Abstract
Dry bubble, caused by Lecanicillium fungicola, is one of the most detrimental diseases affecting button mushroom cultivation. In a previous study, we demonstrated that breeding for resistance to this pathogen is quite challenging due to its quantitative inheritance. A second-generation hybrid progeny derived from an intervarietal cross between a wild strain and a commercial cultivar was characterized for L. fungicola resistance under artificial inoculation in three independent experiments. Analysis of quantitative trait loci (QTL) was used to determine the locations, numbers, and effects of genomic regions associated with dry-bubble resistance. Four traits related to resistance were analyzed. Two to four QTL were detected per trait, depending on the experiment. Two genomic regions, on linkage group X (LGX) and LGVIII, were consistently detected in the three experiments. The genomic region on LGX was detected for three of the four variables studied. The total phenotypic variance accounted for by all QTL ranged from 19.3% to 42.1% over all traits in all experiments. For most of the QTL, the favorable allele for resistance came from the wild parent, but for some QTL, the allele that contributed to a higher level of resistance was carried by the cultivar. Comparative mapping with QTL for yield-related traits revealed five colocations between resistance and yield component loci, suggesting that the resistance results from both genetic factors and fitness expression. The consequences for mushroom breeding programs are discussed.
INTRODUCTION
Dry bubble, caused by the fungal pathogen Lecanicillium fungicola (Preuss) Zare et Gams (formerly Verticillium fungicola) is one of the most detrimental diseases that affect cultures of Agaricus bisporus (Lange) Imbach worldwide. Undifferentiated spherical mass (dry bubble), stipe blowout, and spotty cap (17, 21) characterize L. fungicola infection, leading to yield decreases and severe economic losses. Today, control of L. fungicola relies on prophylactic measures and the use of fungicides. However, drastic specifications for the application of chemicals for pest and disease control in mushroom farms, together with the emergence of fungicide tolerance, lead to the consideration of mushroom breeding for resistance to disease as an environmentally sustainable and effective way to limit pathogen epidemics.
Potential sources of resistance to dry bubble have been identified in wild types of A. bisporus, but no complete resistance has been found (6, 15). Thus, wild strains from the Sonoran desert belonging to A. bisporus var. burnettii showed a high level of tolerance for L. fungicola infection (18). In a recent study, the analysis of segregation data for dry-bubble resistance in progeny derived from an A. bisporus var. bisporus × A. bisporus var. burnettii intervarietal cross suggested polygenic inheritance of the partial resistance to L. fungicola carried by A. bisporus var. burnettii (8). The introgression of such complex genetic resistance into new button mushroom cultivars through conventional breeding remains challenging. The recent development of molecular markers and linkage maps (7, 10) provides efficient tools to individualize the quantitative trait loci (QTL) underlying complex traits and could be very helpful for monitoring breeding programs through marker-assisted selection.
In the present study, we investigated the genetic architecture of the partial resistance to L. fungicola in A. bisporus var. burnettii through QTL mapping. The use of wild germ plasm as a source of resistance to disease in crossbreeding may reduce agronomic fitness and hamper genetic progress due to unfavorable traits. Thus, we also analyzed the relationships between the QTL of resistance to dry bubble found here and those related to other crop production traits described in the companion paper (9). Consequences for breeding management and opportunities for marker-assisted selection in the button mushroom are discussed. Also, the present results provide a deeper understanding of A. bisporus-L. fungicola interaction.
MATERIALS AND METHODS
Fungal strain and phenotypic data collection.
The fungal materials, experiments, disease trait evaluations, and statistical analyses of phenotypic data have all been previously described (8). Briefly, 89 second-generation hybrids obtained by crossing the homokaryotic offspring (Hi) of the hybrid JB3-83 × U1-7 (H) with the homokaryon U1-2 (4, 14) were evaluated for resistance to dry bubble under artificial inoculation in three independent experiments referred to here as H0419, H0420, and H0441. The phenotypic data and their statistical analyses were described previously (8). From these data, four pertinent variables were retained for QTL analyses. The two symptoms typifying dry-bubble disease, spotted mushroom and bubble formation, were treated separately, since it has been demonstrated that bubble differentiation and the appearance of necrotic lesions are two independent phenomena. The cumulative data after two flushes reflected the final level of resistance of the hybrids, and consequently, the variables PB2F and PS2F, which stand for the percentage of cumulative weight for the first two flushes of harvested mushrooms showing bubbles and spotty cap, respectively, were used. High values for these two variables expressed a high level of susceptibility to dry bubble. The latent period (LP) in days between the first picking day of healthy mushrooms and the day of appearance of the first symptom was retained for QTL analysis. The longer the LP was, the more resistant the hybrids were. The area under the disease progress curve calculated for the bubble symptom (AUDPCb), inversely proportional to the level of resistance, was also included as the fourth variable. Except for LP, data were corrected for normality by transformation, and the transformed data were used in all subsequent analyses.
QTL analysis.
The process of QTL detection is fully described in the companion paper (9). In brief, QTL analyses were based on the genetic linkage map developed by Foulongne-Oriol et al. (10) and were carried out using the composite interval mapping (CIM) module implemented in the QTL Cartographer software (2). A minimum logarithm of the odds (LOD) threshold of 2.5 was chosen to declare a QTL significant. However, in order to compare the three experiments, genomic areas highlighted for one of the experiments were also checked for significance in the other two with a LOD score cutoff value of 2.0. The map location, the LOD score, the percentage of phenotypic variation explained by the QTL, and the additive effect were given by the model. For the LP variable that did not fit well with the analysis of variance (ANOVA) prerequisite, a nonparametric test based on Kruskal-Wallis statistics was also performed. Digenic epistatic interactions between all pairs of loci were tested with a two-way ANOVA. Epistasis was inferred to be significant at a P value of <10−5. For each trait, the proportion of phenotypic variation explained by all the QTL detected (R2t) was estimated by a multiway ANOVA, including all additive effects and possible digenic epistatic effects. The QTL with overlapping LOD-1 support intervals were assumed to be a single QTL if they belonged to the same trait and to be linked or pleiotropic if they represented different traits. Likewise, the colocations between the QTL described here and QTL detected for other crop production traits (9) were also assessed by comparing confidence interval (CI) positions. The probability that QTL colocations occurred by chance was calculated according to the method of Patterson (22), using the hypergeometric probability distribution function, as follows:
where n is the number of common intervals, m is the number of matches between QTL, l is the total number of QTL found for the trait exhibiting the largest number of QTL, and s is the total number of QTL found for the other trait. One interval was defined as 20 centimorgans (cM), which corresponds to the average length of QTL confidence intervals recommended by Patterson (22).
In order to verify the additivity of QTL effects, hybrids were classified according to the number of favorable alleles present at the closest marker for each QTL detected. Regression analysis was used to estimate the disease incidence associated with an increasing number of resistance alleles.
RESULTS
QTL detection for dry-bubble resistance-related traits.
Considering all four of the selected traits (PB2F, PS2F, LP, and AUDPCb), a total of 10, 8, and 12 QTL were detected in H0419, H0420, and H0441, respectively (Table 1). QTL with an average LOD score of 3.5 were detected, and four QTL with a LOD value between 2 and 2.5 were detected. They individually explained between 7.3% (pb2f-I in H0441) and 20.4% (lp-X in H0441) of the phenotypic variance. The total R2 accounted for by all QTL ranged from 19.3% for AUDPCb in H0419 to 42.1% for LP in H0441 and averaged 28.95% over all traits in all experiments.
Table 1.
QTL for dry-bubble resistance traits detected in the second-generation hybrid progeny derived from the cross U1-7 × Jb3-83g
| Trait | H0419 |
H0420 |
H0420 |
H0441 |
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QTL | LG | Markera | Position (cM) | LODb | CI position (cM) | Parental allelec | Additive effect | R2 (%) | R2t (%) | QTL | LG | Marker | Position (cM) | LOD | CI position (cM) | Parental allele | Additive effect | R2 (%) | R2t (%) | QTL | LG | Marker | Position (cM) | LOD | CI position (cM) | Parental allele | Additive effect | R2 (%) | R2t (%) | |
| PB2F | pb2f-I | I | PR005 | 71.6 | 3.20 | 70–74 | Jb3-83 | 9.00 | 11.1 | 36.6 | 28.0 | pb2f-I | I | PR005 | 71.6 | 2.28 | 62–79 | Jb3-83 | 5.9 | 7.3 | 35.2 | |||||||||
| pb2f-II | II | AbSSR65 | 25.5 | 2.75 | 12–38 | Jb3-83 | 3.80 | 9.4 | pb2f-II | II | AbSSR65 | 25.5 | 2.3 | 12–46 | Jb3-83 | 6.12 | 7.4 | |||||||||||||
| pb2f-VIII | VIII | PR025 | 40.2 | 2.52 | 23–49 | U1-7 | 5.08 | 10.5 | ||||||||||||||||||||||
| pb2f-X | X | PR022 | 41.1 | 4.60 | 35–49 | Jb3-83 | 4.90 | 17.9 | pb2f-X | X | EAAMCAu | 35.0 | 3.31 | 29–44 | Jb3-83 | 5.70 | 11.4 | pb2f-X | X | PR022 | 43.1 | 5.00 | 34–51 | Jb3-83 | 8.72 | 18.1 | ||||
| PS2F | 30.3 | 19.4 | ps2f-I | I | PR007 | 70.5 | 2.8 | 62–77 | Jb3-83 | 6.2 | 11.6 | 20.5 | ||||||||||||||||||
| ps2f-III | III | EAGMCAg | 102.2 | 2.52 | 93–122 | Jb3-83 | 3.36 | 8.3 | ||||||||||||||||||||||
| ps2f-VIII | VIII | ECAMCAi | 70.7 | 4.80 | 57–75 | Jb3-83 | 4.80 | 17.7 | ps2f-VIII | VIII | PR031 | 68.4 | 6.20 | 58–72 | Jb3-83 | 4.80 | 19.4 | ps2f-VIII | VIII | PR031 | 70.7 | 3.27 | 62–72 | Jb3-83 | 3.32 | 13.0 | ||||
| ps2f-XIII | XIII | PR003 | 13.2 | 2.80 | 7–18 | Jb3-83 | 3.60 | 9.3 | ||||||||||||||||||||||
| LP | lp-I | I | PR005 | 71.6 | 4e | 68–76 | Jb3-83 | 6.5 | 15.70 | 22.5 | lp-I | I | PR005 | 71.6 | 4.07e | 66–77 | Jb3-83 | 2.80 | 13.8 | 28.0 | lp-I | I | PR005 | 71.6 | 2.7e | 65–80 | Jb3-83 | 2.55 | 7.6 | 42.1 |
| lp-IV | IV | PR099 | 18.2 | 3.6d | 6–21 | U1-7 | 4.80 | 10.0 | ||||||||||||||||||||||
| lp-V | V | PR093 | 5.0 | 2.78e | 0–14 | Jb3-83 | 2.93 | 9.7 | ||||||||||||||||||||||
| lp-IX | IX | PR095 | 39.0 | 2.8e | 31–51 | U1-7 | 2.31 | 9.7 | ||||||||||||||||||||||
| lp-X | X | ECGMACv | 59.4 | 2.9f | 47–67 | Jb3-83 | 3.80 | 12.3 | lp-X | X | ECGMACv | 57.4 | 2.18d | 47–67 | Jb3-83 | 2.02 | 7.5 | lp-X | X | PR022 | 45.12 | 6.34f | 39–55 | Jb3-83 | 4.2 | 20.4 | ||||
| AUDPCb | audpcb-I | I | PR007 | 68.5 | 3.70 | 64–73 | Jb3-83 | 0.76 | 15.1 | 19.3 | 27.1 | audpcb-I | I | PR007 | 68.5 | 3.70 | 61–72 | Jb3-83 | 0.91 | 12.6 | 38.5 | |||||||||
| audpcb-II | II | AbSSR65 | 25.5 | 2.70 | 8–32 | Jb3-83 | 0.48 | 8.4 | ||||||||||||||||||||||
| audpcb-VIII | VIII | EATMACy | 41.0 | 2.80 | 35–50 | U1-7 | 0.39 | 9.4 | ||||||||||||||||||||||
| audpcb-X | X | PR022 | 41.12 | 2.33 | 29–46 | Jb3-83 | 0.37 | 10.1 | audpcb-X | X | PR022 | 45.23 | 2.93 | 35–56 | Jb3-83 | 0.41 | 10.5 | audpcb-X | X | PR022 | 41.12 | 5.6 | 34–49 | Jb3-83 | 0.65 | 19.14 | ||||
Nearest upstream marker to the LOD score peak.
LOD score value at the LOD score peak.
Parental alleles that contribute to increase the trait values.
Kruskal-Wallis test; P < 0.05.
Kruskal-Wallis test; P < 0.01.
Kruskal-Wallis test; P < 10−3.
QTL detected with 2 < LOD score < 2.5 are in italics.
Consistencies between at least two experiments were found for 47% of the QTL. Five QTL were detected in the three experiments, as illustrated for PB2F in Fig. 1, and others were specific to one experiment, such as ps2f-III in H0419, lp-IX in H0420, and ps2f-I in H0441. Thus, for each trait, the total number of genomic regions involved in the phenotypic variation over all the experiments was greater than the number of QTL detected per experiment. For instance, globally, four loci were involved in PB2F trait variation, while up to three QTL were detected per experiment.
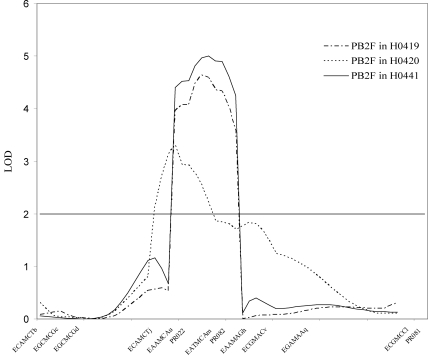
Fig 1.
Plots of LOD scores on LGX calculated by the composite interval mapping module of QTL Cartographer for the PB2F variable in the three experiments. The x axis shows the positions of the molecular marker loci (in cM [Kosambi units]) along the linkage group. The horizontal line shows the LOD-2 threshold.
For most of the QTL, the favorable allele for resistance came from the wild parent, Jb3-83. However, for some QTL, the alleles that contributed to a higher level of resistance were provided by the susceptible parent, U1-7, for instance, pb2f-VIII or LP-IX and LP-IV. No significant epistatic interaction between pairs of markers was detected, whatever the trait and the experiment.
All the QTL detected were grouped into 10 genomic regions distributed over 9 linkage groups (LG). The overlapping of QTL confidence intervals revealed clusters of QTL governing several distinct traits (Table 1 and Fig. 2). Except for the QTL on LGII, found only for PB2F in H0419, The same genomic areas were detected for AUDPCb and PB2F, with quite similar LOD scores and individual effects. On LGI, a genomic region was common to the four traits analyzed. The QTL analyses highlighted another genomic area on LGX, consistently detected in the three experiments, which was involved in PB2F, AUDPCb, and LP.
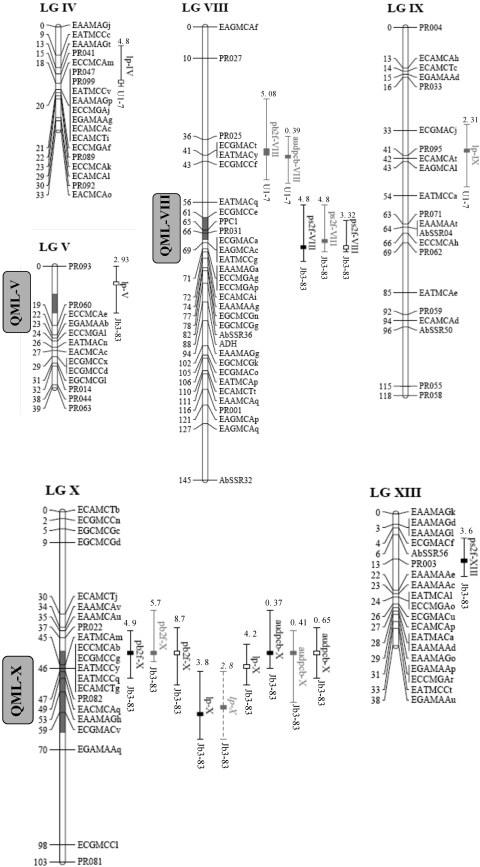
Fig 2.
Map positions of the significant QTL on the A. bisporus linkage map. Only linkage groups for which QTL were found are shown. Positions are given in cM (Kosambi units) to the right of the linkage groups. The QTL nomenclature is described in Materials and Methods. The position of the maximum LOD value is indicated by a box, in black, white, or gray for experiments H0419, H0420, and H0441, respectively. LOD-1 confidence intervals are represented by solid lines for QTL with LOD scores of >2.5 and by dashed lines for QTL with 2 < LOD score < 2.5. The R2 value of each QTL and the parental allele that contributes to the resistance are given at the top and bottom of the confidence intervals, respectively. Gray segments delimited on some linkage groups correspond to QTL for yield-related traits described in the companion paper (9).
Colocation with QTL for mushroom crop production traits.
QTL for various yield-related traits (J1, earliness; Y, yield; N, number of mushrooms per m2; W, average weight per mushroom) and cap color parameters (L, lightness) are described in the companion paper (9). Five overlapping genomic regions controlling both dry-bubble resistance (this study) and the above-mentioned crop production traits were highlighted by comparative mapping (Fig. 2 and 3). These five regions (named here QML, for quantitative multitrait loci) comprised 70% of all the QTL detected in this study. In the same way, 75% of the QTL for production traits were covered by these QML. QTL for LP colocated with QTL for earliness (QML-I, QML-V, and QML-X). The allele of the wild parent, Jb3-83, contributed to increasing the latent period and was associated with the earliest productive hybrids. QTL for PB2F or AUDPCb colocated with QTL for yield (QML-X), number of mushrooms per m2 (QML-II and QML-X), average weight per mushroom (QML-II), and earliness (QML-I and QML-X). The alleles of resistance carried by Jb3-83 were related to an increased number of small mushrooms per m2. On LGVIII, the major QTL involved in the spotty-cap symptom (PS2F) colocated with QTL for the lightness cap color parameter in the vicinity of the PPC1 locus. The brown allele brought by Jb3-83 was associated with a lower percentage of spotty cap. Other QTL for PS2F, as well as for lightness, were also found on LGXIII, but their confidence intervals did not overlap. Therefore, they were considered two independent genomic regions. One QTL for PS2F was also detected in the genomic region involved in earliness on LGI (QML-I). In most cases, these colocations found between resistance QTL and yield-related trait QTL were not fortuitous (P < 0.1), particularly between J1 and LP (P = 0.006) and between PB2F and N (P = 0.02). All these results suggested a complex network of relationships between dry-bubble resistance and yield components.
Fig 3.
Diagram summarizing QML shown in Fig. 2 and defined as overlapping genomic regions between QTL for resistance-related traits described in the present study (PB2F, PS2F, AUDPCb, and LP) and QTL for yield-related traits reported in the companion paper (9). J1, earliness (time in days from casing to first harvest); Y, yield (in kg/m2); N, number of mushrooms/m2; W, average weight/mushroom (in grams); Color, cap color parameters assessed by lightness.
Distribution and number of resistance alleles in progeny.
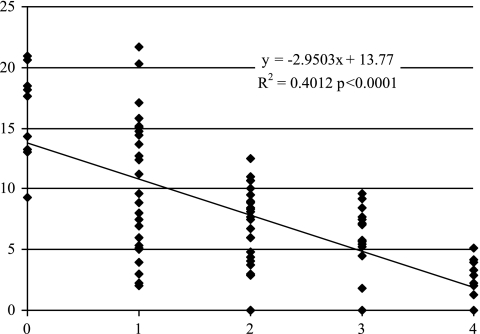
The regression of disease response assessed through the PB2F value according to the number of favorable alleles (up to 4) present at each QTL detected showed a significant (P < 0.0001) slope of −2.9 and a coefficient of determination (R2) of 0.4 for experiment H0419 (Fig. 4). Similar values were observed for experiments H0420 and H0441. The combination of the four alleles allowed the highest level of resistance. Seven hybrids have the favorable alleles at the four resistance loci. This was consistent with what was expected according to the population size and the number of segregating loci [(1/24) × 89 = 5.56]. Furthermore, among these, two are brown and thus also have the favorable allele at the major QTL involved in the spotty-cap resistance trait.
Fig 4.
Regression of PB2F values on the number of favorable resistance alleles present in the four genomic regions detected by QTL analyses in the hybrid progeny in experiment H0419.
DISCUSSION
The QTL analyses performed in the present study made it possible to refine the genetics of the dry-bubble resistance traits. Our results confirmed their polygenic inheritance, as previously suggested (8). Further, the underlying effective factors could be individualized through their map locations and their genetic-effect estimates, while such information remained inaccessible with only phenotypic data. The reliability and accuracy of the QTL detection process are demonstrated and discussed in the companion paper (9). As the phenotypic expression of dry-bubble resistance depends on environmental conditions, the use of independent evaluations is recommended to encompass the various aspects of the resistance. Some QTL were consistent over experiments, suggesting they were essential for resistance expression. Inconsistencies for other QTL could be attributed to differences in environmental conditions that would have influenced either resistance host expression or pathogen development (8). The joint analysis with other important agronomic traits provided further insights into the A. bisporus and L. lecanicillium interaction and prerequisites for multitrait marker-assisted selection in the button mushroom.
Genetic architecture of dry-bubble resistance.
All the resistance components studied presently are polygenically controlled, mainly by a few loci of small effect, as individual R2 values did not exceed 20.5%. The QTL involved in the expression of the two disease symptoms, bubbles (PB2F) and spotty caps (PS2F), were detected in distinct genomic regions, except on LGI. This confirmed that independent mechanisms are involved in the development of these two components of disease development (8).
Up to four genomic regions were involved in PB2F. Similar numbers of QTL were mentioned by Sonnenberg et al. (23), but their locations on the genome and their effects remained unknown. For three of the putative resistance loci identified for PB2F, the allele from the wild-type Jb3 conferred a higher level of resistance, as expected. However, for one locus (on LGVIII), the favorable allele came from the parental cultivar genotype. The susceptibility of U1 to L. fungicola was described as intermediate (8, 16). This genetic inheritance could explain the transgressive phenotypes toward a greater susceptibility observed previously (8).
For resistance to the necrotic-spot symptom, four genomic regions were highlighted with QTL analyses. While the resistance to bubbles was not related to cap color, the confidence interval of major QTL for PS2F surrounded the PPC1 locus, and accordingly, brown genotypes were found to be more resistant to the spotty-cap symptom than the white ones. This result was in agreement with previous observations made by Dragt et al. (6). Indeed, the weakest development of the pathogen L. fungicola, in terms of hyphal growth and sporulation ability, was found in the lesions on an A. bisporus brown strain associated with a deposition of brown pigment in the outer part of the infected cap tissues. Possibly, toxic melanins circumscribe the growth of the pathogen. This reaction reminded us of the plant hypersensitive disease response, as suggested by Berendsen et al. (3). Furthermore, for Pseudomonas tolaassii, another button mushroom pathogen that also produces brown lesions on sporophores, a resistance QTL was found closely linked to the cap color locus (20). Thus, these browning reactions caused by two different pathogens may result from a shared mechanism of resistance based on melanin biosynthesis. Genes involved in this biological pathway could be good candidates for further fine mapping. Molecular markers derived from some of these genes (tyrosinase, laccases, and polyphenol oxidase) are already located on the linkage map (10), but no relation with PS2F traits was detected. The recent release of the reference genome sequence of A. bisporus (http://genome.jgi-psf.org/Agabi_varbisH97_2) will be very helpful in further pursuing this candidate gene approach.
Four clusters of QTL governed distinct resistance traits. These colocations are in agreement with the correlations found between the traits (8). The almost systematic clustering of QTL involved in PB2F and AUDPCb was not surprising, since PB2F was one of the coordinates included in the calculation of the disease curve (8). Our results did not enable us to determine which of these variables was the most informative to describe the inheritance of dry-bubble resistance. Colocation within other resistance traits or between resistance traits and other agronomic traits contributed to explaining the interaction between L. fungicola and A. bisporus. Thus, two QTL for the latent period clustered with the other resistance traits (on LGI and LGX), suggesting that the resistance was tightly associated with the kinetics of the infection. This hypothesis was also supported by the observed colocation between several resistance QTL and earliness QTL, confirming that the relationship between resistance to dry bubble and earliness was not an artifact (8). The earliness was associated with a longer latent period and a higher level of resistance; that is, the earliest hybrids tended to later express fewer disease symptoms. This phenomenon was closely related to the developmental cycle of the hybrids. The colocations between dry-bubble resistance and other agronomic traits indicated that the most resistant hybrids produced numerous small mushrooms early. In such cases, the incidence of severe disease might be reduced due to a delay, in space and time, between the most susceptible host developmental stage and the optimal environmental conditions for L. fungicola infection, leading to escape from disease. This has been commonly reported in plant-pathogen interaction (1, 19). Therefore, we cannot exclude the possibility that, under our experimental conditions, the developmental stage and/or the earliness influenced the disease epidemics and, consequently, QTL detection. However, it is conceivable that the dry-bubble resistance resulted from both genetic factors and fitness expression. To date, our data have not made it possible to estimate which part of each contributes to the resistance. Further studies testing various time points of infection or other mapping populations and not segregating for morphological traits or earliness are needed to disentangle the relationship between resistance and fitness.
Consequences for multitrait selection in mushroom breeding.
The present study demonstrated the polygenic inheritance of partial resistance to dry bubble carried by the wild parental strain, Jb3-83. It also provides pertinent tools to manage selection programs and to further integrate markers into mushroom-breeding schemes. Our results prove that the use of wild types fairly distant from cultivated varieties as sources of resistance for strain improvement could be quite challenging. Thus, unfavorable linkage between resistance and yield components has been clearly demonstrated through QTL comparative mapping. Special emphasis should be placed on LGX, on which QTL have been detected consistently and confidently for either resistance or yield-related traits. These colocations may reflect linkage or pleiotropic effects. The significant results of the hypergeometric tests demonstrated that these colocations did not occur by chance and argued in favor of pleiotropic effects rather than linkage (22). Only fine mapping within this genomic region will help us to solve this question. Consequently, the introgression of the resistance of the wild strain into a commercial genotype would be accompanied by lower agronomic performance. An acceptable compromise may be achieved, as proposed by Sonnenberg et al. (23). The backcross breeding strategy, based on a progressive return to an elite genetic background in each generation, seemed to be rather inappropriate for several reasons. First, the introgression of several QTL simultaneously appeared to be quite difficult without a large population size (11, 12). Second, breaking linkage drag, that is, searching for recombinants between the target QTL and the undesirable trait, even with the help of molecular markers, would be quite arduous, requiring the genotyping and phenotyping of unmanageable numbers of hybrids. If the pleiotropic effect is confirmed, this appears all the more illusive. Third, in practice, the use of backcrosses with the same genetic background as recurrent parents is crippling in the button mushroom due to inbreeding depression (24). Other breeding strategies inspired by allogamous plant breeding seem more suitable to design further crosses in the button mushroom. For example, the QTL-pyramiding approach appears to be promising for multitrait selection in mushrooms. It consists of combining by crosses several QTL from multiple parental strains into a single genotype. Such genotype building is greatly facilitated by molecular markers (5). In this context, finding favorable alleles for disease resistance in commercial strains, as we have shown in the present study, is promising for mushroom strain improvement, since it will accelerate breeding progress. Furthermore, it will be interesting to identify the other possible loci involved in the resistance of the wild strain, Jb3, carried by its second nuclear component. Sonnenberg et al. (23) have demonstrated that the wild strain used for their QTL mapping carried two significant QTL in one homokaryon and five QTL in the other. Recurrent selection and multiway hybrid construction (13) could also be envisaged for button mushroom breeding.
The sum of the data available on A. bisporus, either in genetics or in genomics, makes the button mushroom a model for other mushroom crops. The use of QTL information in combination with whole-genome sequences opens new opportunities for understanding the physiological bases of the studied traits. As the interaction with L. fungicola greatly disturbed the cellular machinery and fungal development, the analysis of the genomic loci underlying the various components of resistance will provide possible tracks for functional genomics.
To our knowledge, this is the most complete and integrative analysis of crop production trait genetics in an edible basidiomycete. This approach can be readily applied to other fungal systems. Validation of the QTL identified in this study is essential prior to implementing a breeding program, since these results are based on only one population. Complementary investigations are in progress to confirm these QTL in other genetic backgrounds.
ACKNOWLEDGMENTS
The French Ministry of Agriculture is acknowledged for financial support through a CASDAR grant (project no. 2005-310). This research was also supported by INRA and CTC under joint contract.
Footnotes
Published ahead of print 13 January 2012
REFERENCES
- 1. Arraiano LS, et al. 2009. Contributions of disease resistance and escape to the control of Septoria tritici blotch of wheat. Plant Pathol. 58:910–922. [Google Scholar]
- 2. Basten CJ, Weir BS, Zeng ZB. 2004. QTL cartographer, version 1.17. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- 3. Berendsen RL, et al. 2010. Lecanicillium fungicola: causal agent of dry bubble disease in white-button mushroom. Mol. Plant Pathol. 11:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Callac P, et al. 1998. Evidence for PPC1, a determinant of the pilei-pellis color of Agaricus bisporus fruitbodies. Fungal Genet. Biol. 23:181–188 [DOI] [PubMed] [Google Scholar]
- 5. Castro AJ, et al. 2003. Pyramiding and validation of quantitative trait locus (QTL) alleles determining resistance to barley stripe rust: effects on adult plant resistance. Crop Sci. 43:2234–2239 [Google Scholar]
- 6. Dragt JW, Geels FP, Rutjens AJ, van Griensven LJ. 1995. Resistance in wild types of Agaricus bisporus to the mycoparasite Verticillium fungicola var. fungicola. Mushroom Sci. 14:679–683 [Google Scholar]
- 7. Foulongne-Oriol M, et al. 2011. Comparative linkage mapping in the white button mushroom Agaricus bisporus provides foundation for breeding management. Curr. Genet. 57:39–50 [DOI] [PubMed] [Google Scholar]
- 8. Foulongne-Oriol M, Rodier A, Rousseau T, Largeteau ML, Savoie JM. 2011. Quantitative genetics to dissect the fungal-fungal interaction between Lecanicillium fungicola and the white button mushroom Agaricus bisporus. Fungal Biol. 115:421–431 [DOI] [PubMed] [Google Scholar]
- 9. Foulongne-Oriol M, Rodier A, Rousseau T, Savoie J-M. 2012. Quantitative trait locus mapping of yield-related components and oligogenic control of the cap color of the button mushroom, Agaricus bisporus. Appl. Environ. Microbiol. 78:2422–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foulongne-Oriol M, Spataro C, Cathalot V, Monllor S, Savoie JM. 2010. An expanded genetic linkage map of an intervarietal Agaricus bisporus var. bisporus × A. bisporus var. burnettii hybrid based on AFLP, SSR and CAPS markers sheds light on the recombination behaviour of the species. Fungal Genet. Biol. 47:226–236 [DOI] [PubMed] [Google Scholar]
- 11. Hospital F. 2009. Challenges for effective marker-assisted selection in plants. Genetica 136:303–310 [DOI] [PubMed] [Google Scholar]
- 12. Hospital F. 2005. Selection in backcross programmes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:1503–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hospital F, Goldringer I, Openshaw S. 2000. Efficient marker-based recurrent selection for multiple quantitative trait loci. Genet. Res. 75:357–368 [DOI] [PubMed] [Google Scholar]
- 14. Imbernon M, Callac P, Gasqui P, Kerrigan RW, Velcko AJ. 1996. BSN, the primary determinant of basidial spore number and reproductive mode in Agaricus bisporus, maps to chromosome I. Mycologia 88:749–761 [Google Scholar]
- 15. Largeteau ML, Baars JJ, Juarez del Carmen S, Regnault-Roger C, Savoie JM. 2005. Wild strains of Agaricus bisporus: a source of tolerance to dry bubble disease, p 77–87 In Pisabarro AG, Ramirez L. (ed), Proceedings of the Genetics and Cellular Biology of Basidiomycetes VI, Pamplona, Spain. Universidad Publica de Navarra, Navarra, Spain: [Google Scholar]
- 16. Largeteau ML, Mata G, Savoie JM. 2004. Verticillium fungicola var. fungicola affects Agaricus bisporus cultivation in Mexico. FEMS Microbiol. Lett. 236:191–196 [DOI] [PubMed] [Google Scholar]
- 17. Largeteau ML, Regnault-Roger C, Savoie JM. 2007. Verticillium disease of Agaricus bisporus: variations in host contribution to total fungal DNA in relation to symptom heterogeneity. Eur. J. Plant Pathol. 118:155–164 [Google Scholar]
- 18. Largeteau ML, et al. 2004. Agaricus susceptibility to Verticiliium fungicola. Mushroom Sci. 16:515–523 [Google Scholar]
- 19. Le May C, Ney B, Lemarchand E, Schoeny A, Tivoli B. 2009. Effect of pea plant architecture on spatiotemporal epidemic development of ascochyta blight (Mycosphaerella pinodes) in the field. Plant Pathol. 58:332–343 [Google Scholar]
- 20. Moquet F, Desmerger C, Mamoun M, Ramos-Guedes-Lafargue MR, Olivier JM. 1999. A quantitative trait locus of Agaricus bisporus resistance to Pseudomonas tolaasii is closely linked to natural cap color. Fungal Genet. Biol. 28:34–42 [DOI] [PubMed] [Google Scholar]
- 21. North LH, Wuest PJ. 1993. The infection process and symptom expression of Verticillium disease of Agaricus bisporus. Can. J. Plant Pathol. 15:74–80 [Google Scholar]
- 22. Patterson AH. 2002. What has QTL mapping taught us about plant domestication? New Phytol. 154:591–608 [DOI] [PubMed] [Google Scholar]
- 23. Sonnenberg ASM, Baars JJP, Hendrickx PM, Kerrigan RW. 2005. Breeding mushroom: state of the art. Acta Edulis Fungi 12(Suppl):163–173 [Google Scholar]
- 24. Xu J. 1995. Analysis of inbreeding depression in Agaricus bisporus. Genetics 141:137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]