Abstract
Post-translational modification by ubiquitin-like proteins (UBLs) is a predominant eukaryotic regulatory mechanism. The vast reach of this form of regulation extends to virtually all eukaryotic processes that involve proteins. UBL modifications play critical roles in controlling the cell cycle, transcription, DNA repair, stress responses, signaling, immunity, plant growth, embryogenesis, circadian rhythms, and a plethora of other pathways. UBLs dynamically modulate target protein properties including enzymatic activity, conformation, half-life, subcellular localization, and intermolecular interactions. Moreover, the enzymatic process of UBL ligation to proteins is itself dynamic, with the UBL moving between multiple enzyme active sites and ultimately to a target. This review highlights our work on how the dynamic conformations of selected enzymes catalyzing UBL ligation help mediate this fascinating form of protein regulation.
Keywords: Dorothy Crowfoot Hodgkin, ubiquitin, protein dynamics, ubiquitin-like proteins
Introduction
A primary mechanism of eukaryotic protein regulation involves their post-translational modification. An abundance of such modifications exist, ranging from attachment of small chemical groups such as phosphate, acetyl, or methyl moieties to ligation of entire proteins in the ubiquitin-like family (UBLs). In higher eukaryotes, there are over a dozen structurally related UBLs that become covalently attached to other proteins, generally via an isopeptide bond between the UBL's C-terminus and a lysine side-chain on the target, although UBLs can also be ligated to protein N-termini, and Ser, Thr, or Cys residues. Numerous types of UBL modifications provide potential targets with an enormous assortment of distinct destinies. For example, ubiquitin can become linked to proteins in many different ways. In some cases, attachment of a single ubiquitin can alter a protein's subcellular localization.1–7 In other cases, proteins are modified by polyubiquitin chains, in which multiple ubiquitin molecules become covalently linked to each other.8 Some polyubiquitin chains shorten a target protein's cellular half-life, by directing degradation by the 26S Proteasome.9–18 Other polyubiquitin chains mediate binding to distinct, ubiquitin-linkage-specific recognition proteins, often to modulate signaling pathways.19–25 Ubiquitination regulates most eukaryotic pathways, including cell division,26,27 stress responses,28 DNA repair,29 and transcription.30,31 In addition to ubiquitin, other UBLs such as NEDD8, SUMO, and ISG1532–37 impart unique functional changes to their own distinct targets.
We are interested in how dedicated cascades of E1 activating enzymes, E2 conjugating enzymes, and E3 ligase enzymes direct UBLs to their targets to regulate the cell cycle, autophagy, and other processes (Fig. 1). Over 600 human proteins display sequences indicating that they are components of E1s, E2s, and E3s in UBL ligation pathways. Deregulation of these pathways through mutations, enzyme mis-expression, or aberrant behavior of UBL-modified proteins is associated with numerous pathologies, including neurogedenerative diseases, immune disorders, viral infections, and many cancers. I believe that determining the mechanisms by which enzymes transfer UBLs will be of broad importance, much like studies of protein kinases have influenced our knowledge of signaling pathways and their roles in diseases.
Figure 1.
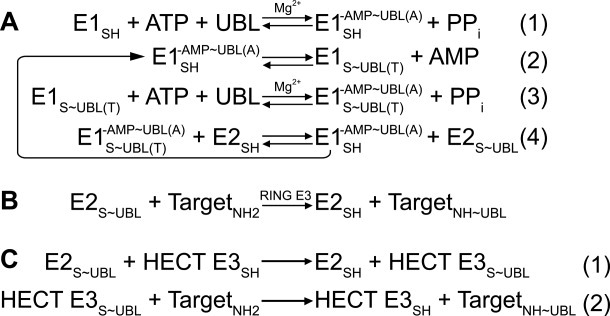
Generalized enzymatic mechanisms of UBL transfer between enzymes and ultimately to a target, based on studies of ubiquitin. ∼ refers to covalent complex; - refers to noncovalent complex. A, Initial steps catalyzed by E1. (1) E1 binds MgATP and a UBL, and catalyzes acyl-adenylation of the UBL's C-terminus. (2) E1 catalytic cysteine attacks the UBL∼AMP intermediate, to form the covalent thioester-linked E1∼UBL intermediate. (3) E1 repeats adenylation reaction on a 2nd UBL molecule, such that E1 binds 2 UBL molecules: UBL(T) is thioester-linked to E1's catalytic cystine; UBL(A) is associated noncovalently at the adenylation site. (4) Doubly-UBL-loaded E1 binds an E2. UBL(T) is transferred from the E1 cysteine to the E2 catalytic cysteine. B, RING E3s (ca. 600 in humans) enhance UBL transfer from E2 to a target. C, HECT E3s (almost 30 in humans) contain a catalytic cysteine, and (1) form a covalent thioester intermediate with a UBL prior to UBL ligation to a target lysine (2). Adapted from Ref. 38.
In addition to the broad physiological importance of UBL modifications,39,40 the biochemical pathway of UBL ligation is captivating in its own right from the perspective of basic protein science.41–43 Modification of a protein with a UBL is a highly dynamic multistep, multienzyme process, which involves transporting an entire protein—the UBL—between multiple enzyme active sites within an E1 enzyme, then transiently onto an E2 enzyme, and ultimately with assistance of an E3 onto a target protein (Fig. 1).44
Between the enormous biological significance and fascinating enzymology, the UBL field is full of stellar scientists, and I am very fortunate to be a part of this exciting arena of protein science. Because this review article stems from my receiving a 2011 Dorothy Crowfoot Hodgkin Award, I will mainly describe my lab's contributions to this vast field. The Hodgkin Award is a tremendous honor, which recognizes the discoveries of numerous trainees and staff in my lab over the years, as well as significant contributions from enriching collaborations and supportive mentors, and enthusiasm, stimulation, and encouragement from wonderful colleagues.
Much of our initial work has focused on the UBL NEDD8, which switches on a family of ∼300 ubiquitin E3s, thereby activating ubiquitin-mediated turnover of presumably thousands of proteins.32–34,45–51 As such, NEDD8 functions as a major regulator of protein homeostasis. Indeed, the NEDD8 pathway was recently hailed as a new target for anticancer therapies.52 In humans, the NEDD8 cascade involves one E1, two E2s, and few E3s and targets.34,53–61 This contrasts with the expansive, hierarchical nature of the human ubiquitin pathway, which involves two E1s, tens of E2s, hundreds of E3s, and thousands of targets with diverse biological functions.26,44,62–74 We are exploiting the minimal nature of the NEDD8 pathway to follow a UBL from beginning to end: how it enters the cascade via E1, moves from enzyme to enzyme to target, and regulates its targets. Sequences of NEDD8 E1-E2-E3 enzymes resemble those for several other UBLs, so our goals are both to understand NEDD8-specific regulation, and to identify general mechanisms of UBL transfer. This latter goal has led us to also investigate specific features of ubiquitin, SUMO, ISG15, and other UBL pathways.
Studies by ourselves and others have revealed that the structures of E1, E2, and E3 enzymes are themselves dynamic, and how plasticity of conformation—be it wholesale domain rotations, local conformational changes, or complete remodeling of portions of structures—helps mediate such a dynamic process in a specific manner. As many recent reviews have discussed the overall structures, mechanisms, and regulation of E1, E2, and E3 enzymes, here I will focus on the structural dynamics and specificity of this process, and provide some recent initial glimpses of how enzyme dynamics can be modulated to influence activity.
Dynamic assembly-line-like UBL activation by E1s
UBL structures have two regions: (1) an N-terminal globular domain adopting a β-grasp fold, which consists of a mixed β-sheet on one side and α-helix on the other, and (2) a flexible C-terminal “tail” ending with a Gly residue.75 Many UBLs are synthesized as precursors with C-terminal extensions, which undergo proteolytic maturation to expose the C-terminal Gly. In all cases, even after maturation, UBLs start out like other proteins, with relatively inert C-termini. Thus, a UBL's C-terminus must be chemically activated for covalent ligation to a target. This is accomplished in a multistep process by a UBL activating enzyme, or “E1.” In general, E1s are specific for only one or a few related UBLs.
E1s are fascinating enzymes, which efficiently select the correct UBL for its pathway, chemically activate the UBL's C-terminus, and coordinate the UBL with the correct downstream pathway.62,64,71,72,76–82 Here I refer to covalent complexes with a tilde (∼), and noncovalent complexes with a hyphen (-). The multistep reaction by which E1s activate UBLs such as ubiquitin, NEDD8, SUMO, and ISG15 is shown in Figure 1(a). First, E1 binds to MgATP and the UBL, and catalyzes UBL C-terminal adenylation. This reaction has two products: (1) the UBL covalently linked by its C-terminus to AMP via a high-energy acyl-phosphate bond, and (2) inorganic pyrophosphate. Second, the E1's catalytic cysteine attacks the UBL C-terminus to produce a transient covalent thioester-linked E1∼UBL complex, concomitant with its liberation from AMP. Third, for ubiquitin and NEDD8, and most likely also for SUMO and ISG15, the E1 next binds and catalyzes adenylation of a second UBL molecule, thus becoming loaded with two UBL molecules asymmetrically. The UBLs are bound at distinct active sites: the first [UBL(T)] is covalently bonded to the E1's cysteine by a thioester linkage. The second [UBL(A)] is associated noncovalently, at the same adenylation active site that initially bound the first UBL. Fourth, the thioester-bound UBL(T) is transferred to a catalytic cysteine on a cognate E2 conjugating enzyme. In this reaction, the E2 interacts physically with E1 to receive the UBL via a thioester transfer reaction. After generation of the E2∼UBL product, the cycle continues for the noncovalently associated UBL(A).
The E1s for ubiquitin and ISG15 exist as single polypeptides that share homology throughout their sequences. The same features are essentially split into two halves in the NEDD8 and SUMO E1s, which exist as heterodimers with one subunit homologous to the N-terminal half of a single polypeptide E1, and the other subunit homologous to the C-terminal half (reviewed in Refs.83 and84). Along with other E1 structures,85–87 our studies of the E1 for NEDD8, alone and in complexes with NEDD8, MgATP, and with NEDD8 E2s provided a structural framework for understanding many E1 functions.61,88–93 The structures revealed three independently-folded globular domains, each specifying distinct E1 activities: (1) an adenylation domain binds the UBL and MgATP and contains the adenylation reaction active site; (2) a distinct domain houses the catalytic cysteine that becomes covalently linked to the UBL C-terminus; and (3) a “ubiquitin-fold domain” (UFD) unexpectedly adopts a ubiquitin-like fold and recruits an E2.
How does the E1 architecture mediate so many binding and catalytic activities? Overall, the three E1 domains are organized so that E1 could function much like an assembly line - binding nucleotide, UBLs, and E2 at different sites - with many moving parts bringing substrates together and directing intermediates down the line for rapid and efficient production of thioester-linked E2∼UBL intermediate complexes (Fig. 2).88
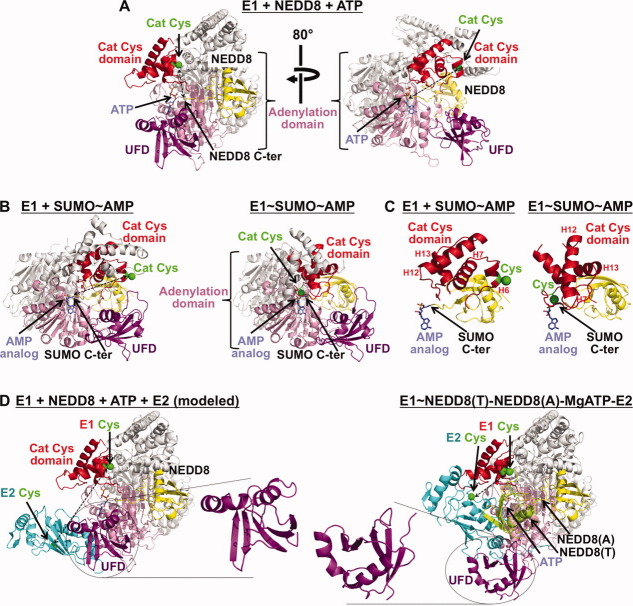
Figure 2.
Dynamic assembly-line like UBL activation by E1s. Some substructures conserved in the E1s for NEDD8, SUMO, and ubiquitin are colored as follows: part of the adenylation domain is shown in pink, part of the catalytic cysteine domain is shown in red with the active site Cys in green with its side-chain as a sphere, and the E2-binding ubiquitin-fold domain (UFD) is shown in purple. Portions of the structures not discussed in the text, undergoing lesser conformational changes, or not conserved among E1s are shown in white and light grey. A, Two views, separated by an 80° rotation around the y-axis, of an E1-NEDD8-ATP complex highlighting domains that are common among E1s and that undergo significant conformational rearrangement during the activation cycle.89 NEDD8 is shown in yellow with its C-terminus approaching the α-phosphate of ATP. A dashed line shows the large gap between the E1 Cys and the UBL C-terminus, suggesting conformational change for forming a thioester-linked E1∼UBL intermediate. B, Chemically-trapped mimics of complexes between SUMO's E1 and SUMO after the adenylation reaction (left) and a transition-state analog for forming the covalent E1∼SUMO intermediate (right).86 Remarkable structural remodeling accompanies this transition, including rotation of the catalytic cysteine domain, dismantling of some substructures such as the helix that contains the catalytic cysteine during the adenylation reaction that does not involve the catalytic Cys, and formation of new catalytic substructures such as the loop containing the catalytic cysteine that is covalently linked to the UBL's C-terminus in the right structure. The structures are oriented by superimposition on the adenylation domain of NEDD8's E1 in the right panel in A. C, Same as B, but close-up views highlighting the reorientation/structural remodeling of the catalytic cysteine domain (red) of SUMO's E1 between the structures representing adenylation of SUMO (yellow) (left) and formation of the E1∼SUMO covalent intermediate (right).86 Helices 6, 7, 12, and 13 from SUMO's E1 are labeled H6, H7, H12, and H13, respectively, at roughly the same relative locations on the two structures, except for H6 which has “melted” into a loop in the structure mimicking the E1∼SUMO∼ AMP intermediate. D, Left, docking model of NEDD8 E2 (cyan) binding to the NEDD8 E1 UFD as in the structure in A, left panel. In docking models of E2 binding to NEDD8 or SUMO E1s, E1 and E2 cysteines (green spheres) face opposite directions and are widely separated (dashed line).87,92 Right, structure of a trapped activation complex containing NEDD8's E1, two NEDD8s [one thioester-linked to E1—lime (T), one noncovalently associated for adenylation—yellow (A)], a catalytically inactive NEDD8 E2 (Ubc12, cyan), and MgATP.93 This structure revealed multiple conformational changes. First, in the double-UBL-loaded E1, the catalytic cysteine domain has returned to its original position and conformation. Second, the UFD (purple, highlighted) has undergone significant rotation such that the E2 cysteine now faces the E1.
The first E1 catalyzed reaction is centered around an ∼45-Å wide platform from the adenylation domain, with MgATP and the UBL globular domain binding at opposite ends. The UBL C-terminal tail spans the intervening distance, and is maintained in a highly ordered extended conformation by numerous UBL-specific contacts to its particular E1 [Fig. 2(a)]. Comparison of the structures of E1 alone and complexed with NEDD8(A) and ATP revealed that both the UBL C-terminal tail and the E1 undergo conformational changes to juxtapose NEDD8's C-terminus with the α-phosphate of ATP, to drive the adenylation reaction.88,89
Although the initial structures provided much insight into how an E1 selects and catalyzes C-terminal adenylation of its particular UBL, they also raised a major topological conundrum because it was unclear how the subsequent reaction could occur. Formation of the E1∼UBL thioester intermediate requires juxtaposition of the E1 catalytic cysteine and UBL C-terminus, despite their separation by an ∼30 Å gap in the E1-NEDD8-ATP structure [Fig. 2(a)].88,89 Thus, the structures implied that a conformational change must accompany the second E1-catalyzed reaction. Indeed, flexibility observed in the loops linking the E1 domains and plasticity of NEDD8's C-terminal tail supported the notion of other dramatic conformational changes driving downstream reactions in the transfer cascade.88,89 Similar properties of SUMO and ubiquitin E1s in complexes with their UBLs, revealed by structures from the Lima and Schindelin labs respectively, suggested that all these E1s utilize some common mechanisms [Fig. 2(b), left].85,87
The structural transformation required for formation of an E1∼UBL thioester intermediate, recently revealed by a brilliant chemical biology collaboration from Christopher Lima and Derek Tan, exceeded all possible expectations and in my opinion is one of the most awesome illustrations of the general beauty and exciting transformability of protein structure.86 Their crystal structure of SUMO's E1 in a chemically trapped complex mimicking formation of the E1∼SUMO thioester linkage showed a 130° relative rotation of the entire domain housing the catalytic cysteine to approach SUMO's C-terminus. Most remarkably, the structure revealed that during formation of a covalent E1∼UBL intermediate, many substructures associated with the adenylation reaction are completely dismantled, and replaced with entirely new substructures required for forming the thioester bond between E1 and the UBL C-terminus.86 For example, this includes the region around the catalytic cysteine, which plays no role in adenylation and during that reaction is in a helix distal from the UBL, but which melts into a repositioned loop for covalent linkage to the UBL [Fig. 2(b,c)].
Ultimately, the UBL bound to the E1 catalytic Cys is transferred to the next enzyme in a cascade, a member of the E2 or UBL conjugating enzyme (UBC) family. We showed that the E2 is recruited by a conserved ubiquitin-fold domain (UFD).88,92 Our structures of complexes between the ubiquitin-fold domains from NEDD8 and SUMO E1s and their respective E2s revealed that E1s recognize their cognate E2s with a concave surface formed by the UFD's twisted β-sheet.61,92,94 However, upon docking an isolated E2-UFD subcomplex onto prior structures of full-length NEDD8 or SUMO E1s,85,88,89,91 an E2 would bind the opposite side and face away from the E1's catalytic cysteine [Fig. 2(d), left]. Thus, significant conformational changes would be required for the E1 and E2 catalytic cysteines to face each other.
Several previous biochemical studies had suggested distinct structural properties for E1 and E2 forms involved in latter steps of UBL activation.64,71,72,77–79 Thus, we determined the structure of a trapped activation complex, containing NEDD8's heterodimeric E1, two NEDD8s [one thioester-linked to E1 (T), one noncovalently-associated for adenylation (A)], a catalytically-inactive NEDD8 E2 (Ubc12), and MgATP.93 The structure revealed (1) that after formation of the E1∼UBL(T) thioester linkage and with E1 also bound noncovalently to UBL(A), the catalytic cysteine-containing domain returns to its original conformation/position, and (2) that a striking E1 conformational change orients the E2. Relative to prior structures of NEDD8's E1, here the E2-associated UFD is rotated ∼120° so that the E1 and E2 catalytic cysteines faced each other [Fig. 2(d), right]. Although the structure still displays a moderate gap between the E1 and E2 cysteines, mutations hindering rotation of the structurally-observed hinge impaired E1-to-Ubc12 NEDD8 transfer, confirming the importance of UFD rotation.92 Furthermore, in the structure, the E2 also interacted with an additional E1 domain, and the NEDD8(T) that is thioester-bound to E1. Thus, the results suggested that transferring the UBL's thioester linkage between successive conjugation enzymes may induce conformational changes and alter interaction networks to drive consecutive steps in UBL cascades. This may enable transient protein-protein interactions to be established and eliminated, so that molecular handoffs can occur between successive components of UBL conjugation cascades.
Interprotein interaction dynamics of E2 enzymes
Following E1-mediated UBL-loading, E2s are generally employed by E3s, which facilitate UBL attachment to substrates by various mechanisms. Structural comparisons of E1-E292 interactions with Pavletich and coworker's prior structures of E2-E3 complexes95,96 suggested overlap between E1 and E3 binding sites on E2s. Our collaboration with Brian Kuhlman examining three distinct sets of E1-E2-E3 enzyme cascades confirmed that some E2s cannot bind their E1 and E3 partners simultaneously.97 The results suggested that E2 interactions with E1 and E3 are highly dynamic, with a common E2 surface shuttling back and forth between E1 to pick of a UBL and an E3 catalytic domain for UBL delivery to the target.92,97
This notion raises the question of how UBL transfer cascades can proceed without the E2 becoming trapped bound to either E1 or E3. One possible answer to this question is that E1s bind free E2s, but release their E2∼UBL products, which are then available to bind E3s.77,78,98 A corollary is that E3 binding to substrate and E2 might partially define mono- or polyubiqutinating enzymes. E3s with high off-rates for substrate and low off-rates for free E2 may catalyze monoubiquitination, whereas E3s with relatively low off-rates for substrate and high off-rates for free E2s may catalyze polyubiquitination. Recently, it has become clear that additional E2 interactions also influence their polyubiquitination activities. These include a variety of types of interactions between E2s and UBLs.99–102 Also, pathway-specific E2-E3 interactions outside of the catalytic domains may tether an E2 to a nonconserved E3 domain during E1-mediated UBL loading, and allowing the UBL-loaded E2's rapid return to a conserved E3 catalytic domain for UBL transfer.103–106
Dynamic conformational control of RING E3 ligases
Like other UBLs, NEDD8 ultimately becomes ligated to its targets. The best-studied targets of NEDD8 are cullin proteins, which are themselves components of ubiquitin cascades.32–34,45–51 The six best-understood human cullin proteins (CUL1-, 2, 3, 4A, 4B, and 5) are subunits of the modular, multisubunit Cullin-RING ligase (CRL) family of ubiquitin E3s. These cullins assemble with a β-strand of one of 2 RBX-family RING proteins (RBX1 and 2), and tens to hundreds of cullin-specific substrate receptor complexes to generate a family of ∼300 distinct CRLs that serve as E3s and mediate ubiquitination of thousands of targets (reviewed in Refs.107–111). Isopeptide linkage of NEDD8's C-terminus to a conserved cullin lysine activates CRL ubiquitinating activity, and it is estimated that 10–20% of all proteasomal degradation is regulated by NEDD8 activation of CRLs.52 Thus, we are interested in understanding both how NEDD8 becomes ligated to cullins, and how NEDD8 ligation activates CRL E3 activity.
We wished to investigate E3-mediated NEDD8 ligation to cullins. Studies from ourselves and others had suggested that RBX proteins are RING-type E3s for NEDD8.54–57,60,61,112 RING E3s are thought to function as adaptors promoting UBL transfer from an E2 cysteine directly to the target lysine [Fig. 1(b)].66–70,73,74 RING E3s contain a RING domain that binds an E2 surface distal from the E2 catalytic cysteine, and a protein-interaction domain that recruits substrates. RING E3s enhance UBL transfer without donating any residues directly to the catalytic center, by bridging the catalytic E2∼UBL intermediate with a target. Indeed, an intact RBX1 RING domain is required for NEDD8 ligation,54–57,60,61 and RBX1 and the NEDD8 E2, Ubc12, interact via surfaces that parallel ubiquitin RING E3-E2 interactions.60,61,112 Further credence to this notion came from our finding of distinct molecular pathways for NEDD8 ligation to different cullin proteins.61 On the basis of a series of systematic analyses defining E2 surfaces that dictate their UBL specificities, we identified a second, previously uncharacterized NEDD8 E2 (UBE2F). In collaboration with Martine Roussel, we found that in vivo Ube2F specifically pairs with Rbx2 to regulate neddylation of CUL5, whereas Ubc12-RBX1 specifically regulates NEDD8ylation of Cullins 1–4.61 Swapping the RBX1 and RBX2 RING domains swapped their preferences for Ubc12 and UBE2F, respectively, in vitro.61 Thus, RBX proteins serve as selective, classic RING-type E3s for the NEDD8 pathway, by recruiting both a particular cullin protein target and a specific NEDD8 E2.
Nonetheless, initial structures raised questions about the mechanism of NEDD8 ligation to cullins. Cullin-RBX complexes have been the subjects of intense crystallographic analyses due to their important functions as ubiquitin E3 ligases. Detailed knowledge of CRL assembly with ubiquitination targets has been established for a subset of model CRLs from studies by Nikola Pavletich's lab, in collaborations with Wade Harper, Stephen Elledge, and Michele Pagano, and by Frank Sicheri and Mike Tyers.113–121 The best-characterized CRL ubiquitin E3s are the SCFs (SKP1-CUL1-Fbox).68–70,73,74,122–125 In human SCFs, one of nearly 70 substrate-recruiting F-box proteins binds interchangeably to SKP1, which in-turn binds CUL1-RBX1.111 In addition to its role as an E3 for NEDD8 ligation to cullins, the RBX1 RING domain recruits the catalytic domains of partner ubiquitin E2s, for ubiquitin ligation to F-box-protein associated targets. A series of studies determined structural bases for F-box protein recruitment to SKP1 and for substrate recruitment to F-box proteins,113,114,116,118,120,126–128 and revealed how the ∼400-residue CUL1 N-terminal domain recruits substrate-binding adaptors (i.e., SKP1-F-box protein) and how a cullin's ∼375-residue C-terminal domain binds RBX1.113,114,116,118–121 Work led by Ning Zheng showed that SCFs and other CRLs generally adopt an overall elongated architecture, with substrate and E2-binding sites at opposite ends, separated by ∼50–60 Å.119,129 Notably, 5 structures of CUL1-, CUL4-, and CUL5-RBX1 complexes showed homologous RBX1 RING domain packing against a cullin's C-terminal region.119,129–131 However, structural models of SCF-E2 complexes raised topological challenges in terms of how CRLs could have UBL ligase activities. In the models, large gaps separated Ubc12's catalytic cysteine and the cullin's NEDD8 acceptor lysine, raising the question of how RBX1 could be an E3 for NEDD8 [Fig. 3(a)].112 Also, in the models a ubiquitin E2's catalytic cysteine an F-box protein-bound substrate are separated by 50–60 Å, raising questions as to how CRLs promote substrate ubiquitination [Fig. 3(a)].119,131
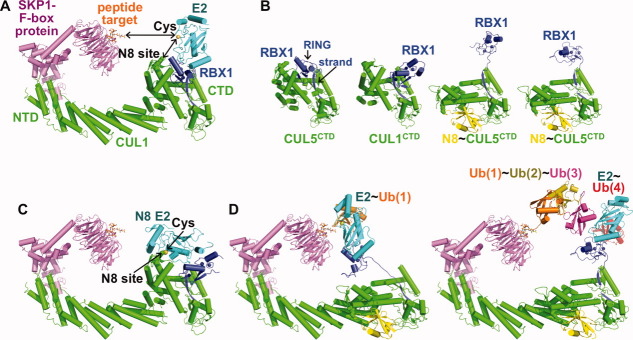
Figure 3.
Examples of RING domain rotation and conformational control of cullin-RING E3 ligases. A, Initial structural model of an SCF CRL (SKP1 and the F-box protein β-TRCP in purple, CUL1 in green, RBX1 in blue) bound to the peptide target (β-catenin phosphopeptide, orange sticks) and a generic E2 (cyan, with catalytic cysteine in yellow). Gaps between E2 cysteine and CUL1's NEDD8 acceptor lysine (“N8 site,” spheres) and to the ubiquitination target (in this case, peptide) are indicated with arrows.96,118,119,130 B, RBX1 RING domain orientational flexibility revealed from comparing structures of CUL5CTD-RBX1 (RBX1 RING in a similar orientation as in CUL1-RBX1 as in panel A),131 CUL1CTD-RBX1 with the RBX1 RING domain in a different orientation,112 and two complexes of NEDD8∼ CUL5CTD-RBX1,131 with cullin C-terminal domains (CTDs) in green, RBX1 in blue, and NEDD8 in yellow. Structures are aligned over the subdomain containing the RBX1 strand. C, Model for juxtaposition of the CUL1 NEDD8 acceptor lysine (N8 site) and NEDD8 E2 catalytic cysteine based on docking NEDD8's E2 Ubc12 (cyan with catalytic Cys in yellow) onto SCFβTRCP as in A but substituted with RBX1 from the CUL1CTD-RBX1 as in B, with the RBX1 RING domain in an orientation poised for NEDD8 ligation.92,96,112,118,119,130 D, Model for ubiquitin (orange) transfer from ubiquitin E2 (cyan) to the ubiquitination peptide target by a NEDD8-activated CRL. The E2 cysteine and peptide target are first brought into proximity via extension/rotation of the RBX1 linker (left), with polyubiquitination (ubiquitins in orange, olive, magenta and red) by a NEDD8-activated CRL, where rotation about the RBX1 linker allows multiple catalytic geometries associated with building a polyubiquitin chain.131
Consistent with the idea of an RBX conformational change for NEDD8 ligation to cullins, we performed disulfide trapping experiments showing that restraining CUL1's C-terminal region and RBX1's RING domain in close proximity specifically impairs NEDD8 transfer a cullin.112 Futhermore, our recent crystal structure of CUL1 C-terminal domain (CTD) bound to RBX1 explained how RBX proteins can be RING E3s for NEDD8 [Fig. 3(b)].112 As in prior structures, RBX1's N-terminal strand recruits the cullin substrate. However, in the new crystal, the RBX1 RING domain has undergone a relative ∼60° rigid-body rotation [Fig. 3(b)],112 and mutations hindering rotation about this hinge diminish CUL1 NEDD8ylation.131 A docking model of CUL1-RBX1-Ubc12 based on this new orientation for RBX1's RING domain revealed RBX1-associated Ubc12 positioned adjacent to a cullin's NEDD8 acceptor lysine [Fig. 3(c)].112 This work presented a RING E3-substrate complex in a conformation poised for conjugation, and showed how RING E3s can bring together an E2 and substrate for UBL transfer.112
We propose that RING domain rotation is generally associated with UBL ligation by the largest family of E3s, and that RBX1 RING domain orientation and flexibility is modulated by additional factors to achieve specific activities.60,112,131 Indeed, through an effort to resolve an apparent paradox, we found that RBX1's orientation can be harnessed to stimulate NEDD8 ligation.60 The seeming paradox was that a different protein called Dcn1 had previously been reported as being a NEDD8 E3.58,59,132 We dissected the roles of RBX1 and Dcn1, and found that they function synergistically via a “dual E3” mechanism.60 Whereas RBX1 functions like a conventional RING E3, Dcn1 functions only in the presence of RBX1's RING domain, to impart specificity for Ubc12∼NEDD8 rather than an E2∼ubiquitin complex, to reduce nonspecific Ubc12∼NEDD8 reactivity, and to bring Ubc12's active site to the cullin target presumably by favoring the rotated orientation of the RBX1 RING domain.60 The two E3s synergize to massively increase catalytic efficiency of UBL transfer to a target. On the basis of structural data and modeling, we propose that the second E3 (Dcn1) restricts the somewhat flexible RBX1 RING-Ubc12∼NEDD8 to a catalytically competent orientation.60
RBX1 RING domain flexibility can also apparently be hindered by CAND1 (Cullin-Associated and Neddylation-Dissociated 1) to inhibit NEDD8 ligation to cullins. Zheng's crystal structure of a CAND1-CUL1-RBX1 complex revealed that CAND1 binds at the interface between the C-terminal portion of CUL1 and RBX1's RING domain.130 This interaction would prevent rotation of RBX1's RING domain into an orientation compatible with NEDD8 ligation.
Finally, RBX1 RING domain orientational flexibility can also be stimulated to promote ubiquitination—and this appears to be a major function of NEDD8.131 Following NEDD8 ligation to the cullin, a CRL binds a ubiquitin-loaded E2, which is the source of ubiquitin to be transferred to a target.104,133 After ligation of the first ubiquitin, subsequent ubiquitin molecules are transferred in a processive manner to build a polyubiquitin chain.104 However, the prior structures raised the question of how an E2 active site could be juxtaposed with the substrate for ubiquitin transfer.118–120,129 It was also unclear how an RBX1-bound E2 could add ubiquitin to the growing end of a polyubiquitin chain.134 Our X-ray crystallographic, SAXS, disulfide crosslinking, limited proteolysis, and enzymatic data explain how CRLs mediate substrate ubiquitination: NEDD8 ligation to a cullin's C-terminal domain stimulates CRL ubiquitin ligase activity apparently by favoring RBX1 RING domain rotational flexibility [Fig. 3(b)].131 This imparts multiple potential catalytic geometries to an associated E2 [Fig. 3(d)].131 This notion is also supported by contemporaneous biochemical studies from Saha and Deshaies135 and Pan and coworkers,136 as well as more recent in vivo and computational studies.137,138
In addition to RBX RING domains adopting different orientations for E3 ligase functions, there is evidence of additional conformational flexibility influencing CRL activities. First, F-box proteins and other CRL ubiquitination substrate receptors may display some flexibility between their cullin- and substrate-binding regions, allowing presentation of subsrates from different orientations.120,139 Furthermore, ubiquitination substrates themselves range from having flexible regions to being completely intrinsically disordered, but with their lysines optimally spaced to receive ubiquitin from a catalytic E2.118,140 Also, some CRLs dimerize, important for E3 ligase activities.113,121,139 What roles may these additional modes of E3 and substrate conformational variability play? In collaboration with Wade Harper and Kevin White, our results suggested that structural flexibility within a dimeric CRL may allow avid interaction with structurally diverse substrates containing two or more CRL-binding motifs, with dimerization having additional roles in catalysis that have yet to be defined.139
It seems likely that we have only touched the surface in terms of understanding structural mechanisms underlying assembly, regulation, and activities of the several hundred human CRLs. I await with great anticipation many future studies elucidating additional forms of regulation exerted at the level of conformational control for CRLs and other RING E3s.
Structural dynamics of HECT E3s
In addition to RING E3s, members of the HECT family form a major class of ubiquitin ligases, with critical roles in cellular regulation. For many of the 28 human HECT E3s, disrupted function is associated with a range of diseases including cancers, high blood pressure, and retroviral infections.141–143 Thus, it is important to understand catalytic mechanisms of HECT E3s.
An ∼40 kDa C-terminal catalytic “HECT domain” participates in ubiquitin transfer in manner distinct from RING E3s [Figs. 1(c) and 4(a)]. The HECT domain binds a thioester-linked E2∼ubiquitin complex, and ubiquitin is transferred from the E2 catalytic cysteine to the HECT domain catalytic cysteine. Ultimately ubiquitin is transferred from the HECT E3 cysteine to a lysine of an associated target, or to ubiquitin linked to the target. Several mechanisms have been proposed for how HECT E3s build a polyubiquitin chain on a target, but the most favored model is that each ubiquitin is transferred sequentially from the HECT catalytic cysteine to a lysine from the previous ubiquitin in the chain.141 Different HECT E3s build polyubiquitin chains with distinct linkages. For example, E6AP links the C-terminus of one ubquitin to Lys48 in another, whereas Rsp5 builds chains via ubiquitin's Lys63.141
Figure 4.
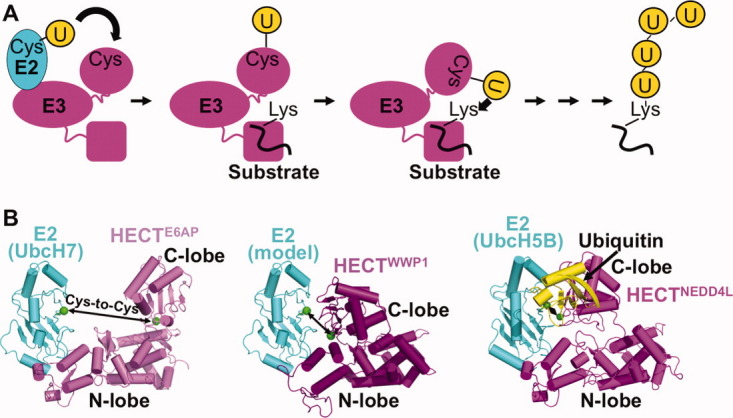
HECT E3 conformational dynamics. A, Schematic view of HECT E3 ubiquitin transfer mechanism. HECT E3s have an N-terminal region of varying sequence/structure containing protein interaction domains responsible for substrate binding and subcellular localization, and a C-terminal HECT domain. The HECT domain contains two “lobes,” an “N-lobe” that binds an E2 and a “C-lobe” that contains the catalytic cysteine. Ubiquitin is first transferred from an associated E2 to the HECT catalytic cysteine, and then to a lysine residue on the target. It is thought that conformational changes accompany many steps in HECT E3-mediated ubiquitin transfer. B, Structural insights into the first HECT E3 function of E2-to-E3 ubiquitin transfer. Shown from left to right are E6AP HECT domain (violet) bound to the E2 UbcH7 (cyan),95 WWP1 HECT domain (purple) with UbcH7 (cyan) modeled,144 and NEDD4L HECT domain (magenta) bound to UbcH5B (cyan)∼ubiquitin (yellow).145 Arrows denote gaps between the E2 and HECT domain cysteines (green). The structures were aligned by superimposing the HECT domain N-lobes.
The Pavletich and Noel labs had shown that HECT domains have two flexibly-tethered “lobes”: (1) the “N-lobe” binds part of an E2 distal from the E2 catalytic cysteine; (2) the “C-lobe” contains the catalytic cysteine, which receives ubiquitin from E2 to form a thioester-linked HECT E3∼ubiquitin complex.95,144 However, the structures showed large gaps between the E2 and HECT active sites, raising questions as to how ubiquitin could be transferred between them [Fig. 4(b)]. To address this question, we determined the crystal structure of a complex between the HECT domain of NEDD4L and the E2 UbcH5B with its active site covalently linked to ubiquitin's C-terminus (UbcH5B∼ubiquitin).145 The structure showed how an E2∼ubiquitin intermediate binds an E3, and revealed a new conformation for the HECT domain, with the E2 and E3 active sites within 8 Å of each other [Fig. 4(b)]. Extensive noncovalent interactions between UbcH5B∼ubiquitin with the HECT domain lead to an overall compact structure, with ubiquitin's C-terminus centrally localized, presumably for transfer between the UbcH5B and HECT domain active sites [Fig. 4(b)].145 It will be exciting to see in the future how HECT E3-mediated substrate ubiquitination, and generation of specific polyubiquitin chains, are driven by distinct HECT E3 conformations.
Additional twists and turns in UBL cascades
In addition to E1-E2-E3 enzyme conformational changes during UBL ligation to targets, many other aspects of UBL pathways are dynamic. Target modification by a UBL is often transient, being reversible by the action of UBL deconjugating enzymes. Interestingly the deubiquitinating enzyme A20 also contains a separate E3 ligase domain, for ubiquitin chain editing: after a polyubiquitin chain linked through ubiquitin's Lys63 is removed, another is rebuilt on the same target, but with Lys48 linkages to mediate a different function.146 Furthermore, protein conformational dynamics play major roles in establishing the functions of UBL modifications. For example, although highly dynamic, ubiquitin chains with different lysine links display different conformations and surface accessibilities, which are distinguished by linkage-specific binding partners that impart unique functionalities.20,24,147 And the best recognized function of ubiquitin—to direct proteins for degradation—relies on remarkable dynamics of the 26S Proteasome, as its subunit composition varies for regulatory purposes, and as it carries out its numerous activities such as binding and recycling particular polyubiquitin chains, unfolding and directing substrates into the catalytic chamber, and mediating substrate proteolysis (e.g., see Refs.11,13, and148–152). It is also noteworthy that two of the first reported small-molecule inhibitors of UBL ligation cascades rely on conformational flexibility of their targets, in one case the NEDD8 E1,153 and in the other a CRL E3.154 These findings inspire great hope that dynamic protein features, especially in UBL systems, will prove useful targets for improving human health. I look forward with great gusto to future studies unveiling what will undoubtedly be a dazzling array of new, unanticipated, and exciting structural mechanisms underlying the dynamic nature of protein regulation by UBLs.
Acknowledgments
The author is grateful to the pioneers in the UBL field for unveiling its exciting enzymology and vast physiological relevance; to the great colleagues for scientific stimulation and friendship; and to the mentors and all members of the lab past and present for allowing her to be a part of their ideas, discoveries, and development, and for making the journey very special and inspiring! And the author also thanks the Protein Society and Dorothy Crowfoot Hodgkin for a tremendous honor.
Editor's Note: Dr. Schulman is the co-recipient of the 2011 Dorothy Crowfoot Hodgkin Award granted in recognition of exceptional contributions in protein science which profoundly influence our understanding of biology.
References
- 1.Hicke L. Protein regulation by monoubiquitin. Nat Rev. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH, Stenmark H. Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys. 2011;40:119–142. doi: 10.1146/annurev-biophys-042910-155404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nature reviews. Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- 4.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 5.Morita E, Sundquist WI. Retrovirus budding. Ann Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 6.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A, Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985;128:1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- 9.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Introduction to intracellular protein degradation. Chem Rev. 2009;109:1479–1480. doi: 10.1021/cr900054t. [DOI] [PubMed] [Google Scholar]
- 11.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Ann Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 13.Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- 15.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson KD, Urban MK, Haas AL. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980;255:7529–7532. [PubMed] [Google Scholar]
- 17.Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. “Protein modifications: beyond the usual suspects” review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 23.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 25.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 26.Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- 27.Goebl MG, Yochem J, Jentsch S, McGrath JP, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 28.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 29.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 30.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 31.Hochstrasser M, Ellison MJ, Chau V, Varshavsky A. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 36.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 38.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Ann Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 39.Hershko A, Ciechanover A, Varshavsky A. Basic medical research award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 40.Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciechanover A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel lecture) Angew Chem Intl Ed. 2005;44:5944–5967. doi: 10.1002/anie.200501428. [DOI] [PubMed] [Google Scholar]
- 42.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture) Angew Chem Intl Ed. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 43.Rose I. Ubiquitin at Fox Chase (Nobel lecture) Angew Chem. 2005;44:5926–5931. doi: 10.1002/anie.200500995. [DOI] [PubMed] [Google Scholar]
- 44.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 45.Amir RE, Iwai K, Ciechanover A. The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. J Biol Chem. 2002;277:23253–23259. doi: 10.1074/jbc.M200967200. [DOI] [PubMed] [Google Scholar]
- 46.Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, Omata M, Tanaka K. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- 48.Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, Palombella VJ. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 51.Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci USA. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 53.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 54.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gray WM, Hellmann H, Dharmasiri S, Estelle M. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell. 2002;14:2137–2144. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 57.Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol Cell Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 59.Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 60.Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciechanover A, Elias S, Heller H, Hershko A. “Covalent affinity” purification of ubiquitin-activating enzyme. J Biol Chem. 1982;257:2537–2542. [PubMed] [Google Scholar]
- 63.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- 64.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–1581. [PubMed] [Google Scholar]
- 65.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 68.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ, Shevchenko A, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 70.Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 71.Haas AL, Bright PM. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J Biol Chem. 1988;263:13258–13267. [PubMed] [Google Scholar]
- 72.Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988;263:13268–13275. [PubMed] [Google Scholar]
- 73.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 74.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 75.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 76.Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci USA. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257:10329–10337. [PubMed] [Google Scholar]
- 78.Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- 79.Haas AL, Warms JV, Rose IA. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry. 1983;22:4388–4394. doi: 10.1021/bi00288a007. [DOI] [PubMed] [Google Scholar]
- 80.Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 81.Wilkinson KD, Smith SE, O'Connor L, Sternberg E, Taggart JJ, Berges DA, Butt T. A specific inhibitor of the ubiquitin activating enzyme: synthesis and characterization of adenosyl-phospho-ubiquitinol, a nonhydrolyzable ubiquitin adenylate analogue. Biochemistry. 1990;29:7373–7380. doi: 10.1021/bi00484a004. [DOI] [PubMed] [Google Scholar]
- 82.Pickart CM, Kasperek EM, Beal R, Kim A. Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1) J Biol Chem. 1994;269:7115–7123. [PubMed] [Google Scholar]
- 83.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Streich FC, Jr, Haas AL. Activation of ubiquitin and ubiquitin-like proteins. Subcell Biochem. 2010;54:1–16. doi: 10.1007/978-1-4419-6676-6_1. [DOI] [PubMed] [Google Scholar]
- 85.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 88.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 89.Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr, Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 90.Souphron J, Waddell MB, Paydar A, Tokgoz-Gromley Z, Roussel MF, Schulman BA. Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1. Biochemistry. 2008;47:8961–8969. doi: 10.1021/bi800604c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 93.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J, Taherbhoy AM, Hunt HW, Seyedin SN, Miller DW, Miller DJ, Huang DT, Schulman BA. Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways. PLoS ONE. 2010;5:e15805. doi: 10.1371/journal.pone.0015805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 96.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 97.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 98.Siepmann TJ, Bohnsack RN, Tokgoz Z, Baboshina OV, Haas AL. Protein interactions within the N-end rule ubiquitin ligation pathway. J Biol Chem. 2003;278:9448–9457. doi: 10.1074/jbc.M211240200. [DOI] [PubMed] [Google Scholar]
- 99.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 100.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 101.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kleiger G, Hao B, Mohl DA, Deshaies RJ. The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J Biol Chem. 2009;284:36012–36023. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 108.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nature Rev. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 109.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 110.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Calabrese MF, Scott DC, Duda DM, Grace CR, Kurinov I, Kriwacki RW, Schulman BA. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat Struct Mol Biol. 2011;18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 114.Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 115.Min JH, Yang H, Ivan M, Gertler F, Kaelin WG, Jr, Pavletich NP. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 116.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 117.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 118.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 119.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 120.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 121.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 122.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 123.Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 124.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 125.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 126.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 127.Mizushima T, Hirao T, Yoshida Y, Lee SJ, Chiba T, Iwai K, Yamaguchi Y, Kato K, Tsukihara T, Tanaka K. Structural basis of sugar-recognizing ubiquitin ligase. Nat Struct Mol Biol. 2004;11:365–370. doi: 10.1038/nsmb732. [DOI] [PubMed] [Google Scholar]
- 128.Mizushima T, Yoshida Y, Kumanomidou T, Hasegawa Y, Suzuki A, Yamane T, Tanaka K. Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc Natl Acad Sci USA. 2007;104:5777–5781. doi: 10.1073/pnas.0610312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 130.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 131.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, Ramanathan Y, Singh B. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 135.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc Natl Acad Sci USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ma B, Tsai CJ, Haliloglu T, Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure. 2011;19:907–917. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boh BK, Smith PG, Hagen T. Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J Mol Biol. 2011;409:136–145. doi: 10.1016/j.jmb.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 139.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 143.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 144.Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 145.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 147.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains—from structures to functions. Nature reviews. Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hanna J, Finley D. A proteasome for all occasions. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 154.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]