance is suggested also to be subject to repression by ACE1 (31). These findings are especially interesting with respect to our result on the influence of PKAC1 on transcript levels of xyr1, ace1, and ace2 as well as those of cel7a/cbh1. On the one hand, the coregulation of xyr1 and cbh1 suggests regulation of cellulase gene expression by phosphorylation of an upstream compound influencing XYR1 abundance. Additional support for this hypothesis comes from the fact that xyr1 and cbh1 show a striking correlation in transcript patterns, not only in wild-type strains but also in mutant strains (43). On the other hand, however, we cannot exclude that this effect is at least in part due to regulation of ace1 and/or ace2, which are upregulated in Δpkac1 and hence may cause the negative effect on xyr1 and consequently cel7a/cbh1 in light. In darkness, transcript levels of all three transcription factors are upregulated in the Δpkac1 mutant, which is not in accordance with these earlier data. We assume that the reason for this discrepancy could be the uncontrolled light conditions applied in the earlier studies. While we do not see considerable alteration in transcript abundances of xyr1, ace1, and ace2 in the wild-type in light and darkness on lactose, their impact on each other cannot automatically be inferred to be unaltered in light and darkness. Clear support for this conclusion comes from the fact that upon deletion of pkac1, transcript levels of xyr1 become light regulated, which cannot be excluded for other factors. The regulatory interactions between xyr1, ace1, and ace2 are unlikely to disappear under controlled light conditions. It thus can be expected that in accordance with the considerable differences of cellulase gene expression on lactose in light and darkness as shown in this study, xyr1, ace1, and ace2 also influence each other differently.
In summary, we could show that the crucial components of the cAMP pathway, adenylate cyclase 1 (ACY1) and cAMP-dependent protein kinase A (PKA), are involved in regulation of cellulase gene expression in T. reesei. Although earlier studies strongly suggested such a function (53, 61), the finding that the regulatory output of PKAC1, but not ACY1, in terms of cellulase gene transcript levels as well as transcript abundance of the respective transcription factors is dependent on the light status is novel. These findings suggest that a comparable function of PKA in stabilization of the photoreceptor complex as observed in N. crassa is likely to be present also in T. reesei. Thereby, a cycle for integration of nutrient signaling (e.g., by heterotrimeric G-proteins) and light response could be established and might be responsible for fine-tuning of the regulatory outputs of both pathways.
Supplementary Material
ACKNOWLEDGMENTS
Our work was supported by grants from the Austrian Science Fund (FWF) (P-20004 and V152-B20) to M.S. D.T. is the recipient of a DOC fFORTE fellowship from the Austrian Academy of Sciences at the Institute of Chemical Engineering, Vienna University of Technology.
Footnotes
Published ahead of print 27 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alspaugh JA, et al. 2002. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aro N, Ilmen M, Saloheimo A, Penttila M. 2003. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aro N, Saloheimo A, Ilmen M, Penttila M. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276: 24309–24314 [DOI] [PubMed] [Google Scholar]
- 4. Atoui A, et al. 2010. Cross-talk between light and glucose regulation controls toxin production and morphogenesis in Aspergillus nidulans. Fungal Genet. Biol. 47: 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker DA, Kelly JM. 2004. Structure, function and evolution of microbial adenylyl and guanylyl cyclases. Mol. Microbiol. 52: 1229–1242 [DOI] [PubMed] [Google Scholar]
- 6. Banno S, et al. 2005. A catalytic subunit of cyclic AMP-dependent protein kinase, PKAC-1, regulates asexual differentiation in Neurospora crassa. Genes Genet. Syst. 80: 25–34 [DOI] [PubMed] [Google Scholar]
- 7. Bölker M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25: 143–156 [DOI] [PubMed] [Google Scholar]
- 8. Carlile MJ. 1965. The photobiology of fungi. Annu. Rev. Plant Physiol. 16: 175–202 [Google Scholar]
- 9. Casas-Flores S, et al. 2006. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot. Cell 5: 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castellanos F, et al. 2010. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet. Biol. 47: 468–476 [DOI] [PubMed] [Google Scholar]
- 11. Choi W, Dean RA. 1997. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Souza CA, Heitman J. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25: 349–364 [DOI] [PubMed] [Google Scholar]
- 13. Fillinger S, Chaveroche MK, Shimizu K, Keller N, d'Enfert C. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44: 1001–1016 [DOI] [PubMed] [Google Scholar]
- 14. Friedl MA, Kubicek CP, Druzhinina IS. 2008. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis. Appl. Environ. Microbiol. 74: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedl MA, Schmoll M, Kubicek CP, Druzhinina IS. 2008. Photostimulation of Hypocrea atroviridis growth occurs due to a cross-talk of carbon metabolism, blue light receptors and response to oxidative stress. Microbiology 154: 1229–1241 [DOI] [PubMed] [Google Scholar]
- 16. Froehlich A, Noh B, Vierstra R, Loros J, Dunlap J. 2005. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell 4: 2140–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Griffioen G, Thevelein JM. 2002. Molecular mechanisms controlling the localisation of protein kinase A. Curr. Genet. 41: 199–207 [DOI] [PubMed] [Google Scholar]
- 18. Gruber F, Visser J, Kubicek CP, de Graaff LH. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18: 71–76 [DOI] [PubMed] [Google Scholar]
- 19. Hickey PC, Jacobson DJ, Read ND, Louise Glass N. 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37: 109–119 [DOI] [PubMed] [Google Scholar]
- 20. Huang G, et al. 2007. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 21: 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ivey FD, Kays AM, Borkovich KA. 2002. Shared and independent roles for a Galpha(i) protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot. Cell 1: 634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamerewerd J, Jansson M, Nowrousian M, Poggeler S, Kuck U. 2008. Three alpha-subunits of heterotrimeric G proteins and an adenylyl cyclase have distinct roles in fruiting body development in the homothallic fungus Sordaria macrospora. Genetics 180: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolarova N, Haplová J, Greík M. 1992. Light-activated adenyl cyclase from Trichoderma viride. FEMS Microbiol. Lett. 93: 275–278 [DOI] [PubMed] [Google Scholar]
- 24. Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. 2009. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol. Biofuels 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larkin M, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- 26. Lee K, Dunlap JC, Loros JJ. 2003. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163: 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee N, D'Souza C, Kronstad J. 2003. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol 41: 399–427 [DOI] [PubMed] [Google Scholar]
- 28. Lengeler KB, et al. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liebmann B, Gattung S, Jahn B, Brakhage AA. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269: 420–435 [DOI] [PubMed] [Google Scholar]
- 30. Mach RL, Schindler M, Kubicek CP. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25: 567–570 [DOI] [PubMed] [Google Scholar]
- 31. Mach-Aigner A, et al. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 74: 6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mandels M, Andreotti RE. 1978. Problems and challenges in the cellulose to cellulase fermentation. Proc. Biochem. 13: 6–13 [Google Scholar]
- 33. Mandels M, Reese ET. 1960. Induction of cellulase in fungi by cellobiose. J. Bacteriol. 79: 816–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mandels M, Reese ET. 1957. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 73: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37: D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Margolles-Clark E, Ihnen M, Penttilä M. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57: 167–179 [Google Scholar]
- 37. Martinez D, et al. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26: 553–560 [DOI] [PubMed] [Google Scholar]
- 38. Mehra A, Baker CL, Loros JJ, Dunlap JC. 2009. Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 34: 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mukherjee M, Mukherjee PK, Kale SP. 2007. cAMP signalling is involved in growth, germination, mycoparasitism and secondary metabolism in Trichoderma virens. Microbiology 153: 1734–1742 [DOI] [PubMed] [Google Scholar]
- 40. Nowrousian M, Duffield G, Loros J, Dunlap J. 2003. The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics 164: 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pall ML. 1981. Adenosine 3′,5′-phosphate in fungi. Microbiol. Rev. 45: 462–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park G, et al. 2011. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot. Cell 10: 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Portnoy T, et al. 2011. Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot. Cell 10: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rauscher R, et al. 2006. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell 5: 447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rocha CR, et al. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12: 3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roelfsema WA, Kuster BFM, Heslinga MC, Pluim H, Verhage M. 2000. Lactose and derivatives. In Ullmann's encyclopedia of industrial chemistry. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany: [Google Scholar]
- 47. Saloheimo A, Aro N, Ilmen M, Penttila M. 2000. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275: 5817–5825 [DOI] [PubMed] [Google Scholar]
- 48. Sands WA, Palmer TM. 2008. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 20: 460–466 [DOI] [PubMed] [Google Scholar]
- 49. Schmoll M. 2008. The information highways of a biotechnological workhorse—signal transduction in Hypocrea jecorina. BMC Genomics 9: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmoll M, Esquivel-Naranjo EU, Herrera-Estrella A. 2010. Trichoderma in the light of day—physiology and development. Fungal Genet. Biol. 47: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmoll M, Franchi L, Kubicek CP. 2005. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell 4: 1998–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmoll M, Kubicek CP. 2003. Regulation of Trichoderma cellulase formation: lessons in molecular biology from an industrial fungus. A review. Acta Microbiol. Immunol. Hung. 50: 125–145 [DOI] [PubMed] [Google Scholar]
- 53. Schmoll M, Schuster A, Silva Rdo N, Kubicek CP. 2009. The G-alpha protein GNA3 of Hypocrea jecorina (anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot. Cell 8: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schuster A, et al. 2012. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol. Biofuels. 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M. 2007. Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genomics 8: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schuster A, Kubicek CP, Schmoll M. 2011. Dehydrogenase GRD1 represents a novel component of the cellulase regulon in Trichoderma reesei (Hypocrea jecorina). Appl. Environ. Microbiol. 77:4553–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schuster A, Schmoll M. 2009. Heterotrimeric G-protein signaling and light response: two signaling pathways coordinated for optimal adjustment to nature. Commun. Integr. Biol. 2: 308–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seibel C, Gremel G, Rdo Nascimento Silva Schuster A, Kubicek CP, Schmoll M. 2009. Light-dependent roles of the G-protein alpha subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei). BMC Biol. 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seiboth B, Pakdaman BS, Hartl L, Kubicek CP. 2007. Lactose metabolism in filamentous fungi: how to deal with an unknown substrate. Fungal Biol. Rev. 21:42–48 [Google Scholar]
- 60. Seidl V, Seibel C, Kubicek CP, Schmoll M. 2009. Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U. S. A. 106: 13909–13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sestak S, Farkas V. 1993. Metabolic regulation of endoglucanase synthesis in Trichoderma reesei: participation of cyclic AMP and glucose-6-phosphate. Can. J. Microbiol. 39: 342–347 [DOI] [PubMed] [Google Scholar]
- 62. Stricker AR, Grosstessner-Hain K, Wurleitner E, Mach RL. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5: 2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stricker AR, Mach RL, de Graaff LH. 2008. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 78: 211–220 [DOI] [PubMed] [Google Scholar]
- 64. Stricker AR, Steiger MG, Mach RL. 2007. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 581: 3915–3920 [DOI] [PubMed] [Google Scholar]
- 65. Stricker AR, Trefflinger P, Aro N, Penttilä M, Mach RL. 2008b. Role of Ace2 (activator of cellulases 2) within the xyn2 transcriptosome of Hypocrea jecorina. Fungal Genet. Biol. 45: 436–445 [DOI] [PubMed] [Google Scholar]
- 66. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 67. Taylor SS, et al. 2005. Dynamics of signaling by PKA. Biochim. Biophys. Acta 1754: 25–37 [DOI] [PubMed] [Google Scholar]
- 68. Thevelein JM, De Winde JH. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918 [DOI] [PubMed] [Google Scholar]
- 69. Tisch D, Kubicek CP, Schmoll M. 2011. New insights into the mechanism of light modulated signaling by heterotrimeric G-proteins: ENVOY acts on gna1 and gna3 and adjusts cAMP levels in Trichoderma reesei (Hypocrea jecorina). Fungal Genet. Biol. 48: 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tisch D, Kubicek CP, Schmoll M. 2011. The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei. BMC Genomics 12: 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tisch D, Schmoll M. 2011. Novel approaches to improve cellulase biosynthesis for biofuel production—adjusting signal transduction pathways in the biotechnological workhorse Trichoderma reesei, p 199–224 dos Santos, Bernardes MA. (ed), Biofuel production—recent developments and prospects. Intech, Rijeka, Croatia: [Google Scholar]
- 72. Trinci APJ. 1984. Regulation of hyphal branching and hyphal orientation, p 23–52 Jennings DH, Rayner AD. (ed), The ecology and physiology of the fungal mycelium. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 73. Wilkie T, Yokoyama S. 1994. Evolution of the G protein alpha subunit multigene family. Soc. Gen. Physiol. Ser. 49: 249–270 [PubMed] [Google Scholar]
- 74. Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119 [DOI] [PubMed] [Google Scholar]
- 75. Zeilinger S, Mach RL, Kubicek CP. 1998. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273: 34463–34471 [DOI] [PubMed] [Google Scholar]
- 76. Zhao W, et al. 2006. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect. Immun. 74: 4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.