Abstract
11-(Dansylamino) undecanoic acid (DAUDA) is a dansyl-type fluorophore and has widely used as a probe to determine the binding site for human serum albumin (HSA). Here, we reported that structure of HSA-Myristate-DAUDA ternary complex and identified clearly the presence of two DAUDA molecules at fatty acid (FA) binding site 6 and 7 of HSA, thus showing these two sites are weak FA binding sites. This result also show that DAUDA is an appropriate probe for FA site 6 and 7 on HSA as previous studied, but not a good probe of FA binding site 1 that is likely bilirubin binding site on HSA.
Keywords: DAUDA, fluorescent fatty acid analogue, crystal structure, human serum albumin
Introduction
Human serum albumin (HSA) plays roles in maintaining the colloid osmotic pressure of plasma and transporting endogenous (including fatty acids, lysophospholipids, bilirubin, metal ions) and exogenous compounds.1 The latter property of HSA is important in determining drug transportation in plasma and thus drug distribution and metabolism. HSA was also thought to play a role in maintaining reducing redox potential in plasma due to the presence of a free cysteine (Cys34),2 which is quite rare for proteins in the oxidizing extra-cellular space. The enzymatic activities of HSA was also reported previously,3,4 and was supported by our structural study showing the presence of the modified residue as a result of esterase activity of HSA.5 The biological functions of HSA are closely related to its huge abundance in plasma (∼640 uM). Structural studies of HSA have provided a wealth of important information on HSA structures.6 HSA folds into three homologous α-helical domains (domain I, domain II, and domain III), which assemble into a heart-shaped molecule. Each domain contains 10 helices and is divided into anti-parallel six-helix and four-helix sub-domains (A and B).7 The X-ray structural studies showed that these two primary drug sites (Sudlow's site I and II)8,9 are located in the subdomain IIA and subdomain IIIA, respectively.10 We and others have shown that Sudlow site I at subdomain IIA is a large binding pocket and can be further divided into three non-overlapping subsites.5,11,12 In addition, a total of seven major fatty acid (FA) binding sites in defatted HSA are also identified mainly through the extensive works by Curry.6,13,14 These FA binding sites distributed across the protein and are available for the binding of exogenous compounds. FA sites 2, 4, and 5 on HSA were proposed to have the high affinity toward FA.15 Lysophospholipids, which has only one acyl chain attached to glycyl backbone, were also found to bind to HSA,16 and such binding was observed in the crystal structure of HSA-lysophosphatidylethanolamine complex,17 suggesting that HSA is an important modulator of lysophospholipids function.
The structural studies of HSA and its complex with different small molecular compounds were pioneered by Carter,10 Curry,6 and others, greatly enhancing our understanding of the ligand binding properties of HSA. Despite these success, crystallographic studies of HSA, however, is not trial and far from a routine procedure. The crystallization of rHSA is not always repeatable and requires major efforts. In addition, HSA is notorious for being difficult to be frozen for X-ray data collection,6 which is the typical practice nowadays. Protein crystals are usually frozen with mother liquor containing some cryoprotectants, typically 20% glycerol or ethylene glycol.18,19 However, these typical cryoprotectant cannot be used for HSA. We have discovered a proper cryoprotectant (5–10% DMSO) to cryo-cool HSA crystals.17
11-(Dansylamino) undecanoic acid (DAUDA) is one of fluorescent FA probes, containing 11 carbon acyl chain and a polarity-sensitive dansyl-type fluorophore.20,21 DAUDA has been used to determine the affinity and localization of compounds with long carbon chains (such as natural FAs, monoacylglycerol, and lysophospholipids) on proteins by monitoring excitation and emission at 345 nm and 543 nm.16,20,21 Dansyl derivatives have been widely used as probes for the study of HSA drug binding pockets for a long time.8,20 Sudlow employed dansylated amino acids as fluorescent marker in drug competitive titration, and identified two primary drug-binding sites on HSA. A previous study showed that DAUDA binds with high affinity (Kd = 8 × 10−7 M) to HSA with 3:1 molar stoichiometry.21 The strongest binding site is distinct from the long-chain FA sites and the other two sites are shared with medium-chain FAs and bilirubin.21 Here, we report that structure of HSA-myristate-DAUDA ternary complex prepared by soaking DAUDA into HSA-myristate crystals. We identified clearly the presence of two DAUDA molecules at position FA6 site and FA7 site of HSA, thus showing these two sites are weak FA binding sites.
Results
Reliability of the rHSA-Myr-DAUDA structure
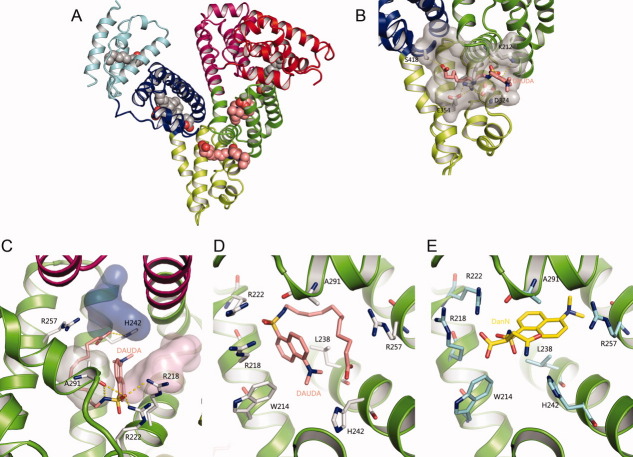
The defatted recombinant human serum albumin (rHSA) crystals are in general quite fragile to allow small molecule ligands to be soaked into them without crystal cracking. Myristate (Myr) is FA with 14 carbon acyl chain and is an important endogenous ligand of HSA. Myristate was identified to stabilize the HSA crystals and HSA protein against denaturation.6,14,22,23 We crystallized HSA in the presence of Myr and soaked the crystals again 10 mM DAUDA into these crystals for 24 h. The resulting rHSA-Myr-DAUDA crystal did not crack, and diffracted to 2.6Å and was in the typical C2 space group.6 After molecular replacement, four myristate molecules was built into the current model based on 2Fobs–Fcalc σ-weighted composite omit map, corresponding to FA site 2, 3, 4, and 5 in PDB entry 1E7G [Fig. 1(A)]. Not much electron density was observed in the FA site 1 located in subdomain IB, consistent with the proposal that this site is a weak FA binding site.13,14 In the FA site 6 and site 7 (Sudlow site I), the electron densities [Fig. 2(A,B)] were much larger than what typically observed for myristate and were modeled with confidence to DAUDA molecules, which has a large dansyl head group compared with myristate. These ligands did not move significantly upon structural refinement, further confirming the right positioning of these ligands. The final structure model was refined to a R factor of 0.251 and a R free of 0.260 and satisfactory geometry with 88.7% of the residues in the most favored Ramachandran region and 2.6% in the additional allowed Ramachandran region, and with root mean square deviations (rmsd) of bond length and bond angles of 0.011Å and 1.8° (Table I), respectively.
Figure 1.
Ligand-binding sites of the HSA–Myr–DAUDA complex. (A) Overview of the crystal structure of HSA-Myr-DAUDA shows DAUDA bound to FA6 and FA7 sites in the presence of myristate. The protein is shown as secondary structures and colored by subdomain. Myristate and DAUDA are shown in a ball representation with atoms colored by atom-type: C-grey for Myr and salmon for DAUDA; O-red; N-blue; S-yellow. This coloring scheme is used throughout this article. (B) Detailed molecular interactions of DAUDA at FA6 site with rHSA. FA6 site is shown semi-transparent gray surface. The key residues that participate in the formation of the trench and locate DAUDA are shown as sticks. Hydrogen bonds are shown in dotted yellow lines. (C) Detailed molecular interactions of DAUDA at FA7 site (subdomain IIA) with HSA. FA7 site can be divided as three subsites: AZT subsite (PDB code 3B9L, light purple), SAL subsite (PDB code 2I30, light grey), and IMN subsite (PDB code 2BXM, pink) and are shown as semi-transparent van der Waals surfaces. (D) Binding model of DAUDA with surround residues at FA7 site (subdomain IIA) on HSA. (E) Binding model of DanN with surround residues at FA7 site (subdomain IIA) on HSA. The HSA residues in contact with DanN are colored as cyan for carbon. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 2.
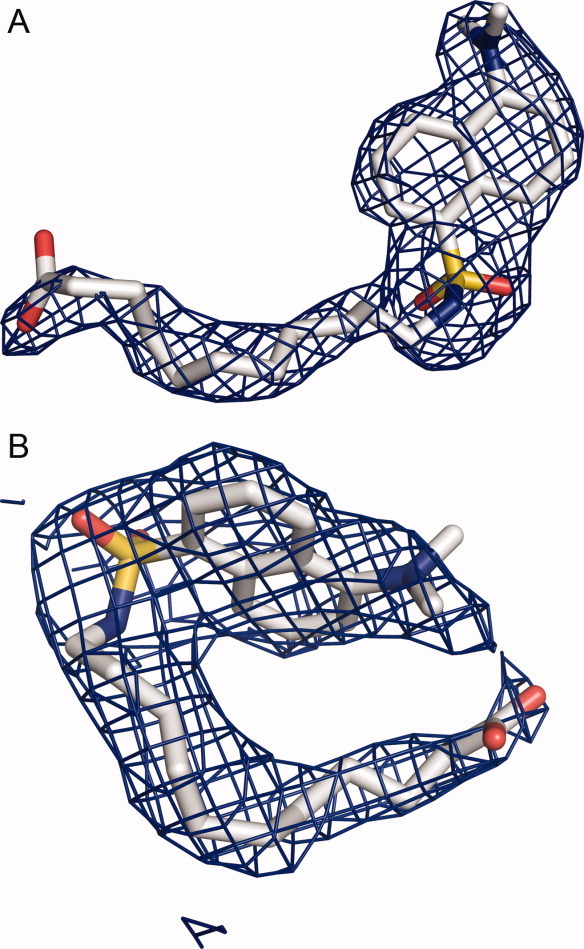
2Fo–Fc electron density omit maps (contoured at 0.5σ) of the HSA–Myr–DAUDA complex at FA6 and FA7 sites were either much larger than what typically observed for myristate and was modeled with confidence to DAUDA molecules (shown in stick), which has a large dansyl head group compared with myristate. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table I.
Data Collection and Structural Refinement Statistics of rHSA-Myr-DAUDA Complex
| Data collection | rHSA-Myr-DAUDA |
|---|---|
| X-ray wavelength (Å) | 1.04 |
| Space group | C2 |
| Cell dimensions (Å) | 176.66, 37.14, 90.20, β = 104.38° |
| Independent reflections | 1177608 |
| Resolution range (Å) | 50.0–2.6 (2.64–2.60)a |
| Completeness (%) | 98.5 (96.1)a |
| Multiplicity (redundancy) | 3.1 (2.3) |
| Rsym(%)b | 8.7 (67.0)a |
| I/σ | 24.9 (2.0)a |
| Refinement statistics | |
| Rwork(%)c | 25.1 (34.2) |
| Rfree(%)d | 26.0 (35.1) |
| Average B values | |
| Protein | 61.82 |
| Myristate | 63.63 |
| DAUDA | 65.65 |
| Root mean square deviation from ideal values | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.8 |
| Percent of residues | |
| in favored region | 88.8 |
| in allowed region | 8.5 |
| and in outlier region | 2.6 |
Values in parentheses are for the outermost resolution shells.
Rsym = Σ| (Ihkl) – (I)|/Σ (Ihkl), where Ihkl is the weighted mean intensity of a given reflection.
Rwork = Σ|Fobs − Fcalc|/ΣFobs, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree is the Rwork calculated using a randomly selected 5% reflection data omitted from the refinement.
DAUDA binding at FA6 site of rHSA
FA6 site is a shallow trench located on the surface of the protein and at the interface between sub-domains IIA and IIB.14 FA6 site was reported to be the binding site for drugs, including halothane, diflunisal, and ibuprofen.12,24 In our structure of rHSA-Myr-DAUDA, a DAUDA molecule was clearly identified at this FA6 site. DUADA6 is surrounded by rHSA residues D324, R209, K351, K212, A213, G328, V325, L327, A350, V216, L347, E354, L331, S480, V482 (ordered by the contact surface area to DAUDA6). The acyl chain of DAUDA sits at the hydrophobic bottom of this trench while both of its charged ends exposed to solvent. The trench was covered up with the surface exposed, opposite-charged residues (domain IIA residue Arg209 and IIB residues Asp324/Glu354) [Fig. 1(B)]. These salt bridges appear important to maintain this FA binding site because hydrogen bonds between opposite-charged residues can be much stronger (4.5 Kcal/mol) than regular hydrogen bonds (2–3 Kcal/mol).25 These salt bridges network could also be a mechanism to regulate the ligand loading and unloading. There is one hydrogen bond between DAUDA 6 and rHSA between the oxygen atom of rHSA Asp324 to the dansyl group (N1) of DUADA. In addition, there is a cation–π interaction between phenyl rings of DUADA 6 interacts with Lys212 [Fig. 1(B)]. All together, the DAUDA has a conformation and position very similar to myristate at this FA6 position (PDB code 1E7G).
DAUDA binding at FA7 site of rHSA
DAUDA was also identified in the rHSA subdomain IIA [Fig. 2(B)], which was also named Sudlow site I.8 Sudlow site I is a large drug-binding site and contains three non-overlapping subsites: a salicylic (SAL) subsite, an indomethacin (IMN) subsite, and a 3′-azido-3′-deoxythymidine (AZT) subsite.5,11,12 FAs are typically not ordered well in the FA7 site of rHSA, and observed as a short crescent of electron density.14 However, in the current ternary HSA-Myr-DUADA structure, the DUADA7 electron density can be clearly defined [Fig. 2(B)]. The acyl chain of DAUDA7 was found to position to hydrophobic SAL subsite of FA7 site, and adopts a binding orientation identical to the myristate molecule in HSA-Myr complex (PDB 1E7G). The AZT and IMN subsites were unoccupied which is similar to the case in FA binding [Fig. 1(C)].
DUADA interacts with HSA residues L238, A291, R218, L219, W214, R222, H242, R257, I290, F211, S287, V241, F223, A215, K199, L260, I264, V293 (ranked by the contact surface area to DUADA7). The NH2 group of Arg222 and Arg218 forms a hydrogen bond with the sulfonyl group of DUADA. And the O atom of Ala291 interacts with the dansyl group (atom N1) of DUADA [Fig. 1(C)]. Moreover, there is a cation–π interaction observed between phenyl rings of DUADA7 and the guanidinium group of Arg218. DAUDA is bulkier than typical FA. Thus, the rHSA His242 side-chain was observed to rotate about 180° away from the Sudlow site I pocket to open up space to accommodate the bulky dansyl group [Fig. 1(C,D)]. However, His242 still maintain a hydrogen bond to the carboxylate head-group of DAUDA.
Discussion
Perturbation of rHSA overall structure by DAUDA
The overall structure of rHSA in the rHSA-Myr-DAUDA complex is similar to the typical rHSA structure in the presence of FA. The overall binding positions and orientation of the DAUDA 6 and DAUDA 7 in rHSA are also quite similar to the binding mode of Myr in rHSA [Fig. 1(B,C)]. These results suggest that DAUDA does not perturb the overall rHSA structure, and thus DAUDA is an appropriate probe for FA binding site 6 and 7 on rHSA.
However, DAUDA molecule contains a dansyl group much larger than typical FA. Thus, some local perturbations to rHSA structure by DAUDA were observed. In the current rHSA-Myr-DAUDA structure, Asp324 is closer to DAUDA ligand at FA6 site than in the rHSA-Myr structure [14] due to the presence of a hydrogen bond to DAUDA dansyl group. Notably, for the DAUDA at the FA7 site, the DAUDA binding causes a 180° rotation of rHSA His242 side chain away from the Sudlow site I pocket, make more space to accommodate the large dansyl group of DAUDA [Fig. 1(D)].
DAUDA does not bind to FA1 site with the current in-crystal displacement method
FA1 site can accommodate molecules with relative large sizes, including hemin, fusidic acid, and bilirubin isomer.26,27 These ligands lie in the centre of the four-helix bundle of sub-domain IB, and their interactions with HSA induces significant conformational changes involving side-chain adjustments of sub-domain IB, especially for ligands containing aromatic rings.12,14,26,27 In addition, this site is exposed to solvent channels in the current crystal form, and thus accessible for in-crystal displacement. However, we observed only some weak electron density at this site that cannot be modeled by a DAUDA molecule, and thus demonstrated no DAUDA in FA1 site. We cannot conclude that DAUDA does not bind to the FA1 site of HSA from this in-crystal displacement study because HSA inside protein crystal lattice has much limited mobility compared with aqueous HSA. The entrance of FA1 site of HSA (residues R117, D183, R186) faces to, and is close to, the other side of the same HSA (domain III), and the mobility may be an important factor for ligands to enter this FA1 site.
A study by Wilton et al. showed that DAUDA is fluorescent probe for bilirubin binding site on HSA.21 Bilirubin is an important endogenous molecule and was identified to have one high-affinity binding site (Kd about 10−8 M) and two additional sites with lower affinities (Kd about 10−6 M) to HSA.28 Displacement studies by Wilton et al. demonstrated that the DAUDA-binding sites were not equivalent to the primary long-chain FA-binding sites on HSA, but corresponded to the bilirubin sites. Curry and coworkers tried to determine the crystal structure of HSA in complex with bilirubin (ratio 1:3) in the absence of FA.26 Surprisingly, they found a bilirubin isomer (4Z,15E-bilirubin-IXα), but not bilirubin, presented in the FA1 binding site. Since the isomer has the size comparable with bilirubin, the intriguing result from this study implies that FA1 would accommodate bilirubin molecule reasonably well, although the exact location of the primary binding site of bilirubin on HSA remains to be established. Thus, further in-solution study is needed to establish if DAUDA can bind at the FA1 site.
Weak FA binding sites on HSA
The packing of the rHSA crystals shows that all FA binding sites are exposed to solvent and accessible for in-crystal displacement. Thus, the success of the current in-crystal reaction reflects that the FA6 and FA7 are weak FA binding sites, at least for the DAUDA replacement. This is consistent with the previous results that FA sites 2, 4, and 5 are the strong binding site, while it also show that FA6 and FA7 are the low-affinity FA binding sites.15,29 The FA in the Sudlow site II is typically difficult to be replaced by a ligand molecule with in-crystal soaking method.6 The best way to find out if DAUDA can replace FA at the Sudlow site II is to co-crystallize pre-formed rHSA-Myr-DAUDA or rHSA-DAUDA complexes, which we had not succeeded yet.
Specificity of dansylated compounds as probes of HSA binding sites
Dansylated amino acids were traditionally used as fluorescent markers for the two primary drug-binding sites on HSA.8,9,30–33 The binding modes of these dansylated amino acids were largely confirmed by a recent structural study by Curry and coworkers.34 They showed that ansyl-l-asparagine (DanN), dansyl-l-glutamate (DanE) and dansyl-l-arginine (DanR) with polar or charged amino acid binds to FA7 (Sudlow site I), while dansyl-l-phenylalanine(DanF), dansyl-l-norvaline (DanNV) and dansylsarcosine (DanSRC) with hydrophobic amino acid are specific for FA3/4 (Sudlow site II).34 An identical feature in these two structures is that the sulfonyl groups are close to the positively charged residues (Arg218 and Arg222) of rHSA. However, the location of the dansyl group of DAUDA in the current structure is different from the dansyl group in the rHSA-DanN structure (PDB 2XVV)34 [Fig. 1(D)]. In the rHSA-DanN structure, it is the dansyl group of DanN that occupies the SAL subsite11 at the bottom of Sudlow site I, while the charged amino acid of DanN points outward and occupies the more polar AZT and IMN subsites. In the rHSA-DAUDA structure, the acyl chain of DAUDA occupies the SAL subsite [Fig. 1(D)], while the dansyl group is sandwiched between Leu238 and Trp214. These differences reflect the large size and the flexible nature of Sudlow site I.
Materials and Methods
Preparation and crystallization of recombinant HSA in complex with myristate and DAUDA
Defatted recombinant HSA, free of FA, was kindly provided by Zhejiang Hisun Pharmaceutical Co. Sodium myristate (Myr) and DAUDA were purchased from Sigma. The rHSA-myristate complex was prepared at molar ratio of 1:10 similar to the previous reports.11,17 The complex was then crystallized by sitting drop vapor diffusion at room temperature by mixing 1 μL of the protein complex with 1ul of reservoir solution containing 28–32% (w/v) polyethylene glycol 3350 in 50 mM potassium phosphate at pH 7.5. Crystals of rHSA-myristate grew spontaneously as clusters of rods after several days. Using these crystals as seeds, diffracting quality crystals were obtained by streak-seeding method. The rHSA-myristate crystals were placed into a crystallization precipitant solution containing a series of DAUDA solution with different concentration for about 24 h. The best concentration of DAUDA was found out to be 10 mM to ensure enough occupancy on rHSA and at the same time not to etch the rHSA-Myr complex crystals.
X-ray data collection
X-ray diffraction data of the crystals were collected at 100 K on the beam line 17U of Shanghai Synchrotron Radiation Facility at a wavelength of 1.04Å. The crystals were soaked briefly in the cryoprotectant solution containing 5% DMSO in the crystallization precipitant solution, and frozen in the liquid-nitrogen stream of the beam line. The diffraction data were indexed and processed using the HKL2000 program.35 The crystal structure of the ternary rHSA-Myr-DAUDA complex was solved by molecular replacement program MOLREP of the CCP4 package using PDB entry 1BJ5, stripped of its ligands, as a search model. The positioned model was initially refined using a rigid body protocol in CNS with the model splitting into its six sub-domains and then refined by energy minimization.36 Model building was performed with COOT.37 To minimize model bias, composite and σ-weighted 2Fo–Fc omit maps, implemented in CNS, were used throughout this work. The structure was analyzed by PyMOL.38 The atomic coordinates were deposited into the PDB (ID: 3TDL).
Conclusion
HSA, which binds with a great number of endogenous and exogenous ligands, has attracted the attention of the pharmaceutical industry for a long time. The introduction and identification of fluorescent FA probes would be a rapid and specific approach to recognize different drugs or compounds binding sites on HSA. The current crystal structure of the HSA-Myr-DAUDA complex revealed that the fluorescent FA probe DAUDA selectively displaced two myristates bound at FA6 and FA7 sites in HSA, and demonstrate that DAUDA is an appropriate probe for these FA binding sites on HSA as previously reported.21 This work also reveal clearly that FA6 and FA7 sites are weak FA binding sites. These sites may be relevant to drug transport and drug metabolism because they may be easier to be replaced by drug molecules.
Acknowledgments
X-ray data were collected at beam line 17U of Shanghai Synchrotron Radiation Facility (SSRF).
Glossary
Abbreviations
- AZT
3′-azido-3′-deoxythymidine
- DAUDA
11-(Dansylamino) undecanoic acid
- FA
fatty acid
- HSA
human serum albumin
- IMN
indomethacin
- SAL
salicylic
References
- 1.Peters T. All about albumbiochemistry, genetics, and medical applications. San Diego: Academic Press; 1996. p. 432. [Google Scholar]
- 2.Fabisiak JP, Sedlov A, Kagan VE. Quantification of oxidative/nitrosative modification of CYS34 in human serum albumin using a fluorescence-based SDS-PAGE assay. Antioxid Redox Signal. 2002;4:855–865. doi: 10.1089/152308602760599016. [DOI] [PubMed] [Google Scholar]
- 3.Burch JW, Blazeryost B. Acetylation of albumin by low-doses of aspirin. Thromb Res. 1981;23:447–452. doi: 10.1016/0049-3848(81)90205-x. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins D, Pinckard RN, Farr RS. Acetylation of human serum albumin by acetylsalicylic acid. Science. 1968;160:780. doi: 10.1126/science.160.3829.780. [DOI] [PubMed] [Google Scholar]
- 5.Huang MD, Yang F, Bian CB, Zhu LL, Zhao GX, Huang ZX. Effect of human serum albumin on drug metabolism: structural evidence of esterase activity of human serum albumin. J Struct Biol. 2007;157:348–355. doi: 10.1016/j.jsb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Curry S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug Metab Pharmacol. 2009;24:342–357. doi: 10.2133/dmpk.24.342. [DOI] [PubMed] [Google Scholar]
- 7.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 8.Sudlow G, Birkett DJ, Wade DN. Characterization of 2 specific drug binding-sites on human-serum albumin. Mol Pharmacol. 1975;11:824–832. [PubMed] [Google Scholar]
- 9.Sudlow G, Birkett DJ, Wade DN. Further characterization of specific drug binding-sites on human-serum albumin. Mol Pharmacol. 1976;12:1052–1061. [PubMed] [Google Scholar]
- 10.He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu L, Yang F, Chen L, Meehan EJ, Huang M. A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography. J Struct Biol. 2008;162:40–49. doi: 10.1016/j.jsb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 13.Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Biol. 1998;5:827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharya AA, Grune T, Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J Mol Biol. 2000;303:721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- 15.Simard JR, Zunszain PA, Ha CE, Yang JS, Bhagavan NV, Petitpas I, Curry S, Hamilton JA. Locating high-affinity fatty acid-binding sites on albumin by x-ray crystallography and NMR spectroscopy. Proc Natl Acad Sci USA. 2005;102:17958–17963. doi: 10.1073/pnas.0506440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thumser AEA, Wilton DC. The binding of natural and fluorescent lysophospholipids to wild-type and mutant rat-liver fatty-acid-binding protein and albumin. Biochem J. 1995;307:305–311. doi: 10.1042/bj3070305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo S, Shi X, Yang F, Chen L, Meehan EJ, Bian C, Huang M. Structural basis of transport of lysophospholipids by human serum albumin. Biochem J. 2009;423:23–30. doi: 10.1042/BJ20090913. [DOI] [PubMed] [Google Scholar]
- 18.Lejon S, Frick IM, Bjorck L, Wikstrom M, Svensson S. Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin. J Biol Chem. 2004;279:42924–42928. doi: 10.1074/jbc.M406957200. [DOI] [PubMed] [Google Scholar]
- 19.Lejon S, Cramer JF, Nordberg P. Structural basis for the binding of naproxen to human serum albumin in the presence of fatty acids and the GA module. Acta Cryst. 2008;F64:64–69. doi: 10.1107/S174430910706770X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thumser AEA, Buckland AG, Wilton DC. Monoacylglycerol binding to human serum albumevidence that monooleoylglycerol binds at the dansylsarcosine site. J Lipid Res. 1998;39:1033–1038. [PubMed] [Google Scholar]
- 21.Wilton DC. The fatty-acid analog 11-(dansylamino)undecanoic acid is a fluorescent-probe for the bilirubin-binding sites of albumin and not for the high-affinity fatty acid-binding sites. Biochem J. 1990;270:163–166. doi: 10.1042/bj2700163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spector AA. Fatty-acid binding to plasma albumin. J Lipid Res. 1975;16:165–179. [PubMed] [Google Scholar]
- 23.Petitpas I, Bhattacharya AA, Twine S, East M, Curry S. Crystal structure analysis of warfarin binding to human serum albumin—anatomy of drug site I. J Biol Chem. 2001;276:22804–22809. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya AA, Curry S, Franks NP. Binding of the general anesthetics propofol and halothane to human serum albumin—high resolution crystal structures. J Biol Chem. 2000;275:38731–38738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- 25.Fersht AR, Shi JP, Knilljones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Hydrogen-bonding and biological specificity analyzed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 26.Zunszain PA, Ghuman J, McDonagh AF, Curry S. Crystallographic analysis of human serum albumin complexed with 4Z,15E-bilirubin-IXalpha. J Mol Biol. 2008;381:394–406. doi: 10.1016/j.jmb.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wardell M, Wang ZM, Ho JX, Robert J, Ruker F, Ruble J, Carter DC. The atomic structure of human methemalbumin at 1.9 angstrom. Biochem Biophys Res Commun. 2002;291:813–819. doi: 10.1006/bbrc.2002.6540. [DOI] [PubMed] [Google Scholar]
- 28.Jacobsen J. Binding of bilirubin to human serum albumin—determination of dissociation constants. FEBS Lett. 1969;5:112. doi: 10.1016/0014-5793(69)80307-8. [DOI] [PubMed] [Google Scholar]
- 29.Simard JR, Zunszain PA, Hamilton JA, Curry S. Location of high and low affinity fatty acid binding sites on human serum albumin revealed by NMR drug-competition analysis. J Mol Biol. 2006;361:336–351. doi: 10.1016/j.jmb.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 30.Chen YM, Guo LH. Fluorescence study on site-specific binding of perfluoroalkyl acids to human serum albumin. Arch Toxicol. 2009;83:255–261. doi: 10.1007/s00204-008-0359-x. [DOI] [PubMed] [Google Scholar]
- 31.Lissi E, Biasutti MA, Abuin E, Leon L. A fluorescence study of human serum albumin binding sites modification by hypochlorite. J Photochem Photobiol B. 2009;94:77–81. doi: 10.1016/j.jphotobiol.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Mathias U, Jung M. Determination of drug-serum protein interactions via fluorescence polarization measurements. Anal Bioanal Chem. 2007;388:1147–1156. doi: 10.1007/s00216-007-1351-7. [DOI] [PubMed] [Google Scholar]
- 33.Nakajou K, Watanabe H, Kragh-Hansen U, Maruyama T, Otagiri M. The effect of glycation on the structure, function and biological fate of human serum albumin as revealed by recombinant mutants. Biochim Biophys Acta. 2003;1623:88–97. doi: 10.1016/j.bbagen.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Ryan AJ, Ghuman J, Zunszain PA, Chung CW, Curry S. Structural basis of binding of fluorescent, site-specific dansylated amino acids to human serum albumin. J Struct Biol. 2011;174:84–91. doi: 10.1016/j.jsb.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38. The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC.