Abstract
The goal of this study was to examine fibril formation by the heterodimeric eukaryotic histones (H2A-H2B and H3-H4) and homodimeric archaeal histones (hMfB and hPyA1). The histone fold dimerization motif is an obligatorily domain-swapped structure comprised of two fused helix:β-loop:helix motifs. Domain swapping has been proposed as a mechanism for the evolution of protein oligomers as well as a means to form precursors in the formation of amyloid-like fibrils. Despite sharing a common fold, the eukaryotic histones of the core nucleosome and archaeal histones fold by kinetic mechanisms of differing complexity with transient population of partially folded monomeric and/or dimeric species. No relationship was apparent between fibrillation propensity and equilibrium stability or population of kinetic intermediates. Only H3 and H4, as isolated monomers and as a heterodimer, readily formed fibrils at room temperature, and this propensity correlates with the significantly lower solubility of these polypeptides. The fibrils were characterized by ThT fluorescence, FTIR, and far-UV CD spectroscopies and electron microscopy. The helical histone fold comprises the protease-resistant core of the fibrils, with little or no protease protection of the poorly structured N-terminal tails. The highly charged tails inhibit fibrillation through electrostatic repulsion. Kinetic studies indicate that H3 and H4 form a co-fibril, with simultaneous incorporation of both histones. The potential impact of H3 and H4 fibrillation on the cytotoxicity of extracellular histones and α-synuclein-mediated neurotoxicity and fibrillation is considered.
Keywords: protein folding intermediates, oligomeric proteins, aggregation, amyloid, domain-swapping
Brief Statement of Significance
Studying the propensity for fibrillation of different histones under varied conditions provides insights into the relative contributions of solubility, stability, and electrostatics in the context of an obligatorily domain-swapped structure. Fibril formation by histones, H3 and H4 in particular, has been implicated in the cytotoxicity of extracellular histones and α-synuclein amyloidogenesis.
The histone fold is a dimerization motif comprised of three α-helices; the long central helix of ∼30 residues is flanked on the N- and C-termini through β-loops to shorter helices of ∼10 residues. Two histone folds dimerize in a head-to-tail orientation, and the juxtaposed β-loops form a parallel β-bridge.1,2 A common function of histone-fold-containing oligomers is the formation of protein–DNA complexes.3 Example macromolecular complexes are the nucleosome core particle (NCP) and TAFs, Tata-Binding-Protein Associated Factors of the TFIID transcription regulation complex, and the SAGA histone acetyltransferase complex.4–6 The NCP, the basic repeating unit of eukaryotic chromatin, consists of ∼150 bp of DNA wrapped in 1.65 turns around eight histone proteins—two H2A-H2B heterodimers and an (H3-H4)2 tetramer. The latter is a dimer of heterodimers that associate via a four-helix bundle formed by the C-termini of the H3 and H3′ histone folds (Fig. 1). The parallel β-bridges formed upon dimerization make significant interactions with the DNA backbone.2,7
Figure 1.
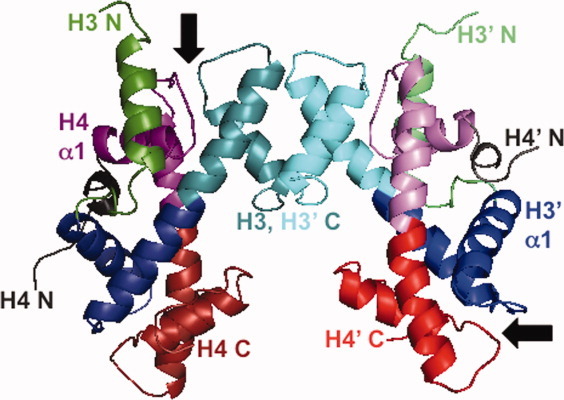
3D domain swapping and the structure of histone oligomers. Ribbon diagram of the (H3-H4)2 tetramer derived from the structure of the nucleosome core particle (1AOI.pdb).2 The H3-H3′ four-helix bundle tetramer interface is at the top of the figure. Some regions of the poorly structured N-terminal tails have been omitted for clarity. The H3 monomers are shown in green (N-terminal tail and helix) and blue to cyan (histone fold); the H4 monomers are shown in gray (N-terminal tail and helix) and purple to red (histone fold). The two of the four β-bridges are indicated by arrows. The protein structure was rendered with the program Mac-PyMol (DeLano Scientific LLC, Portland, OR).
The four eukaryotic histones of the NCP share a common dimerization fold with very similar backbone structures that overlay with root mean square deviation (RMSD) values of 1–2 Å, despite sequence conservation of only 5–10%.8 These histones also contain polypeptide elements that extend beyond the canonical three-helix histone fold. All four histones have poorly structured N-terminal tails that are the sites of regulatory post-translational modifications such as acetylation, methylation, and phosphorylation.9,10 H3 and H2B contain additional N- and C-terminal helices, respectively, while H2A and H4 contain relatively unstructured C-terminal tails. In contrast to the high sequence divergence between the four eukaryotic histones, the individual histones exhibit high sequence identity across species. For example, the H3 histones of Xenopus laevis, used in this study, bovine (used in a previous study of fibrillation)11 and humans are 98% identical, with two differences in 135 residues, and the H4 histones from the three species are identical across 102 residues.
Beyond the impact of histone equilibrium and kinetic folding properties on NCP function, the histones are an interesting model system to investigate protein folding and misfolding. Firstly, the histone fold is a domain-swapped structure. Secondly, histones with similar structures fold by different kinetic mechanisms.12–15
3D domain swapping describes oligomeric structures where an element of structure (ranging from a single α-helix or β-strand to an entire globular domain) is exchanged between monomers such that the swapped segment makes intermolecular interactions in the dimer that are similar to the intramolecular interactions found in the closed monomer.16–19 Domain swapping has been proposed as a mechanism for the evolution of oligomeric structures, and is a plausible hypothesis for the evolution of the dimeric histone fold from a single motif.20 The canonical three-helix histone fold may have arisen by duplication and then fusion of two tandem helix-strand-helix (hsh) motifs (Supporting Information Fig. 1). Because the hsh motifs are fused directly to each other in the long central α-helix, there is no longer a linker or hinge, and thus, the elongated histone fold must dimerize in an obligatorily 3D domain-swapped “closed” structure. As illustrated in Figure 1, there is pseudo-symmetry between the interactions of the N-terminal half of H3 (blue) and the C-terminal half of H4 (red) compared to the N-terminal half of H4 (purple) and the C-terminal half of H3 (cyan).
In addition to mediating appropriate oligomerization, 3D domain swapping has been implicated in the propensity of proteins to aggregate into highly ordered structures such as crystals,21 polymers,22 fibers,23 and fibrils.24–26 Of particular interest is the possible connection between domain swapping and formation of amyloid or amyloid-like fibrils (for review see Ref.27). Amyloid fibrils, formed in vivo by specific proteins, are associated with over 20 protein deposition diseases, such as the Aβ peptide in Alzheimer's, α-synuclein in Parkinson's, β2-microglobulin in dialysis-related amyloidosis, and cystatin C in amyloid angiopathy. Many proteins not associated with amyloidogenic diseases can be induced to form fibrils in vitro, including folded, globular proteins, and intrinsically disordered proteins (for review see Refs.28,29). Despite biological differences, amyloid and amyloid-like fibrils share common features, including a cross-β spine structure.30
It has been proposed that open or “run-away” domain swapping may be a mechanism for protein aggregation, particularly fibrillation.27 The closed dimer of the histone fold is a reciprocal swap, with no open domains lacking a partner. In an open or run-away domain swap, the domains are not exchanged in a reciprocal manner; instead there are open domains that propagate oligomerization and aggregation to satisfy unpaired domains. Runaway domain-swapping leading to amyloid-like fibrils has been observed for T7 endonuclease I26 and an engineered RNase A variant with a highly amyloidogenic insert of 13 Gln residues.25 Some amyloidogeneic proteins adopt domain-swapped structures that have been implicated as precursors for fibril formation, for example, the human prion protein,31,32 β-2 microglobulin,33,34 human stefins A and B, also known as cystatins A and B,35,36 and cystatin C.24,37
A common theme in the conversion of globular proteins to amyloid-like fibrils is the population of a partially unfolded species; the unfolding can be relatively local with most of the protein retaining native structure or more global unfolding to a stable equilibrium or transient kinetic intermediate.38 Fibril formation is generally enhanced by increasing the population of these partially unfolded states, often by destabilizing the native state by mutation or altering the solvent composition, pH, or temperature.
The goal of this study was to exploit the different kinetic folding mechanisms of eukaryotic and archaeal histones to assess the impact of kinetic intermediates on the propensity for the domain-swapped histone fold to form fibrils. The homodimeric hPyA1 archaeal histone from the hyperthermophilic Pyrococcus strain GB-3a (optimal growth at 95°C) folds by a two-state kinetic process at 25°C, with no detectable transient intermediates.13 The hMfB homodimer from the thermophile Methanothermus fervidus (optimal growth at 83°C) shares 85% similarity (57% identity) with hPyA1, but transiently populates a partially folded monomeric species during folding. In contrast, the eukaryotic H2A-H2B and H3-H4 heterodimers populate monomeric and dimeric kinetic intermediates during folding.12,14,15
Previous studies have shown that eukaryotic histones can form amyloid-like fibrils.11,39 These fibrillation studies used a mixture of calf thymus histones containing H2A, H2B, H3, and H4. Thus, the fibrillation propensity of the individual histone sequences was not assessed. This report employs recombinant Xenopus laevis histones and examines the fibrillation of individual histone subunits as well as the eukaryotic heterodimeric pairs.
Results
Fibrillation propensity of different histones
The previously published studies using calf thymus core histones11 served as a guide in choosing conditions to screen for fibril formation: pH 2 and pH 5 with 1M NaCl at room temperature. In these solvent conditions (at 37°C), fibrillation of the bovine histone mixture was detected within 24 h and complete by 60 h. In the current study, the propensity of individual recombinant histones to form fibrils was assessed by starting with histone monomers unfolded in 10 mM HCl with rapid dilution into fibrillation conditions. Fibril formation was detected by the time-dependent increase in ThT FL and an SDS-polyacrylamide gel electrophoresis (PAGE) assay for protein precipitation. Briefly, following centrifugation, the fraction of the protein in the supernatant and pellet were quantitated as a function of time using SDS-PAGE and densitometry (Fig. 2).
Figure 2.
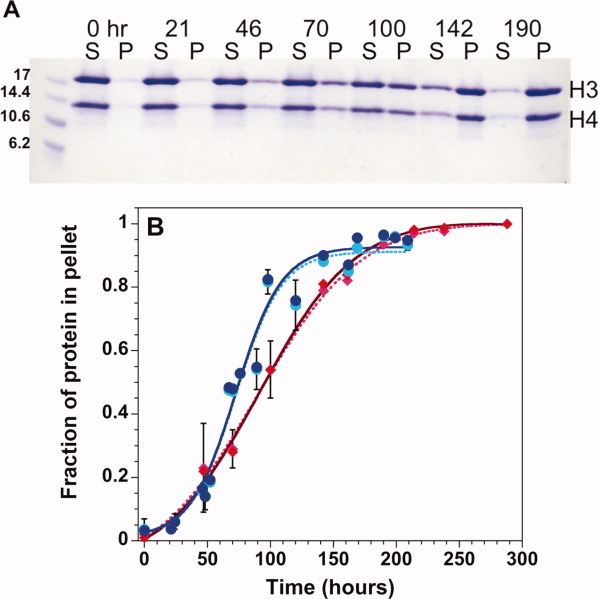
Precipitation gel assay for histone fibrillation. A: Coomassie Blue stained SDS-polyacrylamide gel monitoring the time-dependence of precipitation of H3 + H4 monomers initially unfolded in 10 mM HCl. S, supernatant, P, pellet. B: Densitometer quantitation of the fraction of protein in the pellet for H3 (lighter symbols, dotted line) and H4 (darker symbols, solid line) starting from unfolded H3 + H4 (red diamonds) or from folded tetramer at pH 7.2 (blue circles). The lines represent fits of the data to Eq. (1). Error bars at one standard deviation from multiple experiments are shown or are smaller than the size of the data points. Final conditions: 1M NaCl, 50mM acetic acid/sodium acetate, and 50 mM phosphoric acid/NaH2PO4, pH 5, 23°C and 30 μM each of H3 and H4 monomers. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The unfolded H3 and H4 monomers in isolation form fibrils when shifted to pH 5, and H4 also forms insoluble fibrils at pH 2. Fibrillation of H3 and H4 together was also examined, using two initial conditions: (1) an equi-molar mixture of H3 and H4 monomers unfolded in 10 mM HCl (denoted H3 + H4); and (2) (H3-H4)2 tetramer in a folded state at pH 7.2 (denoted H3-H4). At pH 7.2, the (H3-H4)2 tetramer and H3-H4 dimers are in equilibrium; however, at 200 μM monomer before dilution, the tetramer should be the predominant species.40 At pH values of 5 or lower, the predominant oligomeric state of H3-H4 is the heterodimer.41 Both combinations of H3 and H4 (initially unfolded or folded) readily form fibrils when shifted to acidic conditions with 1M NaCl, with greater efficiency at pH 5 than pH 2 (Table I). However, despite a common fold, other histone dimers exhibited markedly different fibrillation propensities.
Table I.
Representative t50 Values for Fibril Formation by the H3 and H4 Histones
| Conditionsa | Detection methodb | Unfolded H3 | Unfolded H4 | Unfolded H3 + H4 | Folded (H3-H4)2 |
|---|---|---|---|---|---|
| pH 2, 1M NaCl | ThT FL | ∼16 days | <10 min | <6 h | <6 h |
| Gel | Not detectedc | 40 ± 10 min | ∼20 days | ∼5 days | |
| pH 5, 1M NaCl | ThT FL | ∼1 h | <10 min | 26 ± 4 h | 50 ± 5 h |
| Gel | 14 ±1 h | 20 ± 5 min | 98 ± 9 h | 78 ± 7 h |
Final conditions: 50 mM acetic acid/sodium acetate and 50 mM phosphoric acid/NaH2PO4 at 23°C. Protein concentrations were 30 μM monomer for unfolded H3 and H4 (0.34–0.46 mg/ml) or 30 μM of each monomer for unfolded H3 + H4 and folded (H3-H4)2 (0.79 mg/ml). Initial unfolded conditions were 10 mM HCl. Initial conditions for the folded tetramer were 200 mM KCl, 20 mM potassium phosphate, pH 7.2. The values represent the average kinetic response observed in three to five independent replicate experiments. For conditions where the data were collected with sufficient time resolution to determine a t50 value with 10% accuracy for each replicate, the errors at one standard deviation are indicated.
Fibrils were detected by Thioflavin T fluorescence (ThT FL) and a centrifugation/gel-based assay.
After three months, no precipitable fibrils were observed.
For the archaeal homodimers hMfB and hPyA1, no enhancement of ThT FL or fibril precipitation could be detected after incubation up to seven days at pH 5 in 1M NaCl and 60 μM monomer. Additional solvent conditions and cosolutes were explored to promote fibrillation (Supporting Information Table S1). Three general approaches were employed: (1) addition of macromolecular crowding agents, such as sucrose, Ficoll, and poly-ethylene glycol; (2) modulation of ionic strength, type of salt, and pH; and (3) addition of destabilizing cosolutes, such as urea or ethanol. None of these approaches were successful in inducing fibril formation.
The propensity for fibrillation was also examined for the H2A and H2B monomers in isolation and in combination. At pH 5 and 1M NaCl, minor increases in ThT FL were detected: 1.2-fold for H2B, and 2-fold for H2A and H2A + H2B. These changes were small compared to the increases observed for H3 and/or H4 (see below), and no detectable precipitates were observed, even after a month. A series of additional conditions were surveyed for the combination of H2A + H2B (Supporting Information Table S1). Despite small, variable increases in ThT FL, no conditions produced aggregated protein precipitates.
There is clearly no correlation between the population of kinetic folding intermediates and fibril formation. The eukaryotic heterodimers, H2A-H2B and H3-H4, share a multi-state folding pathway,12,14 while hMfB and hPyA1 fold by three- and two-state mechanisms, respectively.13 Yet only H3-H4 readily forms fibrils.
There is no apparent relationship between fibrillation and decreased stability. The pH dependence of the thermal unfolding of H2A-H2B and H3-H4 has been reported, although at lower ionic strengths.41,42 Between pH 4.5 and 5.5, the mid-point of the thermal unfolding transition, TM, for the H3-H4 heterodimer was 50–60°C, depending on the protein concentration, while the TM of H2A-H2B was 10–15°C lower. Thus, H3-H4 is more stable, but forms fibrils much more readily than H2A-H2B or the individual H2A and H2B monomers which are only partially folded even under stabilizing conditions.43 At comparable conditions, the TM values for hMfB and hPyA1 are >80°C.44 Various reports have shown that histones are stabilized by increasing ionic strength.11,41,42,44,45 Yet, decreasing ionic strength (thus stability) did not facilitate fibrillation of hMfB, hPyA1, or H2A + H2B (Supporting Information Table S1) and impeded the fibrillation of H3-H4 (see below). A similar lack of correlation between stability and fibrillation was observed with wild-type and chimeric human stefins.46
The propensity of the histones to form fibrils does correlate with solubility. It was previously known that archaeal histones are highly soluble; during purification, hMfB and hPyA1 remain in solution at 60–70% ammonium sulfate at neutral pH.13 The solubility of the histone monomers and dimers were determined at pH 5 with 0.5M NaCl (Table II), conditions where H3 and H4 monomers did not form fibrils. Under these conditions, the hMfB and hPyA1 monomers as well as H2A + H2B and H3 + H4 associate to form native dimers,41,42,44 with far-UV circular dichroism (CD) spectra comparable to those observed under typical folding conditions (pH 7.2, 200 mM KCl, data not shown). The individual H3 and H4 monomers were difficult to solubilize from a lyophilized state, forming a viscous, cloudy solution with very little soluble protein. Nevertheless, upon dilution from 10 mM HCl to fibrillation conditions, the individual H3 and H4 monomers are initially soluble to at least 30 μM; thus the fibrillation studies described in this report were initiated from a soluble species. In contrast to H3 and H4, the individual H2A and H2B monomers are soluble to mM levels. The H2A-H2B heterodimer and the archaeal histone homodimers also exhibit mM solubility. However, even as a folded heterodimer, H3-H4 is ∼10-fold less soluble than the other histones. Apparently, the relatively low solubility of the H3 and H4 histones, as monomers or heterodimer, facilitates the formation of fibrils.
Table II.
Solubility of Eukaryotic and Archaeal Histones at pH 5, 500 mM NaCla
| Monomers (mM monomer) | ||||
| H3 | H4 | H2A | H2B | |
| negligible | negligible | 10. | 3.6 | |
| Dimers (mM monomer) | ||||
| H3-H4 | ΔN(H3-H4) | H2A-H2B | hMfB | hPyA1 |
| 0.3 | 0.95 | 7 | 5 | 3.7 |
Conditions: 50 mM acetic acid/sodium acetate and 50 mM phosphoric acid/NaH2PO4, 23°C.
Characterization of the histone fibrils by different methods
To confirm that the precipitates identified in the gel-based assay were amyloid-like fibrils, additional methods were employed to characterize the aggregates formed by H3, H4, H3 + H4, and the H3-H4 tetramer, namely ThT FL, FTIR and CD spectroscopies, and electron microscopy.
A hallmark of amyloid-like fibrils is the binding of the benzathiole dye Thioflavin T, resulting in a shift in the excitation and emission maxima and a large increase in fluorescence intensity.47 ThT is generally more selective for fibrils than prefibrillar soluble oligomers or protofibrils, although there are examples of ThT binding to soluble oligomers.48 A complication is that ThT can also bind in hydrophobic cavities of folded proteins.47
As shown in Figure 3, ThT exhibits very little FL in the absence of protein or in the presence of urea-unfolded H3 and H4, conditions where fibril formation is not detected. As an additional control, ThT FL was monitored in the presence of folded (H3-H4)2 at pH 7.2. Significant ThT FL was observed at t = 0, with excitation and emission maximum of 430 and 482 nm, respectively, followed by a slow increase in FL (1.6-fold) that was largely complete by 19 h (Fig. 3). However, there was no indication of fibril formation by precipitation or turbidity.
Figure 3.
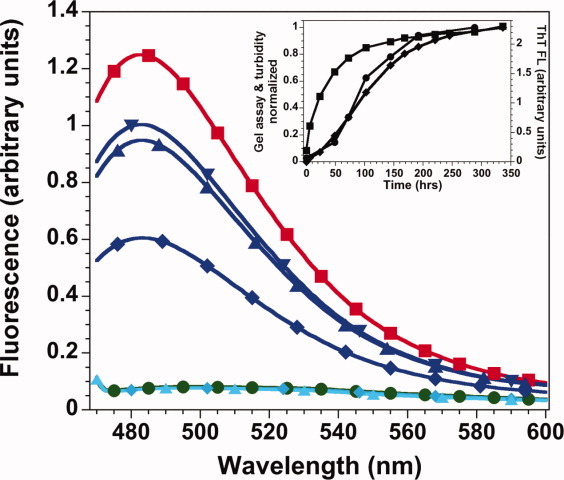
ThT fluorescence spectra. Low intensity, overlapping spectra: ThT alone, green circles; H3-H4 at pH 7.2 in 6M urea (light blue symbols) at t = 0, diamonds, and t = 66 h, triangle. Spectra with (H3-H4)2 at pH 7.2 (dark blue symbols), t = 0, diamond; t = 19 h and t = 66 h, upward and downward triangles, respectively. Spectra of H3-H4 at pH 5, t = 0, red squares. Inset: representative kinetics of fibril formation by H3 + H4. The same sample was monitored by ThT FL, squares, gel precipitation, circles, and turbidity at 500 nm, diamonds. Conditions: 30 μM each of H3 and H4 monomers with 20 μM ThT at pH 7.2, 0.2M KCl, 20 mM potassium phosphate, 0.1 mM EDTA (standard folding conditions) or pH 5, 1M NaCl, 50 mM acetic acid/sodium acetate, and 50 mM phosphoric acid/NaH2PO4, 23°C. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Under fibrillation conditions (1M NaCl, pH 5), higher initial ThT FL was observed in the presence of H3-H4 (Fig. 3) as well as H3 or H4 alone (data not shown). The time-dependent increase in ThT FL for H3-H4 was much slower at pH 5 (Table I) than at pH 7.2 (native conditions). During fibrillation, the ThT FL intensity increased 3- to 4-fold for the individual monomers and 9- to 14-fold for H3 + H4 and H3-H4. The FL emission maxima in the presence of fibrils was 482 nm, but the excitation maxima shifted to 450 nm, as observed for most fibrils.47 Similar ThT FL was observed when fibrils were formed in the presence of ThT or when the dye was added after fibrillation. In summary, there is a substantially greater increase in ThT FL under fibrillation conditions than observed for the native dimer, demonstrating that ThT can bind to the native histone fold, but there is enhanced binding as fibrillation proceeds.
At pH 2 and 5, the rate of increasing ThT FL was more rapid than the detection of protein precipitates by the gel-based assay or turbidity at 500 nm (Table I; Fig. 3 inset). In most conditions, the increase in ThT FL was complete before substantial precipitation was detectable, except for the combinations of H3 and H4 at pH 5. The difference in kinetic response between ThT FL and precipitation suggests that ThT recognizes a soluble protofibril state of the histones. Thus, ThT FL supports the assessment of the precipitated species as fibrils. However, because of the relatively slow binding of ThT to the native fold and the apparent binding to a soluble fibril state, ThT FL was not an optimal probe for monitoring fibrillation kinetics.
FTIR spectroscopy is well-suited for characterization of the secondary structural content of fibrils. The amide I bands between 1700 and 1600 cm−1 reflect the contribution of α-helices, random coil, and different β-sheet structures (parallel or anti-parallel).48–50 The insoluble pellets formed by the individual H3 and H4 histones and those formed from folded tetramer and unfolded H3 + H4 were examined by FTIR and compared to that of the native tetramer [Fig. 4(A)]. The FTIR maxima for the folded tetramer is ∼1650 cm−1, with a broad shoulder extending down to 1620 cm−1, consistent with helical and random coil structure. The four fibril spectra have two peaks, one centered around 1645 cm−1, and a second peak between 1630 and 1625 cm−1. This latter peak corresponds to β-sheet structure as expected for an amyloid-like fibril. The spectra of the folded tetramer and fibrils in Figure 4 are similar to those that have been reported previously for a mixture of calf thymus histones.11 The spectra of the fibrils from individual H3 and H4 histones are quite similar to each other. In contrast, the fibrils from unfolded H3 + H4 and from folded tetramer have different ratios of the 1645 cm−1 and 1625 cm−1 maxima, suggesting there may be subtle differences in the fibrillar structure or the segments of protein not incorporated into the cross-β fibril structure. Similar variation in FTIR spectra of histone fibrils has been observed in the studies with calf thymus histones.11,39
Figure 4.
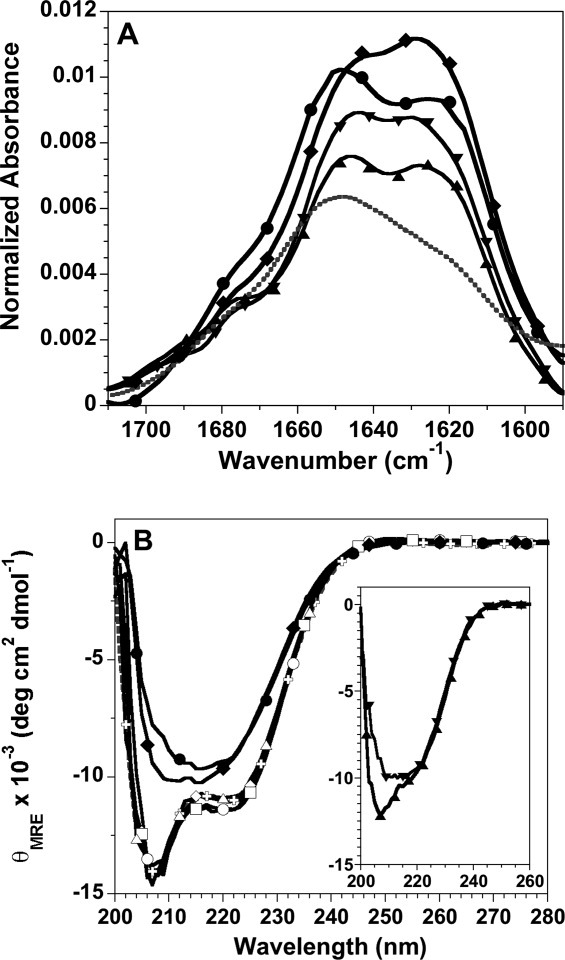
Spectroscopic characterization of histone fibrils. A: FTIR spectra of fibrils formed at pH 5: H3 fibrils, upward triangle; H4 fibrils, downward triangle; fibrils initiated from unfolded H3 + H4, diamonds; fibrils initiated from folded tetramer, circles. For comparison, the spectrum of folded tetramer at pH7.2 is shown as a gray, dotted line. The buffer background spectrum has been subtracted from the protein spectra. B: Far-UV CD spectra normalized to mean residue ellipticity: tetramer at pH 7.2, gray crosses and dotted line; tetramer at pH 5, t = 0 and 196 h, open and closed circles, respectively; initially unfolded H3 + H4 at pH 5, t = 0 and 196 h, open and closed diamonds, respectively; t = 0 spectra for tetramer and H3 + H4 at pH 2, open squares and triangles, respectively. Inset: t = 0 spectra for the individual H3 and H4 monomers, upward and downward triangles, respectively. Fibrillation conditions: 1M NaCl, 50 mM acetic acid/sodium acetate and 50 mM phosphoric acid/NaH2PO4, pH 5 or pH 2, 23°C and 30 μM of the individual H3 and H4 monomers or 30 μM of each monomer for tetramer samples and H3 + H4 initially unfolded in 10 mM HCl.
Far-UV CD is less sensitive than FTIR to β-sheet structure; nonetheless, CD is useful for assessing the degree of helical secondary structure in the species initially populated in fibrillation conditions and monitoring the change in secondary structure during fibril formation [Fig. 4(B)]. The equi-molar mixture of H3 + H4, unfolded in 10 mM HCl, folds rapidly upon dilution into 1M NaCl at pH 2 and pH 5; the initial species under fibrillation conditions exhibit far-UV CD spectra, and thus α-helical content, that are very similar to those of the folded tetramer. The far-UV CD spectra of the H3-H4 oligomer are superimposable at pH 5 and pH 2, where the dimer is the predominant species, and exhibit slightly greater ellipticity at 222 nm compared to the H3 + H4 spectra. The similarity in α-helical content of the (H3-H4)2 tetramer and H3-H4 dimer has been reported previously.12 Therefore, the initial species that form fibrils at pH 2 and pH 5 contain a similar amount of helical secondary structure, regardless of whether the precursory state was unfolded monomers or the folded tetramer. Upon incubation, the far-UV CD spectra change from that expected for an α-helical protein, with double minima at 222 and 208 nm, to spectra that are indicative of β-sheet structure with a broad minimum centered around 216 nm. The time dependent decrease in ellipticity at 222 nm is comparable to the kinetics observed by the precipitation gel-based assay (data not shown). Like the FTIR spectra, the far-UV CD spectra suggest that there may be subtle structural differences in the fibrillar structures formed by folded and unfolded mixtures of the H3 and H4 histones.
The far-UV CD spectra of the H3 monomer at the initiation of fibrillation at pH 5 indicate a combination of random coil and helical structure [Fig. 4(B) inset]. In contrast, the spectra of the H4 monomer are very similar to that of the fibril formed by H3 + H4, suggesting that the isolated H4 monomer adopts β-sheet structure under fibrillation conditions. Pre-formed β-structure in the initial soluble species may contribute to the unusually rapid fibrillation of the H4 monomer (Table I). After fibrillation is complete, the far-UV CD spectra of the fibrils formed by H3 and H4 are comparable to those shown for the combination of H3 and H4 (data not shown).
The macromolecular structure of the histone precipitates was also evaluated by transmission electron microscopy (TEM). Supporting Information Figure 2 shows the structures formed by shifting the (H3-H4)2 tetramer to pH 5 fibrillation buffer conditions. Similar EM images were obtained for fibrils formed by the tetramer at pH 2 and by initially unfolded H3 + H4 at pH 5 and pH 2 (data not shown). The TEM images show that the histone precipitates are not amorphous aggregates. However, the structures do not exhibit the relatively straight morphology of classical amyloid-like fibrils, such as reported previously for fibrillation of the mixture of bovine histones.11 While the fibril diameter is comparable, the morphology of the histone fibrils is more worm-like. Similar EM images have been reported for “immature fibrils” formed by human H2A at elevated temperatures,51 fibrils formed from individual and combinations of calf thymus histones,52 and the small, indistinct fibrils formed at pH 4 by the repeat domain of Pmel17 involved in melanosome structure.53
The H3-H4 heterodimer contains the helical histone fold as well as the poorly structured N-terminal tails that are the sites of a variety of post-translational modifications that regulate chromatin function.9,54 Given that both folded, globular, and intrinsically disordered proteins can form fibrils,28,38 it is of interest to determine whether the folded or disordered regions of H3 and H4 contribute to the fibril structure. Fibrils formed by H3, H4, H3 + H4, and folded tetramer at pH 5 were subjected to limited proteolysis and then analyzed by SDS-PAGE (Supporting Information Fig. 3). The gel mobility of the H4 proteolysis product was identical whether it was from an H4 fibril or a fibril formed in the presence of H3; similarly, the H3 products were the same whether H4 was present or not. This indicates that the same regions of polypeptide were involved in fibrillar structure whether or not both monomers were present. For ease of analysis, the proteolysis products of fibrils from the individual histones were used for further characterization. N-terminal sequencing showed that the H3 proteolysis products started at residue 25, retaining ∼20 residues of the N-terminal tail. Mass spectrometry indicated that the polypeptides extended to residue 127 or 115 (of 135 residues), resulting from cleavage in the center of the third helix of the histone fold or the loop before this helix. The H4 product started at residue 28, removing most of the N-terminal disordered region; the polypeptide extended to residue 84–86 (of 102 residues) in the third helix of the histone fold. Thus, it appears that the majority of polypeptide protected by the fibril structure is helical in the native state (Supporting Information Fig. 4).
In summary, the fibrils formed by H3 at pH 5, H4 at pH 2 and pH 5 as well as the fibrils formed by the combination of H3 and H4, either initially folded or unfolded, share many similar features. The fibrils are positive for ThT FL, form from helical regions of H3 and H4, contain significant β-sheet structure as evidenced by FTIR and far-UV CD spectroscopies and have morphologies consistent with immature fibrils (or protofibrils) as detected by TEM.
Kinetics of fibril formation by the H3 and H4 histones
To elucidate the mechanism of fibril formation by the H3 and H4 histones, individually and in combination, the kinetics of fibrillation were examined under different conditions, using primarily the gel-based precipitation assay. Table I lists the t50 values [Eq. (1) in Methods] for fibrillation at pH 5 and pH 2 at 1M NaCl. The H4 histone forms fibrils rapidly at both pH values; however at pH 5, the lag times are reproducibly shorter and the rate of fibril growth is faster (data not shown). Isolated H3 exhibits enhanced ThT FL at pH 2, but no precipitates were detected after three months. In contrast, precipitating H3 fibrils were observed in less than a day at pH 5. Similarly, the fibrillation kinetics of H3 + H4 and the H3-H4 dimer are faster at pH 5, with shorter lag times and faster rates of fibril growth. Despite similar lag times at pH 5, the growth rate of fibrils from the H3-H4 dimer are marginally but consistently faster than from the initially unfolded H3 + H4 [Fig. 2(B)]. This difference in rates may reflect the subtle differences observed in the far-UV CD and FTIR spectra. The far-UV CD spectra at t = 0 of fibrillation are identical at pH 5 and pH 2 for H3 + H4, and similarly for H3-H4 at pH 2 and 5. Thus, the effect of different pH values on fibrillation kinetics probably does not arise from structural differences between the initial species.
To further examine the effect of pH, fibrillation was monitored for H3-H4 by the gel-based assay and turbidity from pH 2 to pH 9 (Supporting Information Fig. 5). The rate of fibril growth was maximal at pH 4 and 5, with significantly slower fibrillation at more acidic or alkaline pH values. While the rate of fibril growth was similar at pH 4 and 5, the lag time was significantly longer at pH 5 [Supporting Information Fig. 5(A)].This pH-dependence could arise from dynamic differences (e.g., effects on stability and/or unfolding kinetics) or electrostatic effects (increased electrostatic repulsion at acidic pH because of protonation of Glu and Asp residues in highly basic polypeptides). To differentiate between these possibilities, further perturbations were employed, focusing on fibrillation of H3-H4 at pH 5. As noted above, histones are generally stabilized by increasing ionic strength; this has been demonstrated directly for the H3-H4 dimer at pH 5.41 However, decreased ionic strength inhibited fibril formation. The t50 value for fibril formation in 0.75M NaCl was 18 days, and no fibrils were detected after 41 days at 0.5M NaCl. This lack of correlation with decreased stability at lower ionic strength suggests the importance of electrostatic interactions, specifically the screening of electrostatic repulsion with increasing ionic strength.
To test this hypothesis, the positive charge content of the histones was decreased by deleting the poorly structured N-terminal tails of H3 and H4 at the genetic level. The tails are the most charged region of the histones. For example, the 38 and 19 residues of Xenopus laevis H3 and H4 tails are 29 and 37 mol % basic residues, respectively, with no acidic residues. In contrast, the structured regions of H3 and H4 have 8 and 13 mol % excess basic residues. The ΔN-H3/ΔN-H4 tetramer has WT-like stability.40 Although there are many factors determining solubility, a protein's electrostatic properties are an important contributor, with more net charge often correlating with higher solubility (e.g., Refs.55–57). Despite a reduced net positive charge, the ΔN-tetramer is more soluble than the full-length tetramer (Table II). At pH 5 and 1M NaCl, the ΔN-tetramer formed fibrils with a t50 value similar to full-length tetramer. However, in contrast, the ΔN-tetramer readily formed fibrils at 0.5M NaCl, with a t50 value of ∼90 h. This propensity to form fibrils at lower ionic strength confirms the importance of electrostatic repulsion in modulating histone fibril formation.
The t50 values are highly dependent on temperature (Fig. 5). To characterize this temperature dependence, fibrillation kinetics for the H3-H4 heterodimer at pH 5 were examined. The lag time decreases with increasing temperature, but the most significant effect is on the rate of growth. An Arrhenius plot of the growth rates as a function of temperature yielded an activation energy, EA, of 15.4 ± 0.6 kcal mol−1. This value is lower than the activation energies reported for other fibril growth or elongation reactions, for example, 20 kcal mol−1 for α-synuclein58 and 23 kcal mol−1 for the Aβ peptide.59 Interestingly, the enthalpy for unfolding the H3-H4 heterodimer is 15 kcal mol−1 under related conditions (pH 5, 100 mM NaCl, 25°C).41 The ΔH for unfolding may be higher in the more stabilizing condition of 1M NaCl. Nonetheless, the similarity between EA and the ΔH for unfolding suggests that there is substantial unfolding of the H3-H4 dimer as it is added to the growing fibril.
Figure 5.
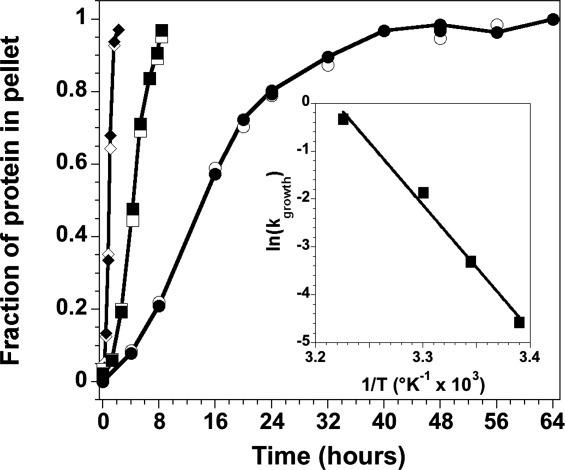
Temperature dependence of fibril growth initiated from the (H3-H4)2 tetramer at pH 5. The data are from the gel precipitation assay, showing the fraction of precipitated protein for H3 (open symbols) and H4 (closed symbols, solid line). 26°C, circles; 30°C, squares; 37°C, diamonds. The lines are drawn to guide the eye and do not represent fits of the data. Inset: Arrhenius plot of the temperature dependence of the rate of fibril growth as determined from a linear fit of the precipitation assay data in the exponential growth time regime. Conditions are described in the legend of Figure 4.
Formation of fibrils containing H3 and H4
The previous studies on bovine histones, containing H2A, H2B, H3, and H4, detected fibril formation by ThT FL, TEM, and FTIR.11,39 It was reported that SDS-PAGE of the insoluble precipitates after the completion of fibrillation indicated that all four histones were present in the fibrils; however, the fibrillation kinetics of the individual histone monomers were not determined in these studies. Therefore, it is not certain whether the histones were forming co-fibrils, with different monomer types present in a single fibril, or if each histone or pair of histones was forming independent fibrils with possibly different kinetics.
In the current study, the gel-based precipitation assay monitors independently the kinetics of incorporation of H3 and H4 into fibrils. When initiated from unfolded H3 + H4 and folded H3-H4, both monomers are incorporated into fibrils with virtually identical kinetics under all buffer conditions examined (e.g., Figs. 2 and 5). Yet in isolation, the H3 and H4 histones exhibit fibrillation kinetics that are distinct from those observed for H3 + H4 and H3-H4 (Table I). For example, at pH 2, H3 does not form fibrils, while H3-H4 and H3 + H4 take days to form fibrils, but H4 fibrillation has a t50 of ∼40 min. These observations suggest that H3 and H4 interact during fibrillation and form a co-fibril, with both monomers being incorporated simultaneously.
To further test the hypothesis of co-fibrillation, the kinetics of H4 fibrillation were examined in the presence of sub-stoichiometric concentrations of H3 and in the presence of ovalbumin (OA) as a control (Fig. 6). The experiments were performed at pH 2 for two reasons: (1) at pH 2, H3 alone does not exhibit fibril formation by ThT FL or the gel-based precipitation assay on the time scale for complete H4 fibrillation; and (2) H4 forms fibrils more slowly at pH 2 than pH 5, simplifying the technical aspects of the gel-based assay. Increasing concentrations of OA accelerated H4 fibril formation; the lag times decrease and the apparent rate of fibril growth is increased by ∼1.6-fold at equi-molar concentrations of H4 and OA. No OA was detected in the protein pellet, indicating that there is no significant interaction between H4 and OA. The OA-dependent acceleration of H4 fibrillation may reflect a small increase in macromolecular crowding or some other non-specific cosolute effect.
Figure 6.
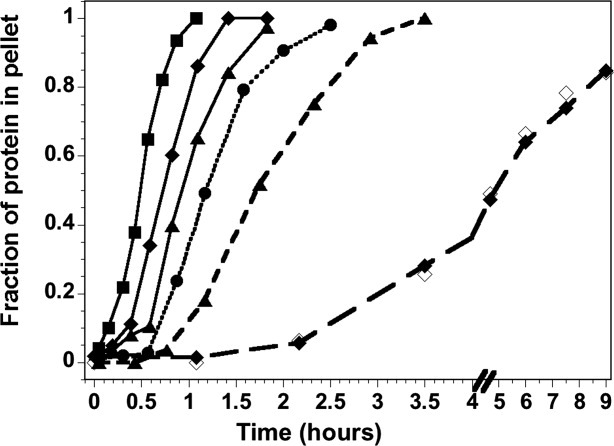
H4 fibrillation kinetics at pH 2 in the presence of ovalbumin (OA) and H3. H4 alone, circles, and dotted line. In the presence of ovalbumin, solid lines; 3 μM OA, triangles; 10 μM OA, diamonds; 30 μM OA, squares. In the presence of 3 μM H3, triangles and short dashed line; 10 μM H3, diamonds and long dashed line with closed and open symbols for H4 and H3, respectively. The lines are drawn to guide the eye and do not represent fits of the data. Conditions are described in the legend of Figure 4.
In contrast, sub-stoichiometric concentrations of H3 (1:10 and 1:3) progressively slow the precipitation of H4. Thus, H3 is not acting as a non-specific crowding agent, and the inhibitory effect is evident even when <10% of the H4 could be in a heterodimeric state. One possible explanation is that the available H3 is associating with H4 to form a heterodimer, which then forms fibrils at a slower rate, with a t50 of several days under these conditions. However, this explanation would predict a rapid precipitation of H4 alone (∼60–70% in the 1:3 ratio), followed by a slower second phase with precipitation of both H3 and H4. This is clearly not observed, as the available H3 precipitates with the same kinetic response as H4 (Fig. 6). The most plausible explanation is that H3 and H4 interact during fibril growth and are incorporated into a co-fibril. It seems unlikely that H4 forms a fibril and H3 is tethered to the precipitate by native-like heterodimeric interactions. In such a scenario, the proteolysis products would likely be different between homo-fibrils of H3 or H4 as compared to fibrils formed with both H3 and H4 present. Instead, as described above, similar sized polypeptides are protected from proteolysis in homo- and co-fibrils.
Discussion
Multiple spectroscopic methods (Figs. 3 and 4) indicate that the insoluble aggregates detected by the gel-based precipitation assay (Fig. 2) for H3 and H4, together or in isolation, are β-sheet rich, amyloid-like fibrils. The protease-resistant regions of the fibrils are predominantly helical structures in the native heterodimer involved in inter-monomer interactions. The enthalpy associated with fibril elongation, ∼15 kcal mol−1 (Fig. 5), is consistent with substantial unfolding of the H3-H4 heterodimer upon addition to the fibril. The kinetics of fibrillation, with concomitant precipitation of H3 and H4 even at substoichiometric ratios (Fig. 6), strongly support the hypothesis that a co-fibril is formed, containing both H3 and H4.
Co-fibrillation and the domain-swapped heterodimer
The formation of a co-fibril by polypeptides with divergent sequences (despite a common fold) is an unusual observation. Protein aggregates and amyloid fibrils formed in vivo generally consist of predominantly a single polypeptide species, despite the complex composition of the protein milieu.60 A few examples of in vitro formation of co-fibrils have been described, mostly employing short peptides. One notable study employed two ∼10 residue peptides derived from the amyloidogenic protein transthyretin, combined at ratios of 1:100, as well as 1% of either transthyretin peptide with bovine insulin.61 The minor components appeared to be randomly incorporated along the length of the fibril. However, it was necessary to employ conditions under which the rate of fibrillation was comparable for the two components. This is in contrast to the co-fibrils formed by H4 and H3, at much higher ratios (1:10, 1:3 and even 1:1), which form readily under conditions (pH 2) where H3 alone does not form fibrils. It seems likely that the ability of H4 to recruit H3 into fibrils reflects the association of H3 and H4 in a domain-swapped structure. It has been noted that successful seeding of fibrillation with pre-formed fibrils requires significant sequence similarity between the components (e.g., Ref.62 and references therein). Consistent with these findings, H4 fibrils failed to induce fibrillation of H2A or H2B (data not shown).
Co-fibrils between H3 and H4 were observed when fibrillation was initiated from both acid unfolded H3 + H4 or from folded tetramer (pH 7.2). The rate of fibril growth is consistently faster when initiated from folded tetramer; the difference is minor at pH 5 (Fig. 2), but quite significant at pH 2 (Table I). We have observed previously that, unlike refolding from GdmCl-unfolded monomers, reconstitution of the H3-H4 heterodimer from acid-unfolded monomers is not always quantitative, with minor amounts of soluble, misfolded aggregate.40 A possible explanation for the difference in fibrillation kinetics is that the presence of misfolded H3-H4 heterodimer upon dilution from 10 mM HCl [suggested by the slightly lower ellipticity at 222 nm, Fig. 4(B)] impedes fibrillation. This explanation is consistent with the hypothesis that the domain-swapped association of native H3-H4 is important for co-fibril formation.
Predictors of fibrillation propensity
To elucidate the contribution of primary sequence to the different fibrillation propensities of the histones, multiple aggregation/amyloid prediction methods were employed: TANGO,63 WALTZ,64 Zyggregator,65,66 and when possible, the structure-based 3D profile ZipperDB database.67 TANGO, WALTZ, and Zyggregator accurately predicted that hMfB and hPyA1 lack regions of sequence prone to aggregation; however, ZipperDB predicted two non-adjacent hexapeptides in α2 of hMfB with high fibrillation propensity. All four methods predicted that sequences in the N-terminus of α2 in H2A and various segments of α1, α2, and αC of H2B were aggregation-prone and amylogenic. The failure to generate fibrils of hMfB, hPyA1, H2A, and/or H2B under a variety of conditions (Supporting Information Table 1) suggests that the overall solubility and possibly electrostatic effects of the entire polypeptide strongly offset the aggregation propensity of local sequences.
WALTZ predicted no amylogenic regions in H3, but other methods identified three aggregation- and fibrillation-prone regions in α2 (Supporting Information Fig. 4). For H4, TANGO, WALTZ, and Zyggregator identified the center to C-terminus of α2 and the α3 helix. For H3 and H4, the predicted sequences in α2 are protected from proteolysis in the fibril, while the H4 α3 sequences extend a couple of residues beyond the protease-resistant segment. Interestingly, there are tertiary and quaternary interactions in the native state between the predicted aggregation/fibrillation-prone sequences (Supporting Information Fig. 4). The H4 segments in the C-terminal region of α2 and α3 dock on each other in the folded state. It is tempting to speculate that such tertiary interactions may facilitate the rapid fibrillation of H4. Similarly, the central portions of the α2 helices of H3 and H4 make inter-monomer contacts via segments of sequence predicted to be aggregation/fibrillation-prone. These results support the speculation that the domain-swapped structure of the H3-H4 heterodimer may facilitate co-fibril formation.
Contribution of solubility and electrostatics
Comparison of the fibrillation propensities of the eukaryotic and archaeal histones and the sequence-based predictions described above suggest that, while domain-swapping may contribute to the fibril formation by H3 and/or H4, the domain-swapped topology of the histone fold and the presence of aggregation/fibrillation-prone sequence elements are not sufficient to facilitate formation of amyloid-like fibrils. The survey of histone folds and conditions in this report indicate that solubility and electrostatics are major determinants of fibrillation propensity.
Several studies have characterized the impact of electrostatics and the content and distribution of charged residues on fibrillation of model peptides68,69 and proteins, for example, ribonuclease Sa (RNase Sa),70 muscle acyl-phosphatase,71 α-synuclein,72 and β-microglobulin.73 Although the details vary regarding the type of charged residue and sequence/structure context, the general trend observed across these diverse systems is that decreased charge facilitates fibril formation. Three observations indicate that electrostatic repulsion mitigates fibrillation by H3-H4 and H3 + H4: (1) the requirement of ionic strengths of > 0.75M for fibrillation by the full-length histones; (2) the alleviation of this requirement for ΔN-H3/ΔN-H4 which readily forms fibrils at an ionic strength of 0.5M; and (3) slower or no fibrillation of H3 and/or H4 at pH 2 compared to pH 5; presumably the acidic residues are more protonated at the lower pH, and thus the overall charge on the protein is greater. The highly basic N-terminal tails of H3 and H4 do not participate in the structure of the fibril, as evidenced by their susceptibility to proteolysis, but their presence clearly impedes fibrillation. A similar inhibitory effect of charged sequences not incorporated into the structural core of the fibril has been observed for the acidic C-terminus of α-synuclein.72
As noted in previous reports, the effects of electrostatics on fibrillation are more complicated than simple net charge.68,69,72 This is apparent as well for the histones in this study. The Xenopus H2A and H2B histones used in this study have lower estimated isoelectric points (pI) than H3 and H4, and the H2A-H2B dimer has lower net basic charge than H3-H4, +34 and +38, respectively. However, the less-charged histones are resistant to fibrillation. The lack of correlation between charge and fibrillation is even more striking for the archaeal histones which have a net charge of +3 and +1 for hMfB and hPyA1, respectively.
Solubility is an important determinant of protein aggregation and fibril formation.55,57 A protein's electrostatic properties influence solubility, but there are additional aspects of protein sequence and structure that dictate solubility.56 Despite the acknowledged importance, there have been relatively few studies directly relating solubility to fibrillation propensity. Using the model system of RNAse Sa and mutants with varying charge, it was shown the pH dependence of fibril formation correlates with changes in the protein's isoelectric point, pI, where it is least soluble.70 A strong correlation between pI and the pH optimum for fibrillation was also noted for several other proteins. Similar conclusions regarding solubility can be drawn for the histones in this study. The more soluble histones (Table II) do not readily form fibrils, even if they are less stable, as is the case for H2A-H2B, relative to H3-H4, under acidic conditions. However, comparison of fibrillation by the full-length and ΔN-H3-H4 tetramers demonstrates the complicated interplay between charge density and solubility. The less charged ΔN-heterodimer is somewhat more soluble, but forms fibrils at lower ionic strength.
Biological significance
There are no known disease states linked directly to histone fibrillation. However, aggregation and/or fibrillation of the nucleosome core histones, particularly H3 and H4, have been implicated in cytotoxicity and α-synuclein amyloidogenesis. Extracellular histones are cytotoxic in vitro to human endothelial and mouse epithelial cell lines at concentrations as low as 20 μg mL−1.74,75 The accumulation of H3 and H4 has been observed in mouse, baboon, and human plasma in inflammatory responses induced by sepsis, with H3 levels reaching ∼15 μg mL−1.74 Intravenous injection of histones into mice at low levels led to symptoms characteristic of sepsis, and higher levels were lethal. It is plausible that this cytotoxicity is related to histone aggregation and precipitation. It is well known that histones induce protein aggregation upon addition to serum and plasma. Fibrinogen is a major component of the histone-associated plasma precipitate75 and associates with H3 through hydrophobic interactions.76 Electron microscopy suggested that some aggregates of histones with fibrinogen or fibrin were rudimentary fibrils.77 Furthermore, during inflammatory responses to sepsis, neutrophils actively make neutrophil extracellular traps (NETs) that associate with bacterial pathogens, and contain DNA and the four core histones.78 Interestingly, these fibrous structures have a diameter of ∼15 nm, about twice the diameter of fibrils formed by calf thymus histones.11
A second biologically significant aspect of the ability of H3 and H4 to form fibrils is their interaction with α-synuclein. Enhanced nuclear localization of α-synuclein, either through genetic manipulation to introduce a nuclear-localization signal or mutations associated with familial Parkinson's disease, contributed to neurotoxicity.79 In tissue culture and in vivo, α-synuclein associates with H3 in the nucleus, leading to reduced H3 acetylation and enhanced neurotoxicity. Furthermore, a mixture of the core histones drastically accelerates the fibrillation of α-synuclein at sub-stoichiometric levels.80
Thus, it seems reasonable to speculate that the ability of H3 and H4 to form fibrils in the test tube may explain their contributions to the cytotoxicity of extracellular histones and the neurotoxicity and fibrillation of α-synuclein.
Materials and Methods
Materials
Thioflavin T was purchased from Sigma (St. Louis, MO). Proteinase K was from Fisher Scientific (Fair Lawn, NJ). All other chemicals were of reagent or molecular biology grade, generally from JT Baker (Philipsburg, NJ) or Fisher Scientific (Fair Lawn, NJ). Recombinant full-length and ΔN constructs of the H3 and H4 histones were over-expressed in E. coli and purified as described previously.40
Preparation and analysis of histone fibrils
Conditions for fibril formation were generally 30 μM monomer in a buffer of 50 mM acetic acid/sodium acetate and 50 mM phosphoric acid/NaH2PO4 at pH 2 or pH 5 with 0.5–1.0M NaCl at 22°C, unless otherwise indicated. Kinetic experiments at different conditions were independently replicated three to five times; the variation in the observed rate was <30%. For experiments that systematically probed variation in an experimental condition (e.g., temperature, pH, or addition of another protein), representative time-dependent data are shown; the relative changes in the kinetic response were very consistent between replicates even if the absolute rates varied by <30%.
The fibrillation reactions were monitored by spectroscopic methods (see below) and SDS-PAGE. To determine the time dependence of fibrillation, aliquots were taken at various times and centrifuged at 16,000g for 2 min. After removal of the supernatant, the pellet was washed with buffer and resuspended in SDS-PAGE loading buffer. Equivalent amounts of supernatant and pellet were applied to a 14% SDS-Tricine polyacrylamide gel.81 After staining with Coomassie Blue, the bands were quantified by densitometry using a Molecular Dynamics Personal Densitometer SI and ImageQuant v4.2 software. The kinetic data for fibril formation were analyzed using an empirical expression for a sigmoidal response39:
| (1) |
The initial and post-fibrillation baselines were defined as (Yi and mit) and (Yf and mft); in general, the slope values were 0, and the Yi and Yf values were 0 and 1, respectively. The apparent rate for fibril growth is k, and the t50 value is the time required for 50% of the fibrils to form.
To determine what regions of the H3 and H4 histones contributed to the fibril core, fibrils were treated with Proteinase K at 20 μg/mL for 10 min at 24°C. Proteolysis was quenched with 1 mM phenylmethylsolfonyl fluoride (PMSF), and the remaining fibril material was collected by centrifugation at 16,000g for 2 min. The molecular weights of the solubilized fibrils were determined with an Applied Biosystems 4800 MALDI TOF/TOF Analyzer. The solubilized fibrils were also applied to a 14% SDS-Tricine polyacrylamide gel and transferred to polyvinylidene fluoride (PVDF) membrane for N-terminal sequencing, using an Applied Biosystems Procise Protein Sequencer.
Determination of solubility
The individual eukaryotic histone monomers and the folded histone dimers were dialyzed into 10 mM HCl then lyophilized. The powder was then resuspended in a buffer of 500 mM NaCl, 50 mM acetic acid/sodium acetate, and 50 mM phosphoric acid/NaH2PO4, pH 5. The solution was equilibrated at room temperature (∼23°C) overnight. After incubation, the samples were centrifuged and the concentration of the soluble fraction monitored by absorbance.
Spectroscopic methods
For ThT FL, 20 μM dye was included in the fibrillation assays. Fluorescence was monitored with an AVIV ATF-105 spectrofluorimeter (excitation at 450 nm and emission at 490 nm) or with a Wallac 1420 VICTOR2 multi-label plate reader (excitation at 450 nm, and emission at 535 nm with a 30 nm bandwidth). Turbidity was monitored at 500 nm in a 1-cm cell, using a Cary 50 Bio UV-visible spectrophotometer. For kinetic experiments, generally performed in conjunction with gel-based assays, spectroscopic measurements were taken at specific intervals, rather than monitored continuously, to avoid artifacts from photo-degradation or instrumental drift. Far-UV circular dichroism spectra were collected in a 1-mm cell with an AVIV 202SF spectrophotometer. FTIR spectra were obtained with a Nicolet 6700 ATF-FTIR, typically averaging 1000 scans per spectra. TEM was performed on a JEOL 1200 EX transmission electron microscope. Samples were prepared on a Formvar coated copper grid and stained with 1% uranylacetate. Magnification was 50,000–100,000×.
Acknowledgments
Authors thank Dr. Kenneth Nash for the use of his FTIR spectrometer and Dr. Mikael Nilsson for technical assistance (Chemistry, WSU). Electron microscopy was performed at the WSU Franceschi Microscopy and Imaging Center with the assistance of Dr. Christine Davitt. Dr. Gerhard Munske and Chongjie Zhu of the WSU Laboratory for Bioanalysis and Biotechnology performed the N-terminal sequencing and mass spectrometry, respectively.
Glossary
Abbreviations
- CD
far-UV circular dichroism
- ΔN
H3 or H4 constructs in which the N-terminal tails have been genetically removed
- FL
fluorescence
- FTIR
Fourier transformed infrared spectroscopy
- hsh
helix-strand-helix
- NCP
nucleosome core particle
- OA
ovalbumin
- PAGE
polyacrylamide gel electrophoresis
- SDS
sodium dodecyl sulfate
- t50
the time required to form 50% of the total observed fibrils
- ThT
Thioflavin T
- transmission electron microscopy
TEM
Supplementary material
References
- 1.Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie X, Kokubo T, Cohen SL, Mirza U, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 5.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, Davidson I, Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 6.Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol. 2001;8:695–700. doi: 10.1038/90408. [DOI] [PubMed] [Google Scholar]
- 7.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan SA, Landsman D. Characterization of sequence variability in nucleosome core histone folds. Proteins. 2003;52:454–465. doi: 10.1002/prot.10441. [DOI] [PubMed] [Google Scholar]
- 9.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 10.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munishkina LA, Fink AL, Uversky VN. Conformational prerequisites for formation of amyloid fibrils from histones. J Mol Biol. 2004;342:1305–1324. doi: 10.1016/j.jmb.2004.06.094. [DOI] [PubMed] [Google Scholar]
- 12.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–1316. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topping TB, Gloss LM. Stability and folding mechanism of mesophilic, thermophilic and hyperthermophilic archael histones: the importance of folding intermediates. J Mol Biol. 2004;342:247–260. doi: 10.1016/j.jmb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 14.Placek BJ, Gloss LM. Three-state kinetic folding mechanism of the H2A/H2B histone heterodimer: the N-terminal tails affect the transition state between a dimeric intermediate and the native dimer. J Mol Biol. 2005;345:827–836. doi: 10.1016/j.jmb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Stump MR, Gloss LM. Mutational analysis of the stability of the H2A and H2B histone monomers. J Mol Biol. 2008;384:1369–1383. doi: 10.1016/j.jmb.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomer ME. Protein folding and three-dimensional domain swapping: a strained relationship? Curr Opin Struct Biol. 2002;12:48–53. doi: 10.1016/s0959-440x(02)00288-9. [DOI] [PubMed] [Google Scholar]
- 19.Rousseau F, Schymkowitz JW, Itzhaki LS. The unfolding story of three-dimensional domain swapping. Structure. 2003;11:243–251. doi: 10.1016/s0969-2126(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 20.Alva V, Ammelburg M, Soding J, Lupas AN. On the origin of the histone fold. BMC Struct Biol. 2007;7:17. doi: 10.1186/1472-6807-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson CL, Benoff B, Berger T, Berman HM, Carey J. E. coli trp repressor forms a domain-swapped array in aqueous alcohol. Structure. 2004;12:1099–1108. doi: 10.1016/j.str.2004.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takehara S, Zhang J, Yang X, Takahashi N, Mikami B, Onda M. Refolding and polymerization pathways of neuroserpin. J Mol Biol. 2010;403:751–762. doi: 10.1016/j.jmb.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 23.Ogihara NL, Ghirlanda G, Bryson JW, Gingery M, DeGrado WF, Eisenberg D. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc Natl Acad Sci USA. 2001;98:1404–1409. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janowski R, Kozak M, Abrahamson M, Grubb A, Jaskolski M. 3D domain-swapped human cystatin C with amyloidlike intermolecular β-sheets. Proteins. 2005;61:570–578. doi: 10.1002/prot.20633. [DOI] [PubMed] [Google Scholar]
- 25.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc Natl Acad Sci USA. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–824. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Abedini A, Raleigh DP. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Phys Biol. 2009;6:015005. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luheshi LM, Dobson CM. Bridging the gap: from protein misfolding to protein misfolding diseases. FEBS Lett. 2009;583:2581–2586. doi: 10.1016/j.febslet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg D, Nelson R, Sawaya MR, Balbirnie M, Sambashivan S, Ivanova MI, Madsen AO, Riekel C. The structural biology of protein aggregation diseases: fundamental questions and some answers. Acc Chem Res. 2006;39:568–575. doi: 10.1021/ar0500618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 32.Hafner-Bratkovic I, Bester R, Pristovsek P, Gaedtke L, Veranic P, Gaspersic J, Mancek-Keber M, Avbelj M, Polymenidou M, Julius C, Aguzzi A, Vorberg I, Jerala R. Globular domain of the prion protein needs to be unlocked by domain swapping to support prion protein conversion. J Biol Chem. 2011;286:12149–12156. doi: 10.1074/jbc.M110.213926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by β-2 microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Sawaya MR, Eisenberg D. β-Microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2011;18:49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenko Kokalj S, Guncar G, Stern I, Morgan G, Rabzelj S, Kenig M, Staniforth RA, Waltho JP, Zerovnik E, Turk D. Essential role of proline isomerization in stefin B tetramer formation. J Mol Biol. 2007;366:1569–1579. doi: 10.1016/j.jmb.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 36.Zerovnik E, Stoka V, Mirtic A, Guncar G, Grdadolnik J, Staniforth RA, Turk D, Turk V. Mechanisms of amyloid fibril formation—focus on domain-swapping. FEBS J. 2011;278:2263–2282. doi: 10.1111/j.1742-4658.2011.08149.x. [DOI] [PubMed] [Google Scholar]
- 37.Wahlbom M, Wang X, Lindstrom V, Carlemalm E, Jaskolski M, Grubb A. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J Biol Chem. 2007;282:18318–18326. doi: 10.1074/jbc.M611368200. [DOI] [PubMed] [Google Scholar]
- 38.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 39.Munishkina LA, Ahmad A, Fink AL, Uversky VN. Guiding protein aggregation with macromolecular crowding. Biochemistry. 2008;47:8993–9006. doi: 10.1021/bi8008399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banks DD, Gloss LM. Equilibrium folding of the core histones: the H3-H4 tetramer is less stable than the H2A-H2B dimer. Biochemistry. 2003;42:6827–6839. doi: 10.1021/bi026957r. [DOI] [PubMed] [Google Scholar]
- 41.Karantza V, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: pH and ionic strength effects on the stability of the (H3-H4)/(H3-H4)2 system. Biochemistry. 1996;35:2037–2046. doi: 10.1021/bi9518858. [DOI] [PubMed] [Google Scholar]
- 42.Karantza V, Baxevanis AD, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: ionic strength and pH dependence of H2A-H2B dimer stability. Biochemistry. 1995;34:5988–5996. doi: 10.1021/bi00017a028. [DOI] [PubMed] [Google Scholar]
- 43.Stump MR, Gloss LM. Mutational studies uncover non-native structure in the dimeric kinetic intermediate of the H2A-H2B heterodimer. J Mol Biol. 2010;401:518–531. doi: 10.1016/j.jmb.2010.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Grayling RA, Sandman K, Edmondson S, Shriver JW, Reeve JN. Thermodynamic stability of archael histones. Biochemistry. 1998;37:10563–10572. doi: 10.1021/bi973006i. [DOI] [PubMed] [Google Scholar]
- 45.Gloss LM, Placek BJ. The effect of salts on the stability of the H2A-H2B histone dimer. Biochemistry. 2002;41:14951–14959. doi: 10.1021/bi026282s. [DOI] [PubMed] [Google Scholar]
- 46.Kenig M, Jenko-Kokalj S, Tusek-Znidaric M, Pompe-Novak M, Guncar G, Turk D, Waltho JP, Staniforth RA, Avbelj F, Zerovnik E. Folding and amyloid-fibril formation for a series of human stefins' chimeras: any correlation? Proteins. 2006;62:918–927. doi: 10.1002/prot.20812. [DOI] [PubMed] [Google Scholar]
- 47.Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindgren M, Hammarstrom P. Amyloid oligomers: spectroscopic characterization of amyloidogenic protein states. FEBS J. 2010;277:1380–1388. doi: 10.1111/j.1742-4658.2010.07571.x. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 50.Cordeiro Y, Kraineva J, Suarez MC, Tempesta AG, Kelly JW, Silva JL, Winter R, Foguel D. Fourier transform infrared spectroscopy provides a fingerprint for the tetramer and for the aggregates of transthyretin. Biophys J. 2006;91:957–967. doi: 10.1529/biophysj.106.085928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aso Y, Shiraki K, Takagi M. Systematic analysis of aggregates from 38 kinds of non disease-related proteins: identifying the intrinsic propensity of polypeptides to form amyloid fibrils. Biosci Biotechnol Biochem. 2007;71:1313–1321. doi: 10.1271/bbb.60718. [DOI] [PubMed] [Google Scholar]
- 52.Wachtel EJ, Sperling R. Low resolution models of self-assembled histone fibers from X-ray diffraction studies. Nucleic Acids Res. 1979;6:139–152. doi: 10.1093/nar/6.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfefferkorn CM, McGlinchey RP, Lee JC. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc Natl Acad Sci USA. 2010;107:21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 55.Pace CN, Trevino S, Prabhakaran E, Scholtz JM. Protein structure, stability and solubility in water and other solvents. Philos Trans R Soc Lond B Biol Sci. 2004;359:1225–1234. doi: 10.1098/rstb.2004.1500. discussion 1234–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trevino SR, Scholtz JM, Pace CN. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J Mol Biol. 2007;366:449–460. doi: 10.1016/j.jmb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trevino SR, Scholtz JM, Pace CN. Measuring and increasing protein solubility. J Pharm Sci. 2008;97:4155–4166. doi: 10.1002/jps.21327. [DOI] [PubMed] [Google Scholar]
- 58.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 59.Kusumoto Y, Lomakin A, Teplow DB, Benedek GB. Temperature dependence of amyloid beta-protein fibrillization. Proc Natl Acad Sci USA. 1998;95:12277–12282. doi: 10.1073/pnas.95.21.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ventura S. Sequence determinants of protein aggregation: tools to increase protein solubility. Microb Cell Fact. 2005;4:11. doi: 10.1186/1475-2859-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacPhee CE, Dobson CM. Formation of mixed fibrils demonstrates the generic nature and potential utility of amyloid nanostructures. J Am Chem Soc. 2000;122:12707–12713. [Google Scholar]
- 62.Krebs MR, Morozova-Roche LA, Daniel K, Robinson CV, Dobson CM. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 2004;13:1933–1938. doi: 10.1110/ps.04707004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 64.Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JW, Rousseau F. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 65.Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J Mol Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 66.Tartaglia GG, Vendruscolo M. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev. 2008;37:1395–1401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- 67.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Picotti P, De Franceschi G, Frare E, Spolaore B, Zambonin M, Chiti F, de Laureto PP, Fontana A. Amyloid fibril formation and disaggregation of fragment 1–29 of apomyoglobinsights into the effect of pH on protein fibrillogenesis. J Mol Biol. 2007;367:1237–1245. doi: 10.1016/j.jmb.2007.01.072. [DOI] [PubMed] [Google Scholar]
- 69.Marshall KE, Morris KL, Charlton D, O'Reilly N, Lewis L, Walden H, Serpell LC. Hydrophobic, aromatic, and electrostatic interactions play a central role in amyloid fibril formation and stability. Biochemistry. 2011;50:2061–2071. doi: 10.1021/bi101936c. [DOI] [PubMed] [Google Scholar]
- 70.Schmittschmitt JP, Scholtz JM. The role of protein stability, solubility, and net charge in amyloid fibril formation. Protein Sci. 2003;12:2374–2378. doi: 10.1110/ps.03152903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 72.Hoyer W, Cherny D, Subramaniam V, Jovin TM. Impact of the acidic C-terminal region comprising amino acids 109–140 on α-synuclein aggregation in vitro. Biochemistry. 2004;43:16233–16242. doi: 10.1021/bi048453u. [DOI] [PubMed] [Google Scholar]
- 73.Raman B, Chatani E, Kihara M, Ban T, Sakai M, Hasegawa K, Naiki H, Rao Ch M, Goto Y. Critical balance of electrostatic and hydrophobic interactions is required for β 2-microglobulin amyloid fibril growth and stability. Biochemistry. 2005;44:1288–1299. doi: 10.1021/bi048029t. [DOI] [PubMed] [Google Scholar]
- 74.Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, Taylor FB, Esmon NL, Lupu F, Esmon CT. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15:1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pemberton AD, Brown JK, Inglis NF. Proteomic identification of interactions between histones and plasma proteins: implications for cytoprotection. Proteomics. 2010;10:1484–1493. doi: 10.1002/pmic.200900818. [DOI] [PubMed] [Google Scholar]
- 76.Gonias SL, Pasqua JJ, Greenberg C, Pizzo SV. Precipitation of fibrinogen, fibrinogen degradation products and fibrin monomer by histone H3. Thromb Res. 1985;39:97–116. doi: 10.1016/0049-3848(85)90125-2. [DOI] [PubMed] [Google Scholar]
- 77.Stewart GJ, Niewiarowski S. Aggregation of fibrinogen and its degradation products by basic proteins. An electron microscope study. Thromb Diath Haemorrh. 1971;25:566–579. [PubMed] [Google Scholar]
- 78.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 79.Kontopoulos E, Parvin JD, Feany MB. α-Synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 80.Goers J, Manning-Bog AB, McCormack AL, Millett IS, Doniach S, Di Monte DA, Uversky VN, Fink AL. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42:8465–8471. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 81.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


