Abstract
A dimeric Baeyer-Villiger monooxygenase (BVMO) catalyzing the lactonization of 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-coenzyme A (CoA), a key intermediate in the metabolism of camphor by Pseudomonas putida ATCC 17453, had been initially characterized in 1983 by Ougham and coworkers (H. J. Ougham, D. G. Taylor, and P. W. Trudgill, J. Bacteriol. 153:140–152, 1983). Here we cloned and overexpressed the 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-CoA monooxygenase (OTEMO) in Escherichia coli and determined its three-dimensional structure with bound flavin adenine dinucleotide (FAD) at a 1.95-Å resolution as well as with bound FAD and NADP+ at a 2.0-Å resolution. OTEMO represents the first homodimeric type 1 BVMO structure bound to FAD/NADP+. A comparison of several crystal forms of OTEMO bound to FAD and NADP+ revealed a conformational plasticity of several loop regions, some of which have been implicated in contributing to the substrate specificity profile of structurally related BVMOs. Substrate specificity studies confirmed that the 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid coenzyme A ester is preferred over the free acid. However, the catalytic efficiency (kcat/Km) favors 2-n-hexyl cyclopentanone (4.3 × 105 M−1 s−1) as a substrate, although its affinity (Km = 32 μM) was lower than that of the CoA-activated substrate (Km = 18 μM). In whole-cell biotransformation experiments, OTEMO showed a unique enantiocomplementarity to the action of the prototypical cyclohexanone monooxygenase (CHMO) and appeared to be particularly useful for the oxidation of 4-substituted cyclohexanones. Overall, this work extends our understanding of the molecular structure and mechanistic complexity of the type 1 family of BVMOs and expands the catalytic repertoire of one of its original members.
INTRODUCTION
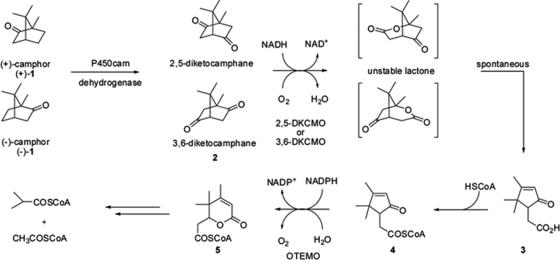
As part of carbon recycling in nature, the microbial metabolism of camphor, a bicyclic terpenoid naturally produced in leaves and wood of the camphor laurel (Cinnamomum camphora) and available as (+) and (−) isomeric forms, was studied by Bradshaw et al. as early as 1959 using a sewage sludge-derived pseudomonad (strain P or C1B), now known as Pseudomonas putida ATCC 17453 (NCIMB 10007) (10). After several elegant studies, including the identification of a large transmissible CAM plasmid controlling camphor metabolism in P. putida (48), the cleavage of the bicyclic ring of camphor was found to involve the participation of three unrelated monooxygenases, one dehydrogenase, and one coenzyme A (CoA) ester synthetase (13–15, 44, 58) (Fig. 1). The flavin adenine dinucleotide (FAD)- and NADPH-dependent 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-mono-oxygenase (OTEMO), sometimes referred to as MO2 (25, 63), was one of the first identified members of the type 1 ring-expand-ing Baeyer-Villiger (BV) monooxygenases (BVMOs) (EC 1.14.13.--), with the prototype and most studied BVMO being the cyclohexanone monooxygenase (CHMO) of Acinetobacter sp. strain NCIMB 9871 (55, 61, 63). Type 1 BVMOs are thus far more frequently found among microorganisms than those of type 2, which require flavin mononucleotide (FMN) as a prosthetic group and NADH as a cofactor (34, 63).
Fig 1.
Catabolic steps of conversion of camphor isomers to acetyl-CoA and isobutyryl-CoA in Pseudomonas putida ATCC 17453. A cytochrome P450-containing enzyme complex (CamCAB) hydroxylates (+)- and (−)-camphor at the 5-exo position to produce 5-exo-hydroxycamphor; upon dehydrogenation (5-exo-hydroxycamphor dehydrogenase [CamD]), the respective diketocamphane is formed. Ring oxygen insertion by the FMN- and NADH-dependent 2,5-diketocamphane monooxygenase for (+)-camphor or 3,6-diketocamphane monooxygenase for (−)-camphor (type 2 BVMOs) produces an unstable lactone that presumably undergoes spontaneous hydrolysis to form 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid (compound 3). The activation of compound 3 by a putative CoA synthetase produces 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-CoA, a substrate for OTEMO (type 1 BVMO), the subject of this study. Cumulative data are from references 30, 44, and 58. COSCoA, carbonyl-CoA; HSCoA, acetyl-CoA.
BVMOs carry out a highly regio- and enantioselective nucleophilic and electrophilic oxygenation of a variety of ketonic substrates. BVMOs are also known to catalyze epoxidations and S- and N-heteroatom oxidations (for recent reviews, see references 4, 18, 29, 33, and 34). The BVMO-catalyzed reaction is recognized by pharmaceutical manufacturers as one of the green chemistry priority research areas in order to reduce the use of chlorinated solvents or strong oxidants (16, 56). The generally accepted mechanism of BVMO based on various kinetic studies indicates that NADPH binds first in the active site and is the last to leave at the end of the catalytic cycle (50, 53, 57). Noncovalently bound FAD is reduced by NADPH to form an enzyme–reduced-FAD (FADH2)–NADP+ complex, which reacts with molecular oxygen to generate a stable flavin-peroxide intermediate. In the presence of a ketone, this reactive intermediate, acting as a nucleophile, attacks the carbonyl carbon of the substrate, leading to the formation of the Criegee intermediate, a characteristic of peracid-catalyzed chemical BV reactions (17). A rearrangement of the Criegee intermediate leads to lactone formation as well as a hydroxyflavin adduct, which, upon hydrolysis, regenerates the oxidized flavin and releases NADP+ at the end of the catalytic cycle. The released NADP+ is then reduced in the cell milieu and returned as NADPH to initiate the next reaction cycle.
The first three-dimensional (3D) structure of a type 1 BVMO, that of a thermostable phenylacetone monooxygenase (PAMO) from Themobifida fusca, showed a two-domain organization, with FAD-binding and NADP-binding domains, in which the active site is positioned in a cleft at the domain interface (35). Importantly, a strictly conserved arginine (R337) was identified to play a critical role in stabilizing the negatively charged flavin-peroxide intermediate. Subsequently, the structures of a rhodococcal CHMO in complex with NADP+ were observed in two conformations, which provided evidence of domain movements and a “sliding” process of the NADP+ during the catalytic cycle (39). In addition, a specific role of the type 1 BVMO signature motif, FXGX3HX3WP (22), in coordinating the domain movement as a possible “atomic switch” was proposed (39).
In this study, we report the dimeric structure of OTEMO from the camphor pathway of P. putida ATCC 17453 bound to FAD alone as well as to both FAD and NADP+. These structures reveal further conformational changes that occur during the BVMO catalytic cycle. In addition, the enzymological characterization of recombinant OTEMO provides new insights into the substrate specificity and biocatalytic spectrum of this enzyme for potential industrial applications.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. putida strain ATCC 17453, referred to as strain PpCam in this study, was purchased from the American Type Culture Collection and grown at 30°C in Luria-Bertani (LB) broth medium (51). Also, Escherichia coli strains were routinely cultured in LB medium, and when necessary, the medium was supplemented with ampicillin (Ap) (100 μg/ml). Strains were maintained on LB medium containing glycerol (50%, vol/vol) at −80°C.
Cloning and genetic manipulations.
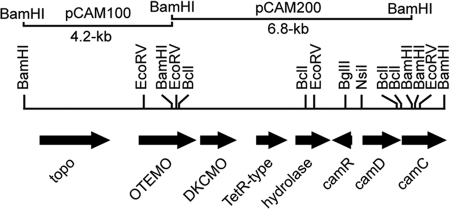
Two degenerate primers that facilitated the cloning of the cyclopentadecanone monooxygenase (CPDMO)-encoding gene (27) were used for the PCR amplification of a ∼1-kb fragment of DNA of PpCam genomic DNA prepared according to a method described previously by Wilson (64). The DNA was labeled by the digoxigenin-11-UTP system according to the manufacturer's instructions (Roche Diagnostics KK) and was used to probe a Southern hybridization of PpCam genomic DNA digested with a number of restriction enzymes. As a result, two BamHI fragments (4.2 kb and 6.8 kb) were probed positive and chosen for cloning into E. coli plasmid pUC19, which had been linearized with BamHI and dephosphorylated. The resulting recombinant plasmids transformed into E. coli XL1-Blue were designated pCAM100 for the 4.2-kb BamHI fragment and pCAM200 for the 6.8-kb BamHI fragment (Fig. 2). DNA sequencing of the cloned inserts was performed by the conventional dideoxy method and analyzed as previously described (26, 27).
Fig 2.
Localizations of the OTEMO-encoding gene and BamHI fragment subclones in an 11-kb region of P. putida ATCC 17453. The identified open reading frames are as follows, from left to right: putative DNA topoisomerase III (topo), OTEMO, 2,5-diketocamphane monooxygenase (DKCMO), a TetR-type regulator, lactone hydrolase, and a camR repressor that regulates the downstream camDCAB operon (camAB) (not shown) (5, 30).
To construct an overexpressing clone of the OTEMO-encoding gene, the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible E. coli pSD80 vector (54) was used to carry the Pfu DNA polymerase-amplified OTEMO-encoding gene using the following respective forward and reverse primers with built-in EcoRI restriction sites (underlined): 5′-CGGAATTCATGAGCAATAGAGCAAAA and 5′-CGGAATTCACGTGCGTTCGCACACTA. The recombinant bacterial strain is designated E. coli BL21[OTEMO].
Site-directed mutagenesis.
The QuikChange II site-directed mutagenesis kit (Agilent Technologies) was used according to the manufacturer's instructions to construct six mutants, each consisting of the following single-amino-acid substitutions (with the corresponding complementary primers in parentheses): Y53A (5′-GGAACCTGGTACTGGAACCGAGCTCCAGGCTGCA and 5′-TGCAGCCTGGAGCTCGGTTCCAGTACCAGGTTCC), Y53F (5′-CCTGGTACTGGAACCGATTTCCAGGCTGC and 5′-GCAGCCTGGAAATCGGTTCCAGTACCAGG), D59A (5′-AGGCTGCAGGCTGGCTACGGAAAGCTACG and 5′-CGTAGCTTTCCGTAGCCAGCCTGCAGCCT), D59N (5′-CCAGGCTGCAGGCTGAATACGGAAAGCTACG and 5′-CGTAGCTTTCCGTATTCAGCCTGCAGCCTGG), R337A (5′-CCCTTCGGTGCTAAGGCCGTGCCGATGGAAAC and 5′-GTTTCCATCGGCACGGCCTTAGCACCGAAGGG), and R337K (5′-GATCATCCCTTCGGTGCTAAGAAGGTGCCGATGGAAACCAATTAT and 5′-ATAATTGGTTTCCATCGGCACCTTCTTAGCACCGAAGGGATGATC). The respective plasmids encoding pSD80[OTEMO] mutant proteins were transformed into E. coli BL21(DE3) and plated onto LB agar plates containing Ap for overnight growth at 37°C and protein expression.
Enzyme assay, substrate specificity, and kinetics.
OTEMO activity was assayed by using a conventional spectrophotometric method by measuring the decrease in the absorbance of NADPH at 340 nm in a reaction mixture (1 ml) containing 50 mM Tris-HCl buffer (pH 9.0), 0.1 mM NADPH, and an appropriate amount of enzyme (10 to 20 mU). The reaction was initiated by the addition of 2 μl of substrate (e.g., 0.2 M 2-n-hexyl cyclopentanone in 2-propanol) to the mixture. Substrates were dissolved in 2-propanol to give a stock concentration of 0.2 M. Depending on the respective enzyme affinity, the final substrate concentrations in the enzyme assay mixtures varied between 0.2 mM (2-n-hexyl cyclopentanone) and 5 mM (methyl cyclohexanones). Specific activity is defined as μmol NADPH (ε = 6.3 liters mmol−1 cm−1) oxidized per min (U) per mg of protein. The protein concentration was determined by a conventional Bradford assay (9a).
Kinetic parameters (Km and kcat) for OTEMO were determined by using a Lineweaver-Burk plot of the Michaelis-Menten equation under steady-state conditions. Results were verified by Eisenthal-Cornish-Bowden direct plots. Initial reaction rates were measured at 25°C in Tris-HCl buffer (50 mM, pH 9.0) by using a total substrate concentration of between 5 μM and 5 mM. The Km for NADPH was estimated with n-hexyl cyclopentanone as a substrate (0.2 mM) in a standard 1-ml assay mixture with NADPH concentrations varying from 2 μM to 100 μM.
Protein purification.
The procedure for protein purification is described in Material SM1 in the supplemental material, together with a description of the sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis. These methods were adapted from methods for the purification of OTEMO from the native PpCam strain reported previously by Ougham et al. (44).
CD spectroscopy and determination of the melting point.
The circular dichroism (CD) spectrum of OTEMO was recorded with a Jasco J-815 spectrometer operating with Spectra Manager software. The temperature was controlled with a Jasco PFD-452S Peltier unit. Purified protein solutions were desalted by using a HiPrep desalting column (26/10; GE Healthcare) previously equilibrated with 20 mM Na-phosphate buffer (pH 7.0). The final protein concentration was adjusted to about 0.1 mg/ml (≈1 μM), and the CD spectrum was recorded at between 190 and 260 nm by using a quartz cuvette (internal diameter [ID] = 0.1 cm). Blanks containing buffer only were prepared and used as a baseline. Temperature-dependent protein unfolding was monitored at 222 nm with thermal profiles ranging from 20°C to 80°C (2°C min−1). Samples were maintained for 5 min at the respective maximum temperatures, and potential protein refolding was monitored under the same conditions as those described above, reversing the thermal profiles. Thermodynamic parameters (melting temperature [Tm], ΔH, ΔS, and ΔG) for the folding/unfolding process were calculated by using Spectra Manager software.
Thermostability of OTEMO.
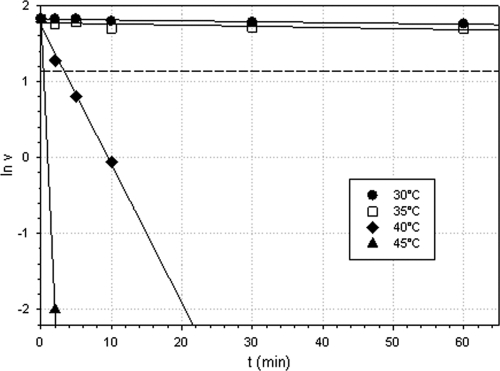
Active enzyme was incubated at different temperatures (30°C, 35°C, 40°C, and 45°C) for different periods (2, 5, 10, 30, and 60 min) and chilled on ice afterwards. Residual enzyme activity was measured spectrophotometrically using 2-n-hexyl cyclopentanone as a substrate. In parallel, purified enzyme was incubated for 10 min at 25°C, 45°C, and 85°C, respectively. After chilling on ice, size-exclusion chromatography was carried out, and the resulting chromatograms were compared.
Chemicals and general procedures.
Unless otherwise stated, all chemicals and reagents, including ketones and chiral lactones, were purchased from Sigma-Aldrich (Mississauga, Canada), Alfar Aesar, TCI America, or Thermo Fisher Scientific and were used without further purification. 2-n-Hexyl cyclopentanone was obtained from Tokyo Chemical Industry Co. Racemic lactone standards were prepared with m-chloroperbenzoic acid as an oxidant according to methods described previously by Meinwald et al. (36). Chiral and enantio-enriched lactone standards were prepared by using whole cells overexpressing CHMO from Rhodococcus sp. strain HI-31 (CHMORhod) (39), CPDMO from Pseudomonas sp. strain HI-70 (27), and cyclopentanone monooxygenase (CPMO) from Comamonas sp. strain NCIMB 9872 (26). Standards were separated by chiral gas chromatography (GC) analysis as described in Material SM2 in the supplemental material. The absolute configuration of samples was determined by chiral GC analysis by coinjection with standards or by a comparison of the optical rotation values of the isolated compounds with data reported in the literature. The structures of isolated compounds from preparative-scale experiments were confirmed by GC/mass spectrometry (MS) and nuclear magnetic resonance (NMR) analyses. 1H and 13C NMR analyses were carried out in CDCl3 on a Bruker (500-MHz) spectrometer. Optical rotations were measured with a Perkin-Elmer 341 polarimeter (courtesy of the Department of Chemistry, University of Montreal). High-resolution mass spectrometry (HRMS) measurements were performed in the positive-ion electrospray (+ESI) mode on a Micromass Waters Q-TOF Ultima Global mass spectrometer equipped with a z-spray ion source, a NanoLockSpray (Waters, Mississauga, Ontario, Canada) source, and an MCP (micro channel plate) detector.
Synthesis of 2-oxo-Δ3-4,5,5-trimethylcyclopentynyl acidic acid and 2-oxo-Δ3-4,5,5-trimethyl cyclopentenylacetyl coenzyme A ester.
2-Oxo-Δ3-4,5,5-trimethylcyclopentynyl acidic acid and 2-oxo-Δ3-4,5,5-trimethyl cyclopentenylacetyl coenzyme A ester compounds were not available commercially and were prepared according to the reaction scheme and procedures shown in Fig. S1 in the supplemental material.
GC/MS analysis of lactones.
Lactones were analyzed on an Agilent Technologies 7890A gas chromatograph coupled to a 5975C quadrupole mass spectrometer and a CTC Analytics Combipal autosampler. One microliter was injected under pulse split (1:10) conditions on a 30-m by 0.25-mm by 0.25-μm HP-5MS capillary column (Agilent). The temperature program was as follows: 70°C for 1.5 min, 65°C/min to 135°C, 5°C/min to 280°C, and 15 min at 280°C. Helium was used as the carrier gas. The injector and the detector were adjusted to 250°C and 280°C. The mass selective detector (MSD) was run in the electron ionization (EI; 70-eV) scan mode between 45 and 500 atomic mass units (amu).
Whole-cell BVMO oxidations on an analytical scale.
A fresh LB agar plate (1.5% [wt/vol] agar) containing Ap was prepared from the stock culture of E. coli BL21[OTEMO], and one colony was transferred into a preculture (20 ml) containing LB medium supplemented with Ap and grown at 30°C at 200 rpm in an orbital shaker overnight. An aliquot of the suspension (2 ml) was used to inoculate LB medium (200 ml) also supplemented with Ap, and the resulting suspension was grown at 30°C at 200 rpm in an orbital shaker. At an optical density at 600 nm (OD600) of 0.5 (∼3 h), protein expression was induced by the addition of IPTG to a final concentration of 1 mM. The cells were allowed to grow for an additional 3 h (OD600 of 2.0 to 2.2), and the cell suspension was divided into 10-ml batches. A solution of substrate in isopropanol (1 M; 30 μl) was added to each batch, and the reaction flask was shaken at 30°C at 200 rpm in an orbital shaker. After 18 h, the cells were pelleted by centrifugation, and the supernatant was extracted with ethyl acetate (10 ml). The layers were separated, and the organic layer was dried over anhydrous sodium sulfate and filtered. The solution obtained was used for GC analysis with the following retention times observed: for chiral GC method A, for 2-ethylcyclohexanone (compound 6), R-6 = 17.86 min, S-6 = 17.60 min, R-6a = 29.81 min, S-6a = 28.83 min; for 2-propylcyclohexanone (compound 7), R-7 = 21.92 min, S-7 = 21.22 min, R-7a = 32.91 min, S-7a = 32.20 min; for 2-methylcyclopentanone (compound 9), R-9 = 14.53 min, S-9 = 14.05 min, R-9a = 36.99 min, S-9a = 37.46 min; for 2,4,4-trimethylcyclopentanone (compound 11), R-11 = 12.59 min, S-11 = 13.11 min, R-11a = 25.49 min, S-11a = 26.06 min; for norcamphor (compound 21), normal lactone (1R)-21a = 45.48 min, normal lactone (1S)-21a = 44.25 min, abnormal lactone (1R)-21b = 45.35 min, abnormal lactone (1S)-21b = 46.11 min; for trans-1-decalone (compound 22), 22 = 30.67 and 30.94 min, normal lactone 22a = 42.65 and 43.76 min, abnormal lactone 22b = 41.23 and 42.09 min; for chiral GC method D (150 min), for 2-phenylcyclohexanone (compound 16), R-8 = 121.13 min, S-8 = 122.45 min, R-8a = 145.05 min, S-8a = 145.19 min; for chiral GC method E (300 min), for 2-hexylcyclopentanone (compound 10), 10 = 47.99 min, R-10a = 123.35 min, S-10a = 122.21 min; for chiral GC method E (Chirasil Dex, 80 min), for bicyclo[3.2.0]hept-2-en-6-one (compound 20), normal lactone (1S,5R)-20a = 38.31 min, normal lactone (1R,5S)-20a = 37.47 min, abnormal lactone (1S,5R)-20b = 36.75 min, abnormal lactone (1R,5S)-20b = 36.20 min.
Whole-cell BVMO oxidation on a preparative scale.
The procedures used for whole-cell BVMO oxidation on a preparative scale are similar to those described above except that after induction, a solution of substrate in isopropanol (1 M; 0.5 to 1.0 ml) was added, and the reaction mixture was shaken (30°C at 200 rpm for 18 h) on an orbital shaker. The cell suspension was centrifuged, and the supernatant was extracted three times with ethyl acetate (200 ml). The organic layers were combined, washed with brine, dried over anhydrous sodium sulfate, and filtered. The organic solvent was removed under reduced pressure, and the crude residue was purified by flash column chromatography (10 g SiO2, ethyl acetate-hexane gradients) to obtain the desired lactones with high purity.
Spectroscopic data.
Spectroscopic data are provided in Material SM3 in the supplemental material.
Crystallization and data collection.
Following purification, OTEMO protein was dialyzed overnight against a buffer consisting of 20 mM HEPES (pH 7.5) and 150 mM NaCl. The dialyzed protein was then concentrated by ultrafiltration to 12.5 mg/ml. For the OTEMO-FAD-NADP complex, NADP (Roche) was added to a final concentration of 5 mM. Crystallization screening was performed by using in-house screens (∼400 conditions) by mixing 0.4 μl protein with 0.5 μl reservoir solution in Intelliplate sitting-drop plates (Art Robbins Scientific). Two crystal forms of the OTEMO-FAD complexes were obtained during optimization by hanging-drop vapor diffusion in 24-well Limbro plates. Prism-shaped (OTEMO-FAD complex form 1) crystals were obtained by mixing 1 μl of protein (12.5 mg/ml) in buffer (20 mM HEPES [pH 7.5], 150 mM NaCl) with 1 μl of reservoir solution (22% [wt/vol] polyethylene glycol 4000 [PEG 4000], 0.1 M HEPES [pH 7.5]). Rod-shaped (OTEMO-FAD complex form 2) crystals were obtained by mixing 1 μl of protein as described above with 1 μl of reservoir solution (22% [wt/vol] PEG 3350, 0.1 M Na/K phosphate buffer [pH 7.5]). In each case, drops were equilibrated over 0.5 ml of reservoir solution.
For data collection, crystals were cryoprotected by transferring them briefly into a reservoir solution supplemented with 15% (vol/vol) ethylene glycol, picked up with a nylon loop, and flash-cooled in an N2 cold stream at 100 K. X-ray diffraction data were collected at the 31-ID (Lilly Research Laboratory Collaborative Access Team [LRL-CAT]) beamline at Advanced Photon Source (APS), Argonne National Laboratory (ANL), or at the CMCF1 beamline at the Canadian Light Source (CLS). Crystals of form 1 belong to space group P21, with a = 59.0, b = 140.0, c = 67.6, and β = 98.4°. Form 2 crystals are also P21, with a = 67.3, b = 92.5, c = 93.4, and β = 103.3°. Cocrystallization with 5 mM NADP using the same reservoir solution as that used for the above-described form 2 crystals led to the OTEMO-FAD-NADP+ complex (NADP+-bound form 1), which has a slightly longer unit cell dimension along the b axis than the FAD-alone crystals, with a = 66.8, b = 95.0, c = 93.1, and β = 102.3°. Additional NADP-bound OTEMO crystals arising from cocrystallization with the substrate but lacking density for this ligand were obtained in the presence of 25% (wt/vol) PEG 8000–0.2 M ammonium sulfate (NADP+-bound form 2), 20% (wt/vol) PEG 8000–0.1 M cacodylate (pH 6.5)–0.2 M ammonium sulfate (NADP+-bound form 3), or 20% (wt/vol) PEG 3350–0.2 M tripotassium citrate (NADP+-bound form 4). Both NADP-bound form 2 and form 4 crystals were in space group P21, with unit cell parameters similar to those of the NADP-bound form 1 crystals. The NADP-bound form 3 crystals belong to space group P212121, with unit cell parameters of a = 61.3, b = 66.2, and c = 282.7, having a Matthew's coefficient of 2.34 Å3/Da, corresponding to a solvent content of 47.4% and diffracted to a 2.8-Å resolution.
Structure determination and refinement.
X-ray diffraction data were integrated and scaled by using the program HKL2000 (43). The structure of the FAD-bound form 1 crystal was obtained by molecular replacement using the program MolRep (59) from the CCP4 suite (12), with the PAMO structure (Protein Data Bank [PDB] accession number 1W4X) being used as the search model. After cycles of restrained refinement using Refmac5 (40), the model was rebuilt by using ARP/WARP (45), followed by alternating cycles of fitting with Coot (21). This structure was subsequently used as the search model for other data sets listed in Table S1 in the supplemental material. All models have good geometry, as analyzed by PROCHECK (32). Data collection and refinement statistics are summarized in Table S1 in the supplemental material.
Accession numbers.
The 10,987-bp DNA sequence of the two BamHI fragments determined in this study have been deposited in the GenBank database under accession number AB671319. Atomic coordinates and structure factors for various OTEMO structures have been deposited in the Research Collaboratory for Structural Bioinformatics PDB (9) under accession numbers 3UOV, 3UOX, 3UOY, 3UOZ, 3UP4, and 3UP5.
RESULTS
Gene context and sequence characteristics of OTEMO.
In addition to the previously established camphor regulatory gene (camR) and the start of the cytochrome P450cam hydroxylase operon (30), three new open reading frames (ORF) and a partial one were found at the 5′ end of the sequenced 6.8-kb BamHI fragment of strain PpCam inserted into E. coli pCAM200. The cloning and sequencing of an adjacent 4.2-kb BamHI fragment of pCAM100 completed a 546-codon ORF that was designated OTEMO, in addition to a possible topoisomerase-encoding gene upstream (Fig. 2). The 3′ end of the OTEMO-encoding gene is 3.794 kb upstream of the 3′ end of the divergently transcribed camR (Fig. 2). OTEMO is likely cotranscribed with the 2,5-diketocamphane monooxygenase (DKCMO) gene 88 bp downstream (our unpublished data).
The predicted 545-amino-acid sequence of OTEMO is typical of the established type 1 BVMO sequences that contain two conserved Rossmann fold fingerprints (16-GAGVTG and 192-GTGATG) and the characteristic BVMO fingerprint (160-FKGESFHSSRWP) (invariant amino acids are underlined). Interestingly, between the second Rossmann fold and the fingerprint sequence, there is an apparent 5- to 6-amino-acid insertion that is absent in the aligned sequences of PAMO and CHMO that shows 40% identity (59% similarity) and 37% identity (57% similarity), respectively (see Fig. S2 in the supplemental material). In a BLAST search (July 2011), OTEMO was found to be most identical (80%) to a predicted 549-residue BVMO (Saro_1480) from the Novosphingobium aromaticivorans DSM 12444 chromosome (GenBank accession number YP_496757.1), followed closely by a 524-residue homolog in Sphingomonas sp. strain SKA58 (accession number ZP_01305158.1). A phylogenetic analysis of experimentally validated BVMO sequences showed that OTEMO is closest to the cluster of the CPMO sequence of Pseudomonas sp. NCIMB 9872 and the BVMOs of Brevibacterium sp. strain HCU (34).
Purification and properties of OTEMO.
Using a three-step chromatography procedure, OTEMO was purified to electrophoretic homogeneity based on the appearance of a single protein band with a relative molecular weight (Mr) of 62,000 in SDS-PAGE gels (see Fig. S3 in the supplemental material). The Mr derived from the sequence is 61,480. Size-exclusion chromatography, however, indicated it to be a homodimeric protein with a molecular weight of 118,000. In all the purification steps (see Table S2 in the supplemental material), the enzyme eluted as a distinct yellow fraction due to the FAD prosthetic group. Up to 250 mg/liter of purified protein could be obtained due to the fact that OTEMO was highly expressed in E. coli BL21(pSD-OTEMO) cells, to as much as 50% of the total soluble protein (not shown).
The pH optimum of the enzyme was found to be 9.0. The enzyme was active for a prolonged time at temperatures below 35°C. However, at temperatures of 40°C and above, the enzyme quickly lost its activity, with estimated half-life (t1/2) values of 3.8 min at 40°C and <0.4 min at 45°C (Fig. 3). The melting temperature (Tm) of the enzyme measured by CD spectroscopy at 222 nm was estimated to be 51°C. The CD spectrum of purified and desalted OTEMO was characteristic of an α-fold protein with typical minima at 208 and 222 nm (not shown). Interestingly, the thermodenaturation of purified OTEMO monitored by CD revealed two protein transitions: one between 34°C and 42°C and the other between 50°C and 70°C (not shown). The dramatic loss of enzyme activity caused by a temperature shift from 35°C to 40°C was attributed to the loss of the FAD prosthetic group at higher temperatures, as demonstrated by size-exclusion chromatography (results not shown).
Fig 3.
Thermostability of OTEMO. Denaturation constants (Kd) were used to calculate the enzyme's half-lives at the respective temperatures.
Substrate specificity and kinetics.
Several C5 and C6 cyclic ketones possessing various substituents and characteristics were tested as possible substrates. OTEMO exhibited the highest affinity (Km, 18 μM) toward CoA-activated 2-oxo-Δ3-4,5,5-trimeth-ylcyclopentenylacetic acid (OT), with a rate of 4 s−1 (Table 1). In contrast, the free acid was a poor substrate, processed with a rate (0.13 s−1) ∼30 times lower than that of its CoA ester. However, OTEMO showed the highest catalytic efficiency (4.3 × 105 M−1 s−1) toward 2-n-hexyl cyclopentanone. Although exhibiting the highest rate constant (13.6 s−1), its apparent affinity (Km = 32 μM) was lower than that of the OTEMO CoA substrate.
Table 1.
Substrate specificity and kinetic parameters of OTEMOa
| Substrate | Mean Km (mM) ± SD | Mean kcat (s−1) ± SD | kcat/Km (s−1 M−1) |
|---|---|---|---|
| OT | ND | 0.13 ± 0.02 | ND |
| OT-CoA ester | 0.018 ± 0.004 | 4.80 ± 0.40 | 2.7 × 105 |
| 2-Oxocyclopentyl acetic acid | ND | 0.11 ± 0.02 | ND |
| 2-Oxocyclopentyl ethyl acetate | 0.31 ± 0.07 | 6.92 ± 1.10 | 2.2 × 104 |
| 2-Oxocyclohexyl ethyl acetate | 0.16 ± 0.02 | 0.90 ± 0.07 | 5.6 ×103 |
| 2-Methyl cyclohexanone | 1.40 ± 0.12 | 5.58 ± 0.16 | 4.0 × 103 |
| 4-Methyl cyclohexanone | 0.60 ± 0.10 | 1.78 ± 0.16 | 3.0 × 103 |
| 2-n-Hexyl cyclopentanone | 0.032 ± 0.001 | 13.60 ± 0.51 | 4.3 × 105 |
OT, 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid; ND, not determined.
2-Oxocyclopentyl ethyl acetate was also a good substrate, as was 2-oxocyclohexyl ethyl acetate. The level of activity toward the former ester was some 70 times higher than that of the free acid (6.9 s−1 versus 0.1 s−1). Methyl-substituted cyclohexanones at the 2- and 4-positions were also relatively good substrates for OTEMO, but unlike the esterified substrates, the Km values were in the mM range.
OTEMO oxidation of selected ketones. (i) 2-Substituted cyclohexanones.
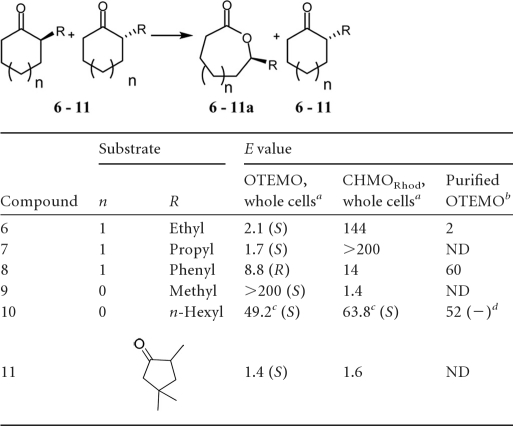
At first, we compared the biocatalytic performance of recombinant OTEMO in whole-cell biotransformations with those of purified OTEMO described in the literature (1, 3, 23–25) (Table 2). Whole cells were used because of the easier handling and the fact that no cofactor regeneration was necessary. The biotransformations were run for 18 h at a substrate concentration of 3 mM (∼0.5 g/liter). E. coli expressing CHMORhod, classified as a typical member of the CHMO family (39), was used as a reference biocatalyst. All of the tested substrates of 2-substituted cyclohexanones were oxidized by OTEMO at a considerable rate. Both enantiomers of 2-ethyl cyclohexanone were oxidized by OTEMO at nearly the same rate although giving a low enantiomeric ratio (E value), a measure for the “selectivity” of an enzymatic resolution (see Table 2 for the calculation). A similar result (E value of 2) was obtained for the oxidation of 2-ethyl cyclohexanone with purified OTEMO (3). A low E value was also observed for 2-propyl cyclohexanone as a substrate with OTEMO. In contrast, CHMORhod proved to be an excellent biocatalyst for the resolution of 2-ethyl and 2-propyl cyclohexanone (E value of >100 to 200), in accordance with data from previous work (39). For 2-phenyl cyclohexanone, both the E values for our recombinant OTEMO and the value reported in the literature for the purified enzyme (3) showed the same trend, although that of the former was lower (E value of 8.8 versus 60). This difference could be due to low or high percent conversions used for the calculations. For obtaining accurate E values, the general rule is to stop the reaction at 50% conversion. The kinetic resolution of 2-methyl cyclopentanones by OTEMO gave an E value of 200, whereas CHMORhod showed no preference for either enantiomer (E value of 1.4). In the case of 2-hexyl cyclopentanone, both OTEMO and CHMORhod showed a preference for the oxidation of the S-enantiomer. The oxidation of 2,4,4-trimethylcyclopentanone by either OTEMO or CHMORhod gave low E values.
Table 2.
Kinetic resolution of racemic ketones
E values were calculated according to a program available at ftp://biocatalysis.uni-graz.at/pub/enantio/.
E values were reported in reference 3 unless otherwise indicated. ND, not determined.
The E value was calculated with percent conversion (c) and percent ee product (eep) using the following equation:E = ln[1 − c(1 + eep)]/ln[1 − c(1 − eep)].
See reference 24. The sign of optical rotation is given in parentheses.
(ii) 4-Substituted cyclohexanones.
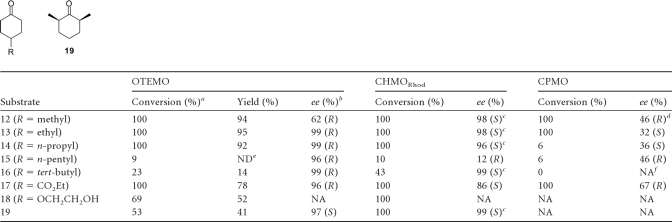
Prochiral ketones using 4-substituted cyclohexanones were tested with recombinant OTEMO (Table 3). The screening of six different 4-substituted cyclohexanones with small to medium substituents at the C-4 position (compounds 12 to 18) in nearly all cases gave enantiopure lactones with an excellent yield. An increase of the steric bulk of the substituent (compounds 15 and 16) led to lower rates of conversion of the starting material; however, the optical purity remained high. The absolute stereoconfiguration of the obtained lactones was determined as R based on optical rotations of isolated compounds as well as comparisons with data obtained from biotransformations with recombinant CHMORhod and also CPMO. This comparison also revealed a clear enantiodivergent behavior of OTEMO from that of CHMORhod. A slight enantiodivergent trend for the oxidation of 4-substituted cyclohexanones is evident for CHMO and CPMO with respect to some of the compounds previously reported (Table 3) (37, 62).
Table 3.
Desymmetrization of prochiral ketones
Interestingly, 2,6-dimethyl cyclohexanone (compound 19) gave the same enantiomer as CHMO, suggesting that CHMO and OTEMO must have similar modes of binding in the vicinity of the keto moiety. On the other hand, the shape of the binding site must differ considerably in order to effectively discriminate between substituents at the C-4 position in cyclohexanones.
(iii) Bicyclic compounds.
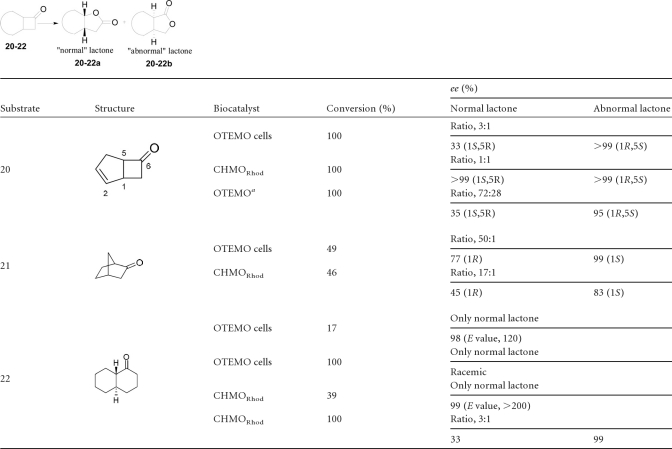
The oxidation of bicyclic compounds (compounds 20 to 22) was investigated (Table 4). Compound 20, bicyclo[3.2.0]hept-2-en-6-one, is probably the most tested substrate for BVMO-mediated oxidations, since it can lead to both the so-called “normal” and “abnormal” lactones. As a result, the whole-cell-mediated oxidation of compound 20 with recombinant OTEMO gave a mixture of normal to abnormal lactone at a ratio of 3:1. Purified OTEMO was reported previously to give a ratio of 2:1 for 100% conversion (24). Recombinant OTEMO oxidized norcamphor (compound 21) nearly exclusively to the normal lactone (ratio, 50:1) at a synthetically usable optical purity when stopped at approximately 50% conversion. CHMORhod gave a lower ratio of 17:1. Upon the kinetic resolution of trans-1-decalone (compound 22), recombinant CHMORhod gave a similar E value of >200, as previously reported (39). At 100% conversion, CHMORhod produced some abnormal lactone. On the other hand, no abnormal lactone was produced by recombinant OTEMO at 100% conversion. An E value of 120 (98% enantiomeric excess [ee]) was obtained for the kinetic resolution of trans-1-decalone (compound 22) at 17% conversion.
Table 4.
Regiodivergent oxidations of bicyclic ketones
See reference 24.
OTEMO structures.
Crystal structures of the OTEMO-FAD and OTEMO-FAD-NADP+ complexes, in different packing environments, were determined by molecular replacement, and the final statistics are presented in Table S1 in the supplemental material. For all the crystal structures reported here, each asymmetric unit contains an OTEMO dimer. The OTEMO-FAD form 1 and form 2 structures consist of residues 6 to 545. Residues S149 to M151 and D390 to T393 are disordered and were not modeled, which is different from the PAMO-FAD complex, where the corresponding residues have been modeled (35). For the OTEMO-FAD-NADP+ structures, the above-described flexible linker region, which connects the FAD and NADP domains, is well defined, except in three subunits (out of all 8 different subunits of the OTEMO-FAD-NADP structures reported here), where residues A391 to T393 remained disordered.
Monomer structure.
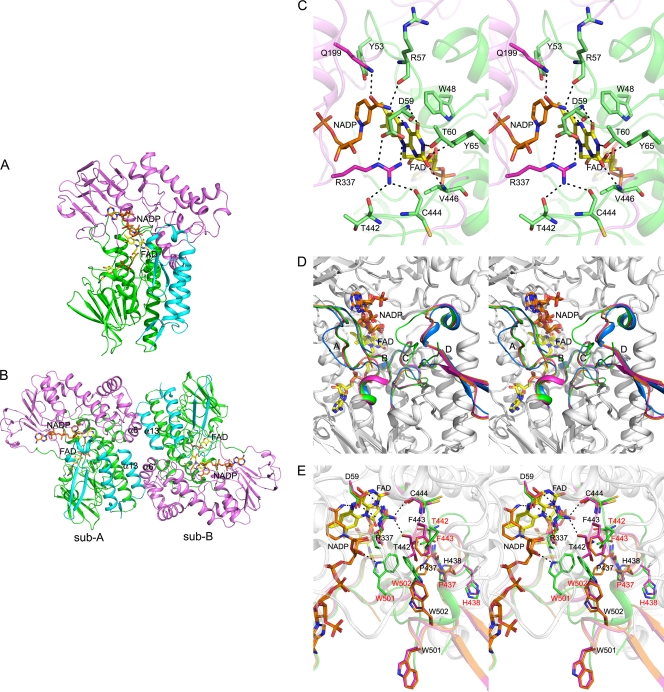
Each OTEMO molecule consists of three domains, with residues 6 to 152 and 391 to 470 forming the FAD-binding domain, residues 153 to 390 forming the NADP-binding domain, and residues 471 to 545 forming the “flap” domain, respectively (Fig. 4A). Connections between the FAD- and NADP-binding domains occur via two linker regions (residues P145 to S152 and D390 to G394). The first linker region becomes ordered upon NADP+ binding, whereas the second linker region is more mobile, as it could not be modeled in some NADP-bound subunits. A flap domain, consisting of a pair of α-helices (α12/α13) and a β-hairpin (β19/β20, i.e., residues 476 to 518), extends from the FAD-binding domain and partially covers the interdomain cleft. The OTEMO crystal structure is closely related to those of PAMO (35) and CHMO (39) (see Fig. S4 in the supplemental material). For example, the closed form of the OTEMO-NADP+ complex (NADP+-bound form 3) shows root mean square deviations of 1.07 Å for 516 C-α atoms with the NADP-bound PAMO (PDB accession number 2YLR) and 1.23 Å for 509 C-α atoms with the closed form of CHMO (PDB accession number 3GWD). All of these BVMOs belong to the FAD/NAD(P)-binding domain superfamily and the FAD/NAD-linked reductase structural family, as classified within the SCOP database (41), a family that includes a variety of dehydrogenases and reductases.
Fig 4.
Crystal structure of OTEMO. (A) Overall structure of the OTEMO monomer, colored by domain, with the FAD-binding domain (residues 6 to 152 and 391 to 470) (green), NADP-binding domain (residues 153 to 390) (magenta), and flap domain (residues 471 to 545) (cyan) indicated. FAD (yellow carbon) and NADP (orange carbon) are shown in a stick representation. This and subsequent depictions of the OTEMO structure were prepared by using the program PyMol (http://www.pymol.org/). (B) Organization of the OTEMO dimer, with the same color scheme as that described above for panel A. The two α-helices, α6 and α13, involved in dimerization are labeled. (C) Stereo view of the FAD- and NADP-binding region within the OTEMO active site (type 1, as summarized in Table 5) (PDB accession number 3UOY). Key active-site residues are labeled. Domains of the OTEMO monomer are colored as described above for panel A. H bonds are shown as black dashed lines. (D) Stereo view of the superposition of different monomers obtained from different crystal forms/crystallization conditions, showing conformational flexibility in the structure. The four regions displaying variations are shown in different colors (types 1, 2, 3, and 4 are shown in blue, orange, magenta, and green, respectively; all other parts having the same conformations are shown in gray) and are indicated by the letters A (residues 145 to 152), B (residues 390 to 394), C (residues 435 to 444), and D (residues 497 to 518). (E) Stereo close-up view of the conformational flexibility of the active-site region shown in panel D. Residues that undergo significant movements are labeled in different colors.
Dimer structure.
Our analysis by size-exclusion chromatography and dynamic light scattering of a sample of purified OTEMO (Mr = 118,000) together with the results of a previous study using equilibrium ultracentrifugation (Mr = 106,000) (44) showed that the enzyme is a dimer in solution. This finding is in contrast to the available data for CHMO and PAMO (35, 39), which indicate that these enzymes function as monomers. A dimeric organization is evident from all of the crystal structures determined in this study (Fig. 4B). The solvent-accessible surface area of the interface is remarkably small, only 1,019 Å2, or 4.5% of the total solvent-accessible area, as computed by using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (31). Contacts between the two subunits of the dimer involve mainly residues from α-helices (α6 and α13) found within the NADP and flap domains, respectively, such that α13 of one subunit contacts α6 of the other subunit. In addition, residues from α3 and α11 of the FAD domain also contribute to dimerization. Hydrogen bonds between the subunits involve R240NH1 to the carbonyl of Y536, and Y63OH to Y536OH, with a number of additional residues forming van der Waals contacts.
Cofactor binding sites. (i) FAD.
Purified OTEMO has a bright yellow color, consistent with the presence of bound FAD. With the exception of the O2′ atom of ribose and the C-8 and N-9 atoms of adenine, the FAD cofactor is completely buried and inaccessible to the solvent. The isoalloxazine ring is sandwiched between the NADP nicotinamide ring on one side (re-side) and the side chains of W48 and Y65 on the other side (si-side) (Fig. 4C). The ribitol moiety is anchored by direct H bonds with Y65OH and T47OG1 and water-mediated H bonds to the amide of V19 and the carbonyl of G45. The pyrophosphate is positioned through H bonds to T20OG1 as well as the main-chain amides of T20 and T47 and several water-mediated interactions. The adenosine portion is anchored by E39 and W50NE1 (ribose) as well as the carbonyl of V112 and water-mediated H bonds to the carbonyls of S395 and R398.
(ii) NADP(H).
The cocrystallization of FAD-bound OTEMO with 5 mM NADP resulted in a structure containing both ligands (Fig. 4A and C). The distance between the C-4 atom of the NADP+, the hydride donor, and the N-5 atom of the FAD acceptor is 5.2 Å. The adenine N-1, N-3, and N-6 atoms of NADP+ participate in water-mediated H bonds to the OTEMO main chain, with the adenine ring stacking against the side chain of R216. The 2′-O-phosphate is positioned via a salt bridge with R216NE and R216NH2, an H bond with T217OG1, and several water-mediated H bonds. The pyrophosphate moiety is anchored via R150NH2 and T196OG1 and to the main-chain amide of T196. The carboxamide of the nicotinamide moiety is involved in two direct H bonds with the protein, one between O-7 and Q199NE2 and the other between N-7 and D59OD2, and the carbonyl of R57 (Fig. 4C). Also, this N-7 atom is within H-bonding distance from the N-5 atom of the FAD isoalloxazine ring.
The OTEMO active-site region revealed in different crystal forms.
We have observed a series of different conformations (Table 5 and Fig. 4D and E) in the OTEMO-FAD-NADP+ structures despite three out of four different crystal forms having almost identical unit cell parameters (see Table S1 in the supplemental material). In the NADP+-bound form 1 crystal, the first linker region connecting the FAD and NADP domains adopts two distinct conformations in the two subunits, resulting in a small difference with respect to the positioning of the NADP adenine ring. In this crystal form, in contrast to the PAMO-NADP+ and the CHMO-NADP+ (closed-form) complexes, no significant conformational changes occur to the segment spanning residues 499 to 518 upon NADP binding, which maintains its β-hairpin structure, resulting in a relatively wide NADP-bound active-site pocket. This is also in contrast to the CHMO-NADP+ “open” form, where the corresponding segment is totally disordered despite the presence of NADP+. While efforts to cocrystallize substrates bound to OTEMO were unsuccessful, a significant rearrangement of the active-site region, corresponding to residues 497 to 518 and 435 to 444, was observed for the other three different NADP+-bound structures obtained from these crystals (NADP+-bound form 2, form 3, and form 4) (Fig. 4D and E). Other notable differences in the structure include residues 390 to 394, which become well defined in these subunits, and differing positions for residues 145 to 152, close to NADP+, which display differing arrangements in the various structures. Considering all these differences, the crystal structures of OTEMO-NADP+ complexes reveal four conformational states (Table 5). In the NADP+-bound form 2 crystal, residues 390 to 394 become well ordered and consequently displace the nearby residues 436 to 439 from their original location to avoid steric clashes. As a result, the active-site pocket becomes less solvent exposed due to a narrowing of the active-site entrance. The NADP+-bound form 3 crystal is in space group P212121, which differs from all other OTEMO crystals (in the P21 space group) (see Table S1 in the supplemental material). In this new form, in addition to the ordering of residues 390 to 394, the segment spanning residues 499 to 518 undergoes a drastic conformational transition, with the C-α of W501 shifting ∼9 Å to form a hydrogen bond with the nicotinamide ribose hydroxyl group. This new conformation adopted by residues 499 to 518 in OTEMO is similar to that observed for the PAMO-NADP+ and CHMO-NADP+ (closed-form) complexes. Interestingly, W501 is the only residue that establishes close contacts with the NADP+ molecule. The immediate consequence of this conformational shift is that the bulky side chain of W502 pushes away the nearby T442 and F443 residues from their positions observed for other OTEMO-NADP+ complexes. Therefore, the segment spanning residues 435 to 444, in close vicinity of the critical residue R337 and the FAD isoalloxazine ring-shaping active-site pocket, displays three alternate conformations in the presence of NADP+ (Table 5). Although the form 4 crystal of the NADP+-bound OTEMO has almost the same unit cell as those of the NADP+-bound form 1 and form 2 crystals, the two subunits in the asymmetric unit adopt two distinct conformations: subunit A is identical to one subunit in form 1, whereas subunit B adopts the same conformation as those observed for form 3. These data show that structural elements forming the boundaries of the active-site pocket in OTEMO are very flexible. As a result, the sizes of the active-site entrance and pocket vary in the different structures presented here.
Table 5.
Conformational flexibility of the OTEMO-FAD-NADP structures observed in different OTEMO subunitsa
| Subunit (PDB accession no.) | Conformation at residues: |
Type | |||
|---|---|---|---|---|---|
| 145–152 | 390–394 | 435–444 | 497–518 | ||
| NADP form 1-A (3UOY) | Open | Disordered | Conformer 1 | Open (β-hairpin) | 1 |
| NADP form 1-B (3UOY) | Closed | Disordered | Conformer 1 | Open (β-hairpin) | 2 |
| NADP form 2-A (3UOZ) | Closed | Ordered | Conformer 2 | Open (β-hairpin) | 3 |
| NADP form 2-B (3UOZ) | Closed | Ordered | Conformer 2 | Open (β-hairpin) | 3 |
| NADP form 3-A (3UP4) | Closed | Ordered | Conformer 3 | Closed | 4 |
| NADP form 3-B (3UP4) | Closed | Ordered | Conformer 3 | Closed | 4 |
| NADP form 4-A (3UP5) | Open | Disordered | Conformer 1 | Open (β-hairpin) | 1 |
| NADP form 4-B (3UP5) | Closed | Ordered | Conformer 3 | Closed | 4 |
The four segments listed here are indicated by the letters A, B, C, and D in Fig. 4D.
A few residues in OTEMO, including W48, N51, Y53, D59, and R337, are proximal to the FAD isoalloxazine ring. In OTEMO, D59 and R337 are ∼4.1 Å and ∼4.4 Å away from the C-4a atom of FAD. Although the available crystal structures of CHMO and PAMO are consistent with the flexibility of this Arg side chain within the active-site pocket (35, 39, 42), superpositions of OTEMO crystal structures in the presence or absence of bound NADP show essentially no change in the position of R337 or other putative active-site residues in the vicinity of the FAD isoalloxazine ring or NADP nicotinamide ring. The slight movement of R337 occurs only in the closed form of the OTEMO-NADP complex (form 3 and form 4 subunit B). In all OTEMO crystal structures, the positively charged guanidinium group of R337 is anchored by the carboxylate group of D59 via salt bridges. It should be noted that due to the slight movement of R337, only one salt bridge interaction is maintained in the closed form of the OTEMO-NADP+ complex, compared to two strong salt bridge interactions observed between R337 and D59 in the other subunits. In addition, the main-chain carbonyls of T442 and C444 also contribute to the positioning of R337 in all OTEMO subunits, with the exception of the OTEMO-NADP+ closed form (NADP+-bound form 3 and form 4 subunit B). Notably, the guanidinium group of R337 is not within H-bonding distance (∼6Å apart) of the carboxamide group of NADP, which instead hydrogen bonds to the N-5 atom of the FAD isoalloxazine ring.
OTEMO mutants and activities.
Guided by the crystal structure, six different mutated OTEMO enzymes containing single-residue changes, consisting of Y53F/Y53A, D59A/D59N, or R337A/R337K, were constructed. Initial activity assays using 2-n-hexyl cyclopentanone as a substrate indicated that all but one of the mutant proteins (Y53F) had lost significant activity. The purified mutant proteins (see Fig. S3 in the supplemental material) indicated that the Y53F variant retained 32% of its activity, whereas both D59 mutants showed only 1 to 3%. This loss of activity was not due to the loss of the prosthetic group, since these mutant proteins were found to have comparable ratios of protein to FAD measured at 280 nm and 435 nm (ratio of 9.5 fro the wild-type protein versus 9 to 9.9 for the rest) for the mutants during purification. No soluble protein was obtained for the Y53A mutant, and it was therefore not analyzed further. The loss of a bulky side chain at this position probably disturbs the local secondary structure required for proper folding and accommodation of the FAD molecule.
The Y53F mutant was further investigated, and its kinetic parameters were compared to those of the wild-type enzyme (Table 6). Interestingly, the catalytic efficiencies of both enzymes appeared to be nearly identical, although the Y53F mutant showed a higher affinity toward OT-CoA while exhibiting a lower kcat value. There was no appreciable difference in the affinities for NADPH between the two proteins, suggesting that the lower catalytic activity of the Y59F mutant may not be due to the lack of a coenzyme interaction.
Table 6.
Comparison of the kinetic parameters of wild-type OTEMO and the Y53F mutanta
| OTEMO protein | Substrate or cosubstrate | Mean Km (μM) ± SD | Mean kcat (s−1) ± SD | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| Wild type | OT-CoA ester | 18 ± 4 | 4.8 ± 0.4 | 2.7 × 105 |
| NADPH | 3.6 ± 0.9 | NA | NA | |
| Y53F | OT-CoA ester | 10 ± 1 | 2.5 ± 0.1 | 2.5 × 105 |
| NADPH | 4.0 ± 1.0 | NA | NA |
OT, 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid; NA, not applicable.
The significant loss of activity for the D59A and D59N mutant enzymes indicates that this residue is very important. In addition to the proper binding or stabilization of NADP+ suggested previously for PAMO (42), its interactions with the R337 present in all OTEMO structures suggest that this charge neutralization is critical for the orientation of R337, the positioning of which is likely pivotal for proper substrate binding and subsequent catalysis.
DISCUSSION
Although regarded as an “unremarkable” enzyme, being a simple dimer carrying FAD as the only detectable nonprotein component (44), OTEMO is unusual in that this monooxygenase requires a CoA ester as opposed to the free 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid as a substrate. Using a recombinantly expressed and purified OTEMO protein, we confirmed this substrate specificity and report a Km (18 μM) toward this ester that is lower than that for 2-n-hexyl cyclopentanone, a substrate exhibiting a ∼3-times-higher turnover rate.
One of the earliest observations for biotransformations catalyzed by partially purified OTEMO (then known as MO2) was its selectivity for monocyclic substrates rather than bicyclic ketone such as bicyclo(2.2.1)heptan-2-one (25). The ability to transform some 2-alkyl cyclopentanones and 2-substituted cyclohexanones, in some cases resulting in excellent enantiomeric excesses (92 to 95%) and enantioselectivities (E values, 52 to 104), has also been reported (3, 23). OTEMO was not particularly effective in the oxidation of 3-substituted cyclobutanones, although in one case with 3-CH2OCH2Ph-cyclobutanone as a substrate, partially purified OTEMO effected a high level of conversion and 90% ee of the R-lactone (23). In terms of providing a stereospecific building block for the asymmetric synthesis of an important pharmaceutical product, R-lipoic acid, OTEMO was most useful for the conversion of 2-(2-acetoxyethyl)cyclohexanone to the R-lactone, resulting in an 83% ee and an E value of ∼17 after only a short incubation (3 h) (1, 2). Partially purified OTEMO was also able to carry out the stereospecific oxygenation of various alkyl aryl sulfides to equivalent S-(−) sulfoxides (8). The “caged” hydrocarbon adamantanone (tricyclo[3.3.13,7]decan-2-one) was tested for biotransformation by intact cells of the camphor-grown PpCam strain, but the exact contribution of OTEMO is not known (52).
Phylogenetic analysis indicated that OTEMO is positioned distinctly from a number of biochemically characterized BVMOs, e.g., Acinetobacter CHMO, but close to the CPMO family (34). Based on a smaller BVMO sequence data set, Mihovilovic and coworkers (38) previously proposed two groups of CHMO and CPMO “family clustering” based on their stereopreferences. The CHMO type generally displayed broad substrate acceptance, whereas the CPMO type was more restrictive. More specifically, members of the CHMO family are known to give S-lactones in high enantiomeric excesses, while members of the CPMO family give R-lactones of moderate optical purity, as also shown in Table 2. CHMO-type enzymes also showed regiodivergent oxidations to both “normal” and “abnormal” lactones, depending on the absolute configuration of the ketone precursors, whereas CPMO-type BVMOs usually yielded abnormal lactone products (11).
In this study, we showed that OTEMO displays a distinct behavior compared to those of both the CHMO and CPMO families, providing unique enantiocomplementarity behavior to BVMOs of the CHMO family. This fact makes OTEMO an extremely useful biocatalyst for the oxidation of 4-substituted cyclohexanones. Now, in combination with CHMO-type BVMOs, both antipodes of chiral lactones derived from prochiral 4-subsituted cyclohexanones are made available. A slight enantiodivergent trend for the oxidation of 4-substituted cyclohexanones was previously observed for CHMO and CPMO (37, 62). The results for recombinant OTEMO oxidation of norcamphor to nearly exclusively the normal lactone (ratio, 50:1) at synthetically usable optical purities showed the apparent superiority of OTEMO over the CHMO-type CHMORhod for this bicyclic substrate. CHMORhod also produced some abnormal lactone (39).
The precise nature of the substrate-binding site in OTEMO and related BVMOs remains largely undefined. An analysis of the OTEMO packing environment within 5 Å of the FAD C-4a atom using the program Voronoia (49) indicated that the more open environment is on the re-side of the FAD ring, against which the nicotinamide ring of NADPH stacks. In the absence of other conformational changes, particularly those involving W48, which stacks closely against the si-side of FAD, it appears that hydride transfer from NADPH, the binding of O2 to form the peroxyflavin intermediate, and the binding of the substrate to form the Criegee intermediate all occur on the re-side of the FAD isoalloxazine ring. This manner of cofactor orientation has become a more frequently observed mode of binding among the solved BVMO structures, as discussed previously by Mirza et al. (39). A structure-based sequence alignment of OTEMO, CHMO, and PAMO (see Fig. S2 in the supplemental material) revealed that key active-site residues are highly conserved among these structures, implying similar modes of substrate binding by each of these enzymes.
A potential role for R337 of PAMO, equivalent to R337 of OTEMO or R329 of CHMO, in the stabilization of the flavin-peroxide or Criegee intermediates was suggested based on the PAMO crystal structure (35). The corresponding R440A mutation in 4-hydroxy-acetophenone monooxygenase (HAPMO) results in an inactive BVMO, although the reduction of the flavin by NADPH is not affected (28, 60). Similarly, the R337A and R337K PAMO mutant enzymes competently form the C-4a peroxyflavin intermediate but cannot effect catalysis with substrates, indicating a key catalytic role (57). In PAMO, D66 (4.7 Å) and R337 (3.7 Å) appear most appropriately positioned relative to the C-4a atom to participate in chemical catalysis. Recent crystallographic studies with PAMO suggested that R337 acts as an anchoring element for the proper binding of the ketone substrate and works in concert with the NADP(H) molecule and that D66 is important (but not essential) for NADPH binding or oxidation (42). In OTEMO, the relative positioning of R337 and D59 in the active site strongly suggests that these two residues work in concert for proper substrate binding or subsequent catalysis. The distances between these two residues observed for OTEMO (∼3.0 Å) are similar to those for both the closed and the open forms of CHMO (∼2.9 Å) but deviate from those in PAMO (35, 42). The closest distance in the latter case is 3.7 Å. Interestingly, the hydrogen-bonding interaction between the guanidinium group of R337 and the carboxamide group of NADP observed for PAMO (oxidized form) is absent from OTEMO, in which these two groups are ∼6 Å apart. However, all of the OTEMO structures are in oxidized forms, because no reducing agent was added during the sample preparation. One possible explanation for this is that these structures represent different snapshots of substrate binding or catalysis. Studies of PAMO suggested that D66 is important for NADPH binding or oxidation. The structure of the PAMO (D66A)-NADP+ complex, however, revealed that in the absence of the D66 carboxylate group, the binding of NADP+ is not disturbed, although the R337 side chain repositions. These results are most consistent with a role of this aspartic acid residue in anchoring R337 for optimal active-site geometry.
Recent studies of PAMO proposed a specific funnel guiding substrate entry and binding (42). No such funnel at the analogous location could be detected in either the OTEMO or CHMO structure (see Fig. S5a in the supplemental material). The very small hole in this region of OTEMO is not sufficient to allow the entry of the substrate. However, surrounded by a cluster of hydrophobic residues (F255, W288, L289, F443, L495, W502, and V521) and also very close to the C-4a flavin atom, this small hole may provide an ideal environment for oxygen diffusion (6). We suggest that the substrate entry path in BVMO enzymes is more likely located at the interface between the two domains, because a small channel was found for all of the structures of OTEMO, CHMO, and PAMO (see Fig. S5b in the supplemental material).
The most striking observation for the structures of OTEMO-NADP+ complexes is the extent of conformational flexibility in the vicinity of the active-site pocket. The structures reported herein could represent snapshots of different stages of substrate binding or release or the adjustment of active-site pocket residues to accept different substrates. The conformational conversion from a β-hairpin to a loop (residues 499 to 518) brings a highly conserved W501, the importance of which has been validated by mutagenesis in CHMO, to the NADP ribose moiety, leading to the closure of the active site. Similar conformational changes for the segment spanning residues 499 to 518 have also been observed for CHMO and PAMO. Therefore, in the “resting state” of these BVMOs, this segment (residues 499 to 518) folds into a β-hairpin that protrudes from the protein surface and functions as a “cork” that will be restructured and inserted back into the “bottle” (enzyme) to shield the active site from solvent upon substrate binding (see Fig. S6 in the supplemental material). The active-site architecture of this closed structure is, however, unlikely compatible with the OT-CoA ester substrate due to the limited space, given that the NADP+ molecule remains bound at the active site. Therefore, the structures of NADP+-bound form 1 and NADP+-bound form 2 (open forms) likely better represent the conformations adapted for the OT-CoA ester substrate. An inspection of the surroundings of the active site, however, does not reveal a readily apparent binding site for the OT-CoA ester in the presence of NADP+, as no other nucleotide-binding motif could be detected. Additionally, a remarkable conformational flexibility was observed for the segment spanning V435 to C444. This is in sharp contrast to both PAMO and CHMO, where the corresponding segments (A435 to S444 for PAMO and L428 to T435 for CHMO) are quite rigid, as shown by all of the available structures. This flexibility likely plays an important role in the reshaping of the active site in the presence of different substrates, which is consistent with the broad substrate profile of OTEMO. An alternative role of this flexibility is to help guide substrate binding and favor subsequent catalysis. In this scenario, the formation and disruption of hydrogen-bonding interactions between two main-chain carbonyl groups in this region (T442 and C444) and the critical R337 in the open and closed forms, respectively, may play an important role for this purpose. The effects of residues 440 to 446 in PAMO (same numbering in OTEMO) have been extensively studied, and an important role of this segment in conferring substrate specificity and enantioselectivity has been well established (46, 47). In addition, a very recent report also identified A435 as contributing to substrate specificity (19). The OTEMO structures reported here reveal for the first time that this region could undergo significant conformational adjustments and provide detailed pictures of different substrate-binding environments. These structural snapshots could also constitute a good template for further protein engineering to expand the scope of substrates used.
The importance of R216 for NADPH binding and the specificity for NADPH over NADH is reflected in the large decrease in the kcat/Km value, resulting from a 3 orders of magnitude increase in the Km observed for the corresponding R217A or R217L mutant PAMO enzymes from T. fusca (20). The corresponding residue in HAPMO, R339, is also critical for NADP+ recognition (28). Structurally, the position of this arginine relative to the cofactor, corresponding to R209 in CHMO, is found to be well conserved. The T218A mutation in PAMO showed little effect on catalytic efficiency (20), consistent with a relatively minor role for T217 of OTEMO in NADPH binding.
Enzymatic analysis using a series of cyclohexanone or cyclopentanone derivatives showed the lowest Km and highest kcat/Km values for several acetate esters of cyclopentanone or cyclohexanone. The important contribution of the ester moiety to reducing the apparent Km of the substrate suggests specific polar interactions between active-site residues and the ester oxygen atoms or the alkyl moiety of the highest-affinity substrate, 2-n-hexyl cyclopentanone. As revealed by the electrostatic potential, the entry into the active-site pocket is remarkably positively charged (see Fig. S7 in the supplemental material). The stereochemical specificity of the reaction as well as the expected attack of the C-4a peroxyflavin at the carbonyl C of the substrate (60) would be consistent with one or more residues specifically interacting with the carbonyl O of the substrate. A candidate residue for this function is Y65, located on the opposite face of the isoalloxazine ring to NADP+.
To date, there has been no structure of a BVMO with a bound substrate that clearly defines the interacting elements (7, 35, 39, 42). From the structure of PAMO in complex with the buffering agent 2-(N-morpholino)ethanesulfonic acid (MES), a low-affinity inhibitor of PAMO, Orru et al. (42) proposed previously that PAMO or BVMOs in general are perhaps mere “oxygen-activating and Criegee-stabilizing” elements that can act on any substrate that diffuses into the substrate-binding pocket. This hypothesis clearly remains challenged by other BVMO structures. The apparent plasticity of BVMOs, orchestrated through multiple domain and loop movements, seems to be key to performing the necessary series of sequential catalytic steps (flavin reduction, oxygen activation, and Criegee intermediate formation) leading to lactone formation and release. Wu and coworkers (65) introduced the concept of induced allostery in the protein engineering of PAMO and observed large domain movements that exposed and reshaped the binding pocket. The possibility that BVMO will be regarded as an intrinsic allosteric enzyme may not be far-fetched.
Supplementary Material
ACKNOWLEDGMENTS
We especially thank Allan Matte for assistance with crystallization and for useful comments on the manuscript.
Contributions from R.S. and M.C. were made possible by grant GSP-48370 from the Canadian Institute of Health Research (CIHR) awarded to M.C. X-ray diffraction data for this study were collected at the CMCF1 beamline, Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the CIHR, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. X-ray diffraction data were also collected at the Lilly Research Laboratory Collaborative Access Team (LRL-CAT) beamline, APS. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. Use of the LRL-CAT beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company, which operates the facility.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adger B, et al. 1995. Application of enzymic Baeyer-Villiger oxidations of 2-substituted cycloalkanones to the total synthesis of R-(+)-lipoic acid. J. Chem. Soc. Chem. Commun. (Camb.) 1995:1563–1564 [Google Scholar]
- 2. Adger B, et al. 1997. The synthesis of R-(+)-lipoic acid using a monooxygenase-catalysed biotransformation as the key step. Bioorg. Med. Chem. 5:253–261 [DOI] [PubMed] [Google Scholar]
- 3. Alphand V, Furstoss R, Pedragosa-Moreau S, Roberts SM, Willetts AJ. 1996. Comparison of microbiologically and enzymatically mediated Baeyer-Villiger oxidations: synthesis of optically active caprolactones. J. Chem. Soc. Perkin Trans. 1996:1867–1872 [Google Scholar]
- 4. Alphand V, Wohlgemuth R. 2010. Applications of Baeyer-Villiger monooxygenases in organic synthesis. Curr. Org. Chem. 14:1928–1965 [Google Scholar]
- 5. Aramaki H, Sagara Y, Hosoi M, Horiuchi T. 1993. Evidence for autoregulation of camR, which encodes a repressor for the cytochrome P-450cam hydroxylase operon on the Pseudomonas putida CAM plasmid. J. Bacteriol. 175:7828–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baron R, et al. 2009. Multiple pathways guide oxygen diffusion into flavoenzyme active sites. Proc. Natl. Acad. Sci. U. S. A. 106:10603–10608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beam MP, Bosserman MA, Noinaj N, Wehenkel M, Jurgen R. 2009. Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthesis pathway. Biochemistry 48:4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beecher J, Richardson P, Willetts A. 1994. Baeyer-Villiger monooxygenase-dependent biotransformations: stereospecific heteroatom oxidations by camphor-grown Pseudomonas putida to produce chiral sulfoxides. Biotechnol. Lett. 16:909–912 [Google Scholar]
- 9. Berman H, et al. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 10. Bradshaw WH, Conrad HE, Corey EJ, Gunsalus IC, Lednicer D. 1959. Microbiological degradation of (+)-camphor. J. Am. Chem. Soc. 1959:5507 [Google Scholar]
- 11. Cernuchova P, Mihovilovic MD. 2007. Microbial Baeyer-Villiger oxidiation of terpenones by recombinant whole-cell biocatalysts—formation of enantiocomplementary regioisomeric lactones. Org. Biomol. Chem. 5:1715–1719 [DOI] [PubMed] [Google Scholar]
- 12. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 13. Conrad HE, Dubus R, Gunsalus IC. 1961. An enzyme system for cyclic ketone lactonization. Biochem. Biophys. Res. Commun. 6:293–297 [DOI] [PubMed] [Google Scholar]
- 14. Conrad HE, Dubus R, Namtvedt MJ, Gunsalus IC. 1965. Mixed function oxidation. II. Separation and properties of the enzyme catalysing camphor lactonization. J. Biol. Chem. 240:495–503 [PubMed] [Google Scholar]
- 15. Conrad HE, Leib K, Gunsalus IC. 1965. Mixed function oxidation. III. An electron transport complex in camphor ketolactonization. J. Biol. Chem. 240:4029–4037 [PubMed] [Google Scholar]
- 16. Constable DJC, et al. 2007. Key green chemistry research areas: a perspective from pharmaceutical manufacturers. Green Chem. 9:411–420 [Google Scholar]
- 17. Criegee R. 1948. Die Umlagerung der Dekalin-peroxydester als Folge von kationischem Sauerstoff. Justus Liebigs Ann. Chem. 560:127–135 [Google Scholar]
- 18. De Gonzolo G, Mihovilovic MD, Fraaije MW. 2010. Recent developments in the application of Baeyer-Villiger monooxygenases as biocatalysts. Chembiochem 11:2208–2231 [DOI] [PubMed] [Google Scholar]
- 19. Dudek HM, et al. 2011. Mapping the substrate binding site of phenylacetone monooxygenase from Thermobifida fusca by mutational analysis. Appl. Environ. Microbiol. 77:5730–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dudek HM, et al. 2010. Investigating the coenzyme specificity of phenylacetone monooxygenase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 88:1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P, Cowtan K. 2004. Coot: model building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Fraaije MW, Kamerbeek NM, van Berkel WJH, Janssen DB. 2002. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 518:43–47 [DOI] [PubMed] [Google Scholar]
- 23. Gagnon R, et al. 1995. Oxidation of some prochiral 3-substituted cyclobutanones using monooxygenase enzymes: a single step method for the synthesis of optically enriched 3-substituted γ-lactones. J. Chem. Soc. Perkin Trans. 1 1995:2527–2528 [Google Scholar]
- 24. Gagnon R, et al. 1994. Biological Baeyer-Villiger oxidation of some monocyclic and bicyclic ketones using monooxygenases from Acetinobacter calcoaceticus NCIMB 9871 and Pseudomonas putida NCIMB 10007. J. Chem. Soc. Perkin Trans. 1 1994:2537–2543 [Google Scholar]
- 25. Grogan G, Roberts S, Wan P, Willetts A. 1993. Camphor-grown Pseudomonas putida, a multifunctional biocatalyst for undertaking Baeyer-Villiger monooxygenase-dependent biotransformations. Biotechnol. Lett. 15:913–918 [Google Scholar]
- 26. Iwaki H, Hasegawa Y, Wang S, Kayser MM, Lau PCK. 2002. Cloning and characterization of a gene cluster involved in cyclopentanol metabolism in Comamonas sp. strain NCIMB 9872 and biotransformations effected by Escherichia coli-expressed cyclopentanone 1,2-monooxygenase. Appl. Environ. Microbiol. 68:5671–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwaki H, et al. 2006. Pseudomonad cyclopentadecanone monooxygenase displaying an uncommon spectrum of Baeyer-Villager oxidations of cyclic ketones. Appl. Environ. Microbiol. 72:2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamerbeek NM, Fraaije MW, Janssen DB. 2004. Identifying determinants of NADPH specificity in Baeyer-Villiger monooxygenases. Eur. J. Biochem. 271:2107–2116 [DOI] [PubMed] [Google Scholar]
- 29. Kayser MM. 2009. ‘Designer reagents’ recombinant microorganisms: new and powerful tools for organic synthesis. Tetrahedron 65:947–974 [Google Scholar]
- 30. Koga H, et al. 1986. camR, a negative regulator locus of the cytochrome P-450cam hydroxylase operon. J. Bacteriol. 166:1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski RA, MacArthur MW, Moss DS, Thornston JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 33. Lau PCK, et al. 2010. Sustained development in Baeyer-Villiger biooxidation technology, p 343–372 In Cheng HN, Gross R. (ed), Green polymer chemistry: biocatalysis and biomaterials. ACS Symposium series, vol 1043 American Chemical Society, Washington, DC [Google Scholar]
- 34. Leisch H, Morley K, Lau PCK. 2011. Baeyer-Villiger monooxygenases: more than just green chemistry. Chem. Rev. 111:4165–4222 [DOI] [PubMed] [Google Scholar]
- 35. Malito E, Alfieri A, Fraaije MW, Mattevi A. 2004. Crystal structure of a Baeyer-Villiger monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 101:13157–13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meinwald J, Tufariello JJ, Hurst JJ. 1964. Fused small-ring compounds. I. Synthesis of some trans-bicyclo[3.2.0]heptanes and trans-bicyclo[4.2.0]octanes. J. Org. Chem. 29:2914–2919 [Google Scholar]
- 37. Mihovilovic MD, Muller B, Stanetty P. 2002. Monooxygenase-mediated Baeyer-Villiger oxidations. Eur. J. Org. Chem. 2002:3711–3730 [Google Scholar]
- 38. Mihovilovic MD, et al. 2005. Family clustering of Baeyer-Villiger monooxygenases based on protein sequence and stereopreference. Angew. Chem. Int. Ed. Engl. 44:3609–3613 [DOI] [PubMed] [Google Scholar]
- 39. Mirza IA, et al. 2009. Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor. J. Am. Chem. Soc. 131:8848–8854 [DOI] [PubMed] [Google Scholar]
- 40. Murshudov GN, Lebedev A, Vagin AA, Wilson KS, Dodson EJ. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 55:247–255 [DOI] [PubMed] [Google Scholar]
- 41. Murzin AG, Brenner SE, Hubbard T, Chothia C. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536–540 [DOI] [PubMed] [Google Scholar]
- 42. Orru R, et al. 2011. Snapshots of enzymatic Baeyer-Villiger catalysis: oxygen activation and intermediate stabilization. J. Biol. Chem. 286:29284–29291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Otwinowski Z, Minor W. 1997. X-ray data collection in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 44. Ougham HJ, Taylor DG, Trudgill PW. 1983. Camphor revisited: involvement of a unique monooxygenase in metabolism of 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid by Pseudomonas putida. J. Bacteriol. 153:140–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463 [DOI] [PubMed] [Google Scholar]
- 46. Reetz MT, Wu S. 2008. Greatly reduced amino acid alphabets in directed evolution: making the right choice for saturation mutagenesis at homologous enzyme positions. Chem. Commun. (Camb.) 2008:5499–5501 [DOI] [PubMed] [Google Scholar]
- 47. Reetz MT, Wu S. 2009. Laboratory evolution of robust and enantioselective Baeyer-Villiger monooxygenases for asymmetric catalysis. J. Am. Chem. Soc. 131:15424–15432 [DOI] [PubMed] [Google Scholar]
- 48. Rheinwald JG, Chakrabarty AM, Gunsalus IC. 1973. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc. Natl. Acad. Sci. U. S. A. 70:885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rother K, Hildebrand PW, Goede A, Gruening B, Preissner R. 2009. Voronoia: analyzing packing in protein structures. Nucleic Acids Res. 37:D393–D395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ryerson CC, Ballou DP, Walsh C. 1982. Mechanistic studies on cyclohexanone oxygenase. Biochemistry 21:2644–2655 [DOI] [PubMed] [Google Scholar]
- 51. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Selifonov SA. 1992. Microbial oxidation of adamantanone by Pseudomonas putida carrying the camphor catabolic plasmid. Biochem. Biophys. Res. Commun. 186:1429–1436 [DOI] [PubMed] [Google Scholar]
- 53. Sheng D, Ballou DP, Massey V. 2001. Mechanistic studies of cyclohexanone monooxygenase: chemical properties of intermediates involved in catalysis. Biochemistry 40:11156–11167 [DOI] [PubMed] [Google Scholar]
- 54. Smith SP, Barber KR, Dunn SD, Shaw GS. 1996. Structural influence of cation binding to recombinant human brain S100b: evidence for calcium-induced exposure of a hydrophobic surface. Biochemistry 35:8805–8814 [DOI] [PubMed] [Google Scholar]
- 55. Stewart JD. 1998. Cyclohexanone monooxygenase: a useful reagent for asymmetric Baeyer-Villiger reactions. Curr. Org. Chem. 2:195–216 [Google Scholar]
- 56. ten Brink GJ, Arends IW, Sheldon RA. 2004. The Baeyer-Villiger reaction: new developments toward greener procedures. Chem. Rev. 104:4105–4124 [DOI] [PubMed] [Google Scholar]
- 57. Torres Pazmiño DE, Baas B-J, Janssen DB, Fraaije MW. 2008. Kinetic mechanism of phenylacetone monooxygenase from Thermobifida fusca. Biochemistry 47:4082–4093 [DOI] [PubMed] [Google Scholar]
- 58. Trudgill PW. 1986. Terpenoid metabolism by Pseudomonas, p 483–525 In Sokatch JR, Ornston LN. (ed), The bacteria: a treatise on structure and function. The biology of Pseudomonas. Academic Press Inc, Orlando, FL [Google Scholar]
- 59. Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022–1025 [Google Scholar]
- 60. Van den Heuvel RHH, et al. 2005. Coenzyme binding during catalysis is beneficial for the stability of 4-hydroxyacetophenone monooxygenase. J. Biol. Chem. 280:32115–32121 [DOI] [PubMed] [Google Scholar]
- 61. Walsh CT, Chen Y-CJ. 1988. Enzymic Baeyer-Villiger oxidations by flavin-dependent monooxygenases. Angew. Chem. Int. Ed. Engl. 27:333–343 [Google Scholar]
- 62. Wang S, Kayser MM, Iwaki H, Lau PCK. 2003. Monooxygenase-catalyzed Baeyer-Villiger oxidations: CHMO versus CPMO. J. Mol. Catal. B Enzym. 22:211–218 [Google Scholar]
- 63. Willetts A. 1997. Structural studies and synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 15:55–62. [DOI] [PubMed] [Google Scholar]
- 64. Wilson K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1–2.4.5 In Ausubel FA, et al. (ed), Current protocols in molecular biology. John Wiley & Sons Inc, New York, NY: [DOI] [PubMed] [Google Scholar]
- 65. Wu S, Acevedo P, Reetz MT. 2010. Induced allostery in the directed evolution of an enantioselective Baeyer-Villiger monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 107:2775–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.