Abstract
Silver toxicity is a problem that microorganisms face in medical and environmental settings. Through exposure to silver compounds, some bacteria have adapted to growth in high concentrations of silver ions. Such adapted microbes may be dangerous as pathogens but, alternatively, could be potentially useful in nanomaterial-manufacturing applications. While naturally adapted isolates typically utilize efflux pumps to achieve metal resistance, we have engineered a silver-tolerant Escherichia coli strain by the use of a simple silver-binding peptide motif. A silver-binding peptide, AgBP2, was identified from a combinatorial display library and fused to the C terminus of the E. coli maltose-binding protein (MBP) to yield a silver-binding protein exhibiting nanomolar affinity for the metal. Growth experiments performed in the presence of silver nitrate showed that cells secreting MBP-AgBP2 into the periplasm exhibited silver tolerance in a batch culture, while those expressing a cytoplasmic version of the fusion protein or MBP alone did not. Transmission electron microscopy analysis of silver-tolerant cells revealed the presence of electron-dense silver nanoparticles. This is the first report of a specifically engineered metal-binding peptide exhibiting a strong in vivo phenotype, pointing toward a novel ability to manipulate bacterial interactions with heavy metals by the use of short and simple peptide motifs. Engineered metal-ion-tolerant microorganisms such as this E. coli strain could potentially be used in applications ranging from remediation to interrogation of biomolecule-metal interactions in vivo.
INTRODUCTION
Silver compounds have long been recognized as microbicidal agents (12). The mechanism of silver toxicity in microorganisms has been attributed to multiple targets. Silver ions interact with thiol (sulfhydryl) groups and other functional groups in enzymes and proteins, disrupting many normal cell processes such as respiration and establishment of proton motive force (12, 32). Silver treatment also results in the release of potassium ions from bacterial cells, suggesting that the cytoplasmic membrane is compromised upon silver exposure (26). In addition, studies have shown silver complexes with nucleic acids, though the contribution of this target to toxicity is unclear (35).
Silver is currently used to antagonize the growth of microorganisms in a variety of medical settings, such as in catheters and on burn wounds (15, 31, 32). In addition to exposure to silver in medical settings, microorganisms encounter silver in the environment, with certain strains developing very high tolerance. Bacteria have adapted to combat the potent bactericidal properties of silver in a variety of ways. Often, microorganisms express dedicated metal efflux pumps, many encoded by plasmid-borne gene cassettes (8, 17, 29). Bacteria also express extracellular or periplasmic silver-binding peptides that bind silver ions; while their roles in resistance are not clear, some of these binding proteins apparently act in concert with efflux pumps (8). One such plasmid-encoded periplasmic binding peptide, SilE, probably acts in concert with a coexpressed efflux system to provide silver resistance (at 0.6 mM Ag+) to a Salmonella strain originally isolated from a hospital burn ward (8, 21, 30). This 143-residue metal-binding protein was purified and characterized as binding silver ions but not copper or cadmium ions, metals closely related to silver on the periodic table (8). Other environmental bacteria have adapted to growth in the presence of toxic silver ions by sequestration of silver in other forms. Pseudomonas stutzeri AG259, isolated from a silver mine, grows in the laboratory in 50 mM silver nitrate and produces periplasmically localized silver crystals up to 200 nm in size (16). For comparison, typical bacterial laboratory strains are sensitive to micromolar levels of silver nitrate. The reduction of toxic silver ions into less-reactive metallic particles is assumed to contribute to P. stutzeri's silver resistance.
Reports of biologically mediated deposition of inorganic nanoparticles such as that observed in P. stutzeri AG259 have spurred research dedicated to identifying and characterizing peptide sequences that bind or precipitate metals. These peptides, typically 7 to 14 amino acids in size, have been identified in cell surface or phage display systems (5, 25) and are similar in principle to biologically derived polypeptides such as SilE that exhibit metal binding activity in vitro (8). Notably, the peptide Ag4 was identified from a combinatorial phage display library as a peptide with affinity for metallic silver particles. Strikingly, the Ag4 peptide can also cause the precipitation of silver particles from a silver nitrate solution (23). When characterized in vitro as a fusion to the Escherichia coli maltose-binding protein (MBP-Ag4), it exhibited nanomolar affinity for a crystalline silver surface (27).
Many studies have investigated the in vitro capabilities of artificially selected inorganic binding peptides (5, 10, 33, 34), but little work has been done to explore any in vivo phenotypes associated with these peptides. We selected the silver-binding peptide AgBP2 from a combinatorial peptide display library and engineered it onto the E. coli maltose-binding protein (MBP-AgBP2). When localized to the periplasm of a laboratory strain of E. coli, it confers tolerance to silver nitrate and results in decoration of cells with electron-dense nanoparticles. These data point toward a novel ability to manipulate bacterial interactions with heavy metals by the use of simple peptide motifs. This peptide has potential applications in bioremediation and nanoparticle synthesis and as a tool to further probe biomolecular interactions with heavy metals within microbes to monitor complex cell physiology.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The E. coli strains and plasmids used in this study are summarized in Table 1. Biocombinatorial selection was performed with E. coli GI826 carrying pFliTrx (Invitrogen). Growth, induction, and selection were carried out as described previously (11). Briefly, cultures were grown at 25°C in an M9 salt minimal medium supplemented with 0.5% glucose, 0.2% Casamino Acids, and ampicillin at 100 μg/ml and induced with tryptophan (100 μg/ml). E. coli HS2019 carrying pMal-p2 or pMal-c2 (New England BioLabs) plasmid derivatives was used for silver tolerance growth experiments. Cultures were grown at 37°C in M63 minimal salt medium (22) supplemented with 0.2% glycerol, 18 amino acids (50 mg/ml, except Cys and Met), and ampicillin at 100 μg/ml. When silver nitrate (AgNO3) (Sigma-Aldrich) was used, it was added from a freshly prepared 10 mM stock in water. When copper sulfate (CuSO4) (Sigma-Aldrich) was used, it was added from a freshly prepared 1 M stock in water. Kanamycin was added from a stock of 30 mg/ml in water.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| HS2019 | MC4100 F−araD139, ΔlacU169, rpsL, thi, ΔmalE444 | 28 |
| HS2019 dsbA::kan | HS2019 with dsbA disrupted by a kanamycin resistance cassette | This study |
| HS2019 cusA-, -B-, or -C::kan | HS2019 with cusA, -B, or -C disrupted by a kanamycin resistance cassette | This study |
| GI826 | F−, lacIq, ampC::PtrpcI, ΔfliC, ΔmotB, eda::Tn10 | Invitrogen |
| Plasmids | ||
| pMal-p2 | Signal sequence MBP cloning vector with MCSa | New England Biolabs |
| pMal-c2 | Signal sequence-less MBP cloning vector with MCS | New England Biolabs |
| pMal-AgBP2p | pMal-p2 with AgBP2 cloned into MCS | This study |
| pMal-AgBP2c | pMal-c2 with AgBP2 cloned into MCS | This study |
| pMal-Ag4p | pMal-p2 with Ag4 cloned into MCS | This study |
| pFliTrx | fliC-thioredoxin peptide fusion vector | Invitrogen |
| pFliTrx-AgBP2 | pFliTrx with the AgBP2 peptide | This study |
| pFliTrx-AgBP30 | pFliTrx with the AgBP30 peptide | This study |
| F′42 | F′lac (JCFL0) | 1 |
MCS, multiple cloning site.
Growth experiments.
Cultures were started by diluting an overnight culture 1:100 into 25 ml of fresh media and were grown with shaking (150 rpm) in 125-ml flasks at 37°C. Cultures grew for 3 h to an optical density at 600 nm (OD600) of 0.2 to 0.3 before addition of 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) to induce expression of MBP derivatives from the pMal plasmids. After 1 h of induction, a sample was collected to determine CFU. For the dose-response experiment, AgNO3 was then added at concentrations from 5 to 100 μM. CFU counts were determined 5 h later. For time course experiments, cultures were grown and induced the same way and AgNO3 was added at a final concentration of 28 μM. CFU counts were determined at various time points thereafter. All CFU counts were done on LB (Luria-Bertani) agar plates supplemented with ampicillin (100 μg/ml) at 37°C. The same procedures were followed to determine culture sensitivity to CuSO4 and kanamycin. Initially, CuSO4 was tested at concentrations from 1 to 20 mM. Concentrations 5 mM and above completely killed strains expressing MBPp and MBP-AgBP2p, while no cell death was observed at 1 mM CuSO4. Therefore, 2 mM CuSO4 was used to test for copper tolerance. Coculture experiments used E. coli HS2019 carrying pMal-p2 derivatives and an F′42 plasmid encoding β-galactosidase. Cultures with and without the F′42 plasmid were grown as described above and mixed 1:1 according to the OD after the 1-h induction. AgNO3 (28 μM) was added, and CFU counts were done on LB agar plates supplemented with ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (50 μg/ml), using a blue-white screen to distinguish between the different populations of bacteria.
Biocombinatorial selection of the silver-binding peptide AgBP2.
The silver-binding peptide AgBP2 was selected from the FliTrx bacterial surface library (Invitrogen) displaying randomized dodecapeptides inserted into the FLITRX chimera bacterial surface flagellin protein in E. coli. Clean 99.9% pure silver foils (Goodfellow Corp.) were used as a target for peptide selection. Four selection rounds were used in the panning experiment for the enrichment of silver-binding clones, as described elsewhere (11). The binding affinity characteristics of 60 isolated clones, a combination of clones from each selection round, were further characterized in fluorescence microscopy experiments. Typically, aliquots of induced cell clones (OD600 = 0.5) were labeled using 7.5 μM SYTO9 nucleic acid fluorescent dye (Molecular Probes) and incubated for 1 h with a silver surface (5 by 5 mm) deposited on glass using a sputter coating system (672 PECS; Gatan, Inc.) to a thickness of 25 nm of deposited metal. After a washing step, bound cells were visualized using a Nikon TE-2000U fluorescence microscope coupled with a Hamamatsu ORCA-ER cooled charged-coupled-device (CCD) camera, METAMORPH software (Universal Imaging), and a fluorescein isothiocyanate (FITC) filter (Chroma Technology Co.) (exciter, 460 to 500; dichroic, 505; emitter, 510 to 560). The binding activities of selected clones were estimated by averaging the number of adhering cells in at least three randomly selected fields; all measurements were carried out at least in duplicate. The clones were classified as weak, moderate, or strong silver binders based on relative cell adherence cutoffs. The cutoff for strong binders was more than 40 cells/mm2 and for weak binders was less than 15 cells/mm2. The group of strong binders consisted of 12 clones, including AgBP2. The group of moderate binders had 25 clones and the group of weak binders 23 clones.
DNA manipulation and plasmid construction.
Plasmid pMal-c2 (New England BioLabs), which encodes a signal sequence-less version of E. coli maltose-binding protein (MBP) followed by a multiple cloning site, a polypeptide linker, and the LacZα peptide, was manipulated as described previously (27) with the following modification. Two phosphorylated primers (Invitrogen) encoding an SSSGGG linker peptide followed by the AgBP2 peptide, together with extensions compatible with the overhangs generated upon SacI and XbaI digestion (5′-CG GGC GGT GGC GAA CAA CTG GGC GTG CGC AAA GAA CTG CGC GGC GTG TGA TGA T-3′ and 5′-CT AGA TCA TCA CAC GCC GCG CAG TTC TTT GCG CAC GCC CAG TTG TTC GCC ACC GCC CGA GCT-3′), were annealed and ligated to SacI- and XbaI-digested plasmid. The protein encoded by this plasmid was named MBP-AgBP2c. The procedure was repeated starting with plasmid pMal-p2 (which is identical to pMal-c2 except for the presence of the MBP signal sequence) to obtain a periplasmically localized version of the fusion named MBP-AgBP2p. The C-terminal sequences of these proteins are shown in Table 2.
Table 2.
Sequences and binding coefficients of MBP proteinsa
| Protein | C-terminal sequence | Kd on silver |
|---|---|---|
| MBP-AgBP2 | … DAQTNSSSGGGEQLGVRKELRGV | 20 nM |
| MBP2 (New England Biolabs) | … DAQTNSSSNNNNNNNNNNLGIEGR | ≥1.9 μM |
| MBP-Ag4 | … DAQTNSSSGGGNPSSLFRYLPSD | 180 nM |
Bold type indicates the added peptide sequences. MBP2 and MBP-Ag4 data are from reference 27.
MBP expression analysis.
During a growth experiment, cells were harvested from MBP derivative-expressing cultures immediately before IPTG induction (OD600 = 0.2) and 1 h after induction (OD600 = 0.4 to 0.5). Cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and MBP derivatives were detected by Western blotting using MBP-specific antiserum (NEB) as the primary antibody and an alkaline-phosphatase-conjugated secondary antibody. Bound antibodies were visualized by exposure to Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (XP).
TEM.
Cultures were grown as in the cell viability growth experiments described above. Culture samples (100 μl) were collected at various time points after silver nitrate exposure. The cells were pelleted at 13,000 × g for 10 min, washed twice in 10 mM magnesium chloride, and resuspended in 100 μl of 10 mM magnesium chloride. A drop (2 μl) of each sample was spotted onto a carbon-coated copper mesh transmission electron microscopy (TEM) grid and dried in a vacuum. The grid was inserted into a 100-kV TEM (FEI Morgagni), and images were recorded using a bottom-mount Gatan CCD camera (4 K by 2 K). High-resolution imaging was performed using the same type of samples with a FEI Tecnai G2 F20 TEM at 200 kV and a through-focus series. Around 200 MBP-AgBP2p cells and 50 MBPp cells were observed.
Protein purification.
MBP-AgBP2 was expressed and purified from the periplasmic osmotic shockate as previously described (24). Briefly, E. coli HS2019 cells harboring the plasmid pMalp2-AgBP2 were grown from a 1:100 dilution of an overnight culture at 37°C in 50 ml of M63 medium as described above. The culture was grown to mid-exponential phase (OD600 = ∼0.4), and protein synthesis was induced by addition of 1 mM IPTG. After 3 h of incubation, the cells were harvested by centrifugation at 6,000 × g for 10 min and resuspended in 25 ml of osmotic shock buffer (20% sucrose, 30 mM Tris-HCl [pH 8], and 1 mM EDTA) at room temperature. The cells were gently rotated at room temperature for 10 min followed by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was discarded, and the resulting pellet was rapidly resuspended in 25 ml of ice-cold double-distilled water (ddH2O). The cells were gently rotated for 10 min at 4°C and collected again at 10,000 × g for 10 min at 4°C. The supernatant from this last step contained the periplasmic fraction, which was loaded onto a 3-ml amylose column (New England BioLabs) equilibrated in buffer A (20 mM Tris-HCl [pH 7.6], 200 mM NaCl, and 1 mM EDTA). Contaminants were removed by washing 10 ml of buffer A through the column, and MBP-AgBP2 was eluted in buffer A supplemented with 10 mM maltose. Protein was dialyzed overnight against a large excess of ddH2O and then concentrated using Amicon Ultra spin columns (Millipore) (30-K molecular weight cutoff [MWCO]).
Quantitative adsorption of the engineered silver-binding protein, MBP-AgBP2.
The equilibrium binding coefficient, Kd, of MBP-AgBP2 on polycrystalline silver was determined using a quartz crystal microbalance (QCM) method, as described elsewhere (7), and compared to previously published values for related proteins. Briefly, a KSV QCM-Z500 microbalance with the temperature controller set to 25°C was converted to a batch liquid cell configuration. Gold quartz crystals (catalog no. 151719-4.95; International Crystal Manufacturing Co.) were coated with 25-nm polycrystalline silver by thermal evaporation (Auto 306; BOC Edwards). After allowing the QCM microbalance to equilibrate with 1 ml of ddH2O in the batch cell, 2-μl additions of stock MBP-AgBP2 (0.5 mg/ml) in ddH2O were successively injected, allowing the QCM microbalance to equilibrate between additions. The equilibrium frequency shifts were recorded and fit to a Langmuir adsorption isotherm to determine the binding coefficient.
RESULTS
Identification of the silver-binding peptide AgBP2.
Silver-binding peptides (AgBPs) with high affinity for a silver surface were identified from the FliTrx combinatorial bacterial surface display library. We evaluated the binding characteristics of AgBP-FliTrx-expressing clones on a silver surface by the use of fluorescence microscopy. This analysis allowed us to identify AgBP peptide sequences exhibiting elevated binding for silver foil when displayed on bacterial cells as flagellar fusion surface proteins. Cells adhered to the silver surface were labeled with fluorescent dye and enumerated. The surface-bound cells expressing AgBP-FliTrx fusion proteins appeared as dispersed bright rods on a dark background. Isolated bacterial clones displaying the AgBP2-FliTrx flagellum demonstrated high binding capacity for a silver surface (2,444 ± 209 cells/mm2) compared to the plasmid-free control parent strain (184 ± 139 cells/mm2) or an AgBP candidate classified as a weak binder (194 ± 67 cells/mm2).
Effect of AgBP2 expression on E. coli silver tolerance.
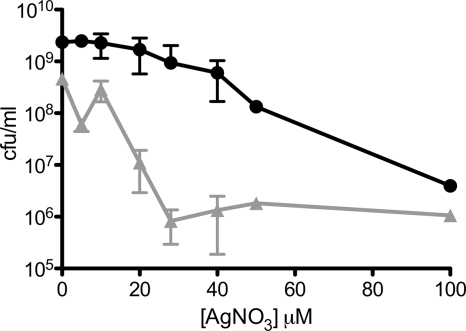
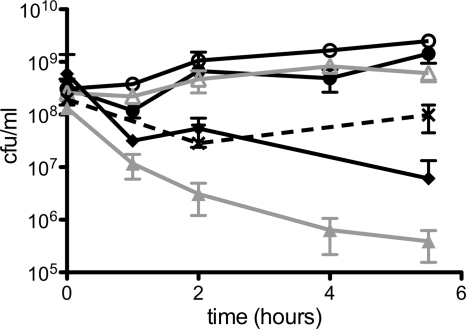
We fused the coding sequence for the AgBP2 silver-binding peptide to the end of the gene encoding the maltose-binding protein (MBP) and expressed both a periplasmic version (MBP-AgBP2p) and a cytoplasmic version (MBP-AgBP2c) of the protein in E. coli strain HS2019. Figure 1 shows that cells expressing MBP-AgBP2p exhibited a higher tolerance for growth in liquid minimal medium supplemented with silver ions relative to control cells expressing periplasmic MBP (MBPp) over a wide range of AgNO3 concentrations. Based on these data, the 28 μM AgNO3 concentration was selected for subsequent experiments. Cultures expressing MBP-AgBP2p, MBP-AgBp2c, and MBPp exposed to 28 μM AgNO3 were compared in a time course experiment. MBP-AgBP2p cultures exposed to silver ions grew as well as MBP-AgBP2p or MBPp cells grown in the absence of silver, whereas MBPp-expressing cultures lost almost 3 logs of cell viability after 5 h of exposure to silver ions. Moreover, cultures expressing cytoplasmic MBP-AgBP2c showed a similar loss of viability upon silver ion exposure. When MBP fused to a different silver-binding peptide (MBP-Ag4 [23, 27]) was expressed in the periplasm, we observed an intermediate but significant and reproducible loss of viability upon silver ion exposure (Fig. 2). The differences in silver ion tolerance between MBP-AgBP2p, MBP-AgBP2c, and MBP-Ag4p cultures cannot be simply attributed to differential expression of the various MBP derivatives, as evidenced by Western blot analysis of cell extracts from 1 h (Fig. 3, lanes 4 to 6) and 2 h (data not shown) after AgNO3 exposure. We observed no difference in protein stability for any of the MBP derivatives. While the overall levels of expression of the different MBP derivatives were not completely equivalent, the most protective MBP-AgBP2p was the least abundant, with steady-state levels that were approximately 75% of the levels of the other relevant tested proteins.
Fig 1.
Silver nitrate dose-response curve for E. coli-expressing MBP-AgBP2p or the MBPp control. HS2019 cultures expressing MBP-AgBP2p (filled circles) or MBPp (filled triangles) were grown in liquid minimal medium and then plated on LB agar for viable cell counts 5 h after AgNO3 exposure. Each concentration was tested in at least duplicate.
Fig 2.
Silver tolerance assay of E. coli HS2019 expressing MBP silver-binding derivatives. Cultures expressing MBP-AgBP2p (filled circles), MBPp (filled triangles), MBP-Ag4p (bold Xs), and MBP-AgBP2c (filled diamonds) with 28 μM AgNO3 or MBP-AgBP2p (open circles) and MBPp (open triangles) without AgNO3 were induced with IPTG for 1 h, and then cell viability in the cultures with or without AgNO3 was measured at the time points indicated. AgNO3 was added at 0 h. Each strain was replicated two to five times. Error bars represent 1 standard deviation.
Fig 3.
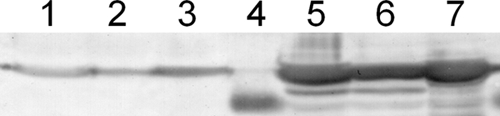
MBP derivative expression analysis. A Western blot analysis of proteins detected with polyclonal MBP antiserum is shown. Data represent cell extracts of MBP-Ag4p (lanes 1 and 5), MBP-AgBP2p (lanes 2 and 6), and MBP-AgBP2c (lanes 3 and 7) cultures before (lanes 1 to 3; OD600 = 0.2) and 1 h after (lanes 5 to 7; OD600 = 0.4 to 0.5) IPTG induction of expression. The predicted size for all proteins is 43 kDa. Lane 4 represents the molecular mass marker at 36 kDa. Equivalent amounts of total cellular protein were loaded per lane.
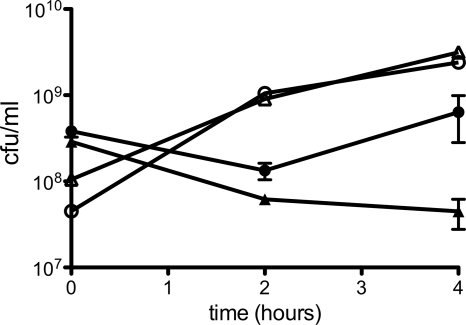
In our system, low levels of MBP-AgBP2p are produced in uninduced cells due to leaky transcription from the plasmid tac promoter. Based on Fig. 3 and additional Western blot analyses, we estimate that protein production is approximately 200 times lower in the absence of IPTG induction than after an hour of induction. Figure 4 shows that this basal amount of protein was sufficient to confer partial tolerance to 28 μM AgNO3, although cell survival was not as robust as with induced cultures.
Fig 4.
Silver tolerance of uninduced E. coli HS2019 expressing MBP silver-binding derivatives. Uninduced cultures expressing MBP-AgBP2p (filled circles) and MBP-AgBP2c (filled triangles) with 28 μM AgNO3 or MBP-AgBP2p (open circles) and MBP-AgBP2c (open triangles) without AgNO3 were plated at indicated time points. AgNO3 was added at 0 h. Experiments using cultures with silver were repeated in triplicate.
The tolerance phenotype of cultures expressing MBP-AgBP2p was limited to cells grown in batch culture. When plated during exponential growth on agar plates (LB supplemented with amplicillin without NaCl) containing 100 μM AgNO3 (9), MBP-AgBP2p-expressing cells did not form colonies overnight at 37°C with or without the inducer IPTG added. Moreover, the MIC of AgNO3 was the same for the MBPp and MPB-AgBP2p cultures (0.625 μg/ml [3.7 μM]). The failure of these cultures to grow on plates containing AgNO3 and their equal AgNO3 MICs support an interpretation of a tolerance, not a resistance, phenotype (2).
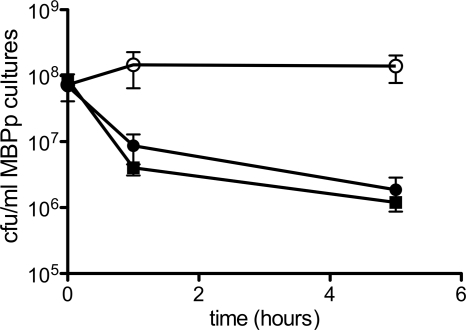
To determine whether MBP-AgBP2p cultures remove silver ions from the media, coculture experiments were performed with MBP-AgBP2p- and MBPp-expressing cultures. Cultures induced for protein expression for 1 h were mixed 1:1 and exposed to AgNO3. We tested cocultures with 5, 10, 20, and 28 μM AgNO3. The 5 μM AgNO3 concentration did not kill MBP-AgBP2p or MBPp cultures in the time course of these experiments. At all concentrations higher than 5 μM AgNO3, MBPp cultures grown either with MBP-AgBP2p cultures or with MBPp control cultures lost cell viability in the presence but not in the absence of AgNO3 (Fig. 5). The fact that MBP-AgBP2p-expressing cultures did not protect the silver-sensitive MBPp cultures from the effects of AgNO3 indicates that the protective effect of MBP-AgBP2p was not solely due to removal of silver ions from the media by the fusion protein.
Fig 5.
Assaying AgBP2-mediated protection from AgNO3 in coculture. HS2019 strains with F′42 pMal-AgBP2p (filled circles) or F′42 pMal-p2 (filled squares) were mixed 1:1 with HS2019 pMal-p2 in the presence of 28 μM AgNO3. HS2019 F′42 pMal-AgBP2p was also mixed 1:1 with HS2019 pMal-p2 without AgNO3 (open circles). The MBP-AgBP2p-expressing strain exhibited the expected silver tolerance in the cocultures. Each coculture experiment was repeated two to five times.
Specificity of AgBP2-mediated tolerance and impact of other host factors.
We tested MBP-AgBP2p for its ability to protect cells from the toxic effects of a different metal salt, CuSO4. Copper and silver belong to the same group of the periodic table, and some metal efflux proteins such as E. coli CusA provide protection to both copper and silver ions (18). If MBP-AgBP2p conferred tolerance to metals in general, we would expect to see protection in the presence of CuSO4. Growth of MBP-AgBP2p cultures matched that of MBPp control cultures in CuSO4, showing no tolerance to growth in 2 mM CuSO4 after 2 h. To test whether MBP-AgBP2p cultures were more tolerant of stress in general rather than specifically tolerant to growth in the presence of silver ions, growth experiments were repeated such as were performed with AgNO3, but kanamycin (30 μg/ml) was added in place of AgNO3. MBP-AgBP2p cultures did not tolerate growth in kanamycin as they had in AgNO3 and showed sensitivity matching that of the control MBPp culture.
Additionally, we tested the contribution of other host proteins to the AgBP2-mediated Ag ion tolerance, starting with the bacterial Dsb-mediated oxidoreduction pathway. We grew MBP-AgBP2p-expressing cells with a dsbA gene disruption in 28 μM AgNO3 and found that DsbA is not required for the AgBP2 tolerance mechanism. Strains with dsbA dsbA+ and dsbA::kan alleles expressing MBP-AgBP2p maintained high CFU counts during growth in AgNO3, while strains expressing MBPp were sensitive to AgNO3. We also found that the CusCBA proteins, which comprise a copper/silver efflux transporter in E. coli (14), do not contribute to AgBP2-mediated silver ion tolerance. We grew MBP-AgBP2p-expressing cells with a cusA, cusB, or cusC gene disruption in 28 μM AgNO3; Cus mutants expressing MBP-AgBP2p remained as tolerant to growth in AgNO3 as the Cus+ parental control.
Transmission electron microscopy of E. coli MBP-AgBP2p cells in silver nitrate.
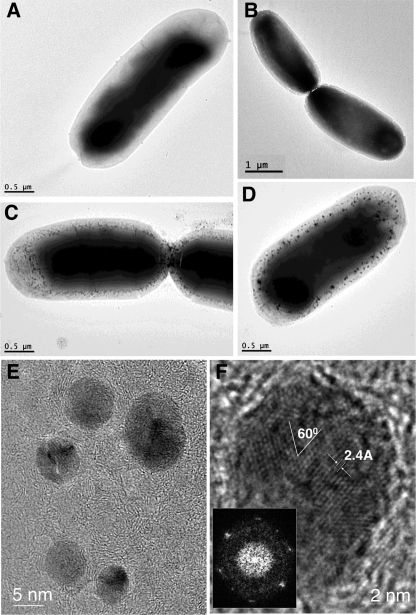
TEM imaging of unstained cells grown in 28 μM AgNO3 for 1 h revealed the presence of electron-dense particles in the periphery of MBP-AgBP2p-expressing cells but not MBPp-expressing cells (Fig. 6A to D). In a representative sample of observed cells, about 63% of MBP-AgBP2p-expressing cells at 1 h after AgNO3 exposure contained these particles whereas none were observed in AgNO3-treated MBPp-expressing control cells. High-resolution imaging in the bacterial septum area revealed that some of these particles were crystalline silver, as evidenced by the crystal lattice formation observed in some of the particles (Fig. 6E and F). The inset in Fig. 6F shows a fast Fourier transform (FFT) pattern recorded using the high-resolution image of the silver nanoparticle, showing angles and lattice spacings matching those of metallic silver. However, not all of the particles were stable under the electron beam, suggesting that some of the electron-dense material might consist of silver salts.
Fig 6.
TEM analysis of whole E. coli cells. (A to D) MBPp (A and B)- or MBP-AgBP2p (C and D)-expressing cells 1 h after AgNO3 exposure. (E and F) High-resolution images of particles from the septum area of MBP-AgBP2p-expressing cells. Angles and lattice spacings match those metallic silver. The inset in panel F represents a fast Fourier transform (power spectrum) of the atomic fringes showing that the particle is a multiply twinned, single crystal and oriented primarily along the <111> zone axis.
We infer that these particles are localized to the bacterial periplasm for several reasons. MBP-AgBP2p works in maltose transport, complementing the malE deletion strain HS2019 to Mal+, which requires MBP in the periplasm (3). Western blot analysis shows that MBP-AgBP2p and MBP-AgBP2c were the same molecular weight (Fig. 3), indicating that MBP-AgBP2p's signal sequence had been processed during export to the periplasm. Also, MBP-AgBP2 must be expressed in the periplasm, not in the cytoplasm, to observe a silver tolerance phenotype (Fig. 2).
Quantitative determination of MBP-AgBP2 affinity for metallic silver.
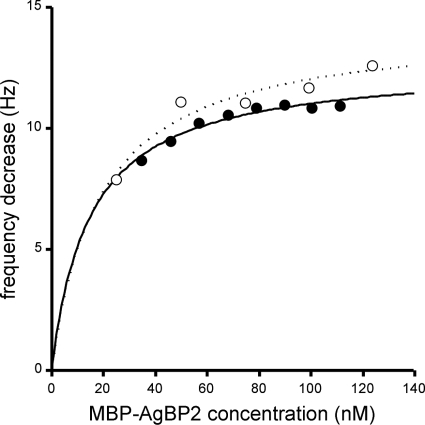
We quantified the affinity of purified MBP-AgBP2 for polycrystalline silver by the use of a QCM method performed with silver-coated QCM crystals. Previously, we reported that MBP-Ag4 has an equilibrium dissociation constant (Kd) of 180 nM for silver whereas an MBP derivative carrying a polyasparagine C-terminal extension of similar length binds the metal with a Kd of only about 2 μM (Table 2) (27). Figure 7 shows that MBP-AgBP2 is a very efficient silver binder, with an affinity for this metal (Kd = ∼20 nM) about 10-fold higher than that of MBP-Ag4.
Fig 7.
MBP-AgBP2 exhibits high affinity for polycrystalline silver. Quartz crystal microbalance (QCM) measurements were performed using two batches of purified protein and two different crystals (open and closed symbols). The lines correspond to the computed Langmuir adsorption isotherms. Calculated equilibrium dissociation constants are 18 and 19 nM.
DISCUSSION
We have identified a combinatorially selected silver-binding peptide that acts in vivo to make a laboratory strain of E. coli silver tolerant. Although many investigations have addressed the in vitro capabilities of combinatorially selected material-binding peptides (6, 10, 13, 20, 34), this is the first demonstration of a de novo-selected peptide conferring a robust in vivo bacterial phenotype. E. coli expressing the periplasmic protein MBP-AgBP2p remained fully viable for several hours in medium with 28 μM AgNO3, while cells expressing the cytoplasmic version of the same protein or the control MBPp lost cell viability within 1 h of AgNO3 exposure (Fig. 1). This observation supports the hypothesis that silver ions exert their toxicity once they have reached either the cytoplasmic membrane or the cytoplasmic compartment, resulting in the release of ions outside the cytoplasmic membrane (26), disruption of proton motive force, and/or malfunction of important intracellular enzymes (12, 32).
This peptide-mediated mechanism of tolerance is unique among the widely distributed mechanisms of metal efflux. Nevertheless, it seems possible that natural bacterial isolates might exploit similar peptide-mediated tolerance as a form of protection from transient metal exposure in a changing environment. This survival strategy would not previously have been easily detected by genetic selections but could contribute significantly to overall fitness in complex environments. In addition, when a binding peptide conferring tolerance can act in concert with, e.g., a specific efflux system, the expression of material-binding peptides might represent the first step in evolution toward true resistance.
MBP-AgBP2p may sequester silver ions in the periplasmic compartment in the form of electron-dense particles, as suggested by TEM analysis (Fig. 6). The reduction of toxic Ag+ into crystalline silver nanoparticles that are less bioavailable may contribute to the tolerance mechanism of the MBP-AgBP2p strain. This mechanism of silver protection is not unprecedented: Pseudomonas stutzeri AG259, isolated from a silver mine, accumulates silver nanoparticles in the periplasm, presumably contributing to its silver resistance phenotype (16). However, if this silver tolerance relies exclusively on sequestration of silver ions, we would expect silver to be removed from the media by MBP-AgBP2p cultures. Coculture experiments indicate that not enough silver is removed from the media by MBP-AgBP2p cultures to protect MBPp cultures from the toxic effects of silver. While this assay may not be sensitive enough to detect small silver concentration decreases, we suggest that the silver tolerance phenotype relies on a mechanism more complicated than simple sequestration.
The Salmonella silver resistance-associated SilE protein has already been shown to specifically bind silver (8) and possesses homology to PcoE, a proposed metal-binding protein of the plasmid-encoded copper resistance system of E. coli (4). However, it has not been determined whether SilE confers silver resistance in the absence of its cognate silver efflux system. Many organisms, like shellfish and diatoms, use peptides to precipitate inorganic materials on or within themselves. Likewise, many bacterial metalloproteins coordinate metals (19). With this information in mind, we have checked for sequences similar to the AgBP2 and Ag4 peptides among currently available genome sequences, but no significant homologues have emerged. Additionally, SilE and PcoE likely rely on several conserved His residues to coordinate metal ions, which are completely lacking in the AgBP2 and Ag4 peptides.
While many environmentally selected bacteria are resistant to high (millimolar) concentrations of toxic metals, we have shown that AgBP2 specifically confers a low level (micromolar) of tolerance of silver. When we grew MBP-AgBP2p-expressing cultures with CuSO4, no broader metal tolerance was observed. In addition, the MBP-AgBP2p cultures are not broadly resistant to environmental stress and do not have significant differences in general membrane permeability, as tested by sensitivity to kanamycin. Moreover, the genes most likely to contribute to silver tolerance (dsbA, cusCBA) are not involved in AgBP2-mediated silver tolerance.
We also compared AgBP2 to Ag4, a previously characterized silver-binding peptide. In vivo, MBP-AgBP2p conferred a higher level of silver ion tolerance than MBP-Ag4p (Fig. 2). Consistent with this observation, purified MBP-AgBP2 exhibits a 10-fold-higher affinity for polycrystalline silver than MBP-Ag4 (Fig. 7). Thus, the affinity of a metal-binding peptide for its cognate material may correlate with its ability to protect a bacterium from the toxic ionic species.
There has been increased interest in understanding the interactions between in-metal ions and proteins and the toxicity of metal ions. We believe that inorganic binding peptides can be a useful molecular tool for these analyses. Engineered bacterial interactions with metals mediated through specific inorganic binding peptide motifs could have uses in diverse applications in bioremediation and biological synthesis of materials by the use of cells as factories. Bacterial proteins have already been engineered to bind other metals such as gold and cuprous oxide (5, 6, 10). This work opens the possibility that these and other engineered proteins could be used in vivo for facile, biological synthesis of inorganic particles. Likewise, in the future such metal-binding peptides could be used as tags for cellular protein localization studies that are not possible with fluorescent or antibody epitope tags.
ACKNOWLEDGMENTS
This project is mainly supported by an NSF-MRSEC grant at the University of Washington (GEMSEC DMR 0520567) and by an NSF BioMat grant (DMR 0706655; B.T., M.S., C.T.). R.H.S. is an NSF Graduate Research Fellow.
The research was carried out at the facilities of GEMSEC, a member of MRFN, and CNT, a member of NNIN, which are supported by NSF. Additional thanks to Tamir Gonen (University of Washington, Department of Biochemistry) for use of his microscopy center, supported by the Murdock Charitable Trust and the Washington Research Foundation, and to Jimmie Lara (University of Washington, Department of Microbiology) for advice on visualizing cells.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Achtman M, Willetts N, Clark AJ. 1971. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bayles KW. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5: 721–726 [DOI] [PubMed] [Google Scholar]
- 3. Bordignon E, Grote M, Schneider E. 2010. The maltose ATP-binding cassette transporter in the 21st century—towards a structural dynamic perspective on its mode of action. Mol. Microbiol. 77: 1354–1366 [DOI] [PubMed] [Google Scholar]
- 4. Brown N, Barrett S, Camakaris J, Lee B, Rouch D. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17: 1153–1166 [DOI] [PubMed] [Google Scholar]
- 5. Brown S. 1997. Metal-recognition by repeating polypeptides. Nat. Biotechnol. 15: 269–272 [DOI] [PubMed] [Google Scholar]
- 6. Dai HX, et al. 2005. Nonequilibrium synthesis and assembly of hybrid inorganic-protein nanostructures using an engineered DNA binding protein. J. Am. Chem. Soc. 127: 15637–15643 [DOI] [PubMed] [Google Scholar]
- 7. Grosh C, Schwartz D, Baneyx F. 2009. Protein-based control of silver growth habit using electrochemical deposition. Cryst. Growth Des. 9: 4401–4406 [Google Scholar]
- 8. Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5: 183–188 [DOI] [PubMed] [Google Scholar]
- 9. Gupta A, Phung LT, Taylor DE, Silver S. 2001. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology 147: 3393–3402 [DOI] [PubMed] [Google Scholar]
- 10. Hall Sedlak R, et al. 2010. An engineered DNA-binding protein self-assembles metallic nanostructures. Chembiochemistry 11: 2108–2112 [DOI] [PubMed] [Google Scholar]
- 11. Hnilova M, et al. 2008. Effect of molecular conformations on the adsorption behavior of gold-binding peptides. Langmuir 24: 12440–12445 [DOI] [PubMed] [Google Scholar]
- 12. Jung W, et al. 2008. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 74: 2171–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kacar T, et al. 2009. Directed self-immobilization of alkaline phosphatase on micro-patterned substrates via genetically fused metal-binding peptide. Biotechnol. Bioeng. 103: 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim EH, Nies DH, McEvoy MM, Rensing C. 2011. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J. Bacteriol. 193: 2381–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klasen H. 2000. Historical review of the use of silver in the treatment of burns. I. Early uses. Burns 26: 117–130 [DOI] [PubMed] [Google Scholar]
- 16. Klaus T, Joerger R, Olsson E, Granqvist CG. 1999. Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. U. S. A. 96: 13611–13614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li XZ, Nikaido H, Williams KE. 1997. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J. Bacteriol. 179: 6127–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long F, et al. 2010. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma Z, Jacobsen F, Giedroc D. 2009. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109: 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mao C, et al. 2003. Viral assembly of oriented quantum dot nanowires. Proc. Natl. Acad. Sci. U. S. A. 100: 6946–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McHugh GL, Moellering RC, Hopkins CC, Swartz MN. 1975. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet i: 235–240 [DOI] [PubMed] [Google Scholar]
- 22. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Naik RR, Stringer SJ, Agarwal G, Jones SE, Stone MO. 2002. Biomimetic synthesis and patterning of silver nanoparticles. Nat. Mater. 1: 169–172 [DOI] [PubMed] [Google Scholar]
- 24. Neu H, Heppel L. 1965. Release of enzymes from Escherichia coli by osmotic shock and during formation of spheroplasts. J. Biol. Chem. 240: 3685–3692 [PubMed] [Google Scholar]
- 25. Sarikaya M, Tamerler C, Jen AK, Schulten K, Baneyx F. 2003. Molecular biomimetics: nanotechnology through biology. Nat. Mater. 2: 577–585 [DOI] [PubMed] [Google Scholar]
- 26. Schreurs W, Rosenberg H. 1982. Effect of silver ions on transport and retention of phosphate by Escherichia coli. J. Bacteriol. 152: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sengupta A, et al. 2008. A genetic approach for controlling the binding and orientation of proteins on nanoparticles. Langmuir 24: 2000–2008 [DOI] [PubMed] [Google Scholar]
- 28. Shuman HA. 1982. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J. Biol. Chem. 257: 5455–5461 [PubMed] [Google Scholar]
- 29. Silver S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27: 341–353 [DOI] [PubMed] [Google Scholar]
- 30. Silver S, Gupta A, Matsui K, Lo JF. 1999. Resistance to ag(i) cations in bacteria: environments, genes and proteins. Met. Based Drugs 6: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silver S, Phung L. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50: 753–789 [DOI] [PubMed] [Google Scholar]
- 32. Slawson R, Vandyke M, Lee H, Trevors J. 1992. Germanium and silver resistance, accumulation, and toxicity in microorganisms. Plasmid 27: 72–79 [DOI] [PubMed] [Google Scholar]
- 33. Tamerler C, et al. 2006. Materials specificity and directed assembly of a gold-binding peptide. Small 2: 1372–1378 [DOI] [PubMed] [Google Scholar]
- 34. Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. 2000. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 405: 665–668 [DOI] [PubMed] [Google Scholar]
- 35. Yakabe Y, Sano T, Ushio H, Yasunaga T. 1980. Kinetic studies of the interaction between silver ion and deoxyribonucleic acid. Chem. Lett. 4: 373–376 [Google Scholar]